Significance
Enteric pathogens have to gauge the intestinal environment and adapt their metabolism and virulence strategies to establish themselves within the host. Here we show using a high-throughput screen that several metabolic pathways intertwine with virulence gene expression in enterohemorrhagic Escherichia coli. This screen identified transcriptional regulators that respond to fluctuations in amino and fatty acids as playing an important role in virulence gene expression both in vitro and during mammalian infection. Our study has fundamental implications for how differential gut metabolite compositions may affect disease outcome and susceptibility to pathogens.
Keywords: enterohemorrhagic E. coli, EHEC, cutR, fadL
Abstract
The gut metabolic landscape is complex and is influenced by the microbiota, host physiology, and enteric pathogens. Pathogens have to exquisitely monitor the biogeography of the gastrointestinal tract to find a suitable niche for colonization. To dissect the important metabolic pathways that influence virulence of enterohemorrhagic Escherichia coli (EHEC), we conducted a high-throughput screen. We generated a dataset of regulatory pathways that control EHEC virulence expression under anaerobic conditions. This unraveled that the cysteine-responsive regulator, CutR, converges with the YhaO serine import pump and the fatty acid metabolism regulator FadR to optimally control virulence expression in EHEC. CutR activates expression of YhaO to increase activity of the YhaJ transcription factor that has been previously shown to directly activate the EHEC virulence genes. CutR enhances FadL, which is a pump for fatty acids that represses inhibition of virulence expression by FadR, unmasking a feedback mechanism responsive to metabolite fluctuations. Moreover, CutR and FadR also augment murine infection by Citrobacter rodentium, which is a murine pathogen extensively employed as a surrogate animal model for EHEC. This high-throughput approach proved to be a powerful tool to map the web of cellular circuits that allows an enteric pathogen to monitor the gut environment and adjust the levels of expression of its virulence repertoire toward successful infection of the host.
The gastrointestinal (GI) tract is a complex environment, where the availability of metabolites and signaling molecules changes in different microenvironments, and is affected by microbiota composition, host physiology, and pathogenic insults (1). Enteric pathogens employ various metabolic and virulence strategies to outcompete and/or exploit the resident microbiota to successfully colonize a GI niche. These strategies include utilization of certain carbon and nitrogen sources as preferred nutrients and/or signals, exploitation of the host inflammation, and acquisition of metals, among others (1).
Enterohemorrhagic Escherichia coli (EHEC) colonizes the colon and causes severe diarrhea. EHEC virulence and intestinal colonization, as well of its surrogate murine infection model, Citrobacter rodentium, is regulated by sugar, nitrogen, organic acid, short chain fatty acid, and oxygen availability (2). The locus of enterocyte effacement (LEE) of EHEC encodes a type III secretion system (T3SS) that is essential for virulence. Effectors translocated through this T3SS induce cytoskeletal rearrangements on epithelial cells referred to as attaching and effacing (AE) lesions (3). Expression of the LEE is a significant metabolic burden for EHEC and must be carefully regulated (4). We established a high-throughput method to define these LEE regulatory mechanisms and identified two transcription factors, CutR and FadR, that govern LEE expression.
CutR (also known as YbaO/DecR) is a member of the feast/famine regulatory protein (FFRP) family of transcription factors (5). Lrp, the canonical example of the FFRP family, regulates the LEE in response to butyrate levels (6). Butyrate is the principal microbiota-derived carbon source for colonic enterocytes (7). CutR is a cysteine-responsive transcription factor (8). Free l-cysteine cannot be detected in the cecal contents of adult specific-pathogen-free (SPF) mice (9), but upon infection with C. rodentium, a bloom of l-cysteine is observed (10). Of note, cysteine is important in the maintenance of the mucosal integrity through its luminal redox status (11). In Salmonella, CutR is essential for the transcription of an adjacent cysteine desulfhydrase, cdsH, and contributes to the detoxification of cysteine (12). CutR was identified as an activator of the yhaOM locus in the E. coli K-12 strain BW25113 (8), and directly binds the yhaO (dslT) promoter in a cysteine-dependent manner. YhaO is a HAAAP family amino acid transporter of d- and l-serine (13), which activates expression of the LEE through YhaJ, a LysR-type transcription factor that directly binds the LEE regulatory region to drive its expression.
FadR is a member of the GntR family of transcriptional regulators. FadR maintains a balance of expression of long chain fatty acid (LCFA) synthesis and catabolism, activating expression of genes required for the former while repressing those of the latter. FadR activity is regulated by the products of FadD, the enzyme facilitating the first step of β-oxidation, conversion of LCFAs to Acyl-CoA derivatives. FadR–DNA interactions are disrupted by binding to long Acyl-CoA molecules. LCFAs influence the virulence of the enteric pathogens Vibrio cholerae and Salmonella enterica serovar Typhimurium. In V. cholera, FadR indirectly activates the expression of the master virulence regulator toxT, which activates the expression of cholera toxin and toxin-coregulated pilus (14). FadR also participates in ToxT regulation by activating the expression of fabA, encoding an enzyme required for unsaturated fatty acid (UFA) synthesis. The UFA linoleic acid directly binds to ToxT to disrupt binding to DNA (15, 16). In S. Typhimurium, disruption of fadD decreases expression of the Salmonella pathogenicity island (SPI)-1 T3SS transcriptional activator, hilA (17). Exogenous LCFAs down-regulate hilA levels through a mechanism that is dependent upon the outer membrane LCFA transport protein, FadL, while being independent of FadR. DNA binding of the hilA activator, HilD, can be abolished by the presence of the LCFA oleate (18).
