Significance
The 2013–2014 silent polio epidemic in Israel was a setback to global eradication efforts because Israel had previously been certified as polio-free by the World Health Organization. Fortunately, Israel has a robust environmental surveillance program that detected the epidemic and allowed rapid mobilization of a vaccine campaign before any cases of acute flaccid paralysis. This kind of silent (caseless) epidemic will be increasingly common as we approach global eradication, demonstrating the need for both enhanced environmental surveillance and an accompanying inference framework to translate environmental data into public health metrics. We incorporate environmental data into a population-level disease transmission model, generating insights into the epidemiology of the outbreak. This framework can be used to guide future interventions.
Keywords: poliovirus, silent transmission, mathematical model, environmental surveillance, vaccination
Abstract
Israel experienced an outbreak of wild poliovirus type 1 (WPV1) in 2013–2014, detected through environmental surveillance of the sewage system. No cases of acute flaccid paralysis were reported, and the epidemic subsided after a bivalent oral polio vaccination (bOPV) campaign. As we approach global eradication, polio will increasingly be detected only through environmental surveillance. We developed a framework to convert quantitative polymerase chain reaction (qPCR) cycle threshold data into scaled WPV1 and OPV1 concentrations for inference within a deterministic, compartmental infectious disease transmission model. We used this approach to estimate the epidemic curve and transmission dynamics, as well as assess alternate vaccination scenarios. Our analysis estimates the outbreak peaked in late June, much earlier than previous estimates derived from analysis of stool samples, although the exact epidemic trajectory remains uncertain. We estimate the basic reproduction number was 1.62 (95% CI 1.04–2.02). Model estimates indicate that 59% (95% CI 9–77%) of susceptible individuals (primarily children under 10 years old) were infected with WPV1 over a little more than six months, mostly before the vaccination campaign onset, and that the vaccination campaign averted 10% (95% CI 1–24%) of WPV1 infections. As we approach global polio eradication, environmental monitoring with qPCR can be used as a highly sensitive method to enhance disease surveillance. Our analytic approach brings public health relevance to environmental data that, if systematically collected, can guide eradication efforts.
Since the beginning of eradication efforts in 1988, incidence of polio has decreased by more than 99% (1), and, as of 2017, polio was endemic in only three countries. The presence of polio is often detected only through acute flaccid paralysis (AFP) surveillance. However, because paralytic polio occurs only in a small fraction of infections (2, 3) and that fraction is shrinking as vaccination coverage improves, AFP will not be sensitive enough to trigger the fast and robust responses needed as we approach global eradication. The 2013–2014 polio outbreak in Israel (4), which had no cases of AFP, underscores the need for fast, sensitive methods of polio detection. Here, we describe the epidemiology of this outbreak using methods we developed to transform environmental quantitative polymerase chain reaction (qPCR) time-series data into an epidemic trajectory.
The discovery of polio transmission in Israel received significant attention (5, 6), as Israel had been previously certified as a “polio-free” country by the World Health Organization. Laboratory genetic testing identified the virus as the South Asian strain of wild poliovirus type 1 (WPV1) (7), and subsequent phylogenetic analysis has suggested that an introduction from Pakistan in late 2012 (8) diverged into Egypt, Israel, and Syria (5, 8). Routine environmental surveillance detected the presence of the virus in Israel in late May 2013, and retrospective analysis showed that the virus had entered southern Israel in February 2013 (4). Prior analysis indicated that Rahat, the largest predominantly Bedouin city (9), sustained significant transmission during the outbreak (10). Israel launched a supplementary vaccination campaign with bivalent oral polio vaccine (bOPV) at the beginning of August 2013 (4), and the outbreak ended in early 2014. OPV contains live poliovirus, and vaccinated individuals can, therefore, transmit the vaccine strains, effectively transmitting protection against WPV strains. The vaccine virus, however, has the potential to mutate to a virulent form that can cause paralysis, known as vaccine-associated paralytic polio (VAPP) (11), and lead to circulating vaccine-derived poliovirus (cVDPV) (12). Thus, countries usually switch from OPV to inactivated polio vaccination (IPV) when the risk of VAPP occurring is greater than the risk of WPV-associated AFP. For countries with high rates of vaccination and good water, sanitation, and hygiene (WASH) practices, the risk of acquiring wild poliovirus is relatively low. Accordingly, Israel switched to solely using IPV in 2005 (4). IPV provides protection against paralysis by developing humoral immunity (13), but it does not prevent fecal–oral polio transmission, making control with IPV alone difficult in areas with poor WASH conditions. Because IPV allows transmission but suppresses AFP, early detection of polio outbreaks becomes more difficult under these conditions (14). Undetected outbreaks and silent circulation place unvaccinated individuals at risk for AFP, particularly in areas where WASH infrastructure is poor (15).
Environmental surveillance that tests for the presence of pathogens in the environment is an important tool for detecting silent circulation because it allows detection of low levels of poliovirus before AFP cases appear (1). Israel has maintained a sewage-based environmental surveillance system for polio since 1989, with monthly composite samples taken at sentinel sites. With the onset of the 2013–2014 silent epidemic, these efforts were intensified with more frequent sampling and expanded to multiple locations across Israel, first in both Bedouin and non-Bedouin cities and towns in southern Israel and later nationwide (7, 16).
Although the standard for environmental surveillance for poliovirus has been qualitative tissue culture challenges, the intensified Israeli surveillance also used quantitative plaque assays as well as a rapid, direct real-time qPCR assay that provided fast, sensitive poliovirus concentration data. The qPCR approach, unlike quantitative plaque assays, could easily distinguish between WPV and OPV strains (7, 16, 17). However, it has been unclear how to best use these qPCR data to inform the outbreak epidemiology (10, 18). Environmental surveillance programs are being expanded around the globe, both for polio and for other pathogens, so developing an inference framework that can translate these data into public health outcomes is crucial, particularly as case data will be sparse or nonexistent as we near eradication. Here, we analyze the outbreak in Rahat, the epicenter of the epidemic, by developing and implementing methods that use qPCR data within an outbreak modeling framework to estimate the epidemic trajectories.
Data
Environmental Surveillance.
Environmental surveillance samples were taken at the mouth of the Rahat sewage treatment plant by an automatic, in-line sampler that collected and pooled samples over a 24-h period (16). The first WPV1-positive sample was collected on March 11, 2013, and this date is designated time (note that the beginning of the outbreak is some unknown time before , as there was likely a delay between initial infection and the first detection of pathogens in the sewage). WPV1, OPV1, and OPV3 strains were separately quantified. Data are available approximately weekly, and our analysis considers samples analyzed through the end of December 2013 (SI Appendix). Some sewage samples are associated with multiple data points: Genetic material was isolated in duplicate (or more) and separately assayed. These qPCR results are treated as distinct data points.
Vaccination Campaigns.
