Significance
Parkinson’s disease (PD) is the second most common neurodegenerative disorder in the world. Several common and rare genetic risk variants associated with PD pathogenesis have been identified, predominantly in persons of European descent, but the genetic contributions to familial PD are largely unknown for Han Chinese. Here, we present a trio-based study to explore the association between de novo-altered genes and early onset PD in Han Chinese. We found that the 12 genes with de novo mutations were biologically connected to each other and likely to be disease-risk genes. Further analyses using two independent cohorts revealed that NUS1 harbored more rare nonsynonymous variants, and subsequent functional studies on Drosophila proved its potential link to PD pathogenesis.
Keywords: Parkinson’s disease, exome sequencing, de novo mutations, disease-risk gene, neurodegenerative disorders
Abstract
Whole-exome sequencing has been successful in identifying genetic factors contributing to familial or sporadic Parkinson’s disease (PD). However, this approach has not been applied to explore the impact of de novo mutations on PD pathogenesis. Here, we sequenced the exomes of 39 early onset patients, their parents, and 20 unaffected siblings to investigate the effects of de novo mutations on PD. We identified 12 genes with de novo mutations (MAD1L1, NUP98, PPP2CB, PKMYT1, TRIM24, CEP131, CTTNBP2, NUS1, SMPD3, MGRN1, IFI35, and RUSC2), which could be functionally relevant to PD pathogenesis. Further analyses of two independent case-control cohorts (1,852 patients and 1,565 controls in one cohort and 3,237 patients and 2,858 controls in the other) revealed that NUS1 harbors significantly more rare nonsynonymous variants (P = 1.01E-5, odds ratio = 11.3) in PD patients than in controls. Functional studies in Drosophila demonstrated that the loss of NUS1 could reduce the climbing ability, dopamine level, and number of dopaminergic neurons in 30-day-old flies and could induce apoptosis in fly brain. Together, our data suggest that de novo mutations could contribute to early onset PD pathogenesis and identify NUS1 as a candidate gene for PD.
Parkinson’s disease (PD) is the second most common neurodegenerative disorder possessing clinical characteristics of resting tremors, bradykinesia, rigidity, and a positive response to dopamine replacement therapy, among others. The classic pathological alterations in PD are the loss of dopaminergic neurons and cytoplasmic inclusions of protein aggregates known as “Lewy bodies” (1). The prevalence of PD is ∼1–2% in people over the age of 60 y and up to 4% in people older than 80 y (2). Although the detailed underlying mechanisms for PD are still unclear, aging, environmental, and genetic factors are major contributors. Since SNCA was identified as the first PD-causative gene in 1997 (3), significant efforts have been made to identify the genetic factors that cause PD, and several genes are recognized as being associated with PD.
Most PD patients arise randomly, but about 10–15% of PD patients have familial PD (4), and another 5–10% have early onset Parkinson’s disease (EOPD, defined as age <40 y at onset) (5). Genetic factors, such as monogenetic causative genes, may play a more crucial role in EOPD than in late-onset PD (6, 7). Although many causative and susceptibility variants have been identified, the definitive genetic contributions to EOPD remain unknown (7, 8).
De novo mutations are reported to associate with neurodevelopmental and neurodegenerative diseases [e.g., autism spectrum disorders (9, 10), intellectual disabilities (11, 12), schizophrenia (13, 14), amyotrophic lateral sclerosis (15), and PD (16)] by whole-exome or whole-genome sequencing. In addition, several studies reported de novo mutations in the established causative genes in individual patients with neurodegenerative diseases such as Alzheimer’s disease (17), progressive supranuclear palsy (18), and PD (19). All these studies indicate the potentially important role(s) of de novo mutations for neurodegenerative diseases.
We performed a two-stage analysis to identify candidate PD genes with deleterious mutations. First, we sequenced the exomes of 19 trios (a patient with EOPD and the patient’s parents) and 20 quads (a patient with EOPD, the patient’s parents and a patient’s healthy sibling) affected by EOPD to identify the genes with deleterious de novo mutations. Second, 12 functionally relevant candidate genes were selected from the trio-based study and were screened on 1,852 patients with sporadic PD and 1,565 controls. NUS1, which is the only gene with nominally significant P value (P = 0.03), was further replicated in another 3,237 PD patients and 2,858 controls. We found that NUS1 harbors significantly more rare nonsynonymous variants in PD patients than in controls (P = 1.01E-5, odds ratio = 11.3). The PD-related phenotypes were observed by knocking down the orthologous gene of NUS1 in Drosophila. We therefore identify NUS1 as a candidate PD gene.