Here we designed a high-throughput screen to establish a dataset of transcription regulators and metabolic pathways that affect LEE gene regulation. Notably CutR, YhaO, and FadR intersect and converge to regulate the LEE in EHEC and C. rodentium. CutR and FadR regulation also occurs during mammalian infection, with cutR and fadR C. rodentium mutants being attenuated for pathogenesis. This links redox, amino acid, and fatty acid metabolism with virulence gene expression in an enteric pathogen. In summary our findings indicate that a complex web of metabolic interactions intersects with virulence regulation to promote enteric disease.
Results
Survey of E. coli Core Genome Metabolic and Transcription Pathways That Influence LEE Gene Expression.
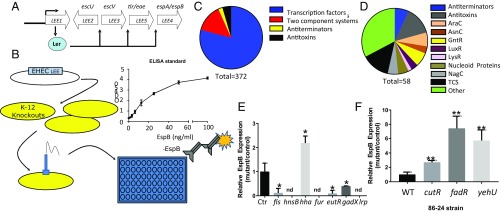
The LEE contains 41 genes, the majority of which are organized in five operons (LEE1–5). The first gene within LEE1 encodes the Ler transcriptional activator of all LEE genes (Fig. 1A). LEE4 encodes the EspB protein that is part of the T3SS translocon (3, 19). We developed an ELISA-based approach for evaluating the expression of EspB (Fig. 1B). The pJAY1512 LEE encoding cosmid was transformed (20) into a subset of the E. coli strains from the BW25113 Keio knockout library (21), generating a library of LEE-expressing K-12 strains. This library included 372 strains deficient in transcription factors, transcription antiterminators, two-component systems, and the DNA-binding type II antitoxin proteins (Fig. 1C and SI Appendix, Table S3). This library was screened in cysteine-supplemented DMEM under anaerobic conditions, as oxygen availability is an important factor governing LEE expression (22), and the pathways controlling LEE expression under anaerobic conditions have been understudied.
Fig. 1.
Screen for LEE regulators. (A) Schematic of the LEE operon arrangement. The LEE1-encoded Ler activates expression of all LEE operons. (B) Schematic of the screen for LEE regulators. BW25113 K-12 deletion strains were transformed with the LEE-encoding cosmid, pJAY1512. Strains were grown in DMEM under anaerobic conditions in 96-well plate format and evaluated by ELISA for EspB production. (Inset) Validation of EspB-directed ELISA procedure by addition of recombinant EspB to culture supernatants of BW25113. (C) Class makeup of the BW25113 deletion strains included in the screen. (D) Makeup of gene families found in the screen with a twofold cutoff in change in EspB production. Statistical significance was calculated as ANOVA with Dunnett’s post hoc test. (E) K-12 knockouts identified in genes previously known to regulate the LEE. (F) EspB ELISA of EHEC (86-24) deletion strains used for validation of screen results. *P < 0.05, **P < 0.01; nd, results below limit of detection.
This screen yielded 58 strains that met the cutoff criteria of a twofold change in EspB expression (Fig. 1D and SI Appendix, Table S3). As a testament to the validity of the screen, we also identified several factors previously described to control LEE gene expression such as Fis, HNS, Hha, EutR, Fur, GadX, and Lrp (23) (Fig. 1E). The majority of the genes involved in LEE regulation were in the other category. The second most common was antiterminators, followed by two-component systems. Transcription factors of the AraC and GntR family were found at similar numbers, and the least common class of transcription factors to regulate the LEE was LysRs (Fig. 1D and SI Appendix, Table S3). From these previously uncharacterized LEE regulators, three promising targets were selected for validation and further investigation: CutR, a FFRP family transcriptional regulator; FadR, a GntR family transcription factor; and YehU, a two-component system histidine sensor kinase. CutR is a cysteine-sensitive activator of the d-serine transporter YhaO, a previously described LEE-controlling protein (8, 13). FadR is the master regulator of fatty acid synthesis and degradation, which has been well described with regards to metabolism, but has not been characterized as having a virulence-related function in pathogenic E. coli. YehU is a peptide and amino acid-sensitive histidine sensor kinase known to activate the expression of yjiY (24), that encodes a transporter that is important during avian pathogenic E. coli infections (25). We generated deletion mutants of these genes in the 86-24 strain of EHEC. Upon rescreening these mutants by ELISA, we find that all three genes contribute to EspB regulation as suggested by our screen in K-12 (Fig. 1F).
CutR, FadR, and YehU in C. rodentium.
To investigate whether the role of these transcription factors in virulence regulation translated into in vivo phenotypes, we employed the C. rodentium murine infection model. C. rodentium is an AE lesion-forming pathogen that is extensively used as a surrogate organism for a mouse model of EHEC infection. C. rodentium contains the LEE and forms AE lesions on murine colonocytes (26). We generated ΔcutR, ΔfadR, and ΔyehU strains of DBS770, a C. rodentium strain harboring the Shiga toxin encoding phage (27). Shiga toxin (Stx) is responsible for the hemolytic uremic syndrome in EHEC infections (3). The C. rodentium Stx model more closely resembles all of the facets of EHEC infection (27).
