Significance
BRCA1 plays a key role in homology-directed repair (HDR) in S/G2-phase cells. It remains unclear why BRCA1 mutation carriers develop cancer predominantly in breast and ovarian tissues. We revealed that a physiological concentration (10 nM) of estrogens efficiently induce TOP2β-dependent DSBs in the absence of BRCA1 in breast cancer cells arrested in G1 phase. This genotoxicity was confirmed also in G0/G1-phase epithelial cells of mouse mammary glands. These findings indicated that BRCA1 contributes to DSB repair independent of HDR. Our data suggested that BRCA1 promotes the removal of TOP2 adducts from DSBs by the nucleolytic activity of MRE11 for subsequent DSB repair by nonhomologous end-joining. This function of BRCA1 may help explain the female-organ-specific carcinogenesis of BRCA1-mutation carriers.
Keywords: BRCA1, estradiol (E2), topoisomerase II, HBOC syndrome, breast cancer
Abstract
Women having BRCA1 germ-line mutations develop cancer in breast and ovary, estrogen-regulated tissues, with high penetrance. Binding of estrogens to the estrogen receptor (ER) transiently induces DNA double-strand breaks (DSBs) by topoisomerase II (TOP2) and controls gene transcription. TOP2 resolves catenated DNA by transiently generating DSBs, TOP2-cleavage complexes (TOP2ccs), where TOP2 covalently binds to 5′ ends of DSBs. TOP2 frequently fails to complete its catalysis, leading to formation of pathological TOP2ccs. We have previously shown that the endonucleolytic activity of MRE11 plays a key role in removing 5′ TOP2 adducts in G1 phase. We show here that BRCA1 promotes MRE11-mediated removal of TOP2 adducts in G1 phase. We disrupted the BRCA1 gene in 53BP1-deficient ER-positive breast cancer and B cells. The loss of BRCA1 caused marked increases of pathological TOP2ccs in G1 phase following exposure to etoposide, which generates pathological TOP2ccs. We conclude that BRCA1 promotes the removal of TOP2 adducts from DSB ends for subsequent nonhomologous end joining. BRCA1-deficient cells showed a decrease in etoposide-induced MRE11 foci in G1 phase, suggesting that BRCA1 repairs pathological TOP2ccs by promoting the recruitment of MRE11 to TOP2cc sites. BRCA1 depletion also leads to the increase of unrepaired DSBs upon estrogen treatment both in vitro in G1-arrested breast cancer cells and in vivo in epithelial cells of mouse mammary glands. BRCA1 thus plays a critical role in removing pathological TOP2ccs induced by estrogens as well as etoposide. We propose that BRCA1 suppresses tumorigenesis by removing estrogen-induced pathological TOP2ccs throughout the cell cycle.
Mutations in the BRCA1 gene predispose carriers to a high incidence of breast and ovarian cancer. BRCA1 plays a critical role in homology-directed repair (HDR) of DNA double-strand breaks (DSBs) (1). The HDR pathway is essential for the repair of spontaneously arising DSBs that occur during DNA replication, and prevents the accumulation of mitotic chromosome breaks (2–4). Since HDR plays an essential function in all cycling cells, a major unresolved question in BRCA biology is, why does the phenotype of a defective BRCA1 manifest in such a highly tissue-restricted manner?
DSBs are repaired by two major repair pathways: HDR and nonhomologous end joining (NHEJ) (reviewed in ref. 5). BRCA1 and Rad51 are involved in HDR, while 53BP1, the catalytic subunit of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), Ku70/80, and ligase IV are all involved in NHEJ. HDR is active only in S and G2 phases, while NHEJ is active throughout the cell cycle. The choice of HDR or NHEJ depends on DSB resection, as the formation of 3′ single-strand overhangs at DSB sites by the nucleases CtIP and MRE11 initiates HDR while inhibiting NHEJ (6). The functional interaction between BRCA1 and 53BP1 plays a critical role in this choice in such a manner that BRCA1 facilitates DSB resection while 53BP1 suppresses it, promoting NHEJ (7). This functional interaction is validated by data demonstrating that a defect in BRCA1 in mice causes embryonic lethality although mice deficient in both BRCA1 and 53BP1 are viable (8), showing a rescue of the HDR defect in BRCA1 mutant cells (9). These viable mice manifest constitutively high levels of genomic instability, but why this is the case remains elusive.
Breast and ovary tissues rely on estrogens for their proliferation. Estrogens stimulate cell proliferation through the activated estrogen receptor alpha (ERα), which serves as a transcription factor. Activated ERα recruits topoisomerase II (TOP2)α and TOP2β to some of the ERα target genes, and triggers the initiation of their transcription (reviewed in ref. 10). In addition to the transcriptional initiation, catalyses by TOP2 play a critical role in transcriptional elongation (11), DNA replication, and decatenation of entangled, newly replicated sister chromatids before the separation of mitotic chromosomes (11, 12) (reviewed in ref. 10). TOP2β has been shown to play a role in transcriptional control by steroid hormones, including both androgen and estrogen hormones (13–16).
The TOP2 enzymes resolve DNA catenanes by catalyzing the transient formation of DSBs, which is followed by enzymatic religation of the broken strands. Transient DSB formation allows an intact DNA duplex to pass through the DSB. During such transient DSB formation, TOP2 becomes covalently bound to the 5′ DNA end of the break, forming TOP2–DNA cleavage-complex intermediates (TOP2ccs) (10). Abortive catalysis, a consequence of failing to complete the religation step, causes the formation of pathological stable TOP2ccs. Abortive catalysis has been demonstrated to occur very frequently during physiological cell cycling (17). The exposure to the male hormone dihydrotestosterone causes persistent DSBs in cells, suggesting that pathological TOP2ccs can be induced by the sex hormone (18).
A number of enzymes contribute to the repair of pathological TOP2ccs. The function of such enzymes can be evaluated by measuring cellular sensitivity to etoposide (VP-16), a TOP2 poison, which strongly stabilizes TOP2ccs and causes genome instability (19). When TOP2 fails to religate TOP2ccs, the resulting 5′ adducts, intact TOP2 and its degradation products, need to be removed before DSB repair by NHEJ (10, 18, 20, 21). Pathological TOP2ccs are removed by tyrosyl-DNA phosphodiesterase 2 (TDP2) (22) as well as by endonucleases such as CtIP and MRE11 in yeast and vertebrate cells (23–26). A genetic study of chicken DT40 cells and biochemical studies with Xenopus egg extracts suggest that the physical interaction between CtIP and BRCA1 contributes to the repair of pathological TOP2ccs (24, 27). These observations indicate that DSB resection by BRCA1, CtIP, and MRE11 in HDR generates 3′ single-strand overhangs and thereby removes 5′ single-strand sequences including 5′ adducts. However, it remains unclear whether BRCA1 removes pathological TOP2ccs in G0/G1 phases, when HDR-mediated DSB repair does not work. We recently demonstrated that the nuclease activity of MRE11 is required to prevent the endogenous accumulation of pathological TOP2ccs in the brains of embryonic mice and tissue-culture cells, including G1-phase cells (17). Thus, MRE11 is able to remove TOP2ccs before DSB repair by NHEJ, independent of its function in HDR.
We here report that BRCA1 promotes the removal of 5′ TOP2 adducts from pathological stable TOP2ccs for subsequent NHEJ in G1 phase. BRCA1 is required for efficient recruitment of MRE11 to TOP2cc sites. Remarkably, BRCA1 depletion leads to the marked accumulation of pathological TOP2βccs over time upon a pulse estrogen treatment in G1-arrested breast cells. This study uncovered the strong genotoxicity associated with a physiological concentration (10 nM) of estradiol-17β (E2), whose genotoxicity depends on both activated ERα and TOP2β. BRCA1 depletion also caused the accumulation of E2-induced γH2AX foci in the epithelial cells of mammary ducts as well as in mice deficient in NHEJ, but not in wild-type mice. BRCA1 thus plays a critical role in removing pathological TOP2ccs. We propose that BRCA1 suppresses tumorigenesis by removing estrogen-induced pathological TOP2ccs throughout the cell cycle.
Results
Recruitment of BRCA1 to E2-Induced DNA-Damage Sites in MCF-7 Cells in G1 Phase.
