Significance
FOXP3+ T regulatory cells (Tregs) dampen immune responses in many environments, particularly in tumors, where they contribute to cancer’s resistance to immunologic defenses. This very broad analysis of tumor-infiltrating Tregs has identified a set of genes that are preferentially expressed by these Tregs in different species, tumor models or cohorts, and types or stages of tumors. This striking commonality suggests that there are core mechanisms that tumors use to attract and mold Tregs, whose perturbation should unleash antitumor immunity. Experimental validation by genome editing provides a proof of concept for the relevance of these genes in TITRs.
Keywords: T cell differentiation, immuno-oncology, immunotherapy
Abstract
FoxP3+ T regulatory (Treg) cells are central elements of immunologic tolerance. They are abundant in many tumors, where they restrict potentially favorable antitumor responses. We used a three-pronged strategy to identify genes related to the presence and function of Tregs in the tumor microenvironment. Gene expression profiles were generated from tumor-infiltrating Tregs (TITRs) of both human and mouse tumors and were compared with those of Tregs of lymphoid organs or normal tissues from the same individuals. A computational deconvolution of whole-tumor datasets from the Cancer Genome Atlas (TCGA) was performed to identify transcripts specifically associated with Tregs across thousands of tumors from different stages and locations. We identified a set of TITR-differential transcripts with striking reproducibility between tumor types in mice, between mice and humans, and between different human patients spanning tumor stages. Many of the TITR-preferential transcripts were shared with “tissue Tregs” residing in nonlymphoid tissues, but a tumor-preferential segment could be identified. Many of these TITR signature transcripts were confirmed by mining of TCGA datasets, which also brought forth transcript modules likely representing the parenchymal attraction of, or response to, tumor Tregs. Importantly, the TITR signature included several genes encoding effective targets of tumor immunotherapy. A number of other targets were validated by CRISPR-based gene inactivation in mouse Tregs. These results confirm the validity of the signature, generating a wealth of leads for understanding the role of Tregs in tumor progression and identifying potential targets for cancer immunotherapy.
Regulatory T cells (Tregs) characterized by the transcription factor FoxP3 are critical for maintaining immunologic homeostasis, enforcing tolerance to self, and preventing runaway immune responses (1, 2) in both mice (3, 4) and humans (5). Tregs regulate the activation and differentiation of conventional CD4+ T cells (Tconv), as well as of many other cell lineages within the innate and adaptive immune systems, through a variety of effector mechanisms (reviewed in ref. 6). There is also increasing recognition of the extraimmunologic roles of Tregs in the homeostasis of several tissues, controlling the noxious side effects of inflammation ensuring effective tissue repair and otherwise promoting homeostasis (7, 8).
Tregs are often found at elevated frequency in tumors relative to blood or lymphoid organs in human cancer patients and mouse models (9, 10). For a number of cancers (but not all), a high density of Tregs is correlated with poor prognosis (reviewed in refs. 9, 11, and 12). However, correlative analyses of this nature can be misleading, because the abundance of Tregs in a locale tends to track with the extent of overall immunocyte infiltration. Causal involvement of Tregs in tumor progression was first demonstrated in mice, where their depletion via administration of anti-CD25 antibody inhibited or reversed several tumors (13). These and other studies (14–19) showed that Treg depletion increased the number of CD4+ and/or CD8+ effector T cells (Teffs) in the tumor, associated with robust tumor-specific killing activity. Thus, tumor eradication in these settings was due, at least in part, to removal of Treg-mediated suppression of the antitumor immune response. Similarly, the success of several Food and Drug Administration-approved immunotherapies for cancer (e.g., anti–CTLA-4 and anti–PD-1) may be attributable to their effects on Tregs in addition to promoting Teff killing (14, 20–22).
There are several indications that the phenotypes of TITRs are distinct from their more generic Treg counterparts found in lymphoid organs. For instance, human TITRs express the cell surface receptor NRP1, which is absent from Tregs in blood or lymphoid organs (23). Analyses of tumor-infiltrating Tregs in colorectal and breast tumors showed that they are highly suppressive, expressing various markers associated with “activated Tregs” (aTregs) (19, 24–28). These aTreg characteristics are also overrepresented in tissue Tregs (8).
At present, immunotherapies approved for use in human cancers often have significant side effects. Some of these side effects are of short duration but can be life-threatening and require intensive care management, whereas others lead to long-lasting autoimmune and autoinflammatory pathologies of the types that might be predicted to result from Treg depletion or incapacitation (reviewed in refs. 29 and 30). The ideal therapy to release the inhibition imparted by Treg cells on antitumor immune responses would be tumor-selective in its effect and target TITRs while not affecting Tregs in general to avoid autoimmune consequences or tissue Tregs to avoid homeostatic perturbations in other organ systems.
Our goal was to identify genes differentially expressed in TITRs relative to Tregs found in secondary lymphoid organs and normal nonlymphoid tissues in both human patients and mouse models. We followed a three-pronged strategy: gene-expression profiling was performed on fresh Tregs from human or mouse tumors, and profiles were compared with those of Tregs from lymphoid organs or normal tissue. In parallel, we conducted a bioinformatic analysis of large datasets from whole tumors in TCGA to ferret out transcripts specifically associated with Treg cells across large numbers of tumors. The combined analysis identified a number of candidate genes encoding plausible targets of antitumor immunotherapy. Direct perturbation of some of these genes by CRISPR-based genetic ablation validates our approach and provides exciting leads for cancer immunotherapy.
