Significance
Intersexual conflict over maternal resource allocation to offspring can lead to the evolution of imprinted genes with parent-of-origin–specific expression. However, the precise mechanism involved in the evolution of such imprinted genes is less well understood, and few clear predictions have been presented. We resolve this issue, and, using different populations of a mixed-mating plant, we demonstrate that more outcrossed paternal populations produce larger seeds when crossed with less outcrossed maternal populations, and vice versa. This provides clear support for a “tug-of-war” mechanism operating between maternally and paternally imprinted genes. Such a mechanism can have important consequences for local adaptation in offspring size in the presence of gene flow between populations with different mating systems.
Keywords: Dalechampia, interlocus contest evolution, kinship genomic imprinting, parent–offspring conflict, sexual conflict
Abstract
In polyandrous species, fathers benefit from attracting greater maternal investment toward their offspring at the expense of the offspring of other males, while mothers should usually allocate resources equally among offspring. This conflict can lead to an evolutionary arms race between the sexes, manifested through antagonistic genes whose expression in offspring depends upon the parent of origin. The arms race may involve an increase in the strength of maternally versus paternally derived alleles engaged in a “tug of war” over maternal provisioning or repeated “recognition-avoidance” coevolution where growth-enhancing paternally derived alleles evolve to escape recognition by maternal genes targeted to suppress their effect. Here, we develop predictions to distinguish between these two mechanisms when considering crosses among populations that have reached different equilibria in this intersexual arms race. We test these predictions using crosses within and among populations of Dalechampia scandens (Euphorbiaceae) that presumably have experienced different intensities of intersexual conflict, as inferred from their historical differences in mating system. In crosses where the paternal population was more outcrossed than the maternal population, hybrid seeds were larger than those normally produced in the maternal population, whereas when the maternal population was more outcrossed, hybrid seeds were smaller than normal. These results confirm the importance of mating systems in determining the intensity of intersexual conflict over maternal investment and provide strong support for a tug-of-war mechanism operating in this conflict. They also yield clear predictions for the fitness consequences of gene flow among populations with different mating histories.
Females of many species frequently mate with several males (polyandry), thus opening an arena for intersexual conflict over the allocation of maternal resources (1–3). Indeed, while mothers maximize their fitness by allocating resources equally among offspring (4), fathers will increase their fitness by causing more maternal resources to be invested in their own offspring, at the expense of offspring sired by other males (3, 5–12). Consequently, selection should favor paternally derived alleles that increase nutrient demands on the mother when expressed in offspring (10, 12) and also maternal mechanisms that counteract the effects of paternally derived alleles to ensure an equal allocation of resources among offspring, thus avoiding the commitment of resources beyond the maternal optimum (13, 14). This conflict of interests should result in an evolutionary arms race between sexes over maternal investment in offspring (2, 13–15).
This intersexual arms race can be manifested through imprinted genes with differential expression depending on the parent of origin (12, 16). Although the importance of such imprinted genes has been demonstrated both during endosperm development in angiosperm seeds (17) and in placental activity in mammals (18), the exact mechanism by which these genes interact to control maternal allocation remains debated. Two genetic mechanisms, which we refer to as “tug of war” and “recognition avoidance,” have been proposed to explain the action of imprinted genes in the arms race between the sexes over maternal investment (19). Here, we provide predictions for distinguishing between these two mechanisms when considering the phenotype of offspring produced by crosses among populations with different levels of intersexual conflict over maternal provisioning. We then test these predictions experimentally by using crosses among natural populations of a mixed-mating plant species. Working with natural populations allows us to interpret our results in the broad context of local adaptation.
The tug-of-war mechanism describes a system where alleles at loci promoting offspring growth are expressed in the offspring when paternally derived and silenced when maternally derived, while alleles at growth-suppressing loci are expressed when maternally derived and silenced when paternally derived (7, 10–13). This mechanism can thus be described as a tug of war between maternally and paternally derived alleles over maternal investment, with coevolution leading to an escalation of the number and/or strength of these genes (20).
In the recognition-avoidance mechanism, genes in maternal tissues surrounding the developing embryo, or maternally expressed alleles in the offspring, may have evolved to recognize and control the effects of paternally derived growth-enhancing alleles (14, 19). In such a case, the arms race between the sexes will lead to the evolution of recognition-avoidance tactics, analogous to host–parasite coevolution (21), where paternally derived growth-enhancing alleles evolve to escape recognition, and hence bypass maternal control, while maternal or maternally derived alleles evolve to recognize new growth-promoting alleles (19). An important difference between these two mechanisms is that maternal (or maternally expressed) alleles involved in the coevolutionary process should directly influence resource allocation in the tug-of-war system, while the effect of these alleles in the recognition-avoidance system is only indirect through controlling the effects of paternally expressed growth-promoting genes with which they have coevolved.
At the population level, the intensity of intersexual conflict depends on the degree of relatedness among the offspring from a given mother, which, in turn, depends on the frequencies of multiple paternity and outcrossing (22). Populations that have historically experienced different intensities of intersexual conflict over offspring size through differences in their mating system should either have reached different tug-of-war equilibria or gone to fixation for different recognition-avoidance alleles (22–26). Consequently, crosses between populations that have experienced different intensities of intersexual conflict should perturb the genetic mechanisms that regulate offspring size. Here, we argue that the direction and magnitude of the difference in offspring size resulting from this genetic perturbation will depend on the genetic mechanism involved in the coevolutionary process, and we derive specific predictions allowing us to distinguish between the two mechanisms.