Results
Identification and Validation of de Novo Mutations Associated with EOPD.
We sequenced and analyzed the whole exome of 39 EOPD families (Fig. 1A). On average, 99.13% of the target regions and 93.61% of the National Center for Biotechnology Information Consensus Coding Sequences database were sequenced; 91.63% target regions were covered at least 20-fold (SI Appendix, Table S1), which was sufficient to detect heterozygous de novo mutations. No significant sequencing coverage bias was observed between the parents and offspring (SI Appendix, Fig. S1). No inherited variants were found as likely causing the PD phenotype (Dataset S1). We designed a two-step validation strategy with a spectrum of quality thresholds to guarantee a low false-negative rate of de novo mutations after removing one trio due to its exceedance of Mendelian errors (SI Appendix, Fig. S2). We confirmed 39 de novo single-nucleotide variants (SNVs) and one de novo insertion among the 38 probands (SI Appendix, Table S2) and 19 de novo SNVs and one de novo deletion in the 20 unaffected siblings (SI Appendix, Table S3). Twenty-five (65.8%) probands and 13 (65.0%) siblings carried at least one de novo SNV. The average number of de novo SNVs in each family (probands: 1.03; siblings: 0.95) followed a Poisson distribution (SI Appendix, Fig. S3), and no significant difference was observed between the probands and their siblings (Fisher’s exact test, P = 0.50) (SI Appendix, Table S4) or between the other covariates (SI Appendix, Fig. S4 and Table S5).
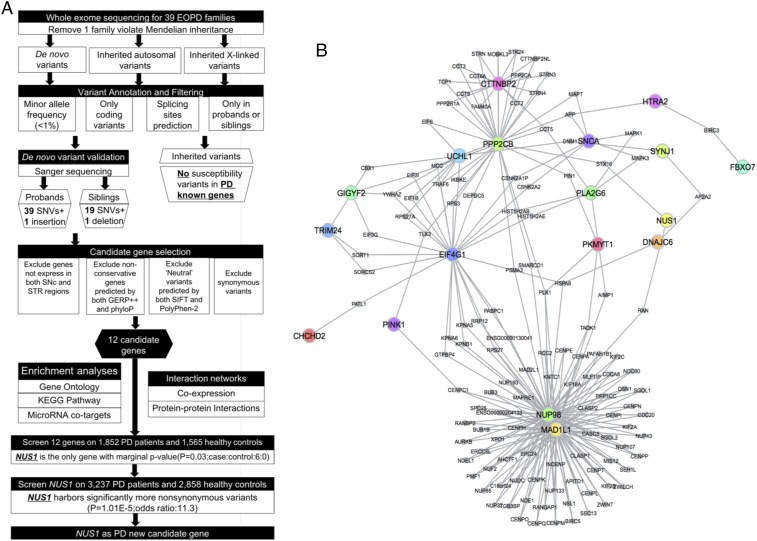
Fig. 1.
Data analysis workflow and the protein–protein interaction networks between candidate and known PD genes. (A) The workflow for identifying PD candidate genes. (B) Protein–protein interaction networks between 12 candidate genes and known PD causative genes.
Potential Pathogenicity of de Novo Mutations Associated with EOPD.