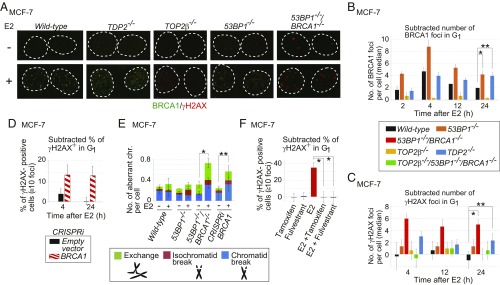
We explored whether TDP2 contributed to the repair of E2-induced DSBs in the MCF-7 human breast adenocarcinoma cell line. We created TDP2−/− MCF-7 cells (SI Appendix, Fig. S1 A and B). SI Appendix, Table S1 provides a list of the gene-disrupted clones analyzed in this study. To exclude the effects of DNA replication and HDR-mediated repair, we enriched G1-phase cells by serum starvation for 24 h (28), added E2, and examined γH2AX foci only in cyclin-A–negative, G1-phase cells (SI Appendix, Fig. S2). Remarkably, the loss of TDP2 caused an ∼70% increase in the number of E2-induced γH2AX foci in G1 phase (Fig. 1A), suggesting the induction of pathological TOP2ccs by E2.
Fig. 1.
E2-induced genomic instability in the BRCA1-deficient MCF-7 cell. (A) BRCA1 and γH2AX foci in serum-starved MCF-7 cells treated with E2 (10 nM) [E2(+)] or DMSO [E2(−)] for 2 h followed by incubation in E2-free medium for 2 h. Dotted lines outline each nucleus. (B and C) The median of the numbers of E2-induced BRCA1 (B) and γH2AX (C) foci in individual MCF-7 cells after 2-h pulse E2 treatment. We subtracted the median number of foci in E2-untreated cells from the median number of foci in E2-treated cells. Actual numbers of foci per cell are shown in SI Appendix, Fig. S3 B and C. Genotypes of the analyzed cells are shown between the graphs. The error bars show SD of three independent experiments. The single and double asterisks in B indicate P < 0.03 and P < 0.02, respectively. The single and double asterisks in C indicate P < 0.04 and P < 0.03, respectively. The P values were calculated by Student’s t test. Representative images of γH2AX foci are shown in SI Appendix, Fig. S3D. (D) Percentage of γH2AX focus-positive cells in CRISPRi-mediated BRCA1-depleted cells. BRCA1-guide RNA (gRNA) vector or empty vector was transiently transfected into MCF-7 cells expressing dCas9-KRAB before E2 treatment (SI Appendix, Materials and Methods and Fig. S4D). A pulse treatment with E2 was done as in B. Focus-positive cells are defined as cells that show at least 10 foci per cell. We subtracted the percentage of untreated focus-positive cells from the percentage of treated focus-positive cells. (E) E2-induced chromosome aberrations in mitotic chromosome spreads. Following 24-h incubation with media containing charcoal-filtered serum, cells were further incubated in media containing charcoal-filtered serum in the absence (−) or presence (+) of E2 (10 nM) for 36 h and then immediately subjected to chromosome analysis. The y axis shows the number of mitotic chromosome aberrations: exchange, isochromatid break (both sister chromatids broken at the same site), and chromatid break (one of two sister chromatids broken). Error bars were plotted for SD from three independent experiments. At least 50 mitotic chromosome spreads were analyzed for each experiment. The single and double asterisks indicate P < 0.02 and P < 0.03, respectively, calculated by Student’s t test. (F) The inhibitory effect of fulvestrant (100 nM) and tamoxifen (1 μM) on E2-induced γH2AX-focus formation in 53BP1−/−/BRCA1−/− cells. Cells were treated with the indicated reagents for 2 h, followed by incubation in drug-free media for an additional 2 h. Error bars indicate SD calculated from three independent experiments. The asterisks indicate P < 0.003, calculated by Student’s t test.
MRE11 as well as TDP2 plays a role in removing pathological TOP2ccs (10). We hypothesized that BRCA1 and MRE11 are both involved in the repair of E2-induced DNA damage in G1 phase, since these two HDR factors collaborate to perform DSB resection and may also play a role in microhomology-mediated end joining even in G1 phase (29, 30). To explore the role played by BRCA1 in E2-induced DNA lesions, we costained γH2AX and BRCA1 foci and analyzed them only in cyclin-A–negative, G1-phase cells (Fig. 1A). To examine the colocalization of the two foci, we scanned individual foci in nuclei and measured the relative intensity of the γH2AX and BRCA1 signals in each focus (SI Appendix, Fig. S3A). Of the foci showing BRCA1 or γH2AX signal, 87% showed both BRCA1 and γH2AX signals (SI Appendix, Fig. S3A, dot plot). Thus, treatment with E2 induces DSBs where BRCA1 accumulates. The recruitment of BRCA1 to DSB sites in G1 cells suggests that BRCA1 is involved in the repair of E2-induced DSBs independent of its function in HDR.
Loss of BRCA1 Causes Prolonged γH2AX-Focus Formation After Pulse Exposure to E2.
We next examined E2-induced γH2AX foci in BRCA1-deficient MCF-7 cells. We disrupted the BRCA1 gene in a 53BP1-deficient background (SI Appendix, Fig. S1 C–F), since inactivation of 53BP1 rescues embryonic lethality in BRCA1-deficient mice (8, 13). The resulting 53BP1−/−/BRCA1−/− cells were able to proliferate with nearly normal kinetics. We did serum starvation for 24 h and examined γH2AX foci only in cyclin-A–negative, G1-phase cells. We monitored γH2AX foci following a 2-h pulse exposure of cells to E2 (10 nM), the concentration equivalent to the serum concentration of pregnant women. This concentration of E2 efficiently induces γH2AX foci in S phase of wild-type MCF-7 cells (31). Remarkably, BRCA1-focus formation was higher in G1-phase 53BP1−/− and TDP2−/− cells than in wild-type cells at 24 h (2-h pulse exposure followed by a 22-h chase period) (Fig. 1B and SI Appendix, Fig. S3B). Prolonged γH2AX-focus formation was seen in 53BP1−/−/BRCA1−/− cells, CRISPRi-mediated BRCA1-depleted cells (SI Appendix, Fig. S4), as well as in TDP2−/− cells at 24 h after addition of E2 (Fig. 1 C and D and SI Appendix, Fig. S3 C and D). These observations suggest that BRCA1 promotes the repair of TOP2ccs, as does TDP2, which eliminates 5′ TOP2 adducts from DSBs (22). Another possibility is that BRCA1 might function upstream of TDP2-dependent repair of TOP2ccs. NHEJ, which is partially dependent on 53BP1 (32), is then able to ligate broken ends in G1 phase.
We investigated the involvement of TOP2 in E2-induced DSB formation by disrupting the TOP2β gene in wild-type and 53BP1−/−BRCA1−/− cells (SI Appendix, Fig. S1 G–I). Expression of TOP2α was down-regulated more than 10 times at 24 h after serum starvation (SI Appendix, Fig. S1J). E2-mediated induction of γH2AX foci was considerably lower in TOP2β−/− and TOP2β−/−/53BP1−/−/BRCA1−/− cells than in wild-type and 53BP1−/−/BRCA1−/− cells, respectively (Fig. 1C and SI Appendix, Fig. S3 C and D). These data suggest that the vast majority of the E2-induced γH2AX foci in serum-starved MCF-7 cells represent TOP2βccs.
To confirm the genotoxicity of E2, we measured chromosomal aberrations in mitotic chromosome spreads following 36 h of continuous exposure of cycling cells to E2 together with serum. The prominent induction of chromosome breaks in 53BP1−/−/BRCA1−/− cells and in BRCA1-depleted cells, but not in wild-type cells, demonstrates that E2 has a strong genotoxic potential in the absence of BRCA1 (Fig. 1E).
E2-Induced DNA Damage Is Dependent on Functional ERs.
We next investigated whether functional ERs are required for E2-induced γH2AX-focus formation in serum-starved MCF-7 cells. Clinically relevant concentrations of inhibitors against the ER, both fulvestrant and tamoxifen (33), completely repressed E2-induced γH2AX-focus formation (Fig. 1F). These observations indicate that activation of the ER by E2 is responsible for the TOP2β-dependent DNA damage.
E2-Induced DNA Damage Is Independent of Ongoing Transcription.