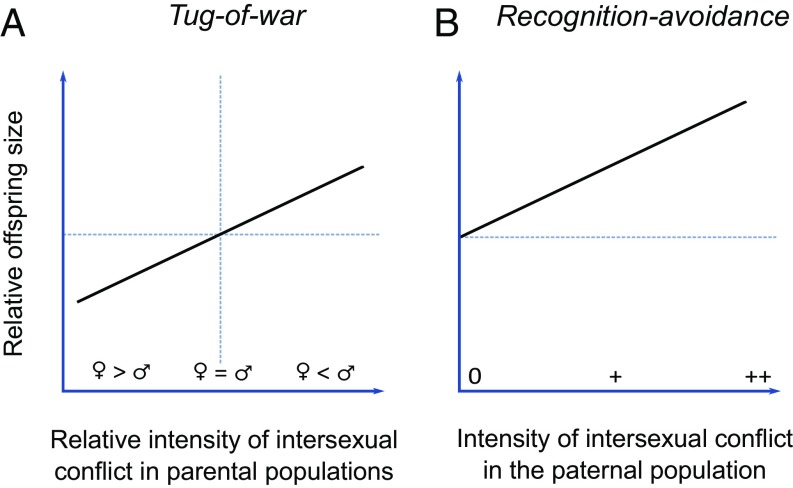
With the tug-of-war mechanism, the divergence in offspring size will depend upon the relative strength of the growth-promoting and growth-suppressing alleles derived from the parental populations. Alleles derived from populations with histories of more intense intersexual conflict are expected to “overpower” alleles from populations with histories of less intense conflict (22, 23, 27, 28). Consequently, crosses involving a maternal population with a history of more intense conflict than the paternal population should produce offspring smaller than the average offspring within the maternal population (growth-suppressing alleles are stronger than growth-enhancing alleles), while crosses in the opposite direction should produce larger offspring (29) (prediction 1; Fig. 1A).
Fig. 1.
Predicted outcomes for hybrid offspring size (solid lines) in crosses between populations relative to the average normal offspring size in the maternal population (horizontal dashed lines) under two different genetic mechanisms. (A) With the tug-of-war mechanism, hybrid offspring are predicted to become larger than the offspring normally produced in the maternal population if the paternal population has experienced more intense intersexual conflict than the maternal population and smaller in the opposite cross-direction. The vertical dashed line represents the point where the intensity of intersexual conflict is similar in both parental populations, and therefore where genes originating from each population have the exact opposite effects on seed size. (B) With the recognition-avoidance mechanism, hybrid offspring should become consistently larger than the offspring normally produced in the maternal populations, and the magnitude of the difference may increase with the intensity of intersexual conflict in the paternal population.
Because any intersexual arms race operating through a recognition-avoidance mechanism will be driven by males evolving new tactics to bypass maternal control, we expect the alleles involved in such a mechanism to be population-specific (14, 19). Consequently, between-population crosses may lead to the failure of maternal (or maternally derived) alleles to recognize “foreign” paternally derived alleles, and thus failure to control their effects on offspring size (19, 25). In the absence of maternal regulation, interpopulation crosses are expected to systematically produce larger offspring than the average offspring within the maternal population (prediction 2; Fig. 1B), unless the paternal population has a long history of strict monogamy or self-fertilization (i.e., with no intersexual conflict over maternal investment). In the latter case, hybrid offspring size should be similar to the average offspring size within the maternal population, because selfish paternal alleles are not expected to have evolved in completely monogamous or selfing populations (22).
These two predictions can be distinguished by the following features. With a tug-of-war mechanism, the difference in size between normal and hybrid offspring should be best explained by the relative intensity of intersexual conflict in the paternal and maternal populations. In contrast, with a recognition-avoidance mechanism, we do not expect the intensity of intersexual conflict in the maternal population to affect the difference in hybrid offspring size. Instead, the magnitude of this difference in offspring size should be explained by the intensity of intersexual conflict in the paternal population alone. Consequently, with a recognition-avoidance mechanism, we do not expect interpopulation crosses to produce offspring smaller than the average offspring normally produced in the maternal population (Fig. 1B). Even if the evolution of offspring size regulation results from a combination of the two mechanisms, the occurrence of such smaller hybrids remains exclusively associated with the tug-of-war mechanism.
Plant mating systems are extremely variable, ranging from functional asexuality to enforced outcrossing through self-incompatibility (30). More than 40% of plant species exhibit mixed mating systems, wherein progeny are produced by a mixture of selfing and outcrossing (31). Populations of mixed-mating species often differ widely in outcrossing rates (32), and hence in the intensity of intersexual conflict over maternal investment. Thus, plants provide ideal experimental systems for testing predictions of intersexual conflict theory. Furthermore, offspring size (seed size) in plants has important fitness consequences because it affects dispersal, germination, and seedling establishment (33–39).
Most previous research assessing seed size in interpopulation crosses in plants seems to support the existence of intersexual conflict over maternal investment, but few studies have explicitly considered the role of the mating system (22). Furthermore, the relative importance of the two genetic mechanisms (tug of war vs. recognition avoidance) in this conflict has rarely been studied, although this knowledge is essential if we want to predict the outcome of interpopulation hybridization. A notable exception is the study by Willi (19), in which support was found for both tug-of-war and recognition-avoidance mechanisms in crosses between predominantly selfing and predominantly outcrossing populations of Arabidopsis lyrata. Given the prevalence of mixed-mating plant species, studies on such species are crucial to achieve a more general understanding of the effect of mating systems on the evolution of intersexual conflict in natural populations. Importantly, the observation of a mating system effect in crosses between populations with subtle differences in their mating systems would underscore the importance of the mating system in the evolution of this conflict. Finally, the predictions we present are general and can be applied to any organism where regulation of offspring size depends on genomic imprinting and where populations vary in the intensity of intersexual conflict over maternal provisioning via, for example, variation in multiple paternity rates.