Among the 39 de novo SNVs in probands, 27 were missense, 1 was nonsense, 2 were located in the canonical splice sites, and 2 (NUP98 and SMPD3) were located in predicted splice sites (SI Appendix, Table S2). Among the 19 de novo SNVs in siblings, 12 were missense, 1 was nonsense, and 1 (FBXL15) was located in a predicted splice site (SI Appendix, Table S3). For small insertions and deletions (indels), one de novo insertion (NUS1) was located in the canonical splice site in probands, and another was a de novo nonframeshift deletion in siblings. The probands tended to have an unexpectedly high ratio of de novo nonsynonymous (NS) to de novo synonymous (S) SNVs (NS:S) of 4.57 (de novo NS SNVs = 32, including missense SNVs, nonsense SNVs, and SNVs in the splice sites; de novo S SNVs = 7), which exceeds the expected value under a random model (NS:S = 2.85) (20) and the ratio observed in siblings (NS:S = 2.80). The difference between de novo mutations in probands and siblings may be an artifact of the small sample size. A collection of 22,866 individual inherited mutations, which were thought to be rare neutral mutations (21), was used for comparison with de novo mutations in probands and siblings as benchmark to evaluate their probability of being genetic risk factors for disease. In probands the NS:S ratio of de novo mutations also was found to be significantly higher than the NS:S ratio of individual inherited mutations. (P = 5.35E-3, Fisher’s exact test). However, no significant difference was observed in siblings (P = 0.20) (SI Appendix, Table S6). Compared with individual mutations, the loss-of-function de novo mutations (including de novo nonsense SNVs, de novo SNVs, and indels from the splice sites) were more likely to be observed in the probands (P = 2.94E-4, Fisher’s exact test) than in the healthy siblings (P = 0.06) (SI Appendix, Table S6). Significant differences in the prediction scores for damaging mutations were observed between the de novo mutations in probands and the individual inherited mutations (Wilcoxon rank sum test: GERP++, P = 0.14; phyloP, P = 0.04; SIFT, P = 2.18E-5; PolyPhen-2, P = 0.03) (SI Appendix, Fig. S5A). However, they were not observed in healthy siblings (Wilcoxon rank sum test: GERP++, P = 0.93; phyloP, P = 1.00; SIFT, P = 0.07; PolyPhen-2, P = 0.99) (SI Appendix, Fig. S5B).
Identification of Functionally Related Candidate Genes by Biological Network.
To determine the potential contribution of these de novo mutations to PD, we chose 12 genes (NUP98, MAD1L1, PPP2CB, PKMYT1, TRIM24, CTTNBP2, NUS1, SMPD3, MGRN1, RUSC2, CEP131, and IFI35) that are expressed in the stratum/substantia nigra region and are potentially functionally relevant to PD pathogenesis (SI Appendix, Table S7A). Seven of these genes (CTTNBP2, MAD1L1, NUP98, NUS1, PKMYT1, PPP2CB, and TRIM24) and 20 known PD genes (Fig. 1B) were found to be involved in a protein–protein interaction network. Seven (NUP98, MAD1L1, PPP2CB, TRIM24, CTTNBP2, NUS1, and MGRN1) were found to be differentially expressed in PD genetic mouse models as well as in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) lesion-induced PD mouse models (SI Appendix, Table S8B). We found additional evidence (SI Appendix, Fig. S6) that these genes are significantly enriched in the targets of hsa-miR-125a-3p (Pcorrected = 6.50E-3) (SI Appendix, Figs. S7 and S8 and Dataset S2), gene coexpression network (Pcorrected = 0.001) (SI Appendix, Fig. S9), GO chromosome (Pcorrected = 6.78E-03) and chromosomal part (Pcorrected = 1.15E-02) (SI Appendix, Fig. S10 A and B) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway [progesterone-mediated oocyte maturation (Pcorrected = 0.03); cell cycle (Pcorrected = 0.03), and oocyte meiosis (Pcorrected = 0.03)] (SI Appendix, Fig. S10 C and D). This suggests that these 12 genes may be involved in similar biological functions and act together in increasing the risk of developing PD.
Analysis of an Independent Case-Control Study Identifies NUS1 as a Candidate PD Gene.
We first screened the 12 candidate genes on 1,852 patients with sporadic PD and 1,565 healthy controls to investigate the burden of rare variants. Molecule molecular inversion probes were designed to capture the exonic regions of the target genes, and their PCR products were sequenced (22). We found no genes harboring significantly more rare nonsynonymous SNVs in patients. Nevertheless, NUS1 was the only gene with a marginally significant P value and with surprisingly full penetrance (case:control = 6:0, P = 0.03) (SI Appendix, Table S7A). It was further screened on additional 3,237 patients by Sanger sequencing and 2,858 controls (SI Appendix, Table S9) whose exomes were sequenced by the HiSeq. 2000 sequencing system and showed a consistent trend (case:control = 20:2, P = 3.2E-4). The replication proved the association between NUS1 and PD (Pcombine = 1.01E-5, odds ratio = 11.3) (Fig. 2 A and B and SI Appendix, Table S7B). None of the 26 mutation carriers in the replication study carried any other known pathogenic mutations for PD, although they displayed typical PD features (Dataset S4).
