Significance
In the 20 years since the discovery of the genetic link between the transcription factor TBX5 and congenital heart defects, few direct targets of TBX5 in cardiac morphogenesis have been identified. In this work, we demonstrate that TBX5 directly regulates canonical Wnt ligands required for initiation of lung development. Lung endoderm forms a Hedgehog signaling source required for morphogenesis of both the lungs and the cardiac inflow septum. Our work expands the role of TBX5 to include a non–cell-autonomous component for atrial septation. We find the mesoderm–endoderm–mesoderm signaling loop initiated by TBX5 is evolutionarily conserved from amphibians to mammals. This work suggests that the evolutionary origin of lungs may have involved the recruitment of cardiac TBX5.
Keywords: lung development, heart development, TBX5, Wnt signaling, Hedgehog signaling
Abstract
Codevelopment of the lungs and heart underlies key evolutionary innovations in the transition to terrestrial life. Cardiac specializations that support pulmonary circulation, including the atrial septum, are generated by second heart field (SHF) cardiopulmonary progenitors (CPPs). It has been presumed that transcription factors required in the SHF for cardiac septation, e.g., Tbx5, directly drive a cardiac morphogenesis gene-regulatory network. Here, we report instead that TBX5 directly drives Wnt ligands to initiate a bidirectional signaling loop between cardiopulmonary mesoderm and the foregut endoderm for endodermal pulmonary specification and, subsequently, atrial septation. We show that Tbx5 is required for pulmonary specification in mice and amphibians but not for swim bladder development in zebrafish. TBX5 is non–cell-autonomously required for pulmonary endoderm specification by directly driving Wnt2 and Wnt2b expression in cardiopulmonary mesoderm. TBX5 ChIP-sequencing identified cis-regulatory elements at Wnt2 sufficient for endogenous Wnt2 expression domains in vivo and required for Wnt2 expression in precardiac mesoderm in vitro. Tbx5 cooperated with Shh signaling to drive Wnt2b expression for lung morphogenesis. Tbx5 haploinsufficiency in mice, a model of Holt–Oram syndrome, caused a quantitative decrement of mesodermal-to-endodermal Wnt signaling and subsequent endodermal-to-mesodermal Shh signaling required for cardiac morphogenesis. Thus, Tbx5 initiates a mesoderm–endoderm–mesoderm signaling loop in lunged vertebrates that provides a molecular basis for the coevolution of pulmonary and cardiac structures required for terrestrial life.
Utilization of atmospheric oxygen revolutionized the ability of vertebrates to adapt to terrestrial life (1). At the center of this revolution are the lungs, a foregut-derived gas-exchange structure (1, 2). The derived cardiovascular system, utilizing pulmonary oxygen, must manage blood from both the body and the lungs simultaneously (2). While most lungfish, amphibians, and reptiles exhibit a three-chambered heart with an atrial septum separating pulmonary and systemic circulation entering the heart (3), the two-sided, four-chambered crocodilian, avian, and mammalian hearts have independently evolved to completely separate pulmonary from systemic circulation (4). The proper development and placement of the cardiac septa are critical for the efficient handling of blood, and defects in these structures comprise common forms of human congenital heart disease.
Recent work has highlighted the common developmental origin of multiple mesodermal derivatives in both the heart and the lung (5, 6). This lateral plate mesodermal population has been termed the “second heart field” (SHF) or “cardiopulmonary progenitors” (CPPs). This population originates dorsal to the cardiac inflow tract and ventral to the anterior foregut and generates multiple structures in the heart, e.g., the atrial septum, and in the lungs, e.g., smooth muscle and vascular endothelium (5, 6). This essential CPP region is labeled by expression of the canonical Wnt signaling ligand Wnt2, the Hedgehog (Hh) signaling-responsive transcription factor Gli1, and the T-box family transcription factor Tbx5 (5, 7–9).