To test whether or not transcriptional elongation causes E2-induced γH2AX foci, we added the RNA polymerase inhibitors 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole and α-amanitin to stop general transcription 3 h before exposure of cells to E2 (SI Appendix, Fig. S5A). In this experiment, we measured γH2AX foci only in cyclin-A–negative G1 53BP1−/−/BRCA1−/− cells. Following serum starvation, the inhibitors had no detectable impact on the E2-induced γH2AX foci (SI Appendix, Fig. S5 B and C). We thus conclude that the E2-induced γH2AX foci seen in G1 phase do not result from transcriptional elongation or the resulting R-loop formation (31, 34–36). The data suggest that E2-induced γH2AX foci may represent TOP2ccs formed at transcriptional regulatory sequences such as the promoter (10, 37, 38).
Delayed Repair of E2-Induced Pathological TOP2βccs at the pS2 Promoter in BRCA1-Deficient Cells.
A previous report showed that ER-mediated transcriptional initiation requires TOP2β-mediated, site-specific DSB formation at the pS2 promoter of MCF-7 cells (13). We detected DSBs by chromatin immunoprecipitation using an α-γH2AX antibody. DSB formation was seen transiently about 10 min after addition of E2 (13). We also detected this site-specific DSB formation at 10 min after addition of E2 in serum-starved wild-type cells, but not in TOP2β−/− cells (SI Appendix, Fig. S5D, Left). We next examined DSB formation at the pS2 promoter after a 2-h exposure to E2 (10 nM) followed by a 10-h chase period (SI Appendix, Fig. S5D, Right). Unrepaired DSBs were detectable in 53BP1−/−/BRCA1−/− cells but not in wild-type cells at 12 h. We conclude that the absence of BRCA1 causes defective repair of TOP2ccs induced by E2 at the pS2 promoter.
Epistatic Relationship Between BRCA1 and the Canonical NHEJ Pathway in the Repair of Etoposide-Induced DNA Damage.
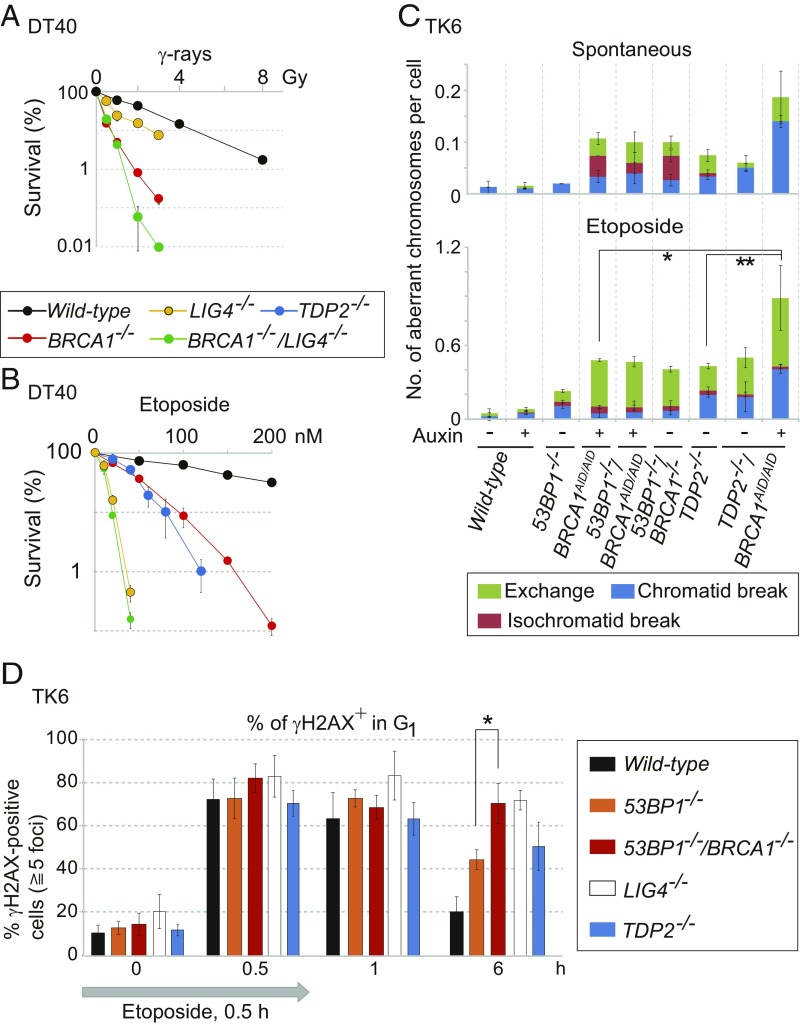
BRCA1’s contribution to the repair of E2-induced DSBs in G1 phase suggests a collaboration between BRCA1 and canonical NHEJ. To investigate this collaboration, we took a genetic approach using the chicken DT40 cell line, since DT40 cells deficient in BRCA1 (BRCA1−/−) are able to proliferate (39). We disrupted the BRCA1 gene in a LIG4−/− background, as described previously (39). We verified gene disruption by genomic PCR and reverse-transcription PCR of the resulting mutant, BRCA1−/−/LIG4−/− cells (SI Appendix, Fig. S6 A and B). To assess the role played by BRCA1 in the removal of 5′ TOP2 adducts from DSBs, we measured cellular sensitivity to etoposide, a TOP2 poison, which strongly stabilizes the TOP2–DNA intermediate (19). As expected, TDP2−/− DT40 cells showed a prominent sensitivity to etoposide (Fig. 2B) (40). DSBs induced by etoposide are repaired mainly by NHEJ, but not by HDR, in DT40 cells (21). The BRCA1−/− cells were also very sensitive to etoposide. Remarkably, the etoposide sensitivity of the BRCA1−/−/LIG4−/− double mutant was very similar to that of LIG4−/− cells (Fig. 2B). This epistatic relationship suggests that BRCA1 collaborates with the canonical NHEJ pathway in the repair of TOP2ccs in DT40 cells in such a manner that BRCA1 promotes the removal of 5′ TOP2 adducts for subsequent NHEJ.
Fig. 2.
Function of BRCA1 is dependent on the canonical NHEJ pathway and independent of TDP2 in the repair of etoposide-induced DNA damage. (A and B) Cellular sensitivity of chicken DT40 cells to γ-rays (A) and etoposide (TOP2 poison) (B) was measured by clonogenic cell-survival analysis. Error bars show SD calculated from three independent experiments. (C) Number of spontaneously arising mitotic chromosome breaks (Upper) and etoposide-induced chromosome breaks (Lower). BRCA1AID/AID cells were treated with auxin (250 μM) (+) or without auxin (−) for 9 h. The cells were treated with etoposide (100 nM) for 9 h (Lower). Cells were incubated with colcemid for the last 3 h. Error bars represent SD of three independent experiments. At least 50 metaphase cells per experiment were examined for the chromosome analysis. The single and double asterisks (Lower) indicate P < 0.03 and P < 0.02, respectively, calculated by Student’s t test. (D) DSB-repair kinetics of TK6 cells in G1 phase following 0.5-h pulse exposure to etoposide (10 nM). Cells were released to fresh medium after the pulse exposure. DSB repair was evaluated by counting the number of γH2AX foci at 0.5, 1, and 6 h after the initiation of the pulse exposure. The percentage of cells with at least five foci per nucleus is shown on the y axis. Error bars were plotted for SD from three independent experiments. The asterisk indicates P < 0.01, calculated by Student’s t test. More than 100 G1 cells (cyclin-A–negative) were analyzed for each experiment. The average numbers of γH2AX foci per cell are shown in SI Appendix, Fig. S8.
BRCA1 Promotes the Removal of 5′ Adducts from DSBs in G1 Phase in the Human TK6 Cell Line.
To investigate the role played by human BRCA1 in the repair of TOP2ccs, we conditionally disrupted the BRCA1 gene in the human lymphoblastoid TK6 cell line (41, 42) by inserting the miniauxin-induced degron (mAID) tag (43, 44) into the C terminus of BRCA1 allelic genes, generating BRCA1AID/AID TK6 cells (SI Appendix, Fig. S7 A–C). The loss of BRCA1 was lethal to the TK6 cells, while the 53BP1−/−/BRCAAID/AID cells were viable even in the presence of auxin (SI Appendix, Fig. S7C). We then generated 53BP1−/−/BRCA1−/− TK6 cells (SI Appendix, Fig. S7 D and G). The 53BP1−/−/BRCA1−/− cells were also able to proliferate, although they showed a substantial number of spontaneously arising chromosome breaks in mitotic chromosome spreads (Fig. 2C, Upper), as seen in mice carrying mutations in both the 53BP1 and BRCA1 genes (8).