Israel conducted two bOPV campaigns in the region. The first began on August 5, 2013 and reached 90% coverage in children under 10 y old. The second began on October 7, 2013 and achieved 53% coverage (10). We assume that the vaccination rate did not vary during the vaccination campaigns and estimated an exponential vaccination rate of 0.074/d for both campaigns from the coverage time-series data (with the vaccination rate set to zero when the campaign was not occurring). We assumed the vaccination take rate was 80% (19).
Mathematical Modeling
Measurement Model: Connecting Concentration and Cycle Threshold.
The presence of poliovirus (WPV1, OPV1, and OPV3 separately) in sewage samples was assessed by quantitative, real-time PCR (7). Quantitative PCR targets a DNA sequence that is copied in each PCR cycle. This method determines the number of amplification cycles needed for the number of copies of the targeted DNA sequence, called the copy number, to reach a threshold number (set by the test operator above the background noise). This number of doubling cycles needed for a sample to achieve the threshold (in essence, become detectable above the level of background noise) is called the cycle threshold (CT), which we denote . A higher CT value thus indicates a lower concentration of virus in the original sample.
Connecting the measured CT data to the concentration of poliovirus in sewage samples requires a measurement model. Each qPCR cycle approximately doubles the copy number of DNA segments (Fig. 1A), so that , where is the threshold copy number, is the poliovirus concentration (i.e., the initial copy number in the sample), is the reaction efficiency [typically 90–110% (20)], and is the CT value (Fig. 1B). Rearranging, we see that is linearly related to the log of the concentration:
| [1] |
We assume (i.e., 100% reaction efficiency or perfect doubling), which simplifies Eq. 1 to
| [2] |
In general, experimental information about the efficiency could be used to change the logarithm base here.
Fig. 1.
(A) Each qPCR amplification cycle doubles the number of targeted DNA segments. (B) qPCR cycles are repeated until the number of copies of the DNA sequence (copy number) crosses the threshold number . The number of cycles needed to cross this threshold is called the cycle threshold (CT), denoted . The initial copy number is denoted .
The laboratory method used to obtain CT values for the 2013 outbreak has been described elsewhere (7, 21–23). The limit of detection in this assay was on the order of 40 cycles, but the experiments were run to 60 cycles to distinguish between negative samples and samples near the limit of detection. Samples with CT greater than 45 were considered to be negative, while samples with CT between 37 and 45 were reanalyzed (more details are available in ref. 21). Here, samples with a CT value of 60 are considered negative for poliovirus. The results were not sensitive to this choice of value under our inference framework (details in Parameter Estimation). Although the threshold level is typically knowable from the experimental setup, we treat it as unknown in this analysis as it is often not recorded. Alternatively, if a quantifying standard is available and run alongside the samples, the CT values can be transformed into concentrations directly for use in the model.
Infectious Disease Model.
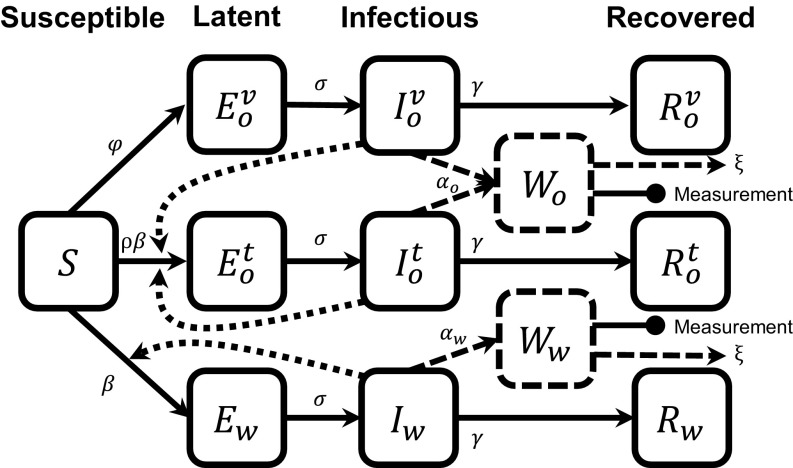
We extend a susceptible, exposed, infectious, and recovered (SEIR) model (SI Appendix) to account for infection by WPV1, vaccination, and subsequent transmission of OPV1 (as part of bOPV) reflected in the 2013 Rahat polio sewage data (Fig. 2, Eqs. 3 and 4, and Table 1). Individuals may be latent, infectious, or recovered with either WPV1 (subscript ) or OPV1 (subscript ) strains. We differentiate between individuals who have the OPV1 strain from vaccination (superscript ) and from transmission (superscript ). OPV3 is modeled analogously to OPV1 (details and equations in SI Appendix).
Fig. 2.
An SEIR-type model for poliovirus, incorporating vaccination and environmental surveillance. The model represents infection by two strains of poliovirus (OPV1 as part of bOPV, with subscript , and WPV1, with subscript ). Individuals infected with OPV1 acquire it through vaccination (superscript ) or transmission (superscript ). Model equations are given in Eqs. 3 and 4, and parameters are in Table 1.
Table 1.
| Parameter | Definition |
| WPV1 transmission rate, 1/d | |
| Ratio of OPV1 to WPV1 transmission rates | |
| Vaccination rate, 1/d | |
| Rate of transition from latent to infectious state, 1/d | |
| Rate of recovery from poliovirus infection, 1/d | |
| Shedding rate for WPV1, copies per sewage volume per day | |
| Shedding rate for OPV1, copies per sewage volume per day | |
| Rate of removal of poliovirus from sewage, 1/d | |
| Copy number threshold, copies per sample volume |
The susceptible compartment includes people who have not previously received OPV and those whose gut immunity has waned. Because Israel transitioned from OPV to IPV in 2005, the target population is predominantly children under age 10 y (24). The size of our target population is unknown and cannot be determined from environmental surveillance alone (and could not be determined even with case data without immunological profiles and movement patterns). Hence, compartments , , , and are scaled to represent the fraction of this (unknown total) population. Our target population consists only of people who are susceptible at the start of the outbreak. Accordingly, there are no recovered individuals in the target population at the start of the outbreak (). Our simulation spans less than 1 y, so we neglect waning immunity and vital dynamics (25).
Susceptible individuals can be vaccinated at a rate , becoming latently infected with the vaccine strain (), or can be infected via transmission, becoming latent with the WPV1 () or OPV1 strain (). Although individuals in any compartment can be vaccinated, we assume that there is no effect on individuals already infected with either strain. Latent individuals progress to the corresponding infectious compartment (, , or ). Infectious individuals are those who can directly transmit pathogens to other individuals, through fecal–oral or oral–oral transmission. Once infectious, individuals shed virus to the sewage compartments ( and ) through their stools. Recovered individuals (, , and ) do not shed and do not transmit virus. We assume that the latent and infectious periods for the WPV1 and OPV1 strains are the same.