Both the ΔcutR and the ΔfadR are attenuated for murine infection compared with WT, and this phenotype could be complemented with cutR and fadR on a plasmid (Fig. 2 A–D and SI Appendix, Fig. S2 D and H) using both infectious doses of 109 (Fig. 2 A–D) and 107 cfu (SI Appendix, Fig. S2 D and H). The ΔyehU is not attenuated for murine infection (SI Appendix, Fig. S1), suggesting that this gene is not critical in this infection model. The attenuation phenotypes of ΔcutR and ΔfadR cannot be explained by an overt difference in bacterial burden, as these strains colonized to levels equivalent to WT throughout the infection in stools at an infectious dose of 109 cfu (SI Appendix, Fig. S2 A and B). However, they did show a small one order of magnitude decrease at day 4 at an infectious dose of 107 cfu, suggesting that at a lower infectious dose, there is a slight decrease in fitness (SI Appendix, Fig. S2 E and I). In terms of tissue colonization WT and ΔcutR colonized the cecum at similar levels, and ΔcutR showed a one order of magnitude decrease in the colonization of the colon (SI Appendix, Fig. S2 F and G). The ΔfadR colonized both cecum and colon at the same levels as WT (SI Appendix, Fig. S2 J and K). This is not due to a growth defect, given that there is no significant difference in generation time for ΔcutR DBS770 (82 ± 8 min) or ΔfadR DBS770 (71 ± 10 min) compared with WT (81 ± 9 min) (SI Appendix, Fig. S3).
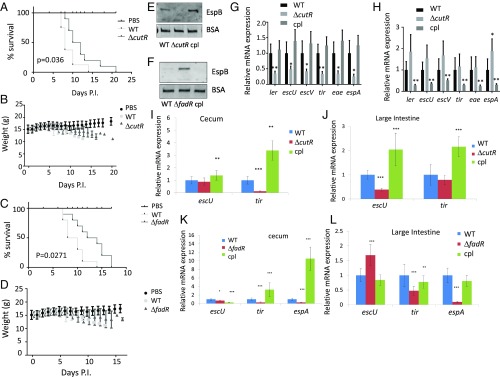
Fig. 2.
Representative screen hits in C. rodentium pathogenesis. All experiments were conducted using conventional mouse feed. Survival curves of C3H/HeJ infected with 109 cfu of WT, (A) ΔcutR, or (C) ΔfadR DBS770 C. rodentium or with PBS control (10 animals per infection group and 8 animals for PBS). Statistical significance calculated by Gehan–Breslow–Wilcoxon test. Weight of animals infected with (B) ΔcutR, or (D) ΔfadR DBS770, WT or mock (PBS). (E) Western for EspB from in vitro culture supernatants from WT, ΔcutR DBS770 and ΔcutR complemented (cultures grown in DMEM). (F) Western for EspB from in vitro culture supernatants from WT, ΔfadR DBS770, and ΔfadR complemented (cultures grown in DMEM). (G) qRT-PCR of C. rodentium LEE mRNAs from in vitro anaerobically grown WT, ΔcutR DBS770, and ΔcutR complemented strains (cultures grown in DMEM). (H) qRT-PCR quantification of C. rodentium LEE mRNAs from in vitro anaerobically grown WT, ΔfadR DBS770, and ΔfadR complemented strains (cultures grown in DMEM). (I) qRT-PCR of C. rodentium LEE mRNAs in murine cecum tissue of animals infected with WT, ΔcutR, and complemented strains. (J) qRT-PCR of C. rodentium LEE mRNAs in murine colon tissue of animals infected with WT, ΔcutR, and complemented strains. (K) qRT-PCR of C. rodentium LEE mRNAs in murine cecum tissue of animals infected with WT, ΔfadR, and complemented strains. (L) qRT-PCR of C. rodentium LEE mRNAs in murine colon tissue of animals infected with WT, ΔfadR, and complemented strains. *P < 0.05, **P < 0.01, ***P < 0.001. P.I., postinfection.
Both CutR and FadR influence LEE expression in C. rodentium in vitro. The ΔcutR has reduced secretion of EspB and LEE mRNA levels compared with WT (Fig. 2 E and G). The ΔfadR secretes higher levels of EspB protein than WT (Fig. 2F) and is a transcriptional repressor of the LEE at the mRNA level, with ΔfadR depicting higher LEE gene expression (Fig. 2H). During murine infection ΔcutR has decreased LEE gene expression in the cecum and the colon (Fig. 2 I and J). The ΔfadR has decreased expression of the LEE genes in the cecum, and dysregulated expression of the LEE genes in the colon, with escU being up-regulated and tir and espA being down-regulated in this mutant (Fig. 2 K and L). It is noteworthy that the LEE4 and LEE5 operons that harbor espA and tir are highly posttranscriptionally regulated by the GlmY–GlmZ sRNAs (28). This indicates that deregulation of LEE expression, whether by decreased or increased expression, affects C. rodentium pathogenesis. Overexpression of the LEE can be detrimental to the pathogen’s virulence because it creates an unnecessary energy burden that decreases fitness (4). Because EHEC has a low infectious dose of 50 cfu (3), it has to efficiently coordinate the right levels of expression of its virulence traits. This is especially important to successfully compete with the dense and highly adapted microbiota for a colonization niche.
We have previously investigated the intestinal metabolic profile of DBS770 infected or uninfected animals (10). Cysteine was the second most increased (155-fold) metabolite in C. rodentium-infected compared with uninfected (PBS control) animals, following antibiotic pretreatment to deplete the resident microbiota. As CutR requires cysteine to function (8), we investigated the course of infection for ΔcutR under depletion of the microbiota to assess whether the bloom of cysteine was microbiota dependent. Following antibiotic treatment, ΔcutR was still attenuated compared with WT, suggesting that changes in the cysteine levels within the intestine are not being dictated by the microbiota (SI Appendix, Fig. S4) and may reflect the host immune responses in attempting to restore mucosal integrity (11). Cysteine levels are increased by antibiotic treatment in the lumen of both cecum and colon, and in the tissues of colon and small intestine, while it remains unchanged in cecum tissues (SI Appendix, Fig. S5). This increase in cysteine is due to the fact that antibiotic treatment per se increases intestinal inflammation, as has been previously reported (29). Expression of most LEE genes in WT C. rodentium is increased during murine infection in mice under a cystine (it is the oxidized stable form of cysteine)-replete versus absent diet (SI Appendix, Fig. S6A). Moreover, ΔcutR is only attenuated for infection when the mice are under a cystine-replete diet (SI Appendix, Fig. S6 B and D). This is because LEE regulation by CutR only occurs in the presence of cysteine. However, in vivo tir regulation was decreased in the presence of cystine, but did not reach statistical significance, because tir is encoded within LEE5 which is highly posttranscriptionally regulated; this discrepancy can be due to that described in ref. 28 (SI Appendix, Fig. S6 C and E). Altogether C. rodentium is sensing cysteine in the gut to up-regulate LEE gene expression increasing its virulence potential, without affecting pathogen expansion.