To evaluate the role played by BRCA1 in the repair of TOP2ccs, we pulse-exposed cells to etoposide and measured the number of chromosome breaks in mitotic chromosome spreads (Fig. 2C, Lower). The auxin-treated BRCA1AID/AID and 53BP1−/−/BRCA1AID/AID cells, as well as the 53BP1−/−/BRCA1−/− cells, showed defective repair of pathological TOP2ccs generated by etoposide. 53BP1−/− cells showed a modest defect, since 53BP1 is largely dispensable for NHEJ (32, 45). We disrupted the TDP2 gene in BRCA1AID/AID cells (SI Appendix, Fig. S7 H and I) and treated the resulting TDP2−/−/BRCA1AID/AID cells with auxin. These cells showed a greater sensitivity to etoposide than did the auxin-induced BRCA1AID/AID cells (Fig. 2C). In summary, human BRCA1 may repair pathological TOP2ccs independent of TDP2, as does MRE11 (17).
To verify the role played by BRCA1 in DSB repair in G1 phase, we monitored the resolution kinetics of γH2AX foci following pulse exposure to etoposide in TK6 cells. We analyzed G1-phase cells by excluding cyclin-A–positive, S/G2-phase cells (Fig. 2D and SI Appendix, Fig. S8). A 30-min pulse exposure to etoposide caused γH2AX-focus increases of several fold, as seen previously (17). By 6 h after exposure, these foci had reduced to almost the background level in wild-type cells (black bar in Fig. 2D), whereas they persisted in the 53BP1−/− and LIG4−/− mutants (orange and white bars in Fig. 2D), both of which are defective in NHEJ. The TDP2−/− cells exhibited the delayed kinetics of γH2AX-focus removal (blue bar in Fig. 2D). Remarkably, the 53BP1−/−/BRCA1−/− cells showed a more prominent delay in DSB repair than did the 53BP1−/− cells (red bar in Fig. 2D). We conclude that BRCA1 promotes the repair of etoposide-induced DSBs in G1 phase. This conclusion, as well as the epistatic relationship between BRCA1−/− and LIG4−/− (Fig. 2B), suggests that BRCA1 may contribute to the repair of pathological TOP2ccs independent of its functioning in HDR. As with the MRE11 nuclease (17), BRCA1 may facilitate the removal of 5′ TOP2 adducts before DSB repair by canonical NHEJ.
Measurement of Stable TOP2ccs by Dot-Blot Analysis.
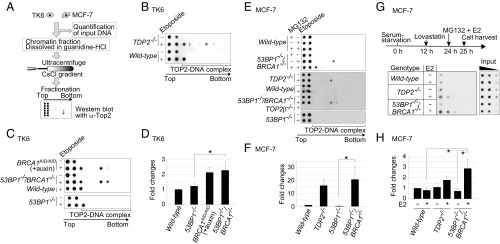
To test the hypothesis that BRCA1 facilitates the removal of 5′ TOP2 adducts, we measured the number of TOP2ccs (TOP2 covalently associated with genomic DNA). To this end, we lysed cells and separated TOP2ccs from free TOP2 in cellular lysates by subjecting them to sedimentation by means of cesium chloride (CsCl)-gradient ultracentrifugation, 100,000 × g (Fig. 3A). We measured the amount of genomic DNA in each cellular lysate (SI Appendix, Table S2). The free TOP2 remained in the top two fractions, whereas the TOP2ccs moved to the lower fractions of the CsCl gradient, which correspond to the migration of chromosomal DNA. TOP2ccs were detected as single or double dots in the middle fractions of the “TOP2–DNA complex” shown at the bottom of the blot (Fig. 3 B, C, and E). Wild-type cells treated with etoposide displayed free TOP2, with a small fraction of TOP2 in the third-lowest fraction (Fig. 3B). Depletion of TDP2, which is known to remove TOP2ccs (22), generated TOP2ccs that were detected in the middle fractions of the CsCl gradient after etoposide treatment (Fig. 3B) (17). In agreement with the increased etoposide sensitivity of 53BP1−/−/BRCA1−/− TK6 cells compared with 53BP1−/− TK6 cells (Fig. 2C, Lower), TOP2ccs were detected in the middle fractions of the CsCl gradient from 53BP1−/−/BRCA1−/− TK6 cells but not 53BP1−/− cells, a result that is similar to TDP2 deletion (Fig. 3 C and D). The BRCA1-depleted BRCA1AID/AID TK6 cells also showed TOP2ccs in the middle fractions of the CsCl gradient (Fig. 3 C and D). In sum, we conclude that BRCA1 promotes the removal of pathological TOP2ccs in the TK6 human B-cell line.
Fig. 3.
Accumulation of TOP2ccs in BRCA1-deficient cells. (A) Schematic of in vivo TOP2cc measurement by immunodetection with α-TOP2 antibody. Genomic DNA (50 μg) (from TK6 and MCF-7) was subjected to sedimentation by CsCl-gradient ultracentrifugation. Genomic DNA from wild-type TK6 cells treated with etoposide (10 μM) for 2 h was included as a control for every dot blot of TK6 cells. The treatment reduced cellular survival by only ∼1% relative to untreated wild-type TK6 cells. Individual fractions were blotted to PVDF filters followed by dot blot using α-TOP2 antibody. The top two fractions include free TOP2 while the bottom fractions include TOP2ccs. (B and C) Dot blot of TOP2 in the indicated TK6 cells treated with etoposide (10 μM) (+) or DMSO (−) for 2 h. BRCA1AID/AID cells were pretreated with auxin for 2 h, and then incubated with etoposide (10 μM) plus auxin for an additional 2 h. (D) Quantification of TOP2cc in the indicated genotypes from C relative to the amount of TOP2cc in wild-type TK6 cells treated with etoposide (10 μM) for 2 h. Each experiment was done independently at least three times. Error bars represent SD. The asterisk indicates P < 0.01, calculated by Student’s t test. (E) TOP2βcc detection in MCF-7 cells. The indicated cells were serum-starved for 24 h and then treated with etoposide (10 μM) for 2 h. The addition of a proteasome inhibitor (MG132) with etoposide caused a signal shift from the middle fractions to the upper-third fraction (Upper). (F) Quantification of etoposide-induced TOP2cc for the indicated genotypes from E, relative to the amount of TOP2cc in wild-type MCF-7. Data are presented as in D. The asterisk indicates P < 0.01, calculated by Student’s t test. (G) Detection of TOP2βcc accumulation by E2 in MCF-7. The diagram (Upper) indicates the experimental design. After incubating MCF-7 cells carrying the indicated genotypes in serum-free medium for 12 h, we added a CDK inhibitor, lovastatin (10 μM), to completely eliminate any cycling cells. We added MG132 and E2 at 24 h after serum starvation and harvested cells at 25 h. MG132 prevents the proteasome degradation of TOP2 at TOP2ccs. Genomic DNA was sheared by sonication. We then conducted immunoprecipitation with α-TOP2β to enrich the TOP2βccs and subjected them to sedimentation by CsCl-gradient ultracentrifugation, as in A. The bottom fractions including TOP2βccs were analyzed by dot blot with α-TOP2β antibody. Threefold serial dilutions of cell lysates were subjected to dot-blot analysis with α–β-actin antibody. (H) Quantification of immunoprecipitated TOP2βccs for the indicated genotypes from G, relative to the amount of immunoprecipitated TOP2cc in wild-type MCF-7 without E2 following normalization with α–β-actin of whole-cell extract (input) as an internal control. Error bars represent SD of three independent experiments. The asterisks indicate P < 0.05, calculated by Student’s t test.