The and compartments model the concentration of WPV1 and OPV1 in sewage where environmental samples are taken, given in copy number per sample volume. Infectious individuals increase the environmental concentration with rates and , which take into account the rate at which virus is shed, the fraction of shed virus that is deposited into the sewage, and the total volume of the sewage. We assume that virus is removed from sewage at rate for both strains, accounting for both sewage flow and pathogen die-off.
Although this model is inspired by other models with environmental compartments, such as the susceptible, infectious, water, and recovered (SIWR) model (26, 27) and the environmental infection transmission system (EITS) model (28, 29), this is not a model of environmentally mediated transmission; i.e., we assume that there is no transmission resulting from ingestion of pathogens from the environment (in this case, sewage). The compartments solely represent reservoirs for environmental surveillance. If the environmental surveillance is analyzed by qPCR, the sample concentration (Eq. 2) corresponds to the concentration of model compartment (Eq. 4) when concentration is given in number of viral genomes per qPCR sample volume.
The equations governing the dynamics of people are
| [3] |
and the equations governing poliovirus concentration in the sewage are
| [4] |
The basic reproductive number, (i.e., the number of secondary infections produced from one infected individual in a completely susceptible population) for this model is the of the WPV1 submodel, .
Identifiability Analysis.
Identifiability analysis is a necessary precursor to parameter estimation from data. Structural identifiability is a theoretical analysis that determines whether model parameters can be uniquely estimated from perfectly observed output trajectories; this analysis reveals inherent barriers to parameter estimation in the model structure. When parameters are not individually identifiable, we can find groups of parameters that can be uniquely determined, called identifiable parameter combinations (30). Once the parameter identifiability is known in the idealized case, practical identifiability analysis then asks whether there are additional barriers to uniquely estimating parameters when real-world data, and the associated variability, are considered (31). Observability is analogous to identifiability but concerns whether the trajectories of state variables, rather than model parameter values, can be recovered from the observed quantities (32, 33).
In this analysis, we are fitting a disease transmission model to a form of data not previously used in this context, i.e., qPCR CT data. It is not a priori clear which infectious disease transmission model parameters are estimable from this kind of data. Hence, we first consider the structural identifiability of this model and the observability of its states, given this kind of data. Second, we use practical identifiability methods to estimate the identifiable parameters or parameter combinations in the context of the Israel polio outbreak. All of the identifiability details and results are in SI Appendix. In brief, we are able to recover the transmission and recovery rates, fractions of the population in each disease stage, and scaled WPV1 and OPV1 concentrations in the sewage. Without additional assumptions or data, we are not able to recover the numbers of individuals in each disease stage, the absolute polio concentrations, or the pathogen shedding or decay rates from the qPCR environmental surveillance data in this modeling framework.
Model Reparameterization.
One important implication of our identifiability analysis is that the concentration of poliovirus can be determined only up to a constant; i.e., we determine the scaled concentrations,
| [5] |
This result is due in part to the ratio in the measurement equation (Eq. 2). The data determine only the ratio , so the actual values of and could be proportionally higher or lower. Moreover, and are in an identifiable combination (details in SI Appendix). After the rescaling, we have
| [6] |
for the full outbreak model. The scaled concentrations take values between 0 and 1 and represent the fraction of the theoretical maximum concentration that would result if the entire population was shedding.
A second implication of the identifiability analysis is that the pathogen decay rate can be arbitrarily large and still fit the data well. Dynamically, this means that and closely follow the values of and , respectively, and
| [7] |
Consequently, we use the measurement equations
| [8] |
to relate the cycle threshold data to the infection prevalence . Even when the individual parameters are not separately identifiable, the parameter combinations and may be identifiable (and are in this analysis).
Simulation.
We fitted the full model (Eq. 3) to the WPV1 and OPV1 qPCR CT data with the scaled measurement equation (Eq. 8); we also fitted to the OPV3 data, as discussed in SI Appendix. At time (March 11, 2013), we assume no vaccination (i.e., , ) and, as previously discussed, that there are no recovered people in the modeled population (). The vaccination rate becomes a fixed, nonzero constant during the two vaccination campaigns, which began August 5 and October 7, 2013 and are modeled as lasting 31 d and 10 d, respectively, and is set to zero at all other times. Parameters and are fixed, whereas parameters , , , and are estimated. The initial condition is estimated. To determine , we simulate a model trajectory with the same parameters but initial conditions , , . is set to the value of at the time when ; i.e., is chosen to be consistent with . All simulations and analyses were done in R (v.3.4.1); we used deSolve for ordinary differential equation model simulation and the David–Fletcher–Powell algorithm in the Bhat package for maximum-likelihood estimation (34, 35).
Parameter Estimation.
It is reasonable to expect lower CT values to be associated with higher variance given that viral particles might be Poisson distributed in sewage samples. Alternatively, higher CT values might have greater variance as differences are magnified and errors are accumulated with each PCR cycle. Regardless, there is no natural variance structure for qPCR CT data, which is a log-transformation of concentration. Further, values are capped at 60, which corresponds in this study to absence of the virus. Consequently, a least-squares approach to parameter estimation is problematic. We found that a sum of absolute differences () approach on the CT scale was a sensible alternative, heuristically resulting in the median trajectory instead of the mean trajectory, eliminating the sensitivity of the results to the choice of 60 as the “absence of poliovirus” value. We confirmed that the choice of the CT value of negative samples does not change the model fit or parameter estimation results, as long as the model trajectory does not cross this value; i.e., setting the negative value to a CT of 45 or higher here gives equivalent results. This approach is equivalent to maximum-likelihood estimation under the assumption that errors are Laplace distributed, and we minimize the negative log-likelihood,
| [9] |
where are the CT data, is the variance of the error distribution of the data, are the modeled CT values, and is the vector of model parameters. We estimate = 0.98 from the multiple qPCR tests of sewage samples collected on certain days.
Uncertainty Quantification.
Likelihood-based 95% CIs for parameter estimates were determined by parameter profile likelihoods (details in SI Appendix). These profile likelihoods are transects through the 95% confidence region of parameter space. We simulated trajectories for each parameter vector in each profile and estimated the 95% CIs for the maximum-likelihood trajectory to be the maximum and minimum at each time point in this set of simulated trajectories.
Results
Polio Epidemic Dynamics in Rahat, Israel, 2013.
Our analysis estimates that peak concentration of WPV1 occurred in late June (Fig. 3A), while the peak concentration of the OPV1 strain occurred shortly after introduction of the bOPV vaccine in August (Fig. 3B), with a moderate rise for WPV1 and a rapid, steep rise for OPV1 following onset of vaccination. We plot CT data on an inverted axis because a higher CT value indicates a lower concentration. Corresponding figures on the linear scale are provided in SI Appendix. Both model fits follow the observed trends in CT values over the outbreak.
Fig. 3.