Here, we assess the role of mating systems in the evolution of intersexual conflict over seed size and test which of the two genetic mechanisms (tug of war vs. recognition avoidance) mediates this conflict. To this end, we analyzed two independent datasets obtained from crosses within and between populations of the mixed-mating plant Dalechampia scandens (Euphorbiaceae) (Fig. 2). The first dataset was obtained from crosses among nine populations spanning a wide range of inferred mating systems (SI Appendix, Table S1). Because the number of crosses performed between each pair of populations was rather small and the design was somewhat unbalanced (SI Appendix, Table S2), we obtained a second dataset from a controlled full-diallel crossing design using four populations (SI Appendix, Tables S3 and S4). Populations of D. scandens differ in their rate of outcrossing, and because pollinators visit several plants per foraging bout (40), the probability of multiple paternity, both within and among fruits, is likely to increase with outcrossing rate. In this system, population-mean herkogamy (i.e., the spatial separation of male and female structures in the inflorescence) correlates positively with outcrossing rate (r = 0.93, n = 4 populations), pollination reliability, and genetic diversity (41). We therefore used the population-mean herkogamy as a proxy for variation in mating system, and hence the intensity of intersexual conflict in each population. Importantly, while outcrossing rates are known to fluctuate between years (42), mean herkogamy is an evolved character, presumably representing the long-term outcome of selection for or against selfing, depending on the long-term average reliability of pollination (43). We estimated the effects of the relative outcrossing rate of the two parental populations and the outcrossing rate of the paternal population alone on interpopulation hybrid seed size to test the first and second predictions, respectively. The effect on hybrid seed size was estimated as percent difference in seed diameter between hybrid seeds and the mean diameter of seeds produced in the maternal population so as to account for the maternal effects commonly observed for this trait (44–48).
Fig. 2.
Blossom inflorescence of D. scandens with the first (terminal) male flower open above the three female flowers. In this species, the shortest distance between anthers and the stigmas affects the outcrossing rate. (Photograph courtesy of C.P.)
Results and Discussion
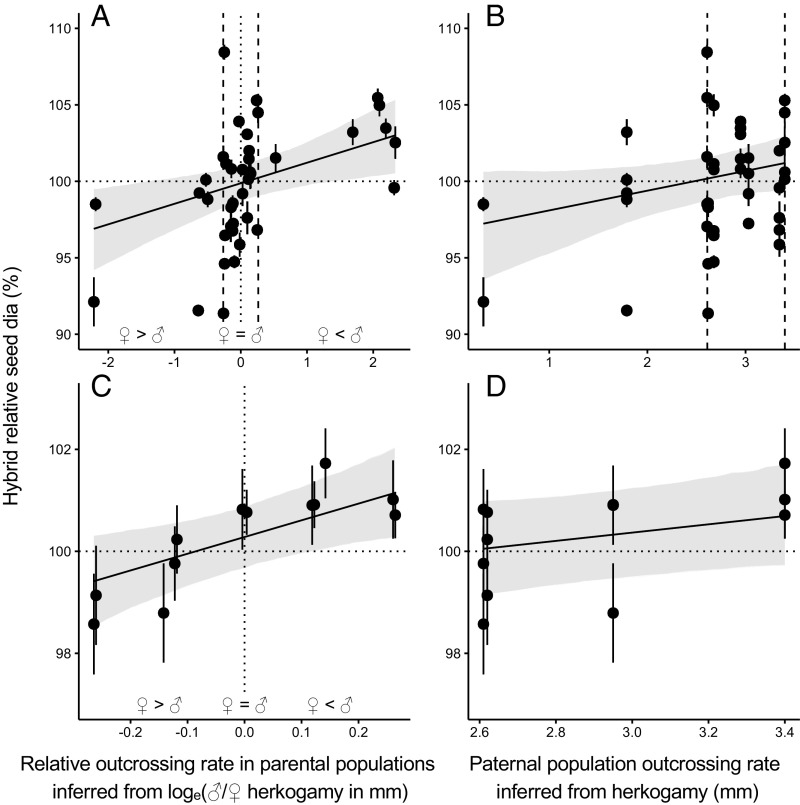
We found no indications of incompatibilities between populations (SI Appendix, Table S9). In both datasets, size differences between seeds resulting from interpopulation crosses and those resulting from within-population (maternal) outcrossing were best explained by the relative outcrossing rates of the two parental populations (Fig. 3 A and C and SI Appendix, Table S5). Hybrid seeds were larger when the inferred historical outcrossing rate of the paternal population exceeded the inferred historical outcrossing rate of the maternal population, and smaller in the opposite case. In the dataset that included crosses among nine populations of D. scandens with a broad range of inferred outcrossing rates (herkogamy ranging from 0.33 to 3.40 mm; SI Appendix, Table S1), hybrid relative seed size increased by 1.34 ± 0.46% (mean ± SE) per unit (log mm) change in the relative outcrossing rate of the parental populations (Fig. 3A). In contrast, the effect of an increase in outcrossing rate of the paternal population alone was weakly supported statistically (slope = 1.29 ± 0.66 %/mm; Fig. 3B and SI Appendix, Table S6B). This pattern was confirmed by the results from the diallel crosses among four populations. Although these populations exhibited a smaller range of inferred outcrossing rates (herkogamy ranging from 2.61 to 3.40 mm; SI Appendix, Table S3), we observed a clear increase in hybrid relative seed size per unit change in the relative outcrossing rate (slope = 3.28 ± 1.19 %/log(mm); Fig. 3C and SI Appendix, Table S5), while the effect of the outcrossing rate of the paternal population on hybrid seed size was limited and weakly supported statistically (slope = 0.81 ± 0.49 %/mm; Fig. 3D and SI Appendix, Table S6B). Finally, between-population crosses did not systematically produce larger seeds compared with within-population crosses, and this was true in the broad dataset (contrast = 0.45 ± 0.57%; SI Appendix, Table S6A) and in the diallel dataset (contrast = 0.11 ± 0.30%; SI Appendix, Table S6A).
Fig. 3.