We next examined TOP2βccs in a serum-starved MCF-7 cell population following pulse exposure to etoposide. As expected, exposure of TOP2β−/− cells to etoposide showed no TOP2βcc signals (Fig. 3E). TOP2βccs were detectable in the middle fractions of the CsCl gradient of 53BP1−/−/BRCA1−/− and TDP2−/− cells, but not in those of 53BP1−/− or wild-type cells (Fig. 3 E and F). These results demonstrate that TOP2βccs accumulate in the absence of BRCA1. Unlike etoposide-treated TK6 cells, no signal was seen in the third-lowest fraction in the etoposide-treated MCF-7 cells. The signals seen in the middle fractions were shifted upward to the third-lowest fraction in the presence of the proteasome inhibitor MG132 (Fig. 3E, compare the fifth and sixth rows). Thus, the third-lowest fraction may contain TOP2ccs containing intact TOP2. The proteasome may have partially degraded the TOP2 included in the TOP2ccs, leading to an increase in the specific gravity of this protein–DNA complex and generating signals in the middle fractions. In conclusion, BRCA1 is important for the removal of the 5′ adducts in G1 phase.
BRCA1 Promotes the Removal of E2-Induced TOP2 Adducts from DSB Ends.
To detect E2-induced TOP2βccs, we treated serum-starved MCF-7 cells with lovastatin to completely inhibit the G1–S transition (46). We then exposed the cells to E2 (100 nM) together with MG132 for 1 h. We enriched TOP2β by immunoprecipitation, followed by the identification of TOP2ccs, as shown in Fig. 3A. The E2 treatment increased the amount of TOP2βccs in 53BP1−/−/BRCA1−/− and TDP2−/− cells by 3.6- and 1.6-fold, respectively, but not in wild-type cells (Fig. 3 G and H). These data suggest that E2-induced γH2AX foci, shown in Fig. 2B, represent TOP2βccs.
BRCA1 Promotes the MRE11-Dependent Removal of 5′ Adducts from Pathological TOP2ccs.
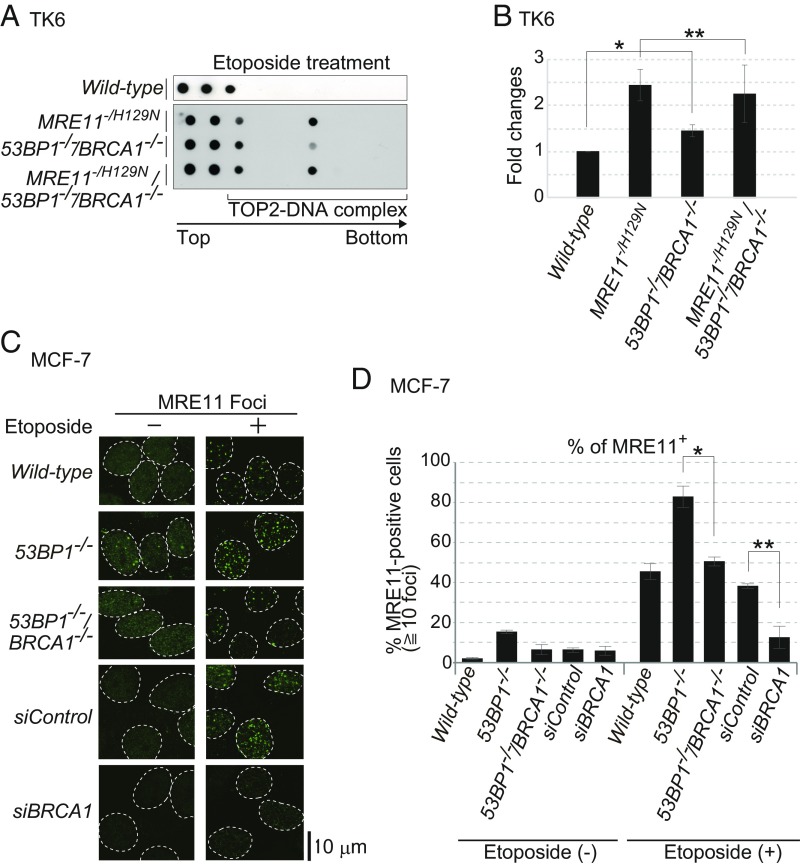
The dominant role played by the nuclease activity of MRE11 in repairing TOP2ccs (17, 47) led us to hypothesize that BRCA1 enhanced the capability of MRE11 to eliminate 5′ TOP2 adducts. To test this hypothesis, we took a genetic approach, comparing etoposide-induced TOP2ccs in MRE11−/H129N, 53BP1−/−/BRCA1−/−, and MRE11−/H129N/53BP1−/−/BRCA1−/− TK6 cells (SI Appendix, Fig. S9A). We analyzed TOP2ccs in MRE11−/H129N/53BP1−/−/BRCA1−/− cells after addition of 4-hydroxytamoxifen (4-OHT), which converts the MRE11+/H129N genotype to MRE11−/H129N by activating the Cre recombinase, and excised the intact MRE11+ allele (48). Note that MRE11−/H129N cells are capable of proliferating exponentially without exhibiting spontaneous mitotic chromosomal breaks for 3 d after addition of 4-OHT (17). We incubated cells with 4-OHT for 3 d, exposing them to etoposide for the last 2 h. Although MRE11−/H129N cells showed more TOP2ccs than did 53BP1−/−/BRCA1−/− cells, MRE11−/H129N and MRE11−/H129N/53BP1−/−/BRCA1−/− TK6 cells showed very similar numbers of TOP2ccs (Fig. 4 A and B). This similarity suggests that BRCA1 contributes to the removal of pathological TOP2ccs possibly through the nuclease activity of MRE11. Supporting this idea, we found DNA damage-induced complex formation between BRCA1 and MRE11 in G1 cells (SI Appendix, Fig. S9C).
Fig. 4.
BRCA1 promotes MRE11-mediated removal of TOP2ccs. (A) MRE11+/H129N/53BP1−/−/BRCA1−/− and MRE11+/H129N TK6 cells were treated with 4-OHT for 3 d to inactivate the wild-type MRE11 allele. Cells were then treated with etoposide (10 μM) for 2 h. Data are shown as in Fig. 3 B and C. (B) Quantification of TOP2cc in the indicated genotypes from A. Data are shown as in Fig. 3D. Error bars were plotted for SD from three independent experiments. The single asterisk indicates P < 0.02 whereas the double asterisk indicates P > 0.3 (not statistically significant difference), calculated by Student’s t test. (C) Etoposide-induced MRE11 foci in serum-starved MCF-7 cells. We examined the indicated genotypes and wild-type MCF-7 cells treated with siRNA targeting BRCA1 (siBRCA1) and control siRNA (siControl). We pulse-exposed cells to etoposide (10 μM) for 30 min. (D) Quantification of MRE11-positive cells with at least 10 foci per nucleus. Error bars were plotted for SD from three independent experiments. The single and double asterisks indicate P < 0.02 and P < 0.03, respectively, calculated by Student’s t test.
To investigate the role played by BRCA1 in MRE11-dependent repair of etoposide-induced DSBs, we performed immunostaining of MRE11 foci following pulse exposure of 53BP1−/− and 53BP1−/−/BRCA1−/− MCF-7 cells to etoposide. We also depleted BRCA1 by siRNA treatment (SI Appendix, Fig. S9B). After serum starvation, we examined only cyclin-A–negative G1 cells. The percentage of MRE11-positive cells was reduced by 68% in the siBRCA1-treated cells, compared with the control siRNA (Fig. 4 C and D). Loss of BRCA1 in 53BP1−/− MCF-7 cells also caused a significant reduction of MRE11 foci, compared with 53BP1−/− cells (Fig. 4 C and D). In summary, BRCA1 may be required for efficient recruitment of MRE11 to pathological TOP2cc sites.
E2 Induces Prominent γH2AX Foci in the Mammalian Epithelial Cells of 53BP1−/−/BRCA1−/− as Well as in NHEJ-Deficient Mice.