PCR CT data and model fits for (A) WPV1 and (B) OPV1 strains in sewage. The PCR detection limit corresponds to about a CT of 40, and experiments were run to 60. The ribbons give the CIs for the maximum-likelihood trajectory using likelihood-based estimates of the 95% confidence parameter region. The gray bars give the approximate time periods of the bOPV campaigns.
Based on these estimated concentration values, our model simulations predict a peak in WPV1 infections around June 26, approximately 6 wk before the bOPV vaccination campaign began on August 5 (Fig. 4). By August 5, 56% (95% CI 9–75%) of the susceptible population was infected with WPV1. Infection from OPV1 was more rapid and peaked in late August, midway through the first vaccination campaign. Most OPV1 infections were from direct vaccination, and there was comparatively little person-to-person transmission of the vaccine strain to susceptible individuals.
Fig. 4.
Modeled fractions of the population that were infected with WPV1, OPV1 through vaccination, and OPV1 through transmission. The ribbons give the CIs for the maximum-likelihood trajectory using likelihood-based estimates of the 95% confidence parameter region. The gray bars give the approximate time periods of the bOPV campaigns.
We estimated five model parameters (Table 2; parameter profiles quantifying the CIs are provided in SI Appendix). From the model fit, we estimate was 1.62 (95% CI 1.04–2.02) for this outbreak.
Table 2.
Fixed and estimated model parameter values
| Parameter | Value (source or 95% CI) |
| Fixed | |
| Ratio of OPV1 to WPV1 transmission | |
| rate, | 0.37 (36) |
| Vaccination rate, | 0.8 0.074/d (10, 19) |
| Rate of progressing from latency, | 0.25/d (37, 38) |
| Estimated | |
| WPV1 transmission rate, | 0.151/d (0.108, 0.174) |
| Recovery rate, | 0.93/d (0.078, 0.108) |
| OPV1 sewage scaling parameter, | 2.02 (1.1, 4.2) 107 |
| WPV1 sewage scaling parameter, | 1.07 (0.03, 1.66) 107 |
| Fraction infected at , | 4.34 (0.39, 14.4) 10−3 |
Comparison with Stool Sample Data.
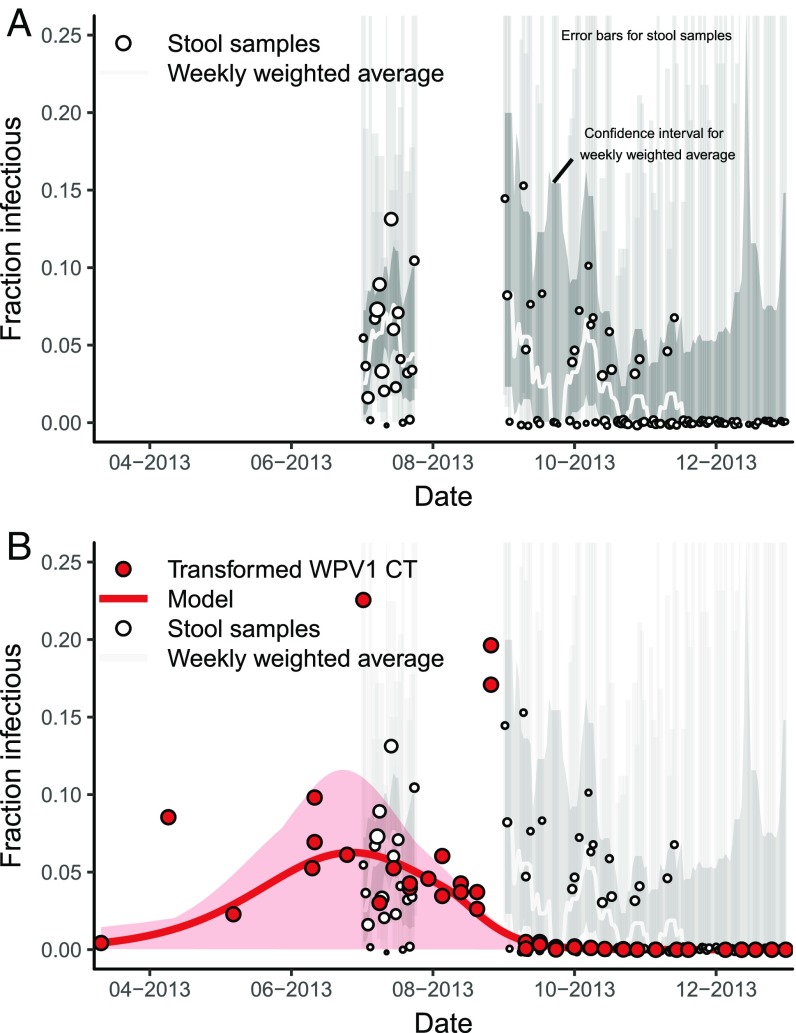
The previous estimate of the trajectory of the Israeli polio epidemic was based on convenience stool samples of children under 10 y old who had not been vaccinated with OPV before the outbreak (10). Stool sampling occurred in two phases—between July 2 and 24 and between September 1 and June 26 of the following year—and resulted in 2,196 nonduplicate samples of children under 10 y old in total. Stools were collected from healthy children in daycares, health clinics, and other convenience locations across the Negev region of southern Israel (24). Children in the first phase were OPV naive but many or most children in the second phase would have been vaccinated in the bOPV campaign. The surveys were intended to detect the presence or absence of WPV1 and estimate poliovirus excretion rates but were not designed to assess infection prevalence [in particular, older children and adults were underrepresented in the original stool sample and were not included in the previous transmission analysis (10)]. The number of samples collected daily varied widely, with a median number of 38 in the first phase and 12 in the second phase. Despite the limitations of the dataset for estimating population-level prevalence, it is the only direct measure of wild poliovirus infection for this epidemic.
The previous modeling analysis of these stool samples indicated that the outbreak peaked in mid-August (10), but there is considerable uncertainty because of the variability and convenience sampling nature of the samples and because there are no samples in the crucial late summer period (Fig. 5A; a plot scaled to show the full data uncertainty is provided in SI Appendix).
Fig. 5.
(A) Prevalence of WPV1 in stool of a convenience sample of under 10-y olds in Southern Israel, where point size is scaled to sample size, with weekly weighted moving average and 95% Clopper–Pearson CIs for the data points and the weekly average. (B) Modeled fraction of infectious people (with 95% CI for the maximum-likelihood estimate) based on environmental surveillance data in Rahat, Israel (WPV1 CT values transformed by the model) and the stool sample prevalence (CIs faded for readability). The environmental surveillance data suggest an earlier peak than the stool samples do. The stool samples and environmental surveillance represent different populations during the same outbreak.
On the other hand, the qPCR environmental sample data represent a different population that experienced the same outbreak (all initially susceptible, not just under 10-y olds, but only those in Rahat, rather than across the Negev). Our analysis of the qPCR environmental surveillance data, which spans the entire period, suggests different outbreak dynamics, with the outbreak peaking approximately 7 wk earlier than previous analysis of the stool samples suggested (Fig. 5B).