Results
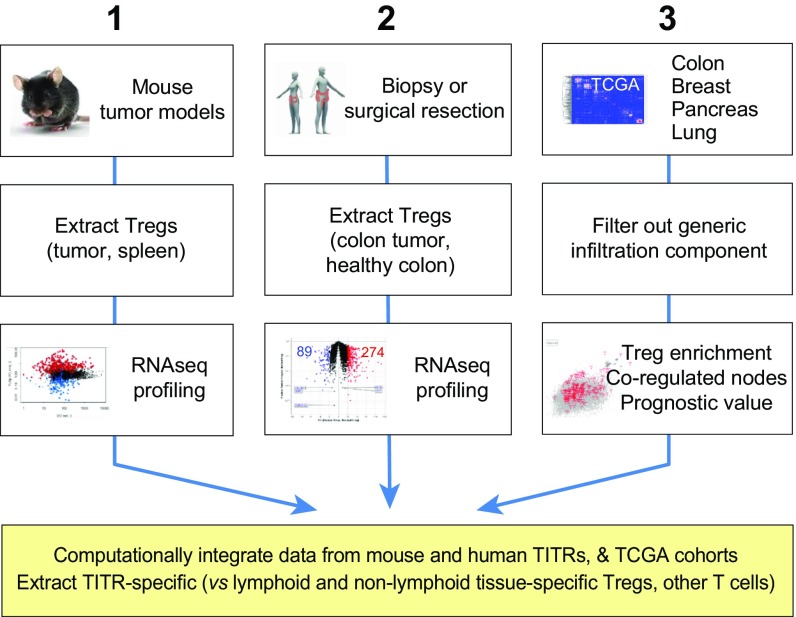
We followed the multipronged approach described in Fig. 1, which provided three orthogonal and cross-validating datasets. First, we purified and profiled TITRs from three transplantable tumors in mice and then compared these profiles with those of Tregs and other lymphocytes from various lymphoid and nonlymphoid organs. Second, we purified and profiled TITRs from patients with colorectal tumors compared with Treg cells from normal colon tissue of the same donors. Both these approaches are aimed at identifying transcripts and pathways that distinguish TITRs from both standard lymphoid organ Tregs and non-lymphoid organ Tregs. Third, we broadened the scope of the data by mining whole-tumor datasets from several different tumor types, generated by TCGA (31; https://cancergenome.nih.gov/), to identify genes whose expression correlated specifically with that of the gene encoding the Treg-defining factor FoxP3. Each of these datasets was analyzed alone, and so their intersection defined high-confidence predictions, conserved across species and tumor types, of gene signatures specifically active in tumor-infiltrating Tregs.
Fig. 1.
Schematic of multipronged work flow. This flowchart describes the generation of our three independent and cross-confirming datasets: (1) Purification and profiling of Treg cells infiltrating three different transplantable tumors in immunocompetent mice; (2) purification of TITR cells from patients with colorectal tumors, and comparison of their gene expression profiles with those of Treg cells purified from normal human colon (many from the same donors); and (3) mining of large datasets from TCGA for genes whose expression correlated with that of the Treg-defining factor FOXP3. Ultimately, these three datasets were combined to identify genes specifically overexpressed in TITRs.
Transcripts Specific to Mouse Tumor Tregs.
We primarily used three mouse models of cancer, namely the transplantable MC38 and CT26 colon carcinomas and B16 melanoma. In each case, tumors were inoculated s.c. into immunocompetent FoxP3IRES-GFP reporter mice (32), such that Treg cells could be sharply and uniquely identified by the fluorescent GFP reporter (Fig. 2A). After establishment and growth of these tumors (21 d), immunocytes were isolated from tumors and spleens, and Tregs were highly purified via cytometric double-sorting (Fig. 2A) for gene expression profiling (in biological triplicates).
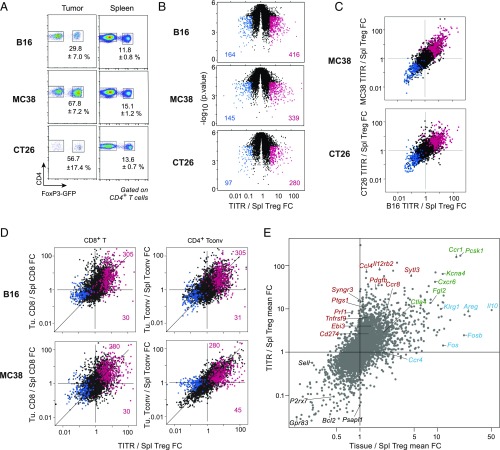
Fig. 2.
Identification of TITR signature. (A) Exemplar gating used for FACS of Tregs from three different mouse tumor models: B16, MC38, and CT26. Data are mean ± SD of Treg population size as a percentage total CD4+ T cells. (B) Volcano plots showing the fold change (FC) in gene expression between TITR and splenic Tregs for each of the mouse tumor models examined. Genes with FC in expression ≥3 (red) or ≤−3 (blue) in TITRs vs. splenic Tregs are highlighted and enumerated. (C) FC × FC plots depicting the FC in expression of genes in tumor vs. spleen for one tumor type vs. another tumor type. (Top) B16 × MC38. (Bottom) B16 × CT26. Additive filtered gene sets [genes with FC in expression ≥3 (red) or ≤−3 (blue)] in TITRs vs. splenic Tregs in each of the three transplantable tumor models are highlighted. (D) Comparison of tumor/spleen FC between CD8+ T or CD4+ Tconv cells and Tregs. The additive TITR gene sets described in C are highlighted. (E) Comparison of transcriptomes between TITRs and tissue-resident Tregs. The mean FC in several tissue Tregs (visceral adipose tissue, injured muscle, and colonic lamina propria) relative to splenic Tregs (x-axis) vs. mean FC in TITR relative to splenic Tregs (average of tumor models noted above; y-axis) is shown.
As illustrated in Fig. 2B, several hundred transcripts were significantly altered in Treg cells of each tumor compared with corresponding splenic Tregs. Some differences were very pronounced, greater than 20-fold. The majority were overexpressed in TITRs relative to spleen Tregs, suggesting inductive events operating in the tumors. Moreover, the vast majority of genes differentially expressed in TITRs from one tumor model were similarly affected in others as well (Fig. 2C and Dataset S1). The finding that the tumor Treg-specific signature is quasi-identical in tumors as different as melanomas and colon carcinomas suggests that these inductive events represent a recurrent response of Tregs to tumor microenvironments.
We then asked which of these differential transcripts were specific to Treg cells or might be activated in all lymphocytes found in the tumor microenvironment. Comparing the tumor/spleen ratio in CD8+ T or Tconv cells with that seen for Tregs (Fig. 2D) showed that many of the transcripts altered in TITRs were also changed in these other intratumoral lymphocytes. This trend was not absolute, however; 10–15% of the transcripts were uniquely induced in TITRs and not in the other T cells tested. To better delineate TITR-specific transcripts, as opposed to those equally expressed in other tumor-infiltrating lymphocytes, we directly compared expression levels among tumor-infiltrating lymphocytes. A large fraction of the transcripts that distinguished TITRs from spleen Tregs (red highlights in SI Appendix, Fig. S1A) were indeed overexpressed in TITRs relative to CD8+ and Tconv cells.
As noted above, there is growing awareness that Tregs have an extraimmunologic role in several tissues, regulating the harmful side effects of inflammation and promoting effective tissue repair and otherwise ensuring tissue homeostasis. Given our goal of identifying molecules specific for TITRs as opposed to Tregs in other locations, and given that some of the transcripts overexpressed in TITRs were reminiscent of nonlymphoid tissue Tregs, we compared the transcriptomes of these two Treg classes. As shown in Fig. 2E, we calculated the mean of fold changes in several tissue Tregs relative to spleen, with data from visceral adipose tissue, injured muscle, and colonic lamina propria (33), and then plotted these vs. the mean of fold changes between TITRs and corresponding splenic Tregs, averaged from the tumor models described above.