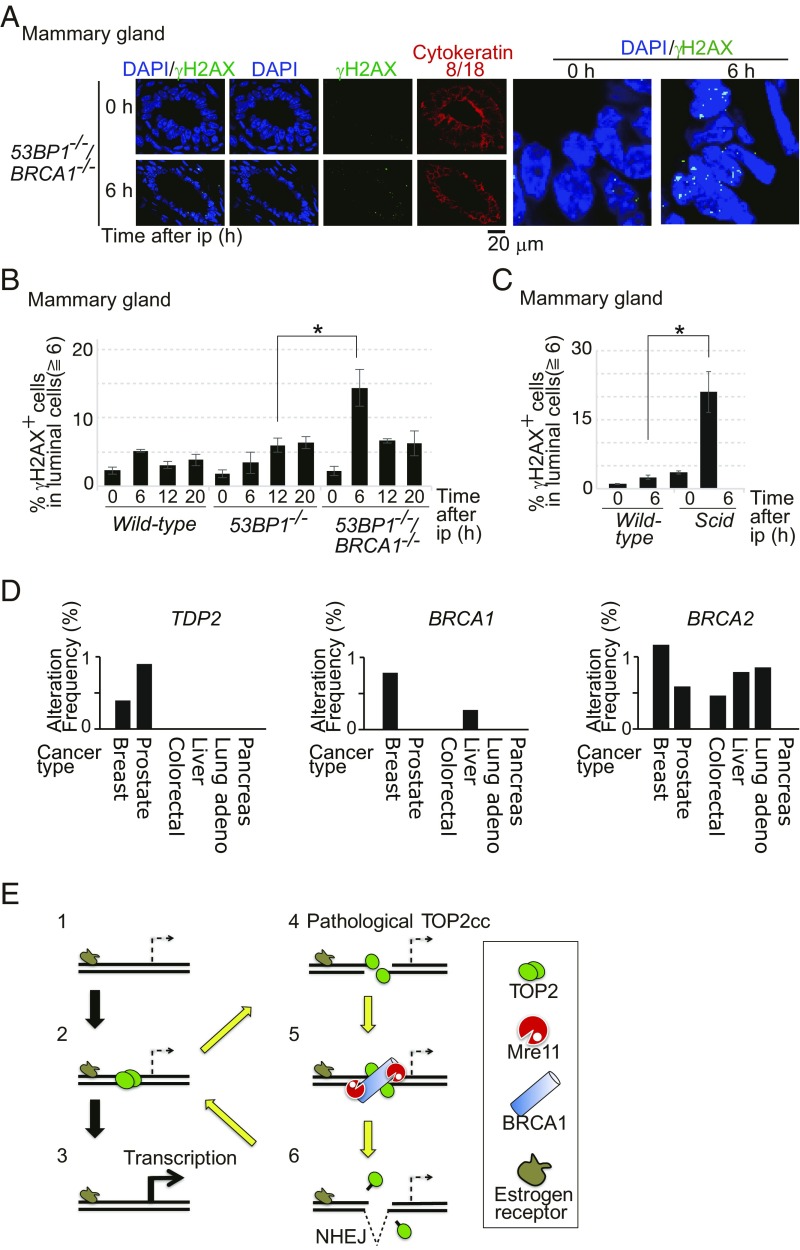
To verify the genotoxicity of E2 in BRCA1-deficient mice, we administered E2 to mice by i.p. injection (ip) and monitored the number of γH2AX foci in the mammary gland. We analyzed luminal epithelial cells in wild-type, 53BP1−/−, and 53BP1−/−/BRCA1−/− mice after ip with E2 (8). Only a few E2-induced γH2AX foci were seen in wild-type mice. In marked contrast, 53BP1−/−/BRCA1−/− mice displayed significant induction of γH2AX focus-positive cells at 6 h after ip with E2 (Fig. 5 A and B and SI Appendix, Fig. S10A). The prominent γH2AX foci are reminiscent of the kinetics of E2-induced γH2AX foci in MCF-7 cells (Fig. 1C). We therefore conclude that BRCA1 significantly prevents genome instability caused by E2 in the mammary luminal epithelial cells of mice.
Fig. 5.
E2 is genotoxic to luminal epithelial cells of mammary glands in mice deficient in BRCA1 or NHEJ. (A) E2 was given to 53BP1−/−/BRCA1−/− mice via i.p. injection. Mammary-gland tissues were isolated at 0 and 6 h after ip, and immunostained with α-γH2AX and α-cytokeratin 8/18 antibodies. Luminal cells, which express the ER, were stained with α–cytokeratin-8/18. (A, Right) Images indicate enlarged view of γH2AX foci (green) on luminal cells. (B) Percentage of γH2AX focus-positive cells in luminal epithelial cells at the indicated time after ip with E2. For a wild-type strain relevant to 53BP1−/− and 53BP1−/−/BRCA1−/− mice, the C57BL6/J strain was used. Representative images of γH2AX foci are shown in SI Appendix, Fig. S10A. The asterisk indicates P < 0.02, calculated by Student’s t test. (C) Data are shown as in B. For a wild-type strain relevant to NHEJ-deficient Scid mice, the C.B.-17/lcr strain was used. The asterisk indicates P < 0.01, calculated by Student’s t test. (D) The search of TCGA database indicates that homozygous deep deletion of the TDP2 gene is found in breast-invasive cancers and prostate cancers but not other types of malignant cancers. The y axis shows the percentage of the indicated cancers carrying homozygous deep deletion of either TDP2, BRCA1, or BRCA2 in 818 breast-invasive (50), 333 prostate (53), 212 colorectal (51), 373 liver (TCGA, provisional), 230 lung (52), and 150 pancreas (TCGA, provisional) cancer samples, registered in TCGA database. Homozygous deep deletion of the TDP2 gene is observed in three cases of the 818 breast-invasive and three cases of the 313 prostate cancer samples, but in neither the colorectal, liver, lung, nor pancreas cancer samples. (E) Proposed model for the effect of E2 on TOP2cc formation in an ER target gene. Exposure of cells to E2 translocates the ER to target genes (step 1) and triggers the transient formation of TOP2ccs in transcriptional regulatory sequences (step 2). Some ER target genes require TOP2-mediated catalysis for their transcriptional control (step 3). Occasional abortive catalysis of TOP2 causes the formation of pathological TOP2ccs, including DSBs covalently associated with the degradative products of TOP2 (step 4, yellow arrow). BRCA1 promotes the recruitment of MRE11 to pathological TOP2cc sites (step 5). MRE11 as well as TDP2 removes TOP2 and its degradative products from DSB ends for subsequent ligation by NHEJ (step 6) in G0/G1 phase.
The 53BP1−/− mice displayed a defect in the repair of E2-induced DSBs, whose defect was less prominent than that seen in 53BP1−/−/BRCA1−/− mice (Fig. 5B). To confirm the role played by NHEJ in the repair of E2-induced DSBs, we examined γH2AX-focus formation at 6 h after ip injection with E2, comparing wild-type and DNA-PKcs–deficient (Scid) mice. Remarkably, Scid mice, but not wild-type mice, displayed numerous γH2AX foci in the individual epithelial cells of the mammary ducts (Fig. 5C and SI Appendix, Fig. S10B). Only less than 1% of epithelial cells was labeled with 5-Ethynyl-2′-deoxyuridine, indicating the formation of γH2AX foci in G0/G1-phase cells (SI Appendix, Fig. S10C). In conclusion, E2 is highly genotoxic in the absence of BRCA1 and NHEJ. The numerous γH2AX foci of the Scid mice displayed in Fig. 5C may explain their susceptibility to mammary adenocarcinoma (49).
If estrogen-dependent formation of pathological TOP2ccs is responsible for oncogenesis of luminal epithelial cells, TDP2 might also play a role in suppressing the genotoxic effect of E2, as does BRCA1. The homozygous deep deletion of the TDP2 gene was seen in 0.4 and 0.8% of 818 breast-invasive carcinomas and 333 prostate cancer patients, respectively, registered in The Cancer Genome Atlas (TCGA) database (50) (Fig. 5D). By contrast, no deep deletion of TDP2 was seen in 212 colorectal, 373 liver, 230 lung, or 150 pancreas cancer patients (50–53). These data support the notion that the sex hormones drive oncogenesis in mammary glands as well as in prostate glands by forming pathological TOP2ccs.
Discussion
We show in this work that BRCA1 plays an important role in eliminating pathological TOP2ccs (Fig. 3), independent of its role in promoting HDR (Fig. 2B). BRCA1 promotes the elimination of pathological TOP2ccs, most likely through nucleolytic processing of MRE11 (Fig. 4). Exposure of cells to E2 induces pathological TOP2βccs, and their efficient eradication requires BRCA1 as well as TDP2 (Fig. 3 G and H). The strong genotoxic effect of E2 in the absence of BRCA1 is also evidenced by numerous chromosome breaks in mitotic chromosome spreads (Fig. 1E) and by the presence of prominent γH2AX foci in murine luminal epithelial cells (Fig. 5 A and B). In summary, BRCA1 promotes genome integrity from abortive TOP2 activity throughout the cell cycle (Fig. 5E). Our data have uncovered a potentially novel molecular mechanism underlying the initiation of cancers restricted to estrogen-regulated tissues, such as breast and ovary tissues, in carriers with germ-line mutations of the BRCA1 gene.