Alternative Vaccination Campaign Timing.
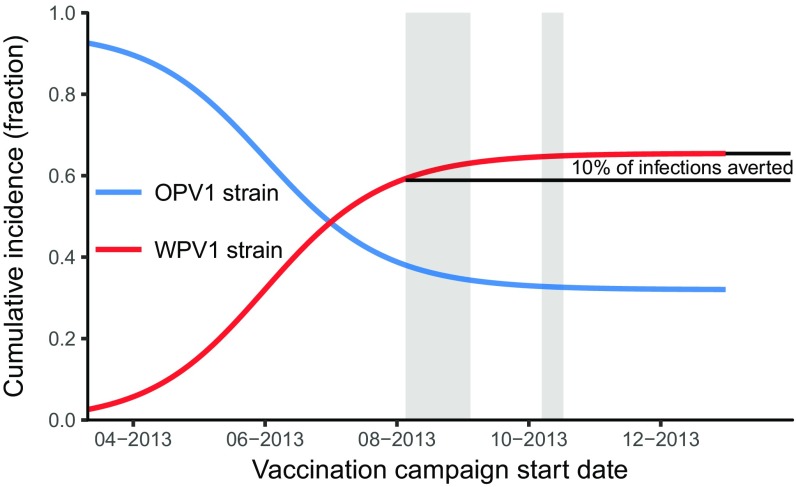
We estimate that the cumulative incidence (i.e., attack ratio) without any vaccination campaign would have been 0.65 (95% CI 0.10–0.80) (Fig. 6). This estimate is consistent with final size calculations using our estimate of = = 1.62 (39, 40). For the true August 5 vaccination start date, we estimate that cumulative incidence was 0.59 (95% CI 0.09–0.77) and that approximately 10% (95%CI 1–24%) of infections were averted by the vaccination campaign. Simulations of earlier implementation of the bOPV campaign resulted in a lower attack ratio.
Fig. 6.
Simulated cumulative incidence (fraction) of the population infected with WPV1 and OPV1 virus strains over the course of the outbreak as a function of the vaccination campaign start date. The relative campaign timing and rates are held constant while the start date changes. The gray bars give the dates of the bOPV campaigns. Earlier vaccination dates result in a larger fraction of individuals infected with the vaccine strain and a smaller fraction with the wild strain.
Discussion
Using high-quality, time-series environmental surveillance data, we provide epidemiological insight into the 2013–2014 polio outbreak in Israel, demonstrating the promise for direct qPCR analysis of environmental samples as a rapid, sensitive means of public health surveillance. To this end, we developed a framework to incorporate qPCR data into a compartmental, deterministic SEIR-type infectious disease transmission model and reanalyzed the 2013–2014 polio outbreak in Israel with environmental surveillance data alone.
Epidemiological Implications.
In the short time between the introduction of WPV1 into Rahat (February–March 2013) and the start of the vaccination campaign (August 2013), Rahat experienced substantial transmission of WPV1. Because of the variability and early sparseness of the PCR CT data, there is considerable uncertainty in the exact epidemic curve; small differences in trajectories through the CT data translate to substantial differences on the population scale. Moreover, while we have captured the uncertainty given our choice of model and likelihood structure, there is additional epistemological uncertainty that is not captured, surrounding both the model structure and the choice of error structure and scale. Nevertheless, while the exact impact of the vaccination campaign is uncertain, our analysis of the data indicates that the epidemic peaked in midsummer and therefore was in decline when vaccination began.
From environmental sampling data in other Bedouin communities in the region, we know that polio transmission was not limited to Rahat, which is the largest predominantly Bedouin community in the region. Indeed, movement patterns likely contributed to sustained regional transmission. Moreover, a large percentage of the Bedouin population in the Negev live in unrecognized communities with little-to-no WASH infrastructure (9). Because these communities have no centralized sewage system, they are outside of the scope of the Israeli environmental surveillance system but likely had high polio transmission. A full regional epidemiological analysis of the spatiotemporal and cultural patterns of the outbreak is a natural next step, which will require an analysis of sewage samples collected throughout Israel.
This epidemic further highlights the complexities and difficulties of polio vaccine policy as we approach eradication. The decision to replace OPV with IPV balances the risk of wild polio importation with the risk of VAPP. Because IPV prevents adverse neural outcomes (AFP) but does not prevent fecal–oral transmission, poliovirus can circulate, particularly when fecal–oral transmission is enhanced by poor WASH infrastructure. Moreover, because IPV prevents AFP, AFP surveillance systems will not catch the outbreak early, allowing silent circulation to put a greater number of unvaccinated people at risk for paralysis. However, use of OPV elevates the risk of strain mutation to a neurovirulent form. Localized poor WASH conditions and the greater proportion of children in Bedouin communities, combined with unexpected outbreaks in neighboring countries, led to the 2013–2014 outbreak in Israel. Fortunately, the early environmental detection and subsequent vaccination campaigns ensured that there were no cases of AFP. Additionally, we cannot know how many cases of VAPP were prevented since the switch from OPV to IPV in 2005 and previous use of IPV followed by OPV (during which periods there were no cases of VAPP).
The model parameter estimates are largely reasonable, although the estimated mean infectious period of 11 d (corresponding to = 0.09) is lower than values reported in the literature, even if much of the population has received IPV (41). This estimate may be a consequence of variable shedding intensity over time, as viral concentrations in stool drop off after the first week of infection (41, 42). Since infectious individuals are measured through their shedding into the sewage, a drop in shedding intensity will be interpreted by the model as a shorter duration of infection; i.e., individuals shedding low levels of the virus will be treated as recovered, even if they still harbor the infection. Thus, we consider the parameter estimate to be an estimate of the duration of peak shedding rather than true infection duration.
Yaari et al. (10) previously analyzed the 2013–2014 Israel polio outbreak using a combination of a subset of stool surveys (24) (from healthy children and infants at daycare centers or routine visits to healthcare clinics) and supplementary vaccination and demographic information to fit a disease transmission model. From these data, the authors estimated the outbreak occurred in mid-August, and they estimated the basic reproduction number for the outbreak to be 1.77 (1.46 –2.30). The environmental and stool sample data suggest different outbreak timing, which has led to the different analyses and interpretations. There are several possible explanations for these differences. The stool sample data may not be a reliable measure of population infection prevalence: These data came from small, convenience samples that were not designed to be fully representative either of the whole population [older children (10+ y old) and adults were underrepresented in the original sample and not included in Yaari et al.’s (10) analysis] or of the likely susceptible population. Alternatively, the stool data may be an accurate, if noisy, reflection of infection prevalence in the under 10-y-old population. In that case, our analysis of the environmental surveillance data suggests an alternative shape with which to interpret the stool data: The data in the first survey may represent the time following peak prevalence rather than a ramp up (Fig. 5B). In some sense it is not surprising that there is a higher prevalence of WPV1 in the stool data than predicted by our analysis since the underlying populations differ: Our analysis is of the initially susceptible population of Rahat (with unknown age distribution), while the stool sample data represent only under 10-y olds sampled from across the region. Finally, because our model may treat people who are still infected but are shedding minimally as recovered, as discussed above, the true epidemic curve may have a longer tail than our analysis suggests, which is consistent with the existence of positive stool samples in October and November.