Mutations in TBX5 have been implicated as the primary genetic cause of Holt–Oram syndrome (HOS), a human syndrome that includes cardiac septal defects (10–14). Previous work has demonstrated that Tbx5 is required in the posterior SHF (pSHF) for atrial septation (7, 9, 15). In addition, Sonic hedgehog (Shh) signaling has been implicated in cardiac septation (7–9, 16). Shh, expressed in the pulmonary endoderm (PE), activates GLI-mediated transcription in the CPPs (7, 8). Shh and Tbx5 genetically interact for cardiac septation, and constitutive activation of Hh signaling in CPPs rescues atrial septal defects caused by reduced Tbx5 dose (7, 9). Furthermore, TBX5 and GLI transcription factors directly cooperate at enhancers for genes required for cardiac septation (7, 9). This has generated a model in which TBX5 and GLI transcription factors directly activate gene expression in the CPPs of the pSHF for cardiac morphogenesis.
CPPs are an important source of signals that induce the pulmonary lineage in the ventral foregut endoderm and contribute directly to the atrial septum and cardiopulmonary vasculature (17–19). An evolutionarily conserved paracrine signaling cascade involving retinoic acid, Hh, Wnt signaling, and bone morphogenic protein (BMP) regulates the induction of pulmonary progenitors from amphibians to mammals (5, 17–21). Tbx5 has been implicated in lung morphogenesis, both alone and in combination with Tbx4 (22). Midgestation conditional deletion of Tbx5 in mouse embryos caused deficiency in lung-branching morphogenesis, and combined deletion of Tbx4 and Tbx5 in allelic combinations caused reduced WNT2 and BMP4 signaling (22).
We report that Tbx5 is required non–cell-autonomously for the initiation of PE and lung formation. We find that Tbx5 is required for the initiation of lung development in both mammals and amphibians but not for the swim bladder (SB) in zebrafish. Furthermore, we show that TBX5 directly drives the lung-inducing ligands Wnt2 and Wnt2b in pSHF CPPs. TBX5-driven mesoderm-to-endoderm canonical Wnt signaling is required for the subsequent endoderm-to-mesoderm Shh signaling required for atrial septation. Tbx5 haploinsufficiency diminishes both mesodermal Wnt2 and endodermal Shh expression, suggesting that atrial septal defects in HOS may be caused in part by diminished Shh signaling rather than solely by a deficiency of a Tbx5-driven cell-autonomous SHF gene-regulatory network (GRN). Tbx5 thereby initiates a mesoderm–endoderm–mesoderm signaling loop, providing a molecular basis for the coevolution of pulmonary and cardiac development.