Intersection of CutR/YhaO and FadR LEE Regulation in EHEC.
Congruent with the C. rodentium results, CutR is also a transcription activator of LEE gene expression in EHEC (Fig. 3A). EspB secretion is decreased in ΔcutR (Fig. 3B), reflecting the overall decreased transcription of the LEE-encoded T3SS (Fig. 3A). However, the levels of EspB in whole cell lysates are increased in ΔcutR (Fig. 3C). The EspB transcript is highly posttranscriptionally regulated in EHEC (30), and the discrepancy in the overall levels of EspB protein versus secreted EspB are probably a reflection of this regulation. This also explains the elevated levels of overall EspB protein in the ELISA performed with the EHEC ΔcutR (Fig. 1F). The cutR gene is located adjacent to the cdsH gene in C. rodentium and S. Typhimurium, which encodes the cysteine desulfhydrase (Fig. 3D). However, this gene is absent in EHEC, in which cutR is immediately preceded by the thiamine pyrimidine pyrophosphate hydrolase, cof. CutR is a cysteine-dependent transcription factor (8) and regulates the expression of the cdsH gene that encodes cysteine desulfhydrase which contributes to the detoxification of cysteine (12). CutR is also a cysteine-dependent transcription factor in the regulation of its previously described target yhaO (8). Therefore, we attempted to determine whether the CutR–LEE regulatory phenotype was dependent upon the presence of cysteine by excluding this amino acid from the growth media (no differences in growth in the presence or absence of cysteine were observed). We observe that the ability of CutR to govern the LEE transcript level in EHEC also requires the presence of cysteine (Fig. 3 E–G), consistent with its dependency on cysteine for transcription activity.
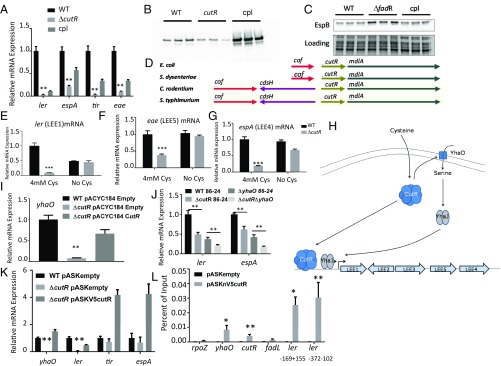
Fig. 3.
CutR regulation of LEE expression in EHEC. (A) qRT-PCR of LEE genes from WT, ΔcutR, and complemented EHEC strains grown anaerobically in the presence of cysteine (cultures grown in DMEM). (B) Western for EspB secreted from WT, ΔcutR, and complemented EHEC strains grown anaerobically in the presence of cysteine (cultures grown in DMEM). (C) Western for EspB from whole cell lysates of WT, ΔcutR, and complemented EHEC strains (cultures grown in DMEM). (D) Schematic of the cutR locus from E. coli, Shigella dysenteriae, C. rodentium, and S. Typhimurium. (E–G) qRT-PCR of LEE transcripts for (E) ler, (F) eae, and (G) espA from WT or ΔcutR grown anaerobically in DMEM either with or without cysteine. (H) Schematic representing a putative mechanism of cutR-dependent LEE regulation. CutR positively regulates the expression of yhaO, a serine transporter that positively regulates LEE expression via the LysR-type transcription factor YhaJ. (I) qRT-PCR of yhaO mRNA from WT, ΔcutR, and complemented strains (cultures grown in DMEM). (J) qRT-PCR to assess LEE expression from ΔcutRΔyhaO double mutant (cultures grown in DMEM). (K) qRT-PCR for complementation of LEE transcriptional phenotype by N-terminally V5-tagged CutR during preparation of cells for ChIP (cultures grown in DMEM). (L) ChIP-qPCR results for empty vector control or N-terminally tagged CutR. Probes are designed to amplify the promoter regions of yhaO (positive control), ybaO (cutR promoter), rpoZ (negative control), fadL, or overlapping fragments of the ler (LEE1) promoter, numbered from the proximal transcriptional start site. *P < 0.05, **P < 0.01, ***P < 0.001.