Several points can be made. First, and as noted previously (27), there was a strong relationship between TITRs and nonlymphoid tissue Tregs, with a strong correlation overall and many genes showing very comparable induction or repression in the two cases (green highlights in Fig. 2E). This correspondence is not surprising, because some of the cues that drive Tregs to normal tissues also might be expected to apply to tumors. Second, there were transcripts with a stronger bias in Tregs within normal tissues (blue in Fig. 2E; e.g., Il10, Fos, Areg) or tumors (red). The latter included Il12rb2, Tnfrsf9, and Cd274. To better separate tissue Treg-specific and TITR-specific transcripts, we used a principal component analysis approach. As illustrated in SI Appendix, Fig. S1B, the first principal component was strongly related to the TITR/spleen differential, but also clearly showed the transcripts that fell off the main line because of overexpression in tissue Tregs (e.g., Il10, Areg, Ctla2a, Ccr5). Furthermore, when we generated new data to compare the transcriptomes of TITR and colon-resident Treg cells, versus splenic Tregs, from MC38 tumor-bearing mice (SI Appendix, Fig. S2), we observed a similar distribution of transcripts as that shown in Fig. 2E. Moreover, when the gene set from Dataset S1 (described below) was highlighted in these data, it was apparent that while certain transcripts were shared by TITRs and tissue Tregs, others were indeed preferentially expressed in TITRs.
These comparisons highlight the numerous transcripts differentially expressed in TITRs from several tumor types relative to generic Treg cells from lymphoid organs, but with significant overlap with other cells (e.g., tissue Tregs, other tumor-infiltrating T cells). Because our intent was to identify a gene set that most specifically characterized TITRs, we selected 139 transcripts based on the following criteria: greater than threefold overexpression in TITRs relative to spleen Tregs for MC38, B16, or CT26 (with P < 10−2); greater than twofold overexpression in TITRs relative to tumor CD4+ or CD8+ T cells; and high values in the principal components distinguishing tumor Tregs from tissue Tregs (Dataset S1). We also discarded a group of differential transcripts typical of tumor-infiltrating myeloid cells, identified using myeloid cell signatures from the ImmGen database; completely eliminating this contamination from the sorts was difficult despite stringent negative controls and flow sorting. Interestingly, our gene selection pathway led us to transcripts encoding proteins already recognized for their role in Treg function and/or costimulation (Ctla4, Tigit, and Tnfrsf9, encoding 4–1BB). We also noted that such genes as Cd274, Icos, and Lag3 narrowly missed the selection criteria. The presence of such genes in our TITR gene set lends credence to their lesser known counterparts in the same gene set and suggests that they might constitute effective targets for Treg modulation.
Transcripts Specific for Human Colorectal Tumor Tregs.
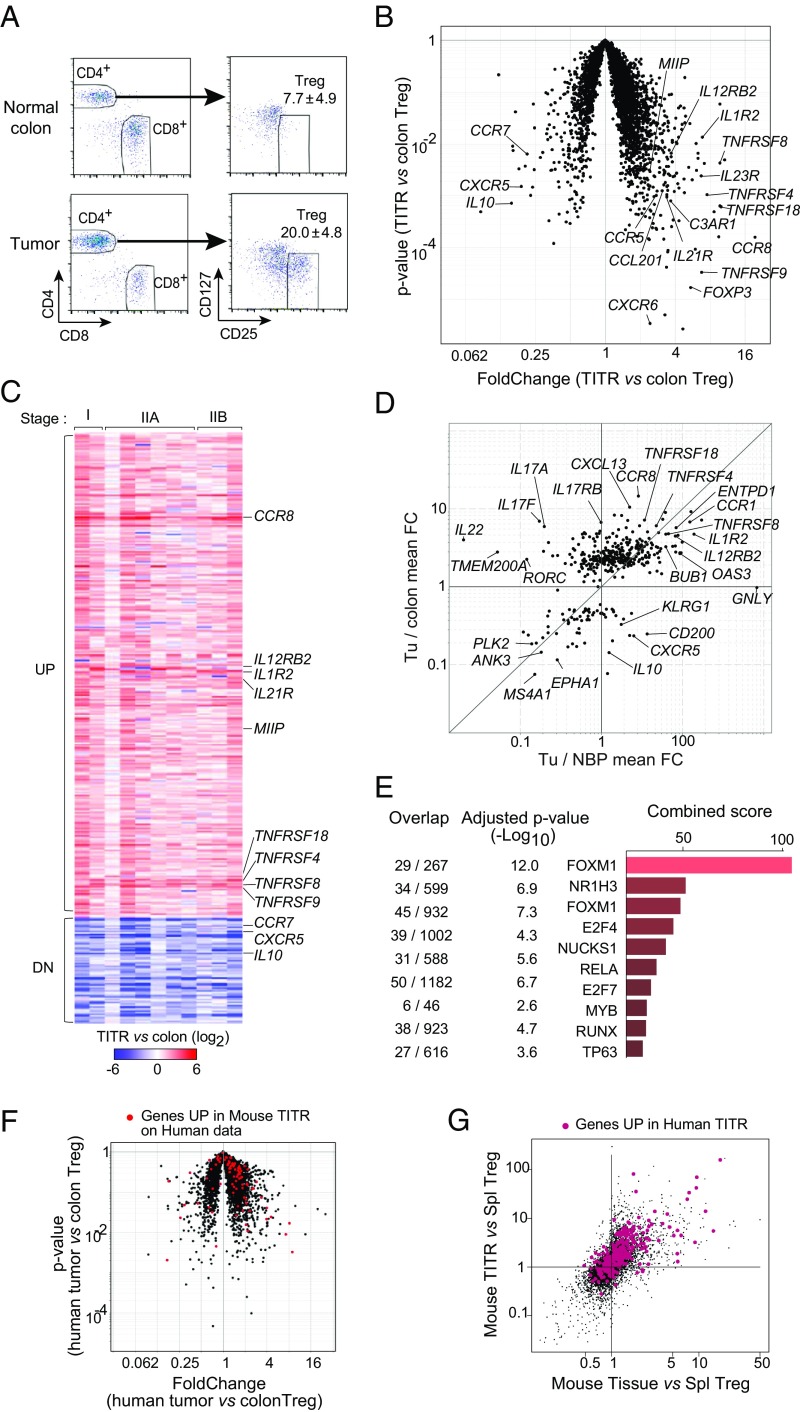
In the second arm of our study, we purified Tregs from 12 surgically resected and cryopreserved human colorectal carcinomas (CRCs) or normal colon tissue, often from the same donors. These tumors spanned a range of stages from early (stage I or IIA) to late (stage IIIB) CRCs. The usual criteria (CD25hiCD127lo) were used to identify and double-sort Tregs to high purity. As in the mouse models, and as previously reported in human tumors (27, 34–37), Tregs constituted a larger fraction of tumor CD4+ T cells than in normal colon (20.0 ± 4.8 vs. 7.7 ± 4.9) (Fig. 3A). RNA sequencing was performed on these purified Treg samples. Differential gene expression analysis comparing all tumor Tregs with all colon Tregs as a group revealed a significant bias in the transcriptome of human TITRs (Fig. 3B). As in the mouse models, these differential transcripts were mostly overexpressed in TITRs relative to Tregs from normal tissues and included several encoding proteins previously recognized to have costimulatory function (i.e., TNFRSF4, TNFRSF9, and TNFRSF18, also known as OX40, 4–1BB, and GITR, respectively). Interestingly, FOXP3 itself was overexpressed relative to colonic Tregs, suggesting that the TITR environment specifically activates the FOXP3 locus. Some transcripts were down-regulated in TITRs, including, surprisingly, genes encoding proteins known to be involved in Treg function, such as CXCR5 and IL10 (38–40). CCR7 is typical of resting Tregs (41), and its reduction points to an increase in activated aTreg phenotypes among TITRs. The drop in IL10 suggests that different suppressor mechanisms may come into play in Tregs from tumors relative to normal colon.