The current study has revealed the strong genotoxicity of the estrogen hormone. Accumulating evidence has indicated that female steroid hormones drive oncogenesis not only as “promoter” but also as “initiator” mutagens. The current study highlights the E2-dependent mutagenicity that is dependent on both ERs and TOP2. Previous studies found that cells deficient in translesion DNA synthesis (TLS) were more sensitive than a wild-type control to 4-hydroxyestradiol, suggesting that 4-hydroxyestradiol may form estrogen–DNA adducts that interfere with DNA replication (54, 55). This idea agrees with the data showing that a short exposure of cycling MCF-10A breast epithelial cells to E2 causes DSBs, mainly in S/G2 phases, even though MCF-10A does not express ERα (56). These studies indicate the mutagenesis is caused by TLS over estrogen–DNA adducts on template strands. The idea that estrogen-DNA adducts interfere DNA replication does not explain carcinogenesis restricted to estrogen-regulated tissues in carriers having BRCA1 mutations, because estrogen–DNA adducts can form in any tissue. Our data show that 10 nM E2 effectively induces DSBs, whose genotoxicity depends on both TOP2β and the ER in G0/G1 phases (Fig. 1 C and F). This ER-dependent genotoxicity can explain the carcinogenesis restricted to estrogen-regulated tissues in carriers having BRCA1 mutations.
The question is, how do TOP2β and the ER cause DSBs outside of S phase? We show that E2-induced R loops (34) are unlikely to cause DSBs in G1-phase MCF-7 cells (SI Appendix, Fig. S5 A–C). We propose that E2 induces pathological TOP2βccs at transcriptional regulatory sequences, including promoter sequences (Fig. 5E), for the following reasons. TOP2β promotes the initiation of transcription by binding to transcriptional regulatory sequences (10). Catalysis of TOP2β is essential for the transcriptional initiation of genes controlled by several nuclear receptors (13, 16). Thus, E2-induced γH2AX foci in BRCA1-deficient cells (Fig. 1 C and D) may result from abortive TOP2βccs formed at transcriptional regulatory sequences in estrogen target genes in G0/G1 phases (Fig. 5E). This idea is verified at least in the pS2 promoter of this ER target gene (SI Appendix, Fig. S5D). In sum, we propose that E2 can cause pathological TOP2βccs at transcriptional regulatory sequences in G0/G1 phases.
Prolonged formation of pathological TOP2ccs suggests that they may cause driver mutations in BRCA1-deficient tumors. One possible scenario is that the activation of ER often causes pathological TOP2ccs (Figs. 1F and 5E) and the loss of BRCA1 might compromise the fidelity of pathological TOP2cc repair, leading to the deletion of transcriptional regulatory sequences important for appropriate responses to the estrogen hormone. In this context, the loss of the TDP2 gene would also decrease the fidelity of TOP2cc repair and might cause deletion of transcriptional regulatory sequences leading to tumorigenesis of breast cancer in response to estrogens. This prediction is verified by the data that homozygous deep deletions of TDP2 were observed in breast malignant tumors (Fig. 5D). The deep deletion is frequently seen also in prostate cancers, indicating that androgens as well as estrogens may be mutagenic in the absence of TDP2. It remains elusive how BRCA1 and MRE11 accurately repair pathological TOP2ccs in collaboration with canonical NHEJ, as endonucleolytic cleavage by MRE11 at the 5′ strands of DSBs can cause deletion of cleaved nucleotides during canonical NHEJ (47). E2-induced TOP2cc sites represent transcriptional regulatory sequences and may be hotspots of nucleotide-sequence deletion in BRCA1-deficient cells. Identifying E2-induced TOP2cc sites in the whole genome would contribute to our understanding of the molecular mechanisms for oncogenesis in BRCA1-deficient mammary epithelial cells.
The loss of BRCA1 may increase the accumulation of mutations to a much greater extent in S phase than in G0/G1 phases through the following mechanisms. First, pathological TOP2ccs may form throughout the cell cycle. Collision between pathological TOP2ccs and replication forks may result in the collapse of replication forks. In addition to this TOP2-dependent genotoxicity, collision between E2-induced R loops and replication forks could cause DSBs in the S phase of BRCA1-deficient cells, since the formation of the R loops is suppressed by BRCA1 (31, 34–36). These collision events may lead to the formation of the DSBs that are repaired by BRCA1-mediated HDR. Collectively, estrogen may have extremely strong genotoxic effects via a TOP2-dependent mechanism as well as via TOP2-independent mechanisms during DNA replication in BRCA1-deficient cells. Multiple roles for BRCA1 in preventing estrogen-induced mutagenesis would explain the carcinogenesis restricted to estrogen-regulated tissues even in carriers with germ-line mutations in the BRCA1 gene.
Since most human BRCA1 mutant breast cancers are basal-like/triple-negative (57, 58), the ER-negative breast stem cell has been suggested to be the “cell of origin” for BRCA1-deficient tumors. However, our current study has suggested that the cell of origin may be ER-positive luminal cells and/or their precursors. The possible scenario is that the extremely high genotoxicity of estrogens in BRCA1-deficient luminal cells can strongly drive their oncogenesis through formation of pathological TOP2ccs. The cell of origin is a highly controversial issue, as ER-positive luminal cells can be dedifferentiated into an ER-negative stem-like state during oncogenesis in humans (59) as well as in mice (60, 61) (reviewed in ref. 62). Once malignant cells are established, the loss of the ER may confer a considerable advantage to malignant cells, because estrogens would no longer generate pathological TOP2ccs. This idea can clearly explain why a majority of BRCA1-deficient breast cancers do not express ER, and may also be relevant to BRCA2-deficient breast cancers (57). It should be noted that a majority of BRCA1-deficient breast cancers, 43 of the 65 cases registered in TCGA, have null mutations of p53 (50, 63). The loss of p53 is also likely to counteract the strong genotoxicity of estrogens in BRCA1-deficient cells. In summary, the idea that activated ER could induce pathological TOP2ccs, which strongly stimulate p53-dependent apoptosis, may explain why BRCA1-deficient cells acquire a growth advantage in the absence of both p53 and ER.
Materials and Methods
All materials and cell lines used in the paper are described in SI Appendix, Tables S1, S3, and S4. Animal studies were conducted in accordance with our institutional guidelines, and the experimental procedures were approved by the Kyoto University Animal Care Committee. The DT40 cell line was cultured as previously described (2). MCF-7 was maintained in Dulbecco’s modified Eagle’s medium (0845964; Gibco) containing FBS (10%; Gibco), penicillin (100 U/mL), and streptomycin (100 μg/mL; Nacalai). Human TK6 B cells were incubated in RPMI 1640 medium (3026456; Nacalai) supplemented with horse serum (5%; Gibco), penicillin (100 U/mL), streptomycin (100 μg/mL; Nacalai), and sodium pyruvate (200 mg/mL; Thermo Fisher). TK6 and MCF-7 mutants were generated by CRISPR/Cas9 gene targeting with a guide RNA designed and cloned into pX330 or pX459 (SI Appendix, Table S3). Details of targeting, clone selection, and screening are given in SI Appendix, Materials and Methods. Details of estrogen injection into mice, preparation of cryosections from mammary-gland tissue, the immunostaining method, TOP2cc detection, Western blot analysis, chromosome analysis, chromatin immunoprecipitation assay, and CRISPRi/siRNA method used for gene silencing are also described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank R. Chapman from Oxford for the genome-editing protocol used with the MCF-7 cells; and J. Haber at Brandeis University; M. Jasin at Memorial Sloan Kettering Cancer Center; L. Zou at Massachusetts General Hospital; and A. Canela, N. Wong, and J. Sam at the NIH for stimulating discussion and critical reading of the manuscript. We are also grateful to M. Kato, A. Kobayashi, and O. Takahashi of KEYENCE and the staffs of the Medical Research Support Center, supported by Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS) from AMED (Grant JP18am0101092), for technical assistance with the cell sorter and confocal microscope. The rat anti-cytokeratin 8/18 antibody developed by Dr. P. Brulet was obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the NIH and maintained at The University of Iowa, Department of Biology. This work was supported by a Grant-in-Aid from the Ministry of Education, Science, Sport and Culture (KAKENHI 23310133) (to K.T.), (KAKENHI 25650006, 23221005, and 16H06306) (to S.T.), and (KAKENHI 16H02953 and 18H04900) (to H.S.). This work was also supported by grants from the Takeda Research and Mitsubishi Foundation (to H.S.) and Japan Society for the Promotion of Science Core-to-Core Program, Advanced Research Networks (to S.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803177115/-/DCSupplemental.