Results of the experimental tests of the tug-of-war and recognition-avoidance mechanisms. The hybrid relative seed size of crosses among populations of D. scandens is regressed against the relative outcrossing rate of parental populations (A and C) to test the tug-of-war hypothesis and against the outcrossing rate of the paternal population (B and D) to test the recognition-avoidance mechanism. Data in A and B are from the crosses among nine populations covering a wide range of outcrossing rates. Data in C and D are from the diallel among four populations. The vertical dashed lines on A and B mark the range in relative outcrossing rate and paternal population outcrossing rate covered by the populations in the diallel experiment. The hybrid relative seed size is the hybrid seed diameter (dia) expressed as a percent of the average seed diameter within the maternal population. Shaded areas represent 95% confidence intervals for the regression models. Regression lines are estimated using individual seed data in mixed-effect models. Each point represents the mean (±SE) relative seed diameter for each cross-combination. The relative outcrossing rate of the parental populations is estimated as: loge (Paternal population herkogamy/Maternal population herkogamy). The outcrossing rate of the paternal population is estimated as the mean herkogamy (millimeters).
Overall, these results support the hypothesis that the intensity of intersexual conflict over maternal investment in seeds increases with outcrossing rate in this self-compatible perennial plant species. More specifically, the results support the tug-of-war hypothesis, wherein uniparentally expressed genes with opposite effects on seed growth have coevolved within populations. Although differences in measured seed diameter between hybrid and within-population seeds were small (ca. 5% and 2% in the first and second datasets, respectively; Fig. 3 A and C), these translate into ca. 14% and 6% differences in seed mass, respectively.
As expected from previous studies (49–52), we found little evidence for additive genetic effects on seed size. Indeed, in both datasets, hybrid relative seed size was not detectably affected by the average seed size in the paternal population relative to the maternal population (crosses between the nine populations: slope = 10.37 ± 13.38 %/log(mm), diallel crosses: β = 3.34 ± 4.12 %/log(mm); SI Appendix, Table S7). This observation further supports our main result that seed size is determined by a more complex mechanism of inheritance.
Inbreeding depression in seed size, measured as the difference in size between seeds produced by selfing versus outcrossing within populations, tended to increase with outcrossing rate in the four diallel populations. Although this result is consistent with the purging of deleterious alleles in the most inbred populations (53) (SI Appendix, Fig. S1), these differences were limited and statistically not significant (SI Appendix, Table S8). Nevertheless, an increase in the size of hybrid seeds relative to the seeds produced by within-population crosses could have resulted from heterosis [i.e., hybrid vigor due to the restoration of heterozygosity in crosses between inbred populations (53–55)]. Although heterosis effects cannot explain the decrease in seed size observed in some hybrid crosses as predicted by the tug-of-war mechanism, it may explain the small upward shift in the intercept of the relationship between hybrid relative seed size and relative outcrossing rate in the diallel data (Fig. 3C).
The present study supports the idea that uniparentally expressed genes with antagonistic effects on seed growth have coevolved within natural populations of D. scandens as an outcome of intersexual conflict over maternal investment. It further suggests that subtle differences in mating systems have resulted in the rapid evolution of genes involved in this conflict. The importance of imprinted genes influencing offspring growth has been well documented in mammals (18), in some crops (56, 57), and in the model organism Arabidopsis thaliana (17, 58–60). However, knowledge derived from artificially selected or highly inbred species is of limited relevance for understanding the evolution of intersexual conflict in a natural context. To our knowledge, only one prior study, that of Willi (19), has performed the kinds of interpopulation crosses between natural populations needed to determine which of the two mechanisms, tug of war versus recognition avoidance, mediates intersexual conflict over maternal resources. Our study differs from that of Willi (19) in several aspects. First, Willi (19) used populations that were either predominantly selfing or predominantly outcrossing (with the exception of one mixed-mating population), while all our populations were mixed-mating populations with relatively small differences in outcrossing rates, as inferred from the limited variation in herkogamy. Second, instead of comparing hybrid seed size with the mid-parent average (which assumes an additive genetic effect as the null hypothesis), we compared the hybrid seed size with the average seed size in the maternal population, therefore accounting for the strong maternal effects generally observed on seed size (e.g., refs. 44–47). Finally, based on the mating systems of the populations involved (22), we made clear predictions regarding the direction of the deviation in hybrid seed size compared with the average seed size in the maternal population. In contrast to Willi (19), we did not observe a consistent increase in hybrid relative seed size when the pollen donor came from another outcrossing population, suggesting that the recognition-avoidance mechanism is of limited importance in our study system. The upward shift in the intercept of the relationship between hybrid relative seed size and relative outcrossing rate could be explained by the fact that maternal plants in our system partly fail to control some of the effects of foreign paternal growth-enhancing alleles; however, as previously mentioned, this could also be explained by heterosis effects. Nevertheless, the general pattern in both datasets suggests that a tug-of-war mechanism is the most prevalent mechanism in these populations.
To conclude, our study on a mixed-mating perennial plant shows that crosses between populations with subtle differences in mating systems yield hybrid seeds that deviate from the within-population equilibrium seed size in the manner predicted by the tug-of-war hypothesis, but not by the recognition-avoidance hypothesis. These results support the idea that maternally and paternally derived alleles with antagonistic effects on seed growth have coevolved within populations, and that antagonistic forces are stronger in more outcrossed populations that have histories of more intense intersexual conflict over maternal investment. This may have important consequences for the maintenance of local adaptation in the presence of gene flow. Indeed, if seed size is locally adapted, depending on environmental conditions and size/number trade-off (4), any gene flow from populations with different outcrossing rates should negatively affect maternal fitness by affecting the size of the seeds produced. However, the fitness of individual seeds (e.g., germination and establishment success) is predicted to increase when the paternal population is more outcrossed but to decrease in the opposite case. Although neither additive genetic effects nor heterosis seemed to explain much of the variation in hybrid seed size, it would still be interesting to investigate systematically how genomic imprinting due to intersexual conflict interacts with local selection pressures on maternal and paternal genetic components of seed size.
Methods
Study Species and Populations.