Fig. 3.
Conservation of TITR signature across species and individual human CRC patients. (A) Exemplar gating used for FACS of Tregs from patient samples. Data are mean ± SD of Treg population size (as a percentage total CD4+ T cells). (B) Transcriptomic profile of human CRC Tregs vs. normal colonic mucosa Tregs. The plot shows FC and P values for the expression of each gene in tumor/normal colonic Tregs. Annotated genes include some known to be involved in Treg activity and/or costimulation. (C) A total of 408 genes (335 up-regulated, 73 down-regulated) that distinguish TITRs from normal colon Tregs were selected. FC (TITRs/normal colon Tregs) values for these modulated genes for individual patients are shown in the heatmap. (D) Overlap of our TITR transcriptome (408 genes) with a recently published Treg dataset from breast cancer. The mean FC in breast cancer TITRs relative to normal breast parenchyma (NBP) Tregs (x-axis) vs. mean FC in CRC TITRs relative to normal colonic Tregs (y-axis) is shown. (E) Top 10 hits from the Enrichr motif analysis of a gene set preferentially up-regulated in TITRs compared with normal colonic Tregs (335 up-regulated genes). The overlap indicates the number of transcripts from our signature/number of transcripts known to be associated with a given transcription factor. (F) Mouse TITR signature highlighted in red on human tumor/normal colonic Treg data. (G) Human TITR transcripts highlighted in red on mouse tumor/tissue Treg comparison (per Fig. 2E).
From these data, we selected a set of 408 genes (355 UP, 73 DN; Dataset S2) that distinguish TITRs from normal-tissue Tregs based on a combination of criteria (mean overall fold change, or high fold change in at least two patients; Methods). Importantly, these differences in gene expression were highly reproducible between individuals (Fig. 3C), irrespective of the stage of CRC examined (namely stage I, IIA, or IIIB), a pattern that was not necessarily expected given the heterogeneity of human genetics and the various tumor stages represented in our samples. To further test the generality of our TITR signature, we compared the overlap with recently published Treg datasets from non–small-cell lung cancer (NSCLC) (28), breast cancer (27), and hepatocellular carcinoma (37). Indeed, there was marked overlap, as illustrated for breast cancer TITRs in Fig. 3D. Thus, the TITR signature is indeed a general one shared among multiple cancer types.
We then investigated the composition of these UP and DN signatures. Gene Ontology analysis was not informative, yielding mainly generic categories. As for the corresponding selection for mouse TITRs, a number of transcripts encoding proteins implicated in aTreg differentiation, such as several members of the TNFR superfamily (GITR, 4.1BB, OX40, and CD30) and chemokine receptors (CCR1, CCR5, and CCR8), were present. Accordingly, a number of these transcripts were associated with the cell cycle (Dataset S2), and Enrichr motif analysis revealed FOXM1 as a transcription factor likely controlling a portion of these TITR-specific transcripts, particularly those associated with the cell cycle (Fig. 3E). The set also included a cluster typical of Th17 cells (IL17A, IL17F, IL22), in keeping with previous results showing RORg-dependent expression of these cytokines in human colon tumors (42).
Finally, we asked whether the genes overexpressed in TITRs within mouse tumors were also differentially represented in human TITRs. We found significant overlap of the tumor-Treg signature between the mouse and human datasets (Fig. 3F); 77 of 90 transcripts that were up-regulated by more than fourfold in TITR vs. splenic Tregs in the mouse were also overrepresented in human TITRs relative to normal colonic Tregs (χ2 P < 10−10). Conversely, highlighting the human TITR up-regulated signature in the mouse tumor/tissue comparison (Fig. 2E) showed a strong bias toward overexpression in tumors, mainly tumor-preferential (Fig. 3G). These results clearly demonstrate conservation of the tumor Treg signature across species and validate the relevance of the mouse results.
Genes Correlated with Tregs in TCGA Tumor Datasets.
In the third arm of this work, the analytical logic was different. FoxP3 is the key transcription factor that defines Tregs and determines much of their transcriptional identity (1). Thus, genes that are specifically expressed in TITRs relative to other tumor-infiltrating immunocytes should exhibit a tight correlation with FOXP3 transcripts across panels of gene expression profiles generated from whole tumors. To this end, we made use of TCGA, a publicly available database of gene expression in 33 types of cancer from more than 11,000 patients. We selected the RNA-sequencing data for four types of cancer, representing different frequencies of mutational load and thus varying likelihoods of antitumor immune response (43): colon, breast, pancreas, and lung, with 285, 1093, 178, and 501 tumor samples, respectively. Our first attempts at correlation with FOXP3 transcripts in these datasets identified numerous genes, many of which were typical of non-Tregs (e.g., Ig transcripts from B cells). This observation indicated that the proportion of Tregs in the tumor samples was parallel to the overall degree of infiltration by immune cells in general, which is known to vary quite widely between individual tumors (9).