A number of assumptions were used in our analysis. First, we assume that sewage trunk lines are well-mixed representations of the population’s sewage as a whole. Consequently, the sewage scaling parameter also accounts for discrepancies between the population captured by the sewage and the true population, like a reporting rate does. Ultimately, microbiologists may need to take an epidemiological perspective when developing their sampling plans for grab samples that are not well mixed. Second, we did not consider the possibility of partial immunity. Individuals with partial immunity would have different transmission rates and would change the number of susceptibles in the population (43). Since we relied on environmental data alone, we do not know the immune status or other characteristics of individuals, such as age. Finally, the assumption that WPV1, OPV1, and OPV3 have the same duration of infectiousness (36) was important for this model, because it allowed the model to use the OPV data to add confidence to the model fit of the decline in WPV1 CT values, which, on its own, is influenced by the negative tests late in the year.
In the prevaccination era, cases of polio were strongly seasonal in temperate areas; the cause of this seasonality is unknown, but may be related to temperature or humidity (3). The average monthly high temperature in this region of Israel ranges from 16.C (F) in January to 33.3C (F) in July, while the relative humidity ranges from 35% in May to 50% in January (https://www.weather-atlas.com/en/israel/beer-sheva-climate). It is unclear whether these climatic variations would substantially impact poliovirus transmission. We modeled seasonal variation in transmission with a periodic function (model fits and parameter estimates are included in SI Appendix). However, the improvement in fit was negligible and did not greatly affect the other parameter estimates, suggesting that seasonality was not an important factor driving transmission. We, therefore, did not include seasonal variation in our final analysis.
Environmental Surveillance and qPCR.
Since the 2013–2014 Israeli epidemic, the World Health Organization and the Global Polio Eradication Initiative have added additional environmental surveillance sites in the remaining polio endemic countries (Nigeria, Afghanistan, and Pakistan) to help detect poliovirus circulation (44, 45). The Israeli epidemic highlighted the value of a highly sensitive environmental surveillance program during the final phases of global eradication. Areas with poor WASH infrastructure—where enteric pathogens such as poliovirus can contribute to sustained transmission (46, 47)—are particularly good candidates for environmental surveillance programs. Environmental surveillance is also well suited to highly vaccinated populations, where paralytic polio is unlikely. Although AFP surveillance remains the standard for polio detection (48), as we approach eradication, silent polio circulation will increasingly be detected through environmental surveillance before AFP cases occur (49, 50). Indeed, early detection is critical to interrupting silent poliovirus circulation by allowing for earlier mobilization of public health responses and vaccination campaigns before the first case of AFP is identified. Environmental surveillance may also be warranted in certified polio-free countries. The recent outbreak of vaccine-derived poliovirus 1 (VDPV1) in Papua New Guinea was detected only by polio case surveillance (51).
However, the current standard method of environmental surveillance of polio is the tissue-culture–based intratypic differentiation (ITD) assay, which is slower than direct qPCR and not quantitative (52, 53). Although presence/absence data are valuable, we should move to quantitative assays as we approach eradication. Because direct qPCR is fast and relatively cheap, the frequency and extent of environmental surveillance could be amplified and used to fit spatial models of transmission. Frequent sampling is essential if we are to understand the underlying dynamics with confidence, as our analysis highlights. With well-planned environmental surveillance, these models may help, for example, to assess the various hypotheses of sustained WPV1 transmission in Pakistan (49). Additionally, initial results suggest that there is a strong correlation between plaque assay and qPCR results, and an analogous modeling and inference framework to incorporate plaque assay data could be developed when those data are available.
Modeling Framework and Identifiability.
Previous environmentally mediated infectious disease modeling work has demonstrated that quantitative environmental surveillance data can improve parameter identifiability and estimation for environmentally mediated infectious disease transmission models when they are used in combination with incidence or prevalence data (27, 29, 54, 55). However, this previous work has considered only simulated environmental surveillance data or environmental surveillance in conjunction with prevalence or incidence data. Since there were no paralytic cases in the 2013–2014 Israeli outbreak, the only case data are stool data that are temporarily limited, are uncertain, and may not be representative of the population at large (24). As a result, this outbreak provided an opportunity to fit a model to time-series PCR data from the environment and an ideal situation to test the potential use of environmental surveillance data alone for model-based prediction and estimation. Because polio transmission will increasingly be detected only through environmental surveillance, a strong modeling and parameter estimation framework for transmission modeling with only environmental surveillance data is necessary. The methods used in this analysis provide such a framework and can be used to understand future disease outbreaks.
The framework presented here may also be modified to examine, for example, surveillance from nonsewage environments (such as environmental water sources, sewage canals, etc.) and could be used in a stochastic setting to examine smaller outbreaks as we approach eradication (potentially using estimation methods better suited for stochastic models, such as refs. 56–58). The methods are also not limited to polio. Many other pathogens can be detected in stool or other environmental reservoirs. There is particular interest in implementing environmental surveillance for panels of enteric pathogens, and methods of detection are being developed (59–61). If the pathogens are transmitted through the environment, the model we developed here will need to be extended to include that transmission pathway (26–29).
Conclusion
Importations of wild and vaccine-derived poliovirus are expected to continue to occur sporadically. Environmental surveillance is particularly useful for detecting silent circulation of the disease in these situations. Our modeling and parameter estimation framework can be used to transform environmental surveillance data into estimates of incidence. Enhancement of environmental surveillance in the remaining polio-endemic countries—and the use of quantitative PCR, in particular—could hasten eradication.
Supporting Information
SI Appendix.
The SI Appendix (i) includes proofs of the identifiability and model reparameterization results, including the practical unidentifiability of the pathogen removal rate; (ii) provides the profile-likelihood plots and parameter estimate CIs; (iii) provides details on the modeling and results for OPV3; (iv) gives model fits on the linear rather than log scale; (v) provides the stool data with the plot scaled to view the full data uncertainty; and (vi) provides details on a model with seasonal variation in polio transmission.
Datasets S1–S4.
The WPV1, OPV1, and OPV3 qPCR CT data used in this analysis are included. The relevant stool sample data for children under 10 y old, available from ref. 10, are included for the reader’s convenience.