References
- 1.Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: The roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonoda E, et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venkitaraman AR. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science. 2014;343:1470–1475. doi: 10.1126/science.1252230. [DOI] [PubMed] [Google Scholar]
- 5.Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Löbrich M, Jeggo P. A process of resection-dependent nonhomologous end joining involving the goddess Artemis. Trends Biochem Sci. 2017;42:690–701. doi: 10.1016/j.tibs.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callen E, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao L, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunting SF, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell. 2012;46:125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pommier Y, Sun Y, Huang SN, Nitiss JL. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol. 2016;17:703–721. doi: 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King IF, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fachinetti D, et al. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell. 2010;39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 14.Sano K, Miyaji-Yamaguchi M, Tsutsui KM, Tsutsui K. Topoisomerase IIbeta activates a subset of neuronal genes that are repressed in AT-rich genomic environment. PLoS One. 2008;3:e4103. doi: 10.1371/journal.pone.0004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haffner MC, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madabhushi R, et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoa NN, et al. Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol Cell. 2016;64:580–592. doi: 10.1016/j.molcel.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Gómez-Herreros F, et al. TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function. Nat Genet. 2014;46:516–521. doi: 10.1038/ng.2929. [DOI] [PubMed] [Google Scholar]
- 19.Pommier Y, Marchand C. Interfacial inhibitors: Targeting macromolecular complexes. Nat Rev Drug Discov. 2011;11:25–36. doi: 10.1038/nrd3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gómez-Herreros F, et al. TDP2 suppresses chromosomal translocations induced by DNA topoisomerase II during gene transcription. Nat Commun. 2017;8:233. doi: 10.1038/s41467-017-00307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maede Y, et al. Differential and common DNA repair pathways for topoisomerase I- and II-targeted drugs in a genetic DT40 repair cell screen panel. Mol Cancer Ther. 2014;13:214–220. doi: 10.1158/1535-7163.MCT-13-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 23.Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aparicio T, Baer R, Gottesman M, Gautier J. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J Cell Biol. 2016;212:399–408. doi: 10.1083/jcb.201504005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makharashvili N, et al. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol Cell. 2014;54:1022–1033. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura K, et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010;6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuella-Martin R, et al. 53BP1 integrates DNA repair and p53-dependent cell fate decisions via distinct mechanisms. Mol Cell. 2016;64:51–64. doi: 10.1016/j.molcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badie S, et al. BRCA1 and CtIP promote alternative non-homologous end-joining at uncapped telomeres. EMBO J. 2015;34:410–424. doi: 10.15252/embj.201488947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biehs R, et al. DNA double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Mol Cell. 2017;65:671–684.e5. doi: 10.1016/j.molcel.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stork CT, et al. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. eLife. 2016;5:e17548. doi: 10.7554/eLife.17548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Difilippantonio S, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stender JD, et al. Structural and molecular mechanisms of cytokine-mediated endocrine resistance in human breast cancer cells. Mol Cell. 2017;65:1122–1135.e5. doi: 10.1016/j.molcel.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatchi E, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell. 2015;57:636–647. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.García-Muse T, Aguilera A. Transcription-replication conflicts: How they occur and how they are resolved. Nat Rev Mol Cell Biol. 2016;17:553–563. doi: 10.1038/nrm.2016.88. [DOI] [PubMed] [Google Scholar]
- 36.Lin YL, Pasero P. Transcription-replication conflicts: Orientation matters. Cell. 2017;170:603–604. doi: 10.1016/j.cell.2017.07.040. [DOI] [PubMed] [Google Scholar]
- 37.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pommier Y, et al. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2) DNA Repair (Amst) 2014;19:114–129. doi: 10.1016/j.dnarep.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qing Y, et al. The epistatic relationship between BRCA2 and the other RAD51 mediators in homologous recombination. PLoS Genet. 2011;7:e1002148. doi: 10.1371/journal.pgen.1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng Z, Cortés-Ledesma F, El Khamisy SF, Caldecott KW. TDP2/TTRAP is the major 5′-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J Biol Chem. 2011;286:403–409. doi: 10.1074/jbc.M110.181016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fowler P, et al. Cadmium chloride, benzo[a]pyrene and cyclophosphamide tested in the in vitro mammalian cell micronucleus test (MNvit) in the human lymphoblastoid cell line TK6 at Covance Laboratories, Harrogate UK in support of OECD draft Test Guideline 487. Mutat Res. 2010;702:171–174. doi: 10.1016/j.mrgentox.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 42.Honma M. Generation of loss of heterozygosity and its dependency on p53 status in human lymphoblastoid cells. Environ Mol Mutagen. 2005;45:162–176. doi: 10.1002/em.20113. [DOI] [PubMed] [Google Scholar]
- 43.Natsume T, Kiyomitsu T, Saga Y, Kanemaki MT. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 2016;15:210–218. doi: 10.1016/j.celrep.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 45.Adachi N, Ishino T, Ishii Y, Takeda S, Koyama H. DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: Implications for DNA double-strand break repair. Proc Natl Acad Sci USA. 2001;98:12109–12113. doi: 10.1073/pnas.201271098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Javanmoghadam-Kamrani S, Keyomarsi K. Synchronization of the cell cycle using lovastatin. Cell Cycle. 2008;7:2434–2440. doi: 10.4161/cc.6364. [DOI] [PubMed] [Google Scholar]
- 47.Deshpande RA, Lee JH, Arora S, Paull TT. Nbs1 converts the human Mre11/Rad50 nuclease complex into an endo/exonuclease machine specific for protein-DNA adducts. Mol Cell. 2016;64:593–606. doi: 10.1016/j.molcel.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 48.Hoa NN, et al. Relative contribution of four nucleases, CtIP, Dna2, Exo1 and Mre11, to the initial step of DNA double-strand break repair by homologous recombination in both the chicken DT40 and human TK6 cell lines. Genes Cells. 2015;20:1059–1076. doi: 10.1111/gtc.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fabre KM, et al. Murine Prkdc polymorphisms impact DNA-PKcs function. Radiat Res. 2011;175:493–500. doi: 10.1667/RR2431.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ciriello G, et al. TCGA Research Network Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550, and correction (2014) 514:262. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizutani A, et al. Extensive chromosomal breaks are induced by tamoxifen and estrogen in DNA repair-deficient cells. Cancer Res. 2004;64:3144–3147. doi: 10.1158/0008-5472.can-03-3489. [DOI] [PubMed] [Google Scholar]
- 55.Yasui M, et al. Mechanism of translesion synthesis past an equine estrogen-DNA adduct by Y-family DNA polymerases. J Mol Biol. 2007;371:1151–1162. doi: 10.1016/j.jmb.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Savage KI, et al. BRCA1 deficiency exacerbates estrogen-induced DNA damage and genomic instability. Cancer Res. 2014;74:2773–2784. doi: 10.1158/0008-5472.CAN-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foulkes WD, et al. Estrogen receptor status in BRCA1- and BRCA2-related breast cancer: The influence of age, grade, and histological type. Clin Cancer Res. 2004;10:2029–2034. doi: 10.1158/1078-0432.ccr-03-1061. [DOI] [PubMed] [Google Scholar]
- 58.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: Different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lim E, et al. kConFab Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 60.Van Keymeulen A, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525:119–123. doi: 10.1038/nature14665. [DOI] [PubMed] [Google Scholar]
- 61.Koren S, et al. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature. 2015;525:114–118. doi: 10.1038/nature14669. [DOI] [PubMed] [Google Scholar]
- 62.Koren S, Bentires-Alj M. Breast tumor heterogeneity: Source of fitness, hurdle for therapy. Mol Cell. 2015;60:537–546. doi: 10.1016/j.molcel.2015.10.031. [DOI] [PubMed] [Google Scholar]
- 63.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.