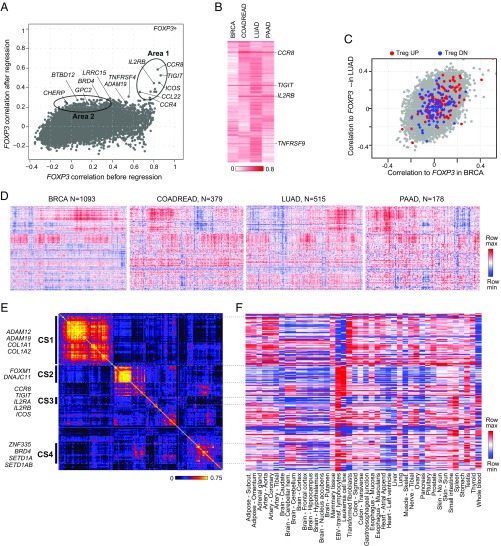
To account for and computationally remove this confounder, we mathematically regressed out the general degree of infiltration, in an approach algorithmically similar to techniques used to estimate the abundance of tumor-infiltrating cells (44, 45). We first curated prototypical signature gene sets corresponding to the major types of immunocytes that can be found in a tumor: B cells, dendritic cells, eosinophils, mast cells, macrophages, neutrophils, NK cells, and T cells (Dataset S3). For each sample, we calculated an average infiltrate index for each cell type, k, using these marker genes, and then used the cell type indices as covariates in a linear model for each gene, i: Yi = β0 + Σβk × cell_indexk + ε). We used the residual of this fit as a measure of gene expression independent of the degree of infiltration into each tumor and then correlated each gene’s residual expression with FOXP3 expression levels, using bootstrap permutation of samples or of signature gene sets to demonstrate the robustness of the procedure and compute 95% confidence intervals (SI Appendix, Fig. S3 and Dataset S4). Fig. 4A displays genome-wide correlations with FOXP3 expression before and after removal of the infiltrate component, averaged across the four tumor types (individual tumor plots in SI Appendix, Fig. S4A). In keeping with our hypothesis, the correlation with FOXP3 expression essentially disappeared for most of the transcripts, although for a minority of genes, the correlation with FOXP3 remained high, with validity supported by disappearance in randomly permuted datasets (SI Appendix, Fig. S4B). These transcripts included some that encode well-known Treg-specific proteins, such as IL2RA or those encoding the costimulatory molecules CTLA4 and ICOS. These transcripts had high initial correlation, which largely persisted in the regressed data (area 1 in Fig. 4A). Another group of transcripts showed low initial correlation with FOXP3 expression, which increased after the correction (area 2). Importantly, these correlation patterns were similar in different types of tumors (Fig. 4B and SI Appendix, Fig. S4), again indicating that the TITR-specific transcriptome was largely shared between human tumor types. In addition, these FOXP3-correlated transcripts were not simply a rediscovery of the classic Treg signature (46); only a limited fraction of the Treg-up signature was positively correlated with FOXP3 expression, as illustrated for the breast and lung tumor datasets in Fig. 4C.
Fig. 4.
Correlation with FOXP3 in TCGA. (A) Correlation with FOXP3 before and after removal of the immune cell component. Genes with stronger correlation to FOXP3 after immune filtrate removal are highlighted. (B) Correlation with FOXP3 after immune infiltrate removal across four TCGA cancer datasets. (C) Canonical up-regulated and down-regulated Treg signature highlighted on postregression correlation with FOXP3 on LUAD vs. BRCA datasets. (D) Variability of the 219 transcripts with the strongest postregression correlation in the four TCGA datasets. (E) Coexpression matrix of 219 transcripts averaged across four tumors. Four gene sets are highlighted. (F) Tissue expression distribution of 219 transcripts in GTEx.
To assess the uniformity of expression of FOXP3-correlated transcripts across individual tumors, we selected 219 transcripts with the greatest postregression correlation and investigated their distribution in the four tumor datasets (Fig. 4D and Dataset S5). Their levels were clearly not uniform across tumors. Some gene modules varied in lockstep between individual tumors in patterns that carried across tumor types, underscoring the robustness of these relationships and hinting at a common and fundamental mechanism of interaction among tumors and the immune system.
In principle, the correlation between these 219 transcripts and FOXP3 transcripts could have three roots: (i) expression in TITRs themselves (the classic Treg-associated transcripts of area 1 clearly belong to this class); (ii) expression connected to mechanisms that draw or retain Tregs into tumors and control their abundance; or (iii) expression resulting from Treg presence and activity. To best resolve the coregulated modules observed in Fig. 4D, we computed a gene-gene correlation matrix from the expression of these 219 genes in each tumor (averaged heatmap in Fig. 4E; individual tumors in SI Appendix, Fig. S5). Several strong modules emerged that were almost identical in all four tumor types, as could be predicted from Fig. 4D. These modules are of very different composition, as evidenced by their expression levels in the tissue datasets from the GTEx database (47) (Fig. 4F). Correlated set (CS) 1 is composed of transcripts primarily encoding collagen or matrix metalloprotease of the ADAM family, which are not expressed in Treg cells or lymphoid cells but are expressed in fibroblasts and connective tissues. CS2 corresponds to a subset of cell-cycle-related transcripts (high in leukemic cells and testis). The previously observed TITR transcripts (e.g., IL2RA, CCR8, TIGIT, CD80, ICOS) are grouped in a more loosely correlated region (CS3). Transcripts of CS4 were less distinctive but included several histone/protein modifiers (e.g., HCFC1, BRD4, SETD1A/B). Thus, correlation with FOXP3 expression brought forth, in addition to TITR-specific transcripts, distinct correlated gene modules whose expression is segregated in many tumors and that may be causally related to Treg presence or activity.
Data Integration.
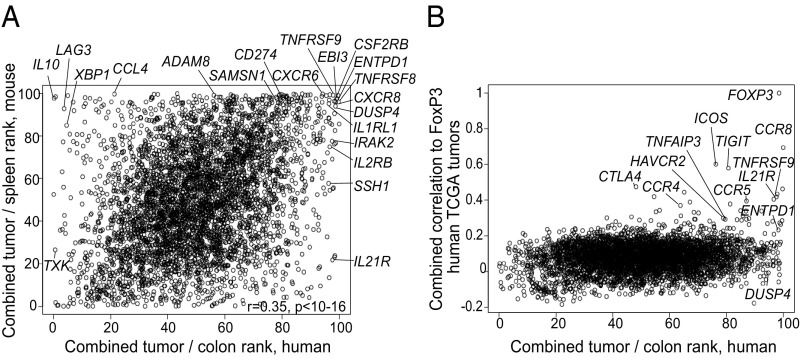
We next integrated these three inputs to compare their outcomes and to select the set of transcripts most specific for TITRs across different tumors. Since the actual data and their distribution differed, we opted to combine the rankings of orthologous genes in the two species rather than the expression metrics. For the human TITR datasets, each gene was ranked according to its expression in TITRs of each tumor sample relative to batch-matched Tregs from normal colon tissue, and these ranks were summed. For the mouse TITR datasets, genes were similarly ranked by their overexpression in TITRs within the three tumor types relative to matched splenic Tregs, and these ranks were also summed. Fig. 5A compares these overall scores for human and mouse TITRs. Consistent with the transspecies conservation of TITR-specific gene expression described above, there was a strong correlation between these rankings (r = 0.34, P < 10−16). A group of transcripts was atop the ranking for tumor specificity in both species, and the gene list selected on the basis of mouse tumor data (Dataset S1) included many transcripts of high rank in the human datasets (SI Appendix, Fig. S6A). Some genes led the ranking for overexpression in human TITRs but not in mice (and vice versa), suggesting that at least some were species-specific (or might be tied to “natural” vs. transplanted tumors). Genes that correlated with FOXP3 in the TCGA data were also high in the rankings for overexpression in human and mouse TITRs (SI Appendix, Fig. S6B, and shown more directly in Fig. 5B). Many of the highest-ranked Treg transcripts also showed the strongest association with FOXP3 transcripts in the TCGA datasets (e.g., CCR8, TNFRSF9, IL21R), although this was not always the case (e.g., DUSP4). From these three orthogonal inputs, also integrating TITR:tissue Treg and TITR:CD8 differential data, we assembled a list of 108 genes most differentially expressed and most closely correlated with tumor Tregs (SI Appendix, Fig. S6C and Dataset S6).