Supplementary Material
Acknowledgments
We thank Ella Mendelson, Danit Sofer, and Merav Weil of the Central Virology Laboratory for their contributions. We also thank the editor and two anonymous reviewers for their insights and comments in revising the manuscript. This project was funded through Models of Infectious Disease Agent Study program within the National Institute of General Medical Sciences of the National Institutes of Health (Grants U01GM110712 and U54GM111274), the National Science Foundation (Grants OCE-1115881 and EAR-1360330), and the World Health Organization (WHO) (Grant 353558 TSA 2014/485861-0). Funding for the Chaim Sheba Central Virology Laboratory was provided by the Israel Ministry of Health and the WHO (Grant 18-TSA-032).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808798115/-/DCSupplemental.
References
- 1.Grassly NC. The final stages of the global eradication of poliomyelitis. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120140. doi: 10.1098/rstb.2012.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Routh J, Oberste S, Patel M. Poliomyelitis. In: Roush SW, Baldy LM, editors. Manual for the Surveillance of Vaccine-Preventable Diseases. Centers for Disease Control and Prevention; Atlanta: 2017. [Google Scholar]
- 3.Nathanson N, Kew OM. From emergence to eradication: The epidemiology of poliomyelitis deconstructed. Am J Epidemiol. 2010;172:1213–1229. doi: 10.1093/aje/kwq320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anis E, et al. Insidious reintroduction of wild poliovirus into Israel, 2013. Euro Surveill. 2013;18:20586. doi: 10.2807/1560-7917.es2013.18.38.20586. [DOI] [PubMed] [Google Scholar]
- 5.Roberts L. Israel’s silent polio epidemic breaks all the rules. Science. 2013;342:679–680. doi: 10.1126/science.342.6159.679. [DOI] [PubMed] [Google Scholar]
- 6.Kopel E, Kaliner E, Grotto I. Lessons from a public health emergency—importation of wild poliovirus to Israel. New Engl J Med. 2014;371:981–983. doi: 10.1056/NEJMp1406250. [DOI] [PubMed] [Google Scholar]
- 7.Shulman LM, et al. Laboratory challenges in response to silent introduction and sustained transmission of wild poliovirus type 1 in Israel during 2013. J Infect Dis. 2014;210:S304–S314. doi: 10.1093/infdis/jiu294. [DOI] [PubMed] [Google Scholar]
- 8.Shulman LM, et al. Molecular epidemiology of silent introduction and sustained transmission of wild poliovirus type 1, Israel, 2013. Euro Surveill. 2014;19:1–8. doi: 10.2807/1560-7917.es2014.19.7.20709. [DOI] [PubMed] [Google Scholar]
- 9.Shmueli DF, Khamaisi R. Bedouin communities in the Negev. J Am Plann Assoc. 2011;77:109–125. [Google Scholar]
- 10.Yaari R, et al. Modeling the spread of polio in an IPV-vaccinated population: Lessons learned from the 2013 silent outbreak in southern Israel. BMC Med. 2016;14:95. doi: 10.1186/s12916-016-0637-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Platt LR, Estivariz CF, Sutter RW. Vaccine-associated paralytic poliomyelitis: A review of the epidemiology and estimation of the global burden. J Infect Dis. 2014;210:S380–S389. doi: 10.1093/infdis/jiu184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns CC, Diop OM, Sutter RW, Kew OM. Vaccine-derived polioviruses. J Infect Dis. 2014;210:S283–S293. doi: 10.1093/infdis/jiu295. [DOI] [PubMed] [Google Scholar]
- 13.Thompson KM, Duintjer Tebbens RJ. Health and economic consequences of different options for timing the coordinated global cessation of the three oral poliovirus vaccine serotypes. BMC Infect Dis. 2015;15:374. doi: 10.1186/s12879-015-1113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson KM, Kalkowska DA, Duintjer Tebbens RJ. Managing population immunity to reduce or eliminate the risks of circulation following the importation of polioviruses. Vaccine. 2015;33:1568–1577. doi: 10.1016/j.vaccine.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swirski S, Hasson Y. 2006. Invisible citizens: Israel government policy toward the Negev Bedouin (Adva Center, Tel Aviv)
- 16.Manor Y, et al. Intensified environmental surveillance supporting the response to wild poliovirus type 1 silent circulation in Israel, 2013. Euro Surveill. 2014;19:20708. doi: 10.2807/1560-7917.es2014.19.7.20708. [DOI] [PubMed] [Google Scholar]
- 17.Kilpatrick DR, et al. Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J Clin Microbiol. 2009;47:1939–1941. doi: 10.1128/JCM.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berchenko Y, et al. Estimation of polio infection prevalence from environmental surveillance data. Sci Transl Med. 2017;9:eaaf6786. doi: 10.1126/scitranslmed.aaf6786. [DOI] [PubMed] [Google Scholar]
- 19.Kalkowska DA, et al. Modeling options to manage type 1 wild poliovirus imported into Israel in 2013. J Infect Dis. 2015;211:1800–1812. doi: 10.1093/infdis/jiu674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Applied Biosystems 2015 Real-time PCR: Understanding Ct, Technical Report. Available at www3.appliedbiosystems.com/cms/groups/mcb_marketing/documents/generaldocuments/cms_053906.pdf. Accessed October 5, 2018.
- 21.Hindiyeh MY, et al. Development and validation of a real time quantitative reverse transcription-polymerase chain reaction (QRT-PCR) assay for investigation of wild poliovirus type 1-South Asian (SOAS) strain reintroduced into Israel, 2013 to 2014. Euro Surveill. 2014;19:1–7. doi: 10.2807/1560-7917.es2014.19.7.20710. [DOI] [PubMed] [Google Scholar]
- 22.Shulman LM, et al. Neurovirulent vaccine-derived polioviruses in sewage from highly immune populations. PLoS ONE. 2006;1:e69. doi: 10.1371/journal.pone.0000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manor Y, et al. Advanced environmental surveillance and molecular analyses indicate separate importations rather than endemic circulation of wild type 1 poliovirus in Gaza district in 2002. Appl Environ Microbiol. 2007;73:5954–5958. doi: 10.1128/AEM.02537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran-Gilad J, et al. Field study of fecal excretion as a decision support tool in response to silent reintroduction of wild-type poliovirus 1 into Israel. J Clin Virol. 2015;66:51–55. doi: 10.1016/j.jcv.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Mayer BT, et al. Successes and shortcomings of polio eradication: A transmission modeling analysis. Am J Epidemiol. 2013;117:1236–1245. doi: 10.1093/aje/kws378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tien JH, Earn DJD. Multiple transmission pathways and disease dynamics in a waterborne pathogen model. Bull Math Biol. 2010;72:1506–1533. doi: 10.1007/s11538-010-9507-6. [DOI] [PubMed] [Google Scholar]
- 27.Eisenberg MC, Robertson SL, Tien JH. Identifiability and estimation of multiple transmission pathways in cholera and waterborne disease. J Theor Biol. 2013;324:84–102. doi: 10.1016/j.jtbi.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Spicknall IH, Koopman JS, Eisenberg JNS. Dynamics and control of infections transmitted from person to person through the environment. Am J Epidemiol. 2009;170:257–265. doi: 10.1093/aje/kwp116. [DOI] [PubMed] [Google Scholar]
- 29.Brouwer AF, et al. Dose-response relationships for environmentally mediated infectious disease transmission models. PLoS Comput Biol. 2017;13:e1005481. doi: 10.1371/journal.pcbi.1005481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cobelli C, DiStefano JJ. Parameter and structural identifiability concepts and ambiguities: A critical review and analysis. Am J Physiol. 1980;239:R7–R24. doi: 10.1152/ajpregu.1980.239.1.R7. [DOI] [PubMed] [Google Scholar]
- 31.Raue A, et al. Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics. 2009;25:1923–1929. doi: 10.1093/bioinformatics/btp358. [DOI] [PubMed] [Google Scholar]
- 32.Griffith EW, Kumar KSP. On the observability of nonlinear systems: I. J Math Anal Appl. 1971;35:135–147. [Google Scholar]
- 33.Hwang M, Seinfeld JH. Observability of nonlinear systems. J Optim Theor Appl. 1972;10:67–77. [Google Scholar]
- 34.Soetaert K, Petzoldt T, Setzer W. Package ‘deSolve’. 2017 Available at https://cran.r-project.org/web/packages/deSolve/deSolve.pdf. Accessed October 5, 2018.