Results
Tbx5-Dependent Transcriptional Profiling of the CPPs.
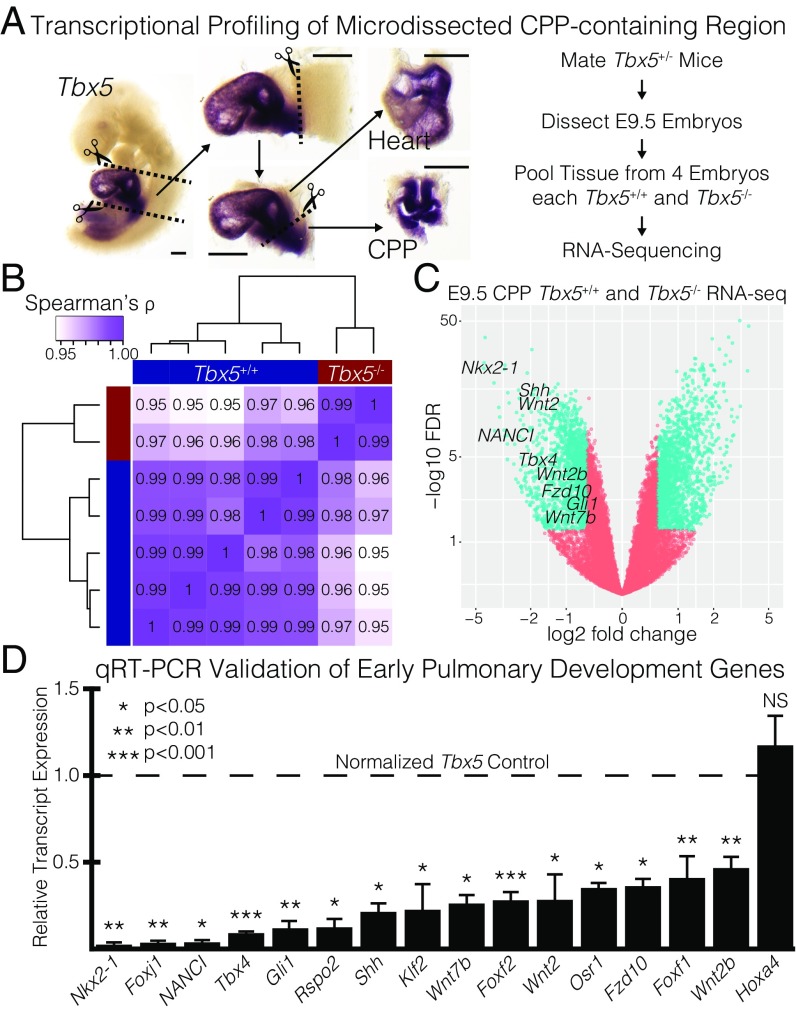
To investigate the role of Tbx5 in CPPs, we performed RNA sequencing (RNA-seq) on microdissected tissue containing the CPPs from Tbx5+/+ and Tbx5−/− mouse embryos at E9.5 (Fig. 1 A and B) (7, 23, 24). Compared with Tbx5+/+ CPPs, 5,486 genes were significantly dysregulated in Tbx5−/− CPPs [false-discovery rate (FDR) <0.05]. We restricted our consideration of genes to those with a magnitude fold change ≥1.5 (SI Appendix, Table S1). This group contained 1,480 down-regulated genes and 1,588 up-regulated genes in the absence of Tbx5 (Fig. 1C). The most significantly down-regulated genes in Tbx5−/− CPPs were transcription factors and signaling factors critical for early lung development. Notably, expression of Nkx2-1 and the long noncoding RNA E030019B13Rik or NANCI, the first markers of PE specification, was extinguished (17, 25–28). Members of the Wnt and Shh signaling pathways, both required for early lung specification and morphogenesis, were also severely down-regulated (Fig. 1C). In addition, we observed 14 other genes among the down-regulated list that have been reported in the literature to be critical for lung development (6, 29–31). As an early role for Tbx5 in lung development has been suggested (22, 32), we validated the significant down-regulation of 15 of 16 “lung development” genes by qRT-PCR in independent samples (Fig. 1D). Together, these data suggested that Tbx5 might occupy a critical position in the GRN for lung induction.
Fig. 1.
Transcriptional profiling of microdissected CPPs identifies a critical role for Tbx5 in pulmonary specification and lung development. (A, Left) Demonstration of microdissection methodology used for embryonic mouse experiments on an E9.5 embryo probed for Tbx5 RNA by ISH. (Scale bars: 0.25 mm.) (Right) Transcriptional profiling strategy used to measure the Tbx5-dependent transcriptome in the CPP-containing tissue by RNA-seq. (B) Spearman’s correlation of RNA-seq replicates. (C) Volcano plot of transcriptional profiling results with significantly dysregulated genes (teal) from the comparison of Tbx5+/+ and Tbx5−/− CPPs. Early markers of the PE and canonical Wnt and Shh signaling are identified. (D) qRT-PCR validation of 16 early lung-development genes that were significantly dysregulated in the RNA-seq.
Tbx5-Dependent Lung Development Is Evolutionarily Conserved.