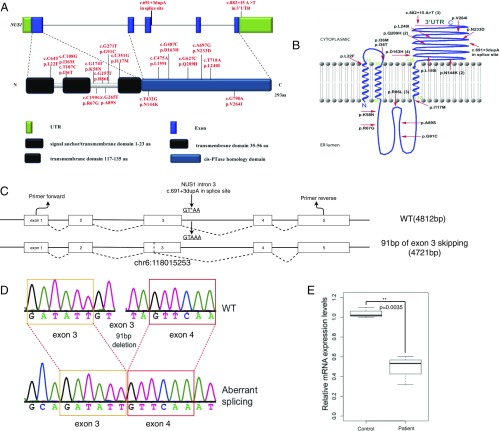
Fig. 2.
The rare variants in NUS1 and analysis of NUS1 c.691+3dupA on the splicing site. (A) The location of rare variants in NUS1. The NgBR domains are identified by Harrison et al. (33). (B) The variant locations on the transmembrane structure of NgBR. The numbers in parentheses represent the mutation recurrence. NgBR exists with two topological conformations. The minor fraction, with its C terminus oriented toward the lumen, is consistent with the regulation of NPC2 stability. The major fraction, with its C terminus oriented toward the cytosol, interacts with human dehydrodolichyl disphosphate synthase (DHDDS, also referred to hCIT), facilitating cis-isoprenyltransferase (cis-IPTase) activity and dolichol biosynthesis (33). (C) Schematic representation of the genomic structure of WT NUS1 and aberrant splicing caused by c.691+3dupA, which introduced a 91-bp deletion from chr6:118015253. (D) Sanger sequencing demonstrated an aberrant splicing event that skipped 91 bp of exon3. (E) Relative gene-expression levels in controls and patients.
We further examined the impact of c.691+3dupA on the splicing and expression of NUS1. Peripheral blood mononuclear cells were isolated from the patient’s peripheral whole blood and from age-matched healthy controls. To determine whether the mutation could alter the splicing, we amplified the cDNA fragment of exon 3 and exon 4 in NUS1. Two bands were presented by dividing PCR products on a 1.2% agarose gel and were sequenced by Sanger sequencing (SI Appendix, Fig. S11). We found an aberrant splicing event that skipped 91 bp of exon3 (Fig. 2 C and D). Furthermore, we determined the relative mRNA expression levels using quantitative RT-PCR. We observed the decrease of NUS1 in the patients (0.4853 ± 0.08473, P = 0.0035) compared with controls (Fig. 2E). These data together demonstrate that c.691+3dupA could cause the missplicing and lead to a significant reduction of NUS1 expression in PD patients.
NUS1 Plays Important Roles in Dopamine Neurons.
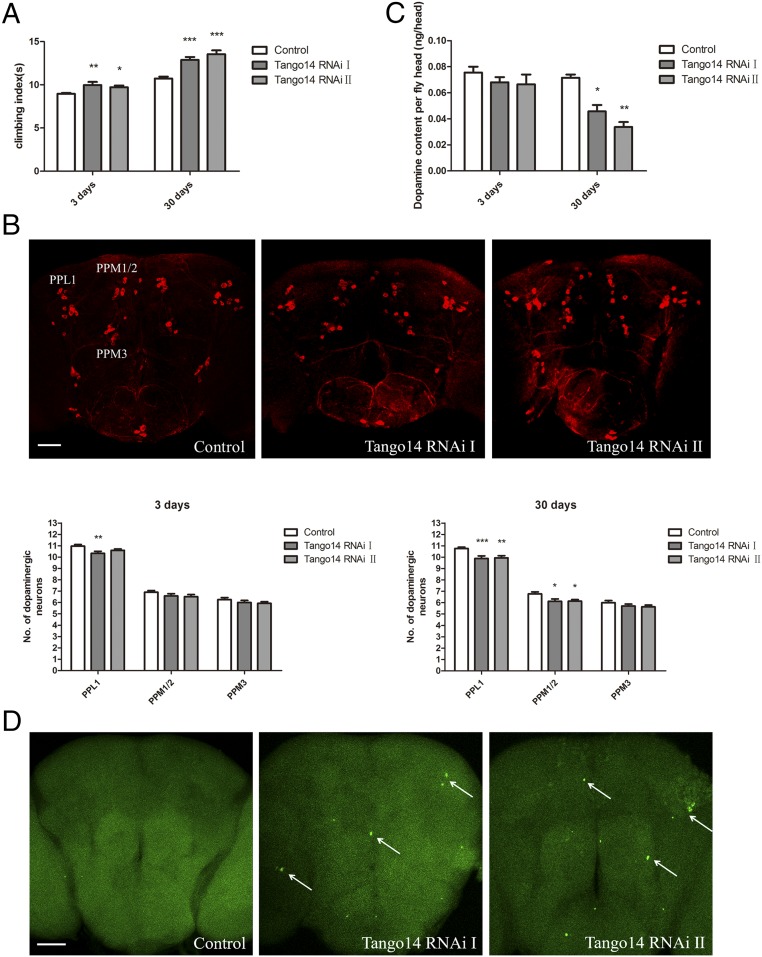
In recent years, Drosophila has been widely used to study the molecular pathogenesis of neurodegeneration disorders such as PD (23, 24). The Drosophila NUS1 is also called “Tango14” and shares 44% similarity with human NUS1 at the amino acid level. We used RNAi-mediated Tango14-knockdown flies to investigate NUS1 roles in vivo (SI Appendix, Fig. S12). The 3-d-old and 30-d-old Tango14 RNAi flies driven by pan-neural Elav-GAL4 showed defects in climbing ability (Fig. 3A). As shown in Fig. 3B, 30-d-old Tango14 RNAi flies induced by the dopaminergic neuron-specific TH-Gal4 driver exhibited a reduction in dopaminergic neurons in the lateral protocerebrum posterior (PPL1) cluster and dorsomedial protocerebral posterior (PPM1/2) cluster (Fig. 3B). We performed HPLC on Drosophila brain extracts and found a dramatic reduction in the brain dopamine levels of 30-d-old flies expressing Tango14 RNAi (Fig. 3C), further confirming that the knockdown of Tango14 leads to dopaminergic dysfunction. A TUNEL assay of 30-d-old flies also demonstrated that Tango14 RNAi treatment led to abnormal apoptotic signals in the brain (Fig. 3D). These data together suggest that NUS1 plays important roles in dopamine neurons and that the loss of NUS1 could lead to neuronal dysfunction that is related to PD.