CutR is an activator of yhaO, which enhances serine import to increase activity of the YhaJ transcription factor that directly activates LEE transcription (13) (Fig. 3 H and I). Therefore, it is conceivable that CutR-dependent LEE transcriptional activation acts through YhaO. We evaluated whether CutR-dependent LEE transcriptional activation occurs through yhaO by generating a double knockout of ΔcutRΔyhaO. This double knockout had an additive effect beyond the single yhaO deletion mutant, suggesting that CutR LEE transcriptional regulation also occurs independently from YhaO (Fig. 3J). YhaO exerts its control over the LEE through the yhaJ-encoded transcription factor that directly activates LEE transcription (13). Therefore, we hypothesized that cutR deficiency might diminish yhaJ expression as it does yhaO, and in turn diminish LEE transcript levels. However, we do not observe a decrease in yhaJ in ΔcutR, suggesting that CutR is not involved in transcriptional regulation of this gene, probably modulating its activity through the levels of imported serine (SI Appendix, Fig. S7). Altogether, these data indicate that there is a CutR–YhaO arm and a CutR–YhaO independent arm to control LEE transcription. These data suggest that CutR may directly regulate transcription of the LEE genes. Because we were unable to purify soluble and folded CutR protein, to evaluate if CutR directly regulates transcription of the LEE, we constructed a tet-inducible V5 N-terminal tagged CutR plasmid for use as a ChIP construct. This construct is capable of complementing the cutR mutant (Fig. 3K). We then performed ChIP-qPCR to evaluate if segments of the LEE1 promoter (that encodes the Ler master regulator of the LEE genes) are capable of interacting with CutR protein in vivo (Fig. 3L). We observed that CutR is capable of interacting with the yhaO promoter as expected, while not interacting with the negative control rpoZ. We observe that CutR interacts with the LEE1 regulatory region, suggesting that direct regulation of ler may be a mechanism by which cutR regulates T3S in EHEC (Fig. 3L).
The CutR regulon has not been thoroughly explored; therefore, we performed transcriptomic studies to evaluate differential gene regulation between ΔcutR and WT EHEC. These studies showed that 121 genes were up-regulated, and 227 genes were down-regulated in ΔcutR (SI Appendix, Fig. S8 A and B and Tables S4 and S5). Notably, many of the up-regulated (10%) and down-regulated (21%) genes fall into a general category of transporters of metabolites, including fatty acids (fadL), polyamines (ydcS), glycine betaine (yehY), arginine (artP), and d/l-serine (yhaO). This suggests that CutR may have a broad function as a regulator of metabolite import. We confirmed the microarray results for a subset of genes via qRT-PCR. In agreement with previous reports, we observe that yhaO is one of the most strongly down-regulated genes in the cutR deletion mutant (SI Appendix, Fig. S8C).
Because there is YhaO-independent CutR regulation of the LEE genes (Fig. 3), we aimed to determine what other CutR-dependent processes may influence the LEE. We reasoned that because the original CutR dependency of the LEE was observed in E. coli K-12, the mechanism of action must be at least partially conserved between K-12 and EHEC. Therefore, we decided to reutilize the LEE cosmid transformed Keio library to evaluate the importance of 304 differentially expressed nonessential K-12 genes on EspB expression. We observe that 12 qRT-PCR-confirmed CutR-regulated genes (SI Appendix, Fig. S9) are capable of influencing EspB expression by at least fivefold. These data suggest that CutR-dependent LEE regulation may be multifactorial.
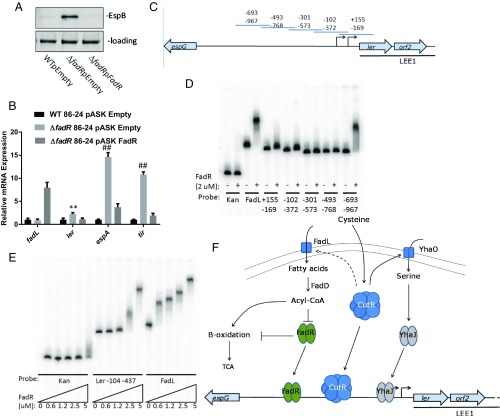
In these studies we observed that CutR activates expression of fadL (SI Appendix, Fig. S8C) that encodes a transporter for fatty acids. These fatty acids are converted to Acyl-CoA, which inhibits the transcription factor FadR (Fig. 4F). In agreement with the C. rodentium data (Fig. 2), FadR acts as a transcriptional repressor of the LEE genes, but this repression is alleviated when CutR activates FadL which increases the levels of Acyl-CoA that inhibit FadR function and repression of LEE transcription (Fig. 4 A and B). These data are congruent with decreased transcription of the LEE ler and tir genes in the fadL mutant (SI Appendix, Fig. S8D). To determine whether FadR may serve as a direct regulator of the LEE, purified recombinant N-terminally His-tagged FadR protein (SI Appendix, Fig. S10) was used on electrophoretic mobility shift assays (EMSAs) with overlapping fragments (Fig. 4C) of the LEE1 promoter. We observe that FadR is capable of interacting with the fadL promoter while not interacting with the kanamycin cassette of pRS551 (negative control). FadR interacts with LEE1 fragments ranging from −967 to −693 bp and −102 to −372 bp upstream of the proximal promoter transcription start site (Fig. 4D). The Citrobacter LEE1 regulatory region lacks the −967 to −693 bp region, as an insertional element has rendered the region significantly shorter than that of EHEC. Therefore, we sought to determine whether FadR is capable of interacting with a more proximal region of Citrobacter’s LEE1. We find that FadR is capable of binding to a fragment of the Citrobacter LEE1 region that is between −437 and −104 bp upstream of the transcriptional start site (Fig. 4E).
Fig. 4.
FadR LEE regulator in EHEC. (A) Western for EspB from supernatants of WT, ΔfadR, and complemented strains (cultures grown in DMEM). (B) qRT-PCR of LEE genes from WT, ΔfadR, and complemented strains (cultures grown in DMEM). (C) Diagram of EHEC LEE1 promoter segments used for EMSA probes. Probes are designed to have ∼80 bp of overlap between segments, extending from 967 bp upstream of the proximal LEE1 transcription start site to 155 bp downstream. (D) EMSA of His-tagged FadR and kanamycin probe (negative control), fadL promoter probe (positive control), and segments of the EHEC LEE1 regulatory region numbered from the proximal transcriptional start site. (E) EMSA of a Citrobacter LEE1 promoter fragment extending from 437 bp upstream to 104 bp upstream of the transcriptional start site. (F) Schematic of model of fadR- and cutR-dependent LEE regulation. **P < 0.01, ##P < 0.0001.