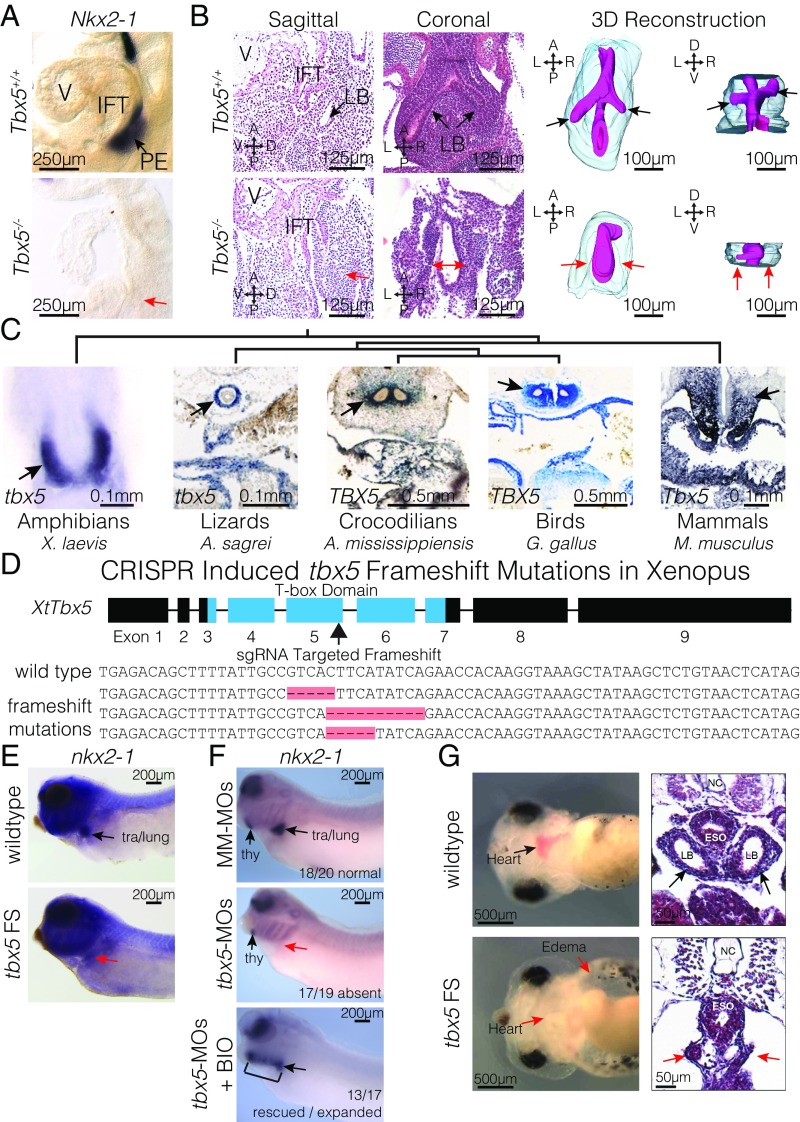
Utilizing the Tbx5−/− mouse embryos, we examined the requirement of Tbx5 for PE specification. The earliest sign of pulmonary induction, Nkx2-1 expression, was absent from the foregut endoderm at E9.5 by in situ hybridization (ISH) (Fig. 2A). Based on sagittal and coronal sections at E10.5, Tbx5−/− mice failed to demonstrate the earliest physical manifestation of lung morphogenesis, the outpouching of the foregut endoderm or lung buds (Fig. 2B). 3D reconstructions highlighting the endoderm further demonstrated the absence of lung bud initiation from the foregut in Tbx5−/− embryos (Fig. 2B).
Fig. 2.
Tbx5 is required for lung development in mice and frogs. (A) RNA ISH for Nkx2-1 in E9.5 Tbx5+/+ and Tbx5−/− embryos. The PE, inflow tract (IFT), and cardiac ventricle (V) are labeled. (B) Histology from both sagittal and coronal perspectives (Left) and 3D reconstruction (Right) of E10.5 lungs from Tbx5+/+ and Tbx5−/− embryos. Black and red arrows point to the lung bud (LB) branches or lack of branches off the foregut. (C) ISH stains of Tbx5 across vertebrate species. Arrows indicate expression in the mesodermal derivatives surrounding the PE. (D, Upper) Strategy for generating biallelic frameshift mutations using sgRNA targeted to the fifth exon of X. tropicalis to disrupt the T-box domain. (Lower) Examples of sequences recovered from tbx5 FS mutants. (E and F) RNA ISH of NF35 tadpoles for nkx2-1 in wild type and tbx5 FS mutants (E) and tadpoles injected with mismatched morpholinos (MM-MOs), tbx5-MOs, or tbx5-MOs cotreated with BIO (F). (G) Live images (Left) and H&E-stained transverse sections (Right) of NF42 tadpoles, depicting anatomical defects induced by CRISPR-mediated mutation of tbx5.
Because of the fundamental role for Tbx5 in lung development in mice, we asked whether this role is conserved across lunged vertebrates. Previously, an evolutionary link across amniotes has been made between Tbx5 expression pattern in the heart and ventricular septation for efficient handling of blood (33). We first examined expression of Tbx5 by ISH in representative species of amphibians (Xenopus laevis), lizards (Anolis sagrei), crocodilians (Alligator mississippiensis), and birds (Gallus gallus). The expression domains of Tbx5 are conserved across each of these species with expression found in the mesodermal derivatives of the lungs in each (Fig. 2C).