Fig. 3.
Characterization of RNAi-mediated Tango14 knockdown in Drosophila. (A) Comparison of the climbing ability of 3-d-old and 30-d-old flies. Control flies are Elav-GAL4/Y, Tango14; RNAi flies are Elav-GAL4/Y;+; Tango14 RNAi/+; n = 3. Error bars indicate mean ± SD. (B, Upper) Whole-mount 30-d-old male brains showing dopaminergic neuron clusters labeled by anti-TH antibody (red). (Scale bar: 50 μm.) (Lower) Graphs show the number of dopaminergic neurons in 3-d-old and 30-d-old flies. Control flies are TH-GAL4/+, Tango14; RNAi flies are TH-GAL4/Tango14 RNAi; n = 15. (C) Amount of dopamine in the heads of 3-d-old and 30-d-old male flies. Control flies are TH-GAL4/+, Tango14; RNAi flies are TH-GAL4/Tango14 RNAi; n = 3. Error bars indicate mean ± SD. (D) TUNEL (green) staining of 30-d-old male fly brains. Control flies are Elav-GAL4/Y, Tango14; RNAi flies are Elav-GAL4/Y;+; Tango14 RNAi/+. The arrows indicated apoptotic signals. (Scale bar: 50 μm.) *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA.
Discussion
Genetic factors are recognized as an important contributor of PD pathogenesis. Genome-wide association studies led to the identification of several common variants associated with PD susceptibility; however, these variants explain only a small proportion of disease heritability (7, 8). Rare variants represent different genetic factors that potentially have large effects that could not be investigated using genome-wide association. In this study, we designed a two-stage sequencing-based strategy to explore and validate EOPD candidate genes by examining rare variants of susceptibility genes detected in PD patients. First, we sequenced the whole exomes of 39 EOPD nuclear families to identify de novo mutations and analyzed their potential pathogenicity. Our data suggest that the dysfunctions are caused by specific de novo mutations rather than by the accumulation of de novo mutations, as is consistent with other human diseases (25, 26). The analyses focus on the de novo mutations with detrimental effects and de novo-altered genes expressed in relevant tissues. Interactome networks boost the power of the sequencing strategy and narrow the candidate gene list (SI Appendix, Fig. S6), assuming they share similar mechanisms (27). For the second stage, we reexamined the 12 functionally related genes in an independent study of sporadic PD cases and controls. This reexamination verified that the de novo-altered gene NUS1 is associated with PD. Furthermore, RNAi experiments on Drosophila revealed that loss of NUS1 caused PD-like phenotypes and abnormal apoptotic cells. A combination of genetic analysis of independent cohorts and functional studies of the candidate genes could help identify the key disease-causing genes.