- 35.Luebeck G, Meza R. Bhat: General likelihood exploration. 2015 Available at https://cran.r-project.org/web/packages/Bhat/Bhat.pdf. Accessed October 5, 2018.
- 36.Duintjer Tebbens RJ, et al. Characterizing poliovirus transmission and evolution: Insights from modeling experiences with wild and vaccine-related polioviruses. Risk Anal. 2013;33:703–749. doi: 10.1111/risa.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul J. World Health Organization Monograph Series No 26. World Health Organization; Geneva: 1955. Epidemiology of poliomyelitis; pp. 9–29. [PubMed] [Google Scholar]
- 38.Krugman S, et al. Immunization with live attenuated poliovirus vaccine. Am J Dis Child. 1961;101:23–29. doi: 10.1001/archpedi.1961.04020020025005. [DOI] [PubMed] [Google Scholar]
- 39.Ma J, Earn DJ. Generality of the final size formula for an epidemic of a newly invading infectious disease. Bull Math Biol. 2006;68:679–702. doi: 10.1007/s11538-005-9047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller JC. A note on the derivation of epidemic final sizes. Bull Math Biol. 2012;74:2125–2141. doi: 10.1007/s11538-012-9749-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Famulare M, Selinger C, McCarthy KA, Eckhoff PA, Chabot-Couture G. Assessing the stability of polio eradication after the withdrawal of oral polio vaccine. PLoS Biol. 2018;16:1–31. doi: 10.1371/journal.pbio.2002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duintjer Tebbens RJ, et al. Review and assessment of poliovirus immunity and transmission: Synthesis of knowledge gaps and identification of research needs. Risk Anal. 2013;33:606–646. doi: 10.1111/risa.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gog J. A status-based approach to multiple strain dynamics. Math Biol. 2002;44:169–184. doi: 10.1007/s002850100120. [DOI] [PubMed] [Google Scholar]
- 44.Asghar H, et al. Environmental surveillance for polioviruses in the global polio eradication initiative. J Infect Dis. 2014;210:S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson Muluh T, et al. Contribution of environmental surveillance toward interruption of poliovirus transmission in Nigeria, 2012-2015. J Infect Dis. 2016;213:131–135. doi: 10.1093/infdis/jiv767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huda TMN, et al. Household environmental conditions are associated with enteropathy and impaired growth in rural Bangladesh. Am J Trop Med Hyg. 2013;89:130–137. doi: 10.4269/ajtmh.12-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown J, Cairncross S, Ensink JH. Water, sanitation, hygiene and enteric infections in children. Arch Dis Child. 2013;98:629–634. doi: 10.1136/archdischild-2011-301528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wassilak SGF, et al. Using acute flaccid paralysis surveillance as a platform for vaccine-preventable disease surveillance. J Infect Dis. 2017;216(Suppl 1):S293–S298. doi: 10.1093/infdis/jiw593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts L. ‘What the hell is going on?’ Polio cases are vanishing in Pakistan, yet the virus won’t go away. Science. 2018 doi: 10.1126/science.aas9789. [DOI] [Google Scholar]
- 50.Cowger TL, et al. The role of supplementary environmental surveillance to complement acute flaccid paralysis surveillance for wild poliovirus in Pakistan – 2011–2013. PLoS ONE. 2017;12:1–16. doi: 10.1371/journal.pone.0180608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.World Health Organization Papua New Guinea confirms poliovirus outbreak, launches response. 2018 Available at www.wpro.who.int/papuanewguinea/mediacentre/releases/20180725/en/. Accessed October 5, 2018.
- 52.Gerloff N, et al. Diagnostic assay development for poliovirus eradication. J Clin Microbiol. 2018;56:1–10. doi: 10.1128/JCM.01624-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manukyan H, et al. Quantitative multiplex one-step RT-PCR assay for identification and quantitation of Sabin strains of poliovirus in clinical and environmental specimens. J Virol Methods. 2018;259:74–80. doi: 10.1016/j.jviromet.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 54.Cortez MH, Weitz JS. Distinguishing between indirect and direct modes of transmission using epidemiological time series. Am Nat. 2013;181:E43–E52. doi: 10.1086/668826. [DOI] [PubMed] [Google Scholar]
- 55.Eisenberg MC, Hayashi MAL. Determining identifiable parameter combinations using subset profiling. Math Biosci. 2014;256:116–126. doi: 10.1016/j.mbs.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 56.Ionides EL, Nguyen D, Atchadé Y, Stoev S, King AA. Inference for dynamic and latent variable models via iterated, perturbed Bayes maps. Proc Natl Acad Sci USA. 2015;112:719–724. doi: 10.1073/pnas.1410597112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ionides EL, Bhadra A, Atchadé Y, King A. Iterated filtering. Ann Stat. 2011;39:1776–1802. [Google Scholar]
- 58.Gamerman D, Lopes HF. Markov Chain Monte Carlo: Stochastic Simulation for Bayesian Inference. Chapman and Hall/CRC; London: 2006. [Google Scholar]
- 59.Liu J, et al. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: A multicentre study. Lancet Infect Dis. 2014;14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]
- 60.Ishii S, et al. Microfluidic quantitative PCR for simultaneous quantification of multiple viruses in environmental water samples. Appl Environ Microbiol. 2014;80:7505–7511. doi: 10.1128/AEM.02578-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nieuwenhuijse DF, Koopmans MP. Metagenomic sequencing for surveillance of food- and waterborne viral diseases. Front Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.