We hypothesized that Tbx5 may be an evolutionarily ancient driver of lung specification. We examined whether the role of Tbx5 in lung specification was evolutionarily conserved in amphibians, the oldest lineage of extant tetrapods (32, 34). Xenopus embryos expressed tbx5 in the heart and in the wnt2b-expressing lateral plate mesoderm surrounding the nkx2-1– and shh-expressing PE (SI Appendix, Fig. S1). We examined the requirement of tbx5 for lung development in Xenopus tropicalis by utilizing CRISPR to induce targeted biallelic frameshift mutations in the fifth exon of tbx5 (termed “tbx5 FS”), causing predicted truncations of the Tbx5 polypeptide due to premature translation termination (Fig. 2D). We observed a loss of nkx2-1 in the foregut endoderm of tbx5 FS embryos compared with controls by ISH of embryos at stage 35, when the lung lineage is being induced (Fig. 2E). We observed an identical loss of nkx2-1 in X. laevis embryos injected with tbx5 morpholinos (MOs) (Fig. 2F). tbx5 FS embryos appear phenotypically similar to previously described tbx5-MO knockdowns at stage 42 (35, 36), including gross edema and loss of blood in the embryonic heart (Fig. 2G). Furthermore, similar to the Tbx5−/− mouse (22), tbx5 FS embryos lacked lung buds as determined by histologic section at stage 42 (Fig. 2 B and G). To further characterize lung development, expression of sftpc, encoding surfactant protein C and a marker of pulmonary epithelium, was examined (6, 37). Sftpc expression was absent from the lung buds of tbx5-MO knockdowns at stage 42 but was rescuable by coinjection with a hormone-inducible version of Tbx5 (GR-Tbx5) (SI Appendix, Fig. S2). Together, these data indicated that Tbx5-dependent PE specification is evolutionarily ancient and is conserved from amphibians to mammals.
The SB of ray-finned fish is an endoderm-derived out-pocket proposed to be a lung homolog and whose development shares many genetic pathways with lung development, including Wnt- and Hh-dependent signaling and transcriptomes (38–44). The role of Tbx5 in heart and limb development of ray-finned fish is conserved with tetrapods (45–47). To address whether Tbx5 is required for SB formation, zebrafish homozygous for the tbx5a mutant heartstrings (hst) allele (45) were examined at 96 h postfertilization (hpf). Similar to clutchmate controls, homozygous hst mutants show SB formation (SI Appendix, Fig. S3). Zebrafish have two copies of the Tbx5 gene, tbx5a and tbx5b (48). To address potential redundancy, we utilized published MOs designed to target tbx5a, tbx5b, or tbx5a and tbx5b (47, 48). In all conditions, early expression of shha in the SB bud at 72 hpf (39) and SB formation at 96 hpf was observed (SI Appendix, Fig. S3). Together, our data suggest that, while Tbx5 is required for the initiation and formation of the lungs, tbx5a/b is not required for the formation of the SB.
Non–Cell-Autonomous Requirement of Tbx5 for PE Specification.
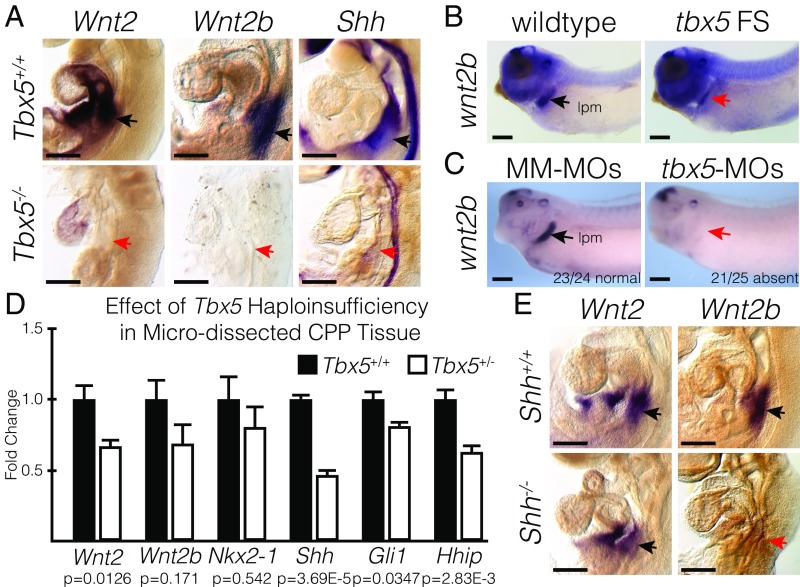
Previous work in mice has demonstrated that Nkx2-1 expression in the foregut endoderm is regulated through canonical Wnt signaling, specifically Wnt2 and Wnt2b. Furthermore, temporal deletion of Tbx5 from cultured embryos demonstrated decreased Wnt2 and Wnt2b expression (22), making the Wnt ligands excellent candidates for direct TBX5 regulation. Wnt2 and Wnt2b are coexpressed with Tbx5 in the SHF mesoderm and are significantly down-regulated in Tbx5−/− CPPs (Fig. 1 C and D) (5, 17, 20). We examined the epistatic requirement for