Crosses were made within and among populations of D. scandens L. (Euphorbiaceae), a mixed-mating perennial vine with a distribution ranging from Mexico to Argentina (61). The bisexual pseudanthial inflorescences (blossoms) comprise 10 male flowers clustered above three female flowers (Fig. 2). Each female flower contains three ovules, so that a blossom can produce up to nine seeds (62). A gland situated above the male flowers secretes a terpenoid resin, which functions as a pollinator reward, attracting apid and megachilid bees that use resin for nest building (61, 63). Two petaloid bracts subtend the flowers and function as an advertisement to attract pollinators (64).
Blossoms are functionally protogynous, with a female phase preceding a bisexual phase during which autonomous selfing can occur (65). Pollinators visit both female-phase and bisexual-phase inflorescences, and they commonly visit multiple plants per foraging bout (40). The rate of autofertility (seed set in the absence of pollinators) declines with increasing distance between anthers and stigmas (herkogamy), a highly evolvable trait that varies among populations (41, 66, 67). In plants, in general, populations with high autofertility rates tend to have low outcrossing rates; thus, herkogamy offers a reliable proxy for variation in mating system (i.e., outcrossing rate) (43). This was confirmed for D. scandens, where variation in outcrossing rates among natural populations was positively correlated with population-mean herkogamy (r = 0.93, n = 4 populations) (41).
The first dataset comprised measurements of seeds produced as part of a larger study of population differentiation in D. scandens, where crosses were performed among nine populations originating from Mexico (SI Appendix, Tables S1 and S2). Because of the unbalanced sampling in this first dataset (missing data for many cross-combinations and few crosses per combination, n = 635 seeds from 86 interpopulation crosses) and the absence of information on covariates that could potentially influence seed size (discussed below), we obtained a second dataset by crossing four populations originating from Veracruz and the Yucatán peninsula in Mexico in a full-diallel design, also including self-pollination (SI Appendix, Tables S3 and S4). All populations differed in average seed size and herkogamy (68) (SI Appendix, Table S1). The populations are interfertile but geographically separated by at least 225 km, so natural gene flow should be extremely rare (40). Population differences in seed size and herkogamy were observed on individuals grown in the greenhouse, and therefore represent genetic differences. Note that the range of herkogamy among the four populations included in the diallel is similar to the range observed among the populations analyzed by Opedal et al. (41), for which the rate of outcrossing ranged from 0.16 to 0.49.
Experimental Design and Measurements.
Crosses for the first dataset were performed between 2007 and 2009 between individuals grown in the greenhouse from field-collected seeds. Between May and July 2016, we used seeds obtained from random crosses within four of the nine populations to grow the plants that we used in the diallel experiment. Hence, the experimental individuals were second-generation greenhouse plants. We used 10 plants per population and distributed them evenly across two tables in a single room in the greenhouse with a 13:11-h light/dark regime and the temperature set at 25 °C during the day and 23 °C at night. We watered the plants each day by flooding the tables with ca. 5 cm of water. Plants were moved weekly to avoid positional effects.
All four populations were crossed in a complete diallel design, with each population used as both a paternal population and a maternal population (SI Appendix, Table S4). This resulted in four sets of within-population crosses and 12 sets of between-population crosses. Within populations, each plant was crossed with two different plants from the same population (outcrossing) and once with itself (geitonogamous selfing). For the between-population crosses, each population was crossed as both a maternal population and a paternal population with each of the three other populations. For each combination of two populations, 10 plants from the maternal population were crossed with three different plants from the paternal population. Hence, each individual was represented three times as a maternal plant and three times as a paternal plant in the crossing design. Crosses (total n = 460) were made from August to December 2016 by one of the authors (A.R.). Blossoms were emasculated and hand-pollinated during the female phase with an ample amount of pollen from a freshly dehisced male flower. Crosses that failed were recorded and repeated. Crosses were performed in a random order to avoid possible confounding factors associated with the timing of pollination and uncontrolled variation in the greenhouse environment. To control for the effect of blossom size on seed size, the peduncle diameter, which correlates with blossom size (50), was measured with digital calipers (0.01-mm precision). Hand-pollinated blossoms were enclosed in empty tea bags to collect seeds after explosive dehiscence.
We counted the number of seeds produced per blossom (seed set) and measured the diameter of each individual seed (seed size) with digital calipers (0.01-mm precision; all measurements were made by A.R.). Repeatability of seed measurements estimated by repeated measurements of one seed per seed set was high (r2 = 0.99, n = 447). Seeds were measured in random order. We used seed diameter as a measure of seed size because seed diameter is less prone than seed mass to vary with time due to water loss. Still, we weighed the seed sets to estimate the allometric relationship between seed diameter and average seed mass. Seed diameter and mass were strongly correlated (r2 = 0.90, n = 428 seed sets), and the allometric exponent was very close to 3 (3.04 ± 0.07) as expected for an allometry between a length and a mass. This allometric relationship indicates that percent differences in seed mass can be accurately estimated by simply multiplying the percent difference in diameter by 3.
Statistical Analyses.
To assess whether the mating system of the parental populations affects seed size in interpopulation crosses, and which of the two genetic mechanisms, tug of war or recognition avoidance, occurs, we compared hybrid seed size with the size of the seeds normally produced in the maternal populations. Maternal effects are expected to strongly influence seed size even in hybrid crosses. Therefore, we expressed hybrid seed size as a percent deviation from the average seed size produced by within-population crosses (excluding selfing) in the maternal population: Hybrid relative seed size = 100 × (Hybrid seed diameter/Mean seed diameter in the maternal population). For the data including nine populations, we calculated the mean seed diameter within each maternal population from the raw data, while for the four populations in the diallel, we estimated this for each maternal population from linear mixed-effect models where paternal population identity was set as a predictor variable with five levels: pollen from another individual in the same population (within-population outcross), pollen from the same plant (selfing), and pollen from each of the three other populations (between-population cross). Effects of peduncle diameter and number of seeds per blossom on seed size were population-specific variables (SI Appendix, Table S8). Therefore, both variables were centered on the mean of the maternal population and included as covariates. Number of seeds per blossom was also allowed to interact with paternal population identity to account for a potential cross-specific size/number trade-off. Paternal plant identity and blossom identity nested within maternal plant identity were set as random factors.