Fig. 5.
Combinatorial data integration. (A) Comparison of overall scores for human (x-axis) and mouse (y-axis) TITRs. Highlighted/annotated are genes either at the top of the ranking in both species, with high scores in the mouse and scores in the top 10% of differential transcripts in human, or highest in the human ranking but not in the mouse ranking. (B) Depiction of overall score for human TITRs (x-axis) vs. average correlation with Foxp3 score derived from the whole-tumor TCGA datasets (y-axis).
Experimental Validation of TITR-Specific Targets.
It was of importance to validate the relevance of these gene sets to TITR physiology, both to understand how they might mechanistically modulate TITR activity and to serve as potential therapeutic targets. We examined the importance of C3AR1, IL12RB2, and IL1RL1 (ST2) using available strains of conventional knockout (KO) mice. MC38 tumors were induced in KO mice and their wild-type (WT) littermates, and tumor growth was measured over time (SI Appendix, Fig. S7). There was no significant difference in tumor growth between the KO and WT littermates in the genes examined, except for the Treg-specific IL1RL1 (ST2) KO group.
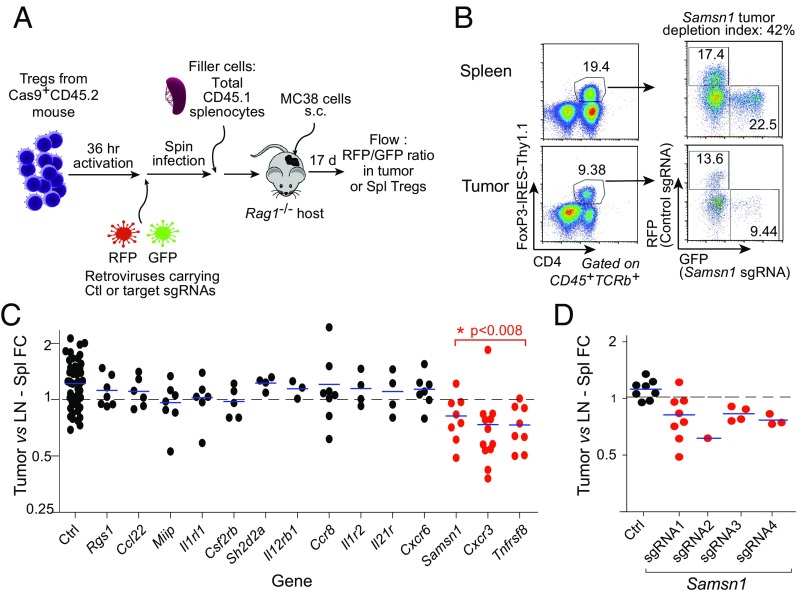
Simultaneously, we performed a systematic in vivo screen of the top-ranked TITR-specific targets by creating loss-of-function (LOF) mutations in the protein-coding regions by adapting the CRISPR/Cas9 system (48) to specifically edit Treg cells and assessing differential representation in the tumor relative to lymphoid organs (Fig. 6A and Dataset S7). Tregs were purified from transgenic mice expressing both the Cas9 protein (48) and the FoxP3-Thy1.1 reporter (49), activated in vitro, and transduced with retroviral vectors encoding the targeting single guide RNAs (sgRNAs) of interest. Pairs of retroviral vectors expressing two different fluorescent proteins (GFP and RFP) were used, which allowed us to label and compare Tregs carrying targeting or control sgRNAs in the same mouse (and thereby increase the informativity of each mouse). Transduced Tregs were then transferred together with “filler” splenocytes (to avoid homeostatically driven expansion) into alymphoid RAG-deficient mice, which concomitantly received MC38 tumor cells. After an interval of 17 d to allow for tumor growth and Treg homing, we analyzed the distributions of Treg cells carrying each of the targeting or control sgRNAs in the tumor relative to lymphoid organs, calculated as a “tumor depletion index” (Fig. 6 B and C). Through DNA amplification and sequencing, we also ensured that the target genes were efficiently edited (estimated as >50% in all cases). The compiled results show that several of the LOF mutations had no impact on TITR proportions, but three of them had an impact: Tnfrsf8 (encodes CD30), Cxcr3 (a chemokine receptor), and Samsn1 (an intracellular signaling adapter). This first Samsn1 LOF result was validated with four independent sgRNAs targeting different regions of Samsn1. Each sgRNA decreased Treg accumulation in the tumors (Fig. 6D), confirming the effect.
Fig. 6.
CRISPR-based Treg KOs. (A) Schematic depiction of protocol to induce LOF mutations in TITR target genes, specifically in Treg cells, using the CRISPR/Cas9 system. (B) Exemplar gating used for determining the tumor depletion index, the ratio of the percentage of GFP or RFP+ Tregs in tumor vs. spleen. (C) Summary of CRISPR-based Treg KO data. The tumor depletion index was significant in the genes highlighted in red. (D) Validation of Samsn1 LOF on Treg accumulation in tumors. Four different sgRNAs showed decreased accumulation of Samsn1 LOF Tregs in the tumors.
Overall, these proof-of-concept results indicate that some of the TITR transcripts highlighted in Dataset S6 encode functionally relevant proteins and might serve as valuable targets for cancer immunotherapy.