tbx5 and canonical Wnt signaling in Xenopus lung specification. We asked if lung specification in tbx5-MO embryos could be rescued by activating the canonical Wnt pathway, using treatment with a glycogen synthase kinase 3 (GSK-3) inhibitor, 6-bromoindirubin-30-oxime (BIO), which stabilizes β-catenin. We found that BIO treatment rescued and expanded nkx2-1 expression in tbx5-MO embryos (Fig. 2E), suggesting that canonical Wnt signaling is downstream of tbx5 in the lateral plate mesoderm of amphibians. Furthermore, we observed that expression of Wnt2 and Wnt2b was extinguished in Tbx5−/− mouse embryos at E9.5 (Fig. 3A) and found by ISH that wnt2b was similarly lost from stage-35 tbx5 FS X. tropicalis and from tbx5-MO–injected X. laevis (Fig. 3 B and C). These observations were consistent with a requirement for Tbx5 upstream of canonical Wnt signaling for pulmonary specification.
Fig. 3.
Tbx5 is required for Wnt2/2b and Shh expression. (A) RNA ISH for Wnt2, Wnt2b, and Shh in E9.5 Tbx5+/+ and Tbx5−/− mouse embryos. Black and red arrows point to positive and negative staining, respectively, in the lung-forming region. (B and C) RNA ISH for wnt2b performed in wild-type or tbx5 FS NF35 tadpoles (B) and in NF35 tadpoles injected with mismatched morpholinos (MM-MOs) or tbx5-MOs (C). The stained region corresponds with the lateral plate mesoderm (lpm). (D) qRT-PCR of microdissected CPP tissue from E9.5 Tbx5+/+ or Tbx5+/− mouse embryos. (E) RNA ISH for Wnt2 and Wnt2b in E9.5 Shh+/+ and Shh−/− mouse embryos. (Scale bars: 250 µm.)
Tbx5 Is Required for Pulmonary Shh Signaling.
Hh signaling from the PE is required for both cardiac and lung morphogenesis (7, 9, 19, 49). Specifically, Shh is expressed in the PE and is required for atrial septation and lung morphogenesis postinduction (8, 50, 51). We predicted that the requirement of Tbx5 for PE specification would also reflect a requirement for pulmonary Shh signaling. We observed by RNA-seq that Shh is dramatically down-regulated in Tbx5−/− embryos, and we observed by ISH that Shh is specifically lost from the foregut/PE at E9.5 (Figs. 1A and 3A). This suggests that the epistatic relationship between Tbx5 and Shh signaling (9) is an indirect feature of the requirement of Tbx5 for pulmonary lung induction.
TBX5 haploinsufficiency in humans results in the congenital HOS, displaying radial forelimb and congenital heart defects, most commonly atrial septal defects (10–12). Shh expression in the PE is required for morphologic development of the atrial septum (8, 16), and we have demonstrated a genetic interaction between Tbx5 and Shh (7, 9). We therefore hypothesized that a quantitative decrement in Tbx5 would result in diminished Wnt and Shh signaling, contributing to the Tbx5-haploinsufficient phenotype. We examined the gene-expression level of the canonical Wnts, PE specification, and Hh signaling in mouse embryos with Tbx5 haploinsufficiency (Fig. 3D). In the CPPs of Tbx5+/− embryos, we observed a significant down-regulation of Wnt2 (0.67 ± 0.05 SEM, P = 0.0126) but not of Wnt2b or Nkx2-1 (0.69 ± 0.15 SEM, P = 0.1707 and 0.80 ± 0.15 SEM, P = 0.5420, respectively). However, a significant down-regulation of Shh (0.46 ± 0.04 SEM, P = 3.693E-05) and the canonical Hh targets Gli1 and Hhip (0.81 ± 0.04 SEM, P = 0.0347 and 0.63 ± 0.05 SEM, P = 0.0028, respectively) was observed in Tbx5+/− embryos compared with controls. Thus, Tbx5 haploinsufficiency caused a decrement of both Shh expression in the PE and Shh signaling reception in CPPs.