To test the tug-of-war model (Fig. 1A), we quantified the relative outcrossing rate of the two parental populations inferred from their mean herkogamy (41) as: Relative outcrossing rate = loge (Paternal population herkogamy/Maternal population herkogamy). This index is symmetrical around zero and will take positive values when the paternal population is more outcrossed than the maternal population and negative values in the opposite case. We fitted a linear mixed-effects model with hybrid relative seed size as the response variable and relative outcrossing rate as the predictor variable. We also included blossom identity nested within maternal plant identity, nested within maternal population identity, and paternal plant identity nested within paternal population identity as random factors. For the diallel data, we also included mean-centered peduncle diameter in interaction with maternal population identity and mean-centered seed number in interaction with maternal and paternal population identities as covariates.
To test the recognition-avoidance model (Fig. 1B), we fitted two different linear mixed-effects models testing two distinct predictions. If hybridization leads to failed maternal recognition of foreign paternal allele products, seeds from interpopulation crosses are expected to be larger than seeds produced by within-population crosses. We tested this prediction by fitting a linear mixed-effects model with hybrid relative seed size as the response variable and cross type (between- vs. within-population) as the predictor variable. In addition, hybrid relative seed size should increase with the outcrossing rate of the paternal population because growth-promoting alleles from more outcrossed populations are expected to have stronger effects. To test this second prediction, we fitted a linear mixed-effects model using hybrid relative seed size as the response variable and the mean herkogamy (as a proxy for outcrossing rate) of the paternal population as the predictor variable. In both models, random effects and covariates were specified as above.
Finally, we tested for a model where seed size determination follows a simple additive genetic inheritance pattern. In this case, hybrid relative seed size should depend on the relative seed size in the two parental populations. We quantified relative average seed size of the two parental populations (excluding selfed seeds) as: Relative seed size = loge (Paternal population seed diameter/Maternal population seed diameter). We then fitted a linear mixed-effects model with hybrid relative seed size as the response variable and relative seed size as the predictor variable. Random effects and covariates were specified as above.
All statistical analyses were conducted in R version 3.3.3 (69), and linear mixed-effects models were fitted using the lme4 package (70).
Supplementary Material
Acknowledgments
We thank Yvonne Willi for comments on an earlier version of this work, three anonymous reviewers for their constructive comments, and Grete Rakvaag for maintaining the plants in the greenhouse. This work was supported by the Research Council of Norway through its Centres of Excellence funding scheme (Project 223257).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 11354.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810979115/-/DCSupplemental.
References
- 1.Arnqvist G, Rowe L. Sexual Conflict. Princeton Univ Press; Princeton: 2013. [Google Scholar]
- 2.Chapman T. Evolutionary conflicts of interest between males and females. Curr Biol. 2006;16:R744–R754. doi: 10.1016/j.cub.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 3.Trivers RL. Parent-offspring conflict. Am Zool. 1974;14:249–264. [Google Scholar]
- 4.Smith CC, Fretwell SD. Optimal balance between size and number of offspring. Am Nat. 1974;108:499–506. [Google Scholar]
- 5.Westoby M, Rice B. Evolution of the seed plants and inclusive fitness of plant tissues. Evolution. 1982;36:713–724. doi: 10.1111/j.1558-5646.1982.tb05437.x. [DOI] [PubMed] [Google Scholar]
- 6.Queller DC. Models of kin selection on seed provisioning. Heredity. 1984;53:151–165. [Google Scholar]
- 7.Haig D, Wilkins JF. Genomic imprinting, sibling solidairity and the logic of collective action. Philos Trans R Soc Lond B Biol Sci. 2000;355:1593–1597. doi: 10.1098/rstb.2000.0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jong TJ, Van Dijk H, Klinkhamer PGL. Hamilton’s rule, imprinting and parent-offspring conflict over seed mass in partially selfing plants. J Evol Biol. 2005;18:676–682. doi: 10.1111/j.1420-9101.2004.00856.x. [DOI] [PubMed] [Google Scholar]
- 9.de Jong TJ, Scott RJ. Parental conflict does not necessarily lead to the evolution of imprinting. Trends Plant Sci. 2007;12:439–443. doi: 10.1016/j.tplants.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Haig D, Westoby M. Parent-specific gene-expression and the triploid endosperm. Am Nat. 1989;134:147–155. [Google Scholar]
- 11.Haig D. The kinship theory of genomic imprinting. Annu Rev Ecol Syst. 2000;31:9–32. [Google Scholar]
- 12.Haig D. Parental antagonism, relatedness asymmetries, and genomic imprinting. Proc Biol Sci. 1997;264:1657–1662. doi: 10.1098/rspb.1997.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkins JF, Haig D. Genomic imprinting of two antagonistic loci. Proc Biol Sci. 2001;268:1861–1867. doi: 10.1098/rspb.2001.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice WR, Holland B. The enemies within: Intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific Red Queen. Behav Ecol Sociobiol. 1997;41:1–10. [Google Scholar]