Discussion
Our goal was to identify genes differentially expressed in TITRs from various origins relative to generic Tregs from secondary lymphoid organs or Tregs from nonlymphoid tissues, as a way to better understand the unique biology of Tregs in tumors and to identify potential targets for immunotherapy. A similar motivation has driven previous studies, but we adopted a broader, multipronged strategy that spanned species, harnessed the large TCGA datasets, and provided proof-of-concept experimental validation. Our complementary approaches also rested on different principles, namely differential expression between Tregs in tumors and nonlymphoid tissues and transcript correlations with FOXP3 expression. This combined strategy allowed us to identify a TITR-specific signature of unexpected constancy. There was a very strong overlap between the transcriptomes of Treg cells infiltrating tumors and nonlymphoid tissues, which perhaps is not surprising, given that some of the signals that permit Tregs to accumulate in tissues might also allow their residence in tumors. These commonalities did present a challenge for defining TITR-specific transcripts, but we were ultimately able to distill a subset of transcripts with marked tumor preference. The power of our strategy was validated by the “rediscovery” of several established targets of tumor immunotherapy, including CTLA4 and TIGIT, with PD-L1 and LAG3 barely missing our thresholds. This validation provides credibility for considering the lesser-known genes in our TITR signature as potential targets for preferentially modulating TITRs.
A striking convergence of TITR signatures was apparent between tumor types in mouse models, between mouse and human TITRs, and between different patients with the same tumor. It also extended to the large sets of tumors in TCGA datasets, for which careful computational parsing uncovered the same core signature of FOXP3-correlated genes in every tumor type tested. In addition, there was a strong overlap with differentially expressed transcripts recently reported in more focused studies of Tregs from human lung, colorectal (28), breast (27), and liver (37) tumors. This convergence implies that a fundamental mechanism must be in play to allow such diverse tumors to elicit the same response. From a practical standpoint, this means that the same pathways might be effectively targeted by immunotherapy to relieve the dampening of antitumor responses by TITRs in many tumor types.
While the bulk of the TITR signature was indeed shared by different cancers, we were able to identify a handful of TITR transcripts specific to CRC when we compared our data with previously reported profiles from breast cancer Tregs (Fig. 3D). These CRC TITR-specific genes included loci encoding cytokines typical of Th17 cells (IL17A, IL17F, and IL22) and RORC, the major driver of Th17 cell differentiation. This observation is consistent with a previous report of RORγ-dependent expression of Th17 cytokines in Tregs from human colon tumors (42), as well as with the microbiome-dependent RORγ+ Tregs that are found electively in the colon (33, 50). Thus, beyond a core TITR signature shared in all tumors, individual locations can imprint an additional local component on TITRs. Similarly, some transcripts were seen to be differential in only one species, such as Il21R, which had a strong differential score in human TITRs but not in mouse TITRs.
Finally, this multipronged exploration, and particularly the TCGA arm, revealed sets of transcript modules that correlated with TITR levels but were not TITR transcripts themselves. It will be interesting to explore the leads provided here, for instance, by assessing how ADAM and collagen gene expression might help Tregs accumulate in the tumors.
Beyond providing a detailed landscape of the strikingly constant phenotype adopted by TITRs in the tumor environment, these explorations have yielded sets of transcripts encoding proteins that might be effective targets for immunotherapy through relieving the brakes that Treg cells impose on antitumor responses. There could be several reasons for the overexpression of specific molecules in TITRs that conditions the actual therapeutic strategy. Some might be overexpressed because they are required by TITRs to survive specifically in this environment; for instance, chemokine receptors are needed for TITRs to accumulate in tumors, and blocking them or reducing their expression could exclude TITRs from tumors. In contrast, other molecules might instead denote negative feedback loops that exist in all biological systems and instead act to limit TITR overexpansion; their elimination actually might benefit TITRs. Still others might be up-regulated in response to stimuli in the tumor microenvironment but play no role in Treg survival or function therein.
Given this diversity, we used parallel genetic approaches to test the significance of the genes revealed by these parallel genomic comparisons. A few could be tested in KO mice, but we leveraged editing by CRISPR/Cas9 to systematically assess how their inactivation affected Treg accumulation in the tumor. This approach was particularly valuable for intracellular targets such as Samsn1, which cannot be easily probed by mAb infusion. In this way, 3 of the 14 targets tested showed a significant effect of editing.
A limitation of the genetic approach with regard to potential therapeutic application is that it is blind to the effects of blocking the encoded protein or of killing TITRs by complement or ADCC mechanisms, which could be revealed by mAb treatment and may have therapeutic value. For example, editing Ccr8 in Tregs did not lower their accumulation in tumors, but our preliminary explorations showed that mAb engagement of CCR8 caused a tumor mass reduction in treated mice. Thus, CCR8 may not be uniquely indispensable for TITR recruitment or survival in tumors (redundancy is not uncommon in chemokine networks), but it might serve as a worthwhile mAb target. Moreover, the other targets for which LOF mutations had no impact on TITR proportions should not necessarily be excluded from exploration as therapeutic targets by other means of perturbation.
In conclusion, this multipronged study has provided numerous perspectives and leads regarding the genomic aspects of Tregs in tumors, pointing to specific transcriptional programs that could be harnessed to lift the immune inhibition by Tregs in multiple cancer contexts.
Methods
Treg cells were purified for RNA sequencing profiling from (i) subcutaneous tumors (CT26, MC38, or B16.F10) induced and subsequently harvested from 6- to 8-wk-old male Foxp3-GFP reporter mice; (ii) matching spleen from the same mice; (iii) surgical resection samples of human CRCs of stage I to IIIB); and (iv) normal colon tissue (matched from the same donor in four cases), collected and analyzed under Institutional Review Board-approved protocols DFCI 12–020, KNUMC 2015–11-005 and HMS CR15-0504–03. Informed consent was obtained after the nature and possible consequences of the study were explained. Computational deconvolution was used to extract Treg-associated transcripts from large expression datasets from four different cancer types (colon, breast, lung, pancreas) obtained from TCGA. CRISPR-based genome editing was performed in mature Tregs from Cas9-expressing transgenic mice, which were subsequently reintroduced into hosts concomitantly challenged with MC38 tumors. More details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. A. Sharpe, M. Pittet, and V. Kuchroo for insightful discussions or materials; K. Hattori, C. Araneo, A. Rhoads, A. Cook, and L. Yang for help with mice, cell sorting, and profiling; and the Broad Technology Labs for library construction and sequencing. This work was funded by the Immune Tolerance Fund at Harvard Medical School and the Blavatnik Biomedical Accelerator at Harvard University. A.M.M. was supported by NIH Grant T32CA079443, and E.K. was supported by a PhD Fellowship from Boehringer Ingelheim Fonds.