Our findings link CutR–cysteine-dependent regulation with YhaO–serine regulation to fatty acid metabolism (FadL and FadR) being converged and interconnected to optimally regulate expression of the LEE virulence genes in EHEC and C. rodentium. These findings illustrate how the bacterial cells read the metabolic landscape of the gut environment to inform themselves on when, how, and where to deploy their virulence armamentarium.
Discussion
Upon horizontal acquisition of a new genetic locus, an organism must control the expression of the genes within the locus. This may be accomplished by silencing gene expression, such as by the H-NS nucleoid-like protein (31). However, to evolve a positive use for a set of acquired genes, the organism must incorporate the locus into the preexisting core genome regulatory architecture. Such incorporation may involve developing the capacity to govern regulators prepackaged within the acquired island. An example of such a system is the LEE1-encoded Ler protein, which is capable of antagonizing the effects of H-NS, enabling the expression of the LEE operons (23). As ler is a lynchpin for the expression of the LEE, it is not surprising that it is under exquisite control by a large number of inputs. Our work represents a method of mapping the core genome components that contribute to these inputs for the LEE, and by extension, to other virulence components of pathogenic E. coli strains. Specifically, we have utilized a K-12 knockout library transformed with a LEE-encoding cosmid to study the conserved E. coli pathways that can influence the production of the T3SS translocon protein, EspB.
We have identified two transcription factors, CutR and FadR, which are known to regulate metabolism in E. coli, but had not yet been identified as contributing to regulation of virulence in EHEC, as key players in enteric virulence. CutR and FadR also intersect with the serine import pump YhaO to inform EHEC of the amino acid, redox, and fatty acid availability within the gut environment. Our results reveal that FadR is mostly a repressor of the LEE that is capable of directly interacting with the LEE1 regulatory regions in EHEC and C. rodentium. CutR functions as an activator of LEE expression, while controlling a network of genes which influence the LEE. Importantly, our transcriptomics analysis for ΔcutR EHEC suggests that this transcription factor may be important for maintaining a network of metabolite transporters. This is interesting from a virulence perspective, as the importance of small molecules in governing LEE expression is being increasingly appreciated.
Our dataset reveals other yet unexplored pathways that may prove important for regulation of T3S in EHEC, comprising a complex cellular web with intersecting circuits that remain to be mapped in detail. The metabolic landscape of the gut is dramatically changed by the presence of certain members of the microbiota and upon enteric infection (10). EHEC is a remarkably efficient pathogen, which is able to establish itself in the host through a very small infectious dose. EHEC’s proficiency to intersect metabolic, signaling, redox, and oxygen sensing may be at the core of its prowess as a successful enteric pathogen.
Materials and Methods
Bacterial Strains and Plasmids.
All strains and plasmids used in this study are listed in SI Appendix, Table S1. Recombinant DNA and molecular biology techniques were performed as previously described (32). All oligonucleotides used are listed in SI Appendix, Table S2. Knockout strains were constructed using lambda red (33). Plasmids pACYC184 (NEB) and pASK IBA32 (IBA Lifesciences) were used as complementation vectors. Cultures were grown in glucose DMEM (Gibco) with or without 4 mM cysteine. Generation times were calculated as [Log2]/slope of semilog plot of OD per time by Graphpad Prism 6. Cultures were grown at 37 °C under strict anaerobic conditions using a ShelLabs Bactron chamber containing 5% H2, 5% CO2, and 90% N2.
EspB Production Screening in K-12.
Keio library K-12 knockout strains were transformed with pAY1512 and grown in 96-well triplicate plates in DMEM with cysteine under anaerobic conditions. Upon reaching stationary phase, growth was halted by transfer to 4 °C and supplementation with 15 mg/mL sodium azide and Sigma Protease Inhibitor Mixture. They were diluted in PBS into Immulon Microtiter plates. EspB levels were determined with ELISA using anti-EspB antisera. Absolute concentrations were interpolated from standard curves from titrations of recombinant EspB protein. The ΔlacA Keio K-12 knockout strain with and without pJAY1512 was used as controls for normalization and background subtraction. All mutants were screened in triplicate; statistical significance was calculated in Graphpad Prism 6 by ANOVA followed by Dunnett’s multiple comparison test.
Chromatin Immunoprecipitation.
ChIP was performed using N-terminally V5-tagged CutR protein, cloned into PCR linearized pASK IBA32 using Gibson cloning with primers described in SI Appendix, Table S2. WT and mutant strains of 86-24 were transformed with pASK empty vector or pASKnV5YbaO. They were grown with supplementation of 12 ng/mL anhydrotetracycline. The cells were harvested and split for use in evaluation of protein expression, mRNA expression, or for ChIP. ChIP samples were treated with 1% formaldehyde for 20 min. Fixed cells were washed, resuspended in 10 mM Tris pH 8, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% RNaseA, and Sigma Protease Inhibitor Mixture and sonicated to fragments of 100–600 bp by seven cycles of 30 s on/60 s off at 95% power on a Qsonica Q125 sonicator. Lysed samples were cleared and quantified by nanodrop. Equivalent loadings of nucleic acids were used for each ChIP replicate and coupled at 37 °C by adding 10 μg of anti-V5 antibody (Abcam) per reaction. These reaction were precipitated with 1.5 mg Protein A Dynabeads and washed in 10 mM Tris pH 8, 500 mM LiCl, 1 mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate. ChIP samples were decrosslinked for 18 h at 65 °C in 10 mM Tris pH 8, 50 mM NaCl, 10 mM EDTA, 1% SDS. Protein was degraded by the addition of 80 μg Proteinase K per reaction and incubation at 55 °C for 4 h, then 95 °C for 10 min. DNA was purified with Qiagen MinElute kits. qRT-PCR was used to evaluate the percent of input of each sample captured during ChIP. Standard curves for each input sample were performed for each probe set (SI Appendix, Table S2) and used to calculate the percent of sequence precipitated.