- 15.Parker GA, Macnair MR. Models of parent-offspring conflict. 4. Suppression–Evolutionary retaliation by the parent. Anim Behav. 1979;27:1210–1235. [Google Scholar]
- 16.Efstratiadis A. Parental imprinting of autosomal mammalian genes. Curr Opin Genet Dev. 1994;4:265–280. doi: 10.1016/s0959-437x(05)80054-1. [DOI] [PubMed] [Google Scholar]
- 17.Vinkenoog R, et al. Genomic imprinting and endosperm development in flowering plants. Mol Biotechnol. 2003;25:149–184. doi: 10.1385/MB:25:2:149. [DOI] [PubMed] [Google Scholar]
- 18.Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- 19.Willi Y. The battle of the sexes over seed size: Support for both kinship genomic imprinting and interlocus contest evolution. Am Nat. 2013;181:787–798. doi: 10.1086/670196. [DOI] [PubMed] [Google Scholar]
- 20.Wilkins JF, Haig D. What good is genomic imprinting: The function of parent-specific gene expression. Nat Rev Genet. 2003;4:359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- 21.Tellier A, Brown JKM. Stability of genetic polymorphism in host-parasite interactions. Proc Biol Sci. 2007;274:809–817. doi: 10.1098/rspb.2006.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brandvain Y, Haig D. Divergent mating systems and parental conflict as a barrier to hybridization in flowering plants. Am Nat. 2005;166:330–338. doi: 10.1086/432036. [DOI] [PubMed] [Google Scholar]
- 23.Kondoh M, Higashi M. Reproductive isolation mechanism resulting from resolution of intragenomic conflict. Am Nat. 2000;156:511–518. doi: 10.1086/303409. [DOI] [PubMed] [Google Scholar]
- 24.Andrés JA, Arnqvist G. Genetic divergence of the seminal signal-receptor system in houseflies: The footprints of sexually antagonistic coevolution? Proc Biol Sci. 2001;268:399–405. doi: 10.1098/rspb.2000.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker GA, Partridge L. Sexual conflict and speciation. Philos Trans R Soc Lond B Biol Sci. 1998;353:261–274. doi: 10.1098/rstb.1998.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pizzari T, Snook RR. Perspective: Sexual conflict and sexual selection: Chasing away paradigm shifts. Evolution. 2003;57:1223–1236. doi: 10.1111/j.0014-3820.2003.tb00331.x. [DOI] [PubMed] [Google Scholar]
- 27.Dawson WD. Fertility and size inheritance in a Peromyscus species cross. Evolution. 1965;19:44–55. [Google Scholar]
- 28.Vrana PB, Guan XJ, Ingram RS, Tilghman SM. Genomic imprinting is disrupted in interspecific Peromyscus hybrids. Nat Genet. 1998;20:362–365. doi: 10.1038/3833. [DOI] [PubMed] [Google Scholar]
- 29.Haig D, Westoby M. Genomic imprinting in endosperm–Its effect on seed development in crosses between species, and between different ploidies of the same species, and its implications for the evolution of apomixis. Philos Trans R Soc Lond B Biol Sci. 1991;333:1–13. [Google Scholar]
- 30.Barrett SCH. Mating strategies in flowering plants: The outcrossing-selfing paradigm and beyond. Philos Trans R Soc Lond B Biol Sci. 2003;358:991–1004. doi: 10.1098/rstb.2003.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodwillie C, Kalisz S, Eckert CG. The evolutionary enigma of mixed mating systems in plants: Occurrence, theoretical explanations, and empirical evidence. Annu Rev Ecol Evol Syst. 2005;36:47–79. [Google Scholar]
- 32.Whitehead MR, Lanfear R, Mitchell RJ, Karron JD. Plant mating systems often vary widely among populations. Front Ecol Evol. 2018;6:38. [Google Scholar]
- 33.Westoby M, Falster DS, Moles AT, Vesk PA, Wright IJ. Plant ecological strategies: Some leading dimensions of variation between species. Annu Rev Ecol Syst. 2002;33:125–159. [Google Scholar]
- 34.Moles AT, et al. A brief history of seed size. Science. 2005;307:576–580. doi: 10.1126/science.1104863. [DOI] [PubMed] [Google Scholar]
- 35.Vaughton G, Ramsey M. Sources and consequences of seed mass variation in Banksia marginata (Proteaceae) J Ecol. 1998;86:563–573. [Google Scholar]
- 36.Westoby M, Leishman M, Lord J. Comparative ecology of seed size and dispersal. Philos Trans R Soc Lond B Biol Sci. 1996;351:1309–1317. [Google Scholar]
- 37.Stebbins GL. Adaptive radiation of reproductive characteristics in angiosperms, II: Seeds and seedlings. Annu Rev Ecol Syst. 1971;2:237–260. [Google Scholar]
- 38.Henery ML, Westoby M. Seed mass and seed nutrient content as predictors of seed output variation between species. Oikos. 2001;92:479–490. [Google Scholar]
- 39.Westoby M, Jurado E, Leishman M. Comparative evolutionary ecology of seed size. Trends Ecol Evol. 1992;7:368–372. doi: 10.1016/0169-5347(92)90006-W. [DOI] [PubMed] [Google Scholar]
- 40.Opedal ØH, et al. Euglossine bees mediate only limited long-distance gene flow in a tropical vine. New Phytol. 2017;213:1898–1908. doi: 10.1111/nph.14380. [DOI] [PubMed] [Google Scholar]
- 41.Opedal ØH, et al. Evolutionary consequences of ecological factors: Pollinator reliability predicts mating-system traits of a perennial plant. Ecol Lett. 2016;19:1486–1495. doi: 10.1111/ele.12701. [DOI] [PubMed] [Google Scholar]
- 42.Eckert CG, Ozimec B, Herlihy CR, Griffin CA, Routley MB. Floral morphology mediates temporal variation in the mating system of a self-compatible plant. Ecology. 2009;90:1540–1548. doi: 10.1890/08-1063.1. [DOI] [PubMed] [Google Scholar]
- 43.Opedal OH, Bolstad GH, Hansen TF, Armbruster WS, Pélabon C. The evolvability of herkogamy: Quantifying the evolutionary potential of a composite trait. Evolution. 2017;71:1572–1586. doi: 10.1111/evo.13258. [DOI] [PubMed] [Google Scholar]