Our results suggest that NUS1 is a candidate PD gene. NUS1 encodes NogoB receptor (NgBR) and localizes primarily to the endoplasmic reticulum. It also interacts with the cholesterol-binding protein Niemann–Pick type C2 (NPC2), a lysosomal protein that is essential for intracellular trafficking of LDL-derived cholesterol (28). Indeed, earlier studies using a limited number of samples did find rare variants of NPC1/2 in PD patients (29, 30). RNAi-mediated disruption of NUS1 or genetic deficiency of NUS1 could lead to a decrease in NPC2 levels, increased intracellular cholesterol accumulation, and a loss of sterol sensing (28). Interestingly, increased cholesterol metabolites were found in degenerative dopaminergic cells with increased α-synuclein, and these metabolites appeared to accelerate α-synuclein aggregation in vitro (31). Consistently, deletion of α-synuclein in mice significantly increased cholesterol in the brain (32). The underlying mechanisms for NgBR involvement in the neurodegenerative diseases remain to be explored in future studies.
Our present study suggests an efficient strategy for exploring the potential effects of de novo mutations associated with human diseases and validating them through genetic and functional studies. In the two independent case-control studies, our finding that the rare NS variants in NUS1 were carried by 26 patients but by only two controls presumed a large effect size of this gene (odds ratio = 11.3). Our data did not replicate the results of Kun-Rodrigues et al. (16) in persons of European descent. This discrepancy may result from the complex mechanisms for de novo mutations and the genetic heterogeneity of PD. In this study, we first pinpointed putative PD-associated genes harboring de novo mutations in EOPD patients, whose disease was more likely to be caused by genetic factors. The 12 selected genes were further screened in patients with sporadic PD, and NUS1 was shown to have certain effects on PD pathogenicity. Because most of the parents of the 26 mutation carriers had died, we could not confirm whether all the rare mutations in NUS1 are indeed de novo. Also, these mutation carriers have sporadic rather than familial PD, suggesting that other, confounding disease risk factors could potentially be involved. Further study should be performed to clarify the mechanism of mutations in NUS1 and to reexamine its effect size using familial cases. Other than NUS1, most candidate genes in the connectome (SI Appendix, Fig. S6) were not replicated in our case-control study. Thus, other independent PD cohorts or more functional experiments are required to verify the pathogenicity of these other candidate genes in the future.
In summary, by combining multistep sequencing strategy with fly genetics, we identified rare de novo mutations associated with neurodegenerative disorders such as PD. Identification of NUS1 as PD-causing gene will further expand our understanding the molecular pathogenesis of PD.
Materials and Methods
The protocol was approved by the Ethics Committee of Central South University, and written informed consent was collected from all the subjects. Details of sample cohort, exome sequencing, variant calls and annotation, gene network analysis, mutation effect analysis, and functional experiments are included in SI Appendix.
Supplementary Material
Acknowledgments
We thank all the patients and family members for their generous participation in this work; BGI-Shenzhen for assistance in exome sequencing and data analysis; Yuanwei Zhang (University of Science and Technology of China) and Jieqiong Tan (School of Life Sciences, Central South University) for their kind help in analyzing the functional impact of splicing mutation; and Prof. Evan E. Eichler for assistance with the data analysis. This work was supported by National Key Plan for Scientific Research and Development of China Grant 2016YFC1306000 and National Natural Science Foundation of China Grant 81430023.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809969115/-/DCSupplemental.