Footnotes
Conflict of interest statement: C.B. co-authored a consortium position paper with Miriam Merad in 2017 that stems from the Human Cell Atlas, a large consortium to which they both belong; they did not collaborate directly on the paper. C.B. is a principal investigator in a bridge funding for a consortium (Immune Cell Atlas), of which Dr. Merad is the Program Director; Dr. Merad’s review of this paper was concluded before this award.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE116347). The plasmids have been deposited on Addgene (IDs 112914 and 116926).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810580115/-/DCSupplemental.
References
- 1.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: Mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benoist C, Mathis D. In: Immune Tolerance. Mathis D, Rudensky A, editors. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2013. pp. 31–44. [Google Scholar]
- 3.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 4.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 5.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 6.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feuerer M, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panduro M, Benoist C, Mathis D. Tissue Tregs. Annu Rev Immunol. 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 10.Roychoudhuri R, Eil RL, Restifo NP. The interplay of effector and regulatory T cells in cancer. Curr Opin Immunol. 2015;33:101–111. doi: 10.1016/j.coi.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 11.deLeeuw RJ, Kost SE, Kakal JA, Nelson BH. The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: A critical review of the literature. Clin Cancer Res. 2012;18:3022–3029. doi: 10.1158/1078-0432.CCR-11-3216. [DOI] [PubMed] [Google Scholar]
- 12.Shang B, Liu Y, Jiang SJ, Liu Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: A systematic review and meta-analysis. Sci Rep. 2015;5:15179. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu J, Yamazaki S, Sakaguchi S. Induction of tumor immunity by removing CD25+CD4+ T cells: A common basis between tumor immunity and autoimmunity. J Immunol. 1999;163:5211–5218. [PubMed] [Google Scholar]
- 14.Quezada SA, Peggs KS, Curran MA, Allison JP. CTLA4 blockade and GM-CSF combination immunotherapy alters the intratumor balance of effector and regulatory T cells. J Clin Invest. 2006;116:1935–1945. doi: 10.1172/JCI27745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liakou CI, et al. CTLA-4 blockade increases IFNgamma-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc Natl Acad Sci USA. 2008;105:14987–14992. doi: 10.1073/pnas.0806075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klages K, et al. Selective depletion of Foxp3+ regulatory T cells improves effective therapeutic vaccination against established melanoma. Cancer Res. 2010;70:7788–7799. doi: 10.1158/0008-5472.CAN-10-1736. [DOI] [PubMed] [Google Scholar]
- 17.Bos PD, Plitas G, Rudra D, Lee SY, Rudensky AY. Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. J Exp Med. 2013;210:2435–2466. doi: 10.1084/jem.20130762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi NS, et al. Regulatory T cells in tumor-associated tertiary lymphoid structures suppress anti-tumor T cell responses. Immunity. 2015;43:579–590. doi: 10.1016/j.immuni.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grinberg-Bleyer Y, et al. NF-κB c-Rel is crucial for the regulatory T cell immune checkpoint in cancer. Cell. 2017;170:1096–1108.e13. doi: 10.1016/j.cell.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simpson TR, et al. Fc-dependent depletion of tumor-infiltrating regulatory T cells co-defines the efficacy of anti-CTLA-4 therapy against melanoma. J Exp Med. 2013;210:1695–1710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peggs KS, Quezada SA, Chambers CA, Korman AJ, Allison JP. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti-CTLA-4 antibodies. J Exp Med. 2009;206:1717–1725. doi: 10.1084/jem.20082492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, et al. PD1 blockade reverses the suppression of melanoma antigen-specific CTL by CD4+ CD25(Hi) regulatory T cells. Int Immunol. 2009;21:1065–1077. doi: 10.1093/intimm/dxp072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Overacre-Delgoffe AE, et al. Interferon-γ drives Treg fragility to promote anti-tumor immunity. Cell. 2017;169:1130–1141.e11. doi: 10.1016/j.cell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darrasse-Jèze G, et al. Tumor emergence is sensed by self-specific CD44hi memory Tregs that create a dominant tolerogenic environment for tumors in mice. J Clin Invest. 2009;119:2648–2662. doi: 10.1172/JCI36628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito T, et al. Two FOXP3+CD4+ T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. 2016;22:679–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 26.Luo CT, Liao W, Dadi S, Toure A, Li MO. Graded Foxo1 activity in Treg cells differentiates tumour immunity from spontaneous autoimmunity. Nature. 2016;529:532–536. doi: 10.1038/nature16486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plitas G, et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Simone M, et al. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity. 2016;45:1135–1147. doi: 10.1016/j.immuni.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michot JM, et al. Immune-related adverse events with immune checkpoint blockade: A comprehensive review. Eur J Cancer. 2016;54:139–148. doi: 10.1016/j.ejca.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 30.Postow MA, Sidlow R, Hellmann MD. Immune-related adverse events associated with immune checkpoint blockade. N Engl J Med. 2018;378:158–168. doi: 10.1056/NEJMra1703481. [DOI] [PubMed] [Google Scholar]
- 31.Grossman RL, et al. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin W, et al. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 33.Sefik E, et al. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu J, et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology. 2007;132:2328–2339. doi: 10.1053/j.gastro.2007.03.102. [DOI] [PubMed] [Google Scholar]
- 35.Salama P, et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009;27:186–192. doi: 10.1200/JCO.2008.18.7229. [DOI] [PubMed] [Google Scholar]
- 36.Tang Y, et al. An increased abundance of tumor-infiltrating regulatory T cells is correlated with the progression and prognosis of pancreatic ductal adenocarcinoma. PLoS One. 2014;9:e91551. doi: 10.1371/journal.pone.0091551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng C, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 38.Murai M, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider MA, Meingassner JG, Lipp M, Moore HD, Rot A. CCR7 is required for the in vivo function of CD4+CD25+ regulatory T cells. J Exp Med. 2007;204:735–745. doi: 10.1084/jem.20061405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung Y, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zemmour D, et al. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat Immunol. 2018;19:291–301. doi: 10.1038/s41590-018-0051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blatner NR, et al. Expression of RORγt marks a pathogenic regulatory T cell subset in human colon cancer. Sci Transl Med. 2012;4:164ra159. doi: 10.1126/scitranslmed.3004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 44.Li B, et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aran D, Hu Z, Butte AJ. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferraro A, et al. Interindividual variation in human T regulatory cells. Proc Natl Acad Sci USA. 2014;111:E1111–E1120. doi: 10.1073/pnas.1401343111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melé M, et al. GTEx Consortium Human genomics: The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liston A, et al. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc Natl Acad Sci USA. 2008;105:11903–11908. doi: 10.1073/pnas.0801506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohnmacht C, et al. The microbiota regulates type 2 immunity through RORγ+ T cells. Science. 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.