- 44.Roach DA, Wulff RD. Maternal effects in plants. Annu Rev Ecol Syst. 1987;18:209–235. [Google Scholar]
- 45.Biere A. Parental effects in Lychnis flos-cuculi. I: Seed size, germination and seedling performance in a controlled environment. J Evol Biol. 1991;4:447–465. [Google Scholar]
- 46.Platenkamp GAJ, Shaw RG. Environmental and genetic maternal effects on seed characters in Nemophila menziesii. Evolution. 1993;47:540–555. doi: 10.1111/j.1558-5646.1993.tb02112.x. [DOI] [PubMed] [Google Scholar]
- 47.Lemontey C, Mousset-Déclas C, Munier-Jolain N, Boutin JP. Maternal genotype influences pea seed size by controlling both mitotic activity during early embryogenesis and final endoreduplication level/cotyledon cell size in mature seed. J Exp Bot. 2000;51:167–175. doi: 10.1093/jexbot/51.343.167. [DOI] [PubMed] [Google Scholar]
- 48.Byers DL, Platenkamp GAJ, Shaw RG. Variation in seed characters in Nemophila menziesii: Evidence of a genetic basis for maternal effect. Evolution. 1997;51:1445–1456. doi: 10.1111/j.1558-5646.1997.tb01468.x. [DOI] [PubMed] [Google Scholar]
- 49.de Jong TJ, Hermans CM, van Der Veen-van Wijk KCAM. Paternal effects on seed mass in Arabidopsis thaliana. Plant Biol (Stuttg) 2011;13(Suppl 1):71–77. doi: 10.1111/j.1438-8677.2009.00287.x. [DOI] [PubMed] [Google Scholar]
- 50.Pélabon C, Albertsen E, Falahati-Anbaran M, Wright J, Armbruster WS. Does multiple paternity affect seed mass in angiosperms? An experimental test in Dalechampia scandens. J Evol Biol. 2015;28:1719–1733. doi: 10.1111/jeb.12692. [DOI] [PubMed] [Google Scholar]
- 51.Pélabon C, et al. Does stronger pollen competition improve offspring fitness when pollen load does not vary? Am J Bot. 2016;103:522–531. doi: 10.3732/ajb.1500126. [DOI] [PubMed] [Google Scholar]
- 52.Galloway LF, Etterson JR, McGlothlin JW. Contribution of direct and maternal genetic effects to life-history evolution. New Phytol. 2009;183:826–838. doi: 10.1111/j.1469-8137.2009.02939.x. [DOI] [PubMed] [Google Scholar]
- 53.Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annu Rev Ecol Syst. 1987;18:237–268. [Google Scholar]
- 54.East EM. Heterosis. Genetics. 1936;21:375–397. doi: 10.1093/genetics/21.4.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheridan PM, Karowe DN. Inbreeding, outbreeding, and heterosis in the yellow pitcher plant, Sarracenia flava (Sarraceniaceae), in Virginia. Am J Bot. 2000;87:1628–1633. [PubMed] [Google Scholar]
- 56.Costa LM, et al. Maternal control of nutrient allocation in plant seeds by genomic imprinting. Curr Biol. 2012;22:160–165, and erratum (2012) 22:1980. doi: 10.1016/j.cub.2011.11.059. [DOI] [PubMed] [Google Scholar]
- 57.Yuan J, et al. Both maternally and paternally imprinted genes regulate seed development in rice. New Phytol. 2017;216:373–387. doi: 10.1111/nph.14510. [DOI] [PubMed] [Google Scholar]
- 58.Spillane C, et al. Positive darwinian selection at the imprinted MEDEA locus in plants. Nature. 2007;448:349–352, and erratum (2007) 450:450. doi: 10.1038/nature05984. [DOI] [PubMed] [Google Scholar]
- 59.Haig D. Kin conflict in seed development: An interdependent but fractious collective. Annu Rev Cell Dev Biol. 2013;29:189–211. doi: 10.1146/annurev-cellbio-101512-122324. [DOI] [PubMed] [Google Scholar]
- 60.Pires ND, et al. Quantitative genetics identifies cryptic genetic variation involved in the paternal regulation of seed development. PLoS Genet. 2016;12:e1005806. doi: 10.1371/journal.pgen.1005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Armbruster WS. Patterns of character divergence and the evolution of reproductive ecotypes of Dalechampia scandens (Euphorbiaceae) Evolution. 1985;39:733–752. doi: 10.1111/j.1558-5646.1985.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 62.Webster GL, Webster BD. The morphology and relationships of Dalechampia scandens (Euphorbiaceae) Am J Bot. 1972;59:573–586. [Google Scholar]
- 63.Armbruster WS. The role of resin in angiosperm pollination–Ecological and chemical considerations. Am J Bot. 1984;71:1149–1160. [Google Scholar]
- 64.Pérez-Barrales R, Bolstad GH, Pélabon C, Hansen TF, Armbruster WS. Pollinators and seed predators generate conflicting selection on Dalechampia blossoms. Oikos. 2013;122:1411–1428. [Google Scholar]
- 65.Opedal ØH, Armbruster WS, Pélabon C. Inbreeding effects in a mixed-mating vine: Effects of mating history, pollen competition and stress on the cost of inbreeding. AoB Plants. 2015;7:plv133. doi: 10.1093/aobpla/plv133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Armbruster WS. Multilevel comparative analysis of the morphology, function, and evolution of Dalechampia blossoms. Ecology. 1988;69:1746–1761. [Google Scholar]
- 67.Hansen TF, Pélabon C, Armbruster WS, Carlson ML. Evolvability and genetic constraint in Dalechampia blossoms: Components of variance and measures of evolvability. J Evol Biol. 2003;16:754–766. doi: 10.1046/j.1420-9101.2003.00556.x. [DOI] [PubMed] [Google Scholar]
- 68.Bolstad GH, et al. Genetic constraints predict evolutionary divergence in Dalechampia blossoms. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130255. doi: 10.1098/rstb.2013.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.R Core Team 2017. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna), Version 3.3.3.
- 70.Bates D, Maechler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;61:1–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.