References
- 1.Lees AJ, Hardy J, Revesz T. Parkinson’s disease. Lancet. 2009;373:2055–2066. doi: 10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 2.de Lau LM, Breteler MM. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006;5:525–535. doi: 10.1016/S1474-4422(06)70471-9. [DOI] [PubMed] [Google Scholar]
- 3.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 4.Verstraeten A, Theuns J, Van Broeckhoven C. Progress in unraveling the genetic etiology of Parkinson disease in a genomic era. Trends Genet. 2015;31:140–149. doi: 10.1016/j.tig.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Schrag A, Schott JM. Epidemiological, clinical, and genetic characteristics of early-onset parkinsonism. Lancet Neurol. 2006;5:355–363. doi: 10.1016/S1474-4422(06)70411-2. [DOI] [PubMed] [Google Scholar]
- 6.Farrer MJ. Genetics of Parkinson disease: Paradigm shifts and future prospects. Nat Rev Genet. 2006;7:306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 7.Keller MF, et al. International Parkinson’s Disease Genomics Consortium (IPDGC); Wellcome Trust Case Control Consortium 2 (WTCCC2) Using genome-wide complex trait analysis to quantify ‘missing heritability’ in Parkinson’s disease. Hum Mol Genet. 2012;21:4996–5009. doi: 10.1093/hmg/dds335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nalls MA, et al. International Parkinson’s Disease Genomics Consortium (IPDGC); Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI); 23andMe; GenePD; NeuroGenetics Research Consortium (NGRC); Hussman Institute of Human Genomics (HIHG); Ashkenazi Jewish Dataset Investigator; Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC); Greek Parkinson’s Disease Consortium; Alzheimer Genetic Analysis Group Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner TN, et al. Genomic patterns of de novo mutation in simplex autism. Cell. 2017;171:710–722.e12. doi: 10.1016/j.cell.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin S, et al. De novo variants in GRIA4 lead to intellectual disability with or without seizures and gait abnormalities. Am J Hum Genet. 2017;101:1013–1020. doi: 10.1016/j.ajhg.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 13.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gulsuner S, et al. Consortium on the Genetics of Schizophrenia (COGS); PAARTNERS Study Group Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chesi A, et al. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat Neurosci. 2013;16:851–855. doi: 10.1038/nn.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kun-Rodrigues C, et al. International Parkinson’s Disease Genomics Consortium (IPDGC) A systematic screening to identify de novo mutations causing sporadic early-onset Parkinson’s disease. Hum Mol Genet. 2015;24:6711–6720. doi: 10.1093/hmg/ddv376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golan MP, et al. Early-onset Alzheimer’s disease with a de novo mutation in the presenilin 1 gene. Exp Neurol. 2007;208:264–268. doi: 10.1016/j.expneurol.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Ogaki K, et al. Analyses of the MAPT, PGRN, and C9orf72 mutations in Japanese patients with FTLD, PSP, and CBS. Parkinsonism Relat Disord. 2013;19:15–20. doi: 10.1016/j.parkreldis.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 19.Puschmann A, et al. A Swedish family with de novo alpha-synuclein A53T mutation: Evidence for early cortical dysfunction. Parkinsonism Relat Disord. 2009;15:627–632. doi: 10.1016/j.parkreldis.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci USA. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu B, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 24.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 25.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahni N, et al. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161:647–660. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison KD, et al. Nogo-B receptor stabilizes Niemann-Pick type C2 protein and regulates intracellular cholesterol trafficking. Cell Metab. 2009;10:208–218. doi: 10.1016/j.cmet.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zech M, et al. Niemann-Pick C disease gene mutations and age-related neurodegenerative disorders. PLoS One. 2013;8:e82879. doi: 10.1371/journal.pone.0082879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kluenemann HH, Nutt JG, Davis MY, Bird TD. Parkinsonism syndrome in heterozygotes for Niemann-Pick C1. J Neurol Sci. 2013;335:219–220. doi: 10.1016/j.jns.2013.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu JP, et al. Cholesterol involvement in the pathogenesis of neurodegenerative diseases. Mol Cell Neurosci. 2010;43:33–42. doi: 10.1016/j.mcn.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Barceló-Coblijn G, Golovko MY, Weinhofer I, Berger J, Murphy EJ. Brain neutral lipids mass is increased in alpha-synuclein gene-ablated mice. J Neurochem. 2007;101:132–141. doi: 10.1111/j.1471-4159.2006.04348.x. [DOI] [PubMed] [Google Scholar]
- 33.Harrison KD, et al. Nogo-B receptor is necessary for cellular dolichol biosynthesis and protein N-glycosylation. EMBO J. 2011;30:2490–2500. doi: 10.1038/emboj.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.