Western Blotting.
For Westerns of secreted proteins, all cultures were grown in DMEM to the same OD600 under anaerobiosis in the presence of cysteine at 37 °C. A total of 10 μg BSA was added to secreted proteins before concentration and loading so that the efficiency of processing was known to be equivalent from sample to sample. Membranes were probed using an anti-EspB antiserum. Whole cell lysates were also run and probed with anti-EspB; loading was evaluated using stain-free settings on a Bio-Rad ChemiDoc imaging system.
Protein Purification and EMSA.
FadR was cloned into the NdeI and BamHI sites of pET28 by Gibson cloning (SI Appendix, Table S2) to create an N-terminal His-tagged construct. This was transformed into NiCO21 (NEB) cells. His-tagged FadR was purified through nickel columns according to manufacturer’s instructions. For EMSA, DNA probes were prepared by PCR (SI Appendix, Table S2 for primers) from genomic templates. Probes were purified by gene electrophoresis and labeled with 32P y-ATP by T4 PNK (NEB). Labeled probes were further purified by Qiagen PCR purification kit. EMSA reactions were prepared as protein diluted into a 2× EMSA buffer (50 mM NaKPO4 pH 7.5, 100 mM NaCl, 1.5 mM DTT, 100 μg/mL BSA, 250 μg/mL sonicated salmon sperm DNA). Binding was resolved on 5% polyacrylamide gels in Tris/borate/EDTA. Gels were dried onto filter paper and exposed to phosphoimager screens and assessed on a GE Typhoon scanner.
qRT-PCR and Microarray.
Samples were grown in triplicate in 50-mL cultures as described above. RNA was purified using an Ambion Bacterial RNA Extraction kit according to manufacturer’s instructions. qPCR was performed with Invitrogen SYBR Green Real-Time PCR Master Mix according to manufacturer’s specifications on an ABI QuantStudio 6 Flex instrument. Data were analyzed by the ΔΔCt method. Statistics were calculated as t test with GraphPad Prism. For microarrays, RNA samples were converted to cDNA and labeled as described in the Affymetrix Gene Expression manual. Samples were hybridized to Affymetrix E. coli 2.0 chips according to manufacturer’s recommendations. Data analysis was done with GeneSpring software, using MAS5 normalization. We report only genes found to be differentially expressed by twofold.
Animal Experiments.
SPF female C3H/HeJ animals were used. We used 8–10 animals per experimental group that were between 4–6 wk of age at the time of experiment. Animals were infected by oral gavage of 109 or 107 cfu of DBS770 or mutants in PBS or PBS alone. Animals were checked daily for survival, weight change, and stools and tissues were collected for analysis of colony-forming units and LEE gene expression. In some experiments, animals were precleared of their microbiota by including in their water 1 g/L ampicillin sodium salt, 1 g/L neomycin trisulfate salt hydrate, 1 g/L metronidazole, and 0.5 g/L vancomycin. This treatment was continued for 10 d, followed by a single day without antibiotics before gavage. In some experiments animals were given a cysteine-containing or a cysteine-absent feed. Statistical significance was determined by Prism 6. Survival was evaluated by Gehan–Breslow–Wilcoxon test. Research involving animals has been approved by the UT Southwestern Institutional Animal Care and Use Committee.
Cysteine Measurements.
Mouse intestinal segments and stools from conventional and antibiotic-treated mice were collected. Tissue and content/stools were homogenized in a sixfold volume of 50:50 methanol:dH20 containing 15 mM DTT using a polytron homogenizer (6× weight of tissue in g = vol of 50:50 MeOH/H20 added; total homogenate volume = 7× weight of tissue). The samples were spun twice and supernatant was collected. An aliquot of each stool supernatant was pooled and diluted 50× in 50:50 methanol:dH20 to make background matrix for preparing the standard curve. A similar diluted homogenate was made for the intestinal tissue standard curve. Standards were made by spiking 100 μL of the diluted homogenate with 2 μL of various concentrations of cysteine prepared fresh from powder. Samples and standards prepared in the 50× diluted pooled homogenate were then mixed 1:1 with crash solution (50:50 MeOH:H20 containing 15 mM DTT, 0.2% formic acid and 100 ng/mL DL-homocysteine d4). Standard concentrations are based on final volume after addition of crash solution so a 2× correction is applied to sample concentrations. After addition of the crash solution, samples were incubated for 10 min and samples were then spun 5′ 16,100 × g, supernatant was collected, and spun a second time. The supernatant was then transferred to an HPLC vial with insert and analyzed by LC-MS/MS. The Qtrap 6500 + analytical conditions used were: buffer A: ddH20 + 0.1% formic acid; buffer B: MeOH + 0.1% formic acid; column: Resteck Ultra IBD column 5 μM, 150 × 4.6 mm; gradient conditions: 0–1.5 min 3% B; 1.5–2.0 min gradient to 100% B; 2.0–3.5 min 100% B; 3.5–3.6 min gradient to 3% B; 3.6–4 min 3% B; flow rate: 1.5 mL/min; ion source/gas parameters: CUR = 30, CAD = 9, IS = 5500, TEM = 600, GS1 = 60, GS2 = 60. Cysteine transition 121.952–75.9; DL-homocysteine-d4 139.96/94.0
Supplementary Material
Acknowledgments
We thank James B. Kaper (University of Maryland Medical School) for the pJAY1512 LEE-encoding cosmid. This work was supported by NIH Grants AI053067, AI77853, AI114511, and AI077613. R.P. was supported through NIH Training Grant 5 T32 AI7520-14.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813451115/-/DCSupplemental.
References
- 1.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cameron EA, Sperandio V. Frenemies: Signaling and nutritional integration in pathogen-microbiota-host interactions. Cell Host Microbe. 2015;18:275–284. doi: 10.1016/j.chom.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 4.Pacheco AR, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokoyama K, et al. Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors. FEMS Microbiol Rev. 2006;30:89–108. doi: 10.1111/j.1574-6976.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- 6.Nakanishi N, et al. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology. 2009;155:521–530. doi: 10.1099/mic.0.023499-0. [DOI] [PubMed] [Google Scholar]
- 7.Roediger WE. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology. 1982;83:424–429. [PubMed] [Google Scholar]
- 8.Shimada T, Tanaka K, Ishihama A. Transcription factor DecR (YbaO) controls detoxification of L-cysteine in Escherichia coli. Microbiology. 2016;162:1698–1707. doi: 10.1099/mic.0.000337. [DOI] [PubMed] [Google Scholar]
- 9.Sasabe J, et al. Interplay between microbial d-amino acids and host d-amino acid oxidase modifies murine mucosal defence and gut microbiota. Nat Microbiol. 2016;1:16125. doi: 10.1038/nmicrobiol.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis MM, et al. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014;16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Circu ML, Aw TY. Intestinal redox biology and oxidative stress. Semin Cell Dev Biol. 2012;23:729–737. doi: 10.1016/j.semcdb.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oguri T, Schneider B, Reitzer L. Cysteine catabolism and cysteine desulfhydrase (CdsH/STM0458) in Salmonella enterica serovar typhimurium. J Bacteriol. 2012;194:4366–4376. doi: 10.1128/JB.00729-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly JP, et al. A highly conserved bacterial D-serine uptake system links host metabolism and virulence. PLoS Pathog. 2016;12:e1005359. doi: 10.1371/journal.ppat.1005359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovacikova G, Lin W, Taylor RK, Skorupski K. The fatty acid regulator FadR influences the expression of the virulence cascade in the El Tor biotype of Vibrio cholerae by modulating the levels of ToxT via two different mechanisms. J Bacteriol. 2017;199:e00762-16. doi: 10.1128/JB.00762-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomson JJ, Plecha SC, Withey JH. A small unstructured region in Vibrio cholerae ToxT mediates the response to positive and negative effectors and ToxT proteolysis. J Bacteriol. 2015;197:654–668. doi: 10.1128/JB.02068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plecha SC, Withey JH. Mechanism for inhibition of Vibrio cholerae ToxT activity by the unsaturated fatty acid components of bile. J Bacteriol. 2015;197:1716–1725. doi: 10.1128/JB.02409-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucas RL, et al. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar typhimurium. J Bacteriol. 2000;182:1872–1882. doi: 10.1128/jb.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golubeva YA, Ellermeier JR, Cott Chubiz JE, Slauch JM. Intestinal long-chain fatty acids act as a direct signal to modulate expression of the Salmonella pathogenicity island 1 type III secretion system. MBio. 2016;7:e02170-e15. doi: 10.1128/mBio.02170-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elliott SJ, et al. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 20.Elliott SJ, Yu J, Kaper JB. The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect Immun. 1999;67:4260–4263. doi: 10.1128/iai.67.8.4260-4263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlson-Banning KM, Sperandio V. Catabolite and oxygen regulation of enterohemorrhagic Escherichia coli virulence. MBio. 2016;7:e01852-16. doi: 10.1128/mBio.01852-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellies JL, Lorenzen E. Enterohemorrhagic Escherichia coli virulence gene regulation. Microbiol Spectr. 2014;2:EHEC-0004-2013. doi: 10.1128/microbiolspec.EHEC-0004-2013. [DOI] [PubMed] [Google Scholar]
- 24.Behr S, Fried L, Jung K. Identification of a novel nutrient-sensing histidine kinase/response regulator network in Escherichia coli. J Bacteriol. 2014;196:2023–2029. doi: 10.1128/JB.01554-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuntufye HN, Lebeer S, Gwakisa PS, Goddeeris BM. Identification of Avian pathogenic Escherichia coli genes that are induced in vivo during infection in chickens. Appl Environ Microbiol. 2012;78:3343–3351. doi: 10.1128/AEM.07677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng W, et al. Dissecting virulence: Systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci USA. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallick EM, et al. A novel murine infection model for Shiga toxin-producing Escherichia coli. J Clin Invest. 2012;122:4012–4024. doi: 10.1172/JCI62746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gruber CC, Sperandio V. Posttranscriptional control of microbe-induced rearrangement of host cell actin. MBio. 2014;5:e01025-e13. doi: 10.1128/mBio.01025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connolly JP, Finlay BB, Roe AJ. From ingestion to colonization: The influence of the host environment on regulation of the LEE encoded type III secretion system in enterohaemorrhagic Escherichia coli. Front Microbiol. 2015;6:568. doi: 10.3389/fmicb.2015.00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navarre WW, et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 33.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.