A Mesoderm–Endoderm–Mesoderm Signaling Loop for Cardiopulmonary Development.
Tbx5 and Shh signaling coordinately control gene expression in the CPPs for cardiac development (7, 9, 49). We asked if Tbx5 and Shh signaling interact to regulate lung development. Previously we showed that Shh reception in CPPs promotes wnt2b expression during lung induction in Xenopus (19). We observed similar results in mice: Wnt2b, but not Wnt2, in CPPs was Shh dependent at E9.5 (Fig. 3E). We further confirmed Wnt2 expression in the embryonic mesoderm of the Smo−/− germline mutant, which ablates all Hh signaling independent of ligand (SI Appendix, Fig. S4). These data suggest that Wnt2 expression is upstream or independent of Shh, while Wnt2b is downstream of Shh.
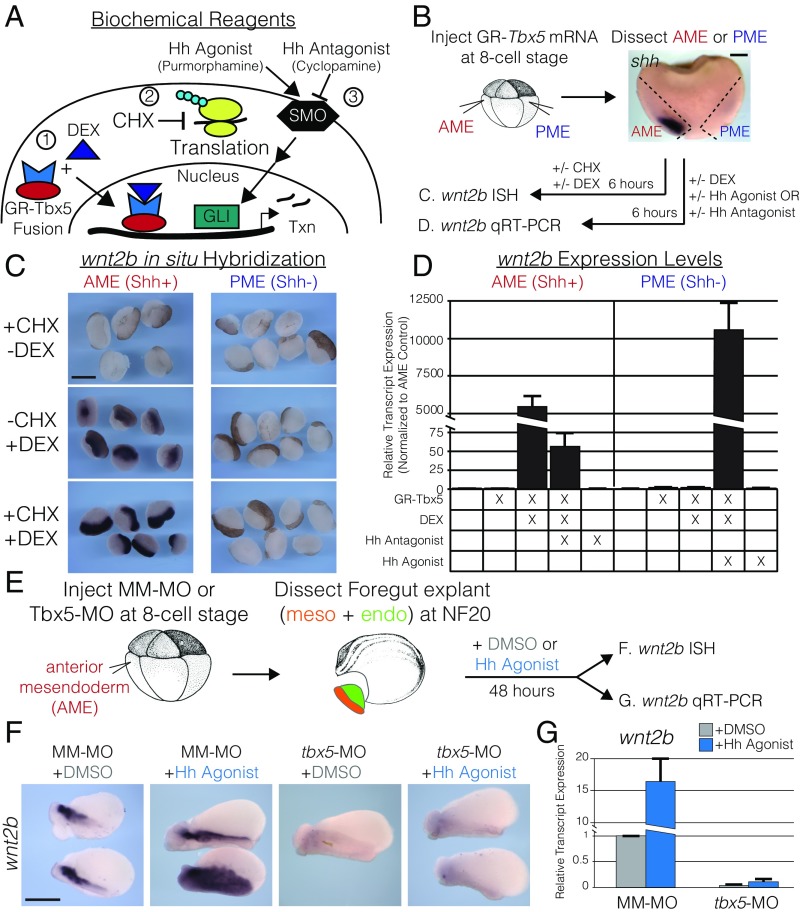
Utilizing a suite of biochemical reagents and the Xenopus model (Fig. 4A), we investigated the interaction between Tbx5 and Shh signaling for wnt2b expression. RNA encoding a fusion protein between Tbx5 and the hormone-inducible region of the glucocorticoid receptor (GR-Tbx5), affording dexamethasone (DEX)-dependent regulation of nuclear import, was injected into the anterior mesendoderm (AME), which has active Hh signaling and gives rise to the foregut (52–55), or into the posterior mesendoderm (PME), which does not have active Hh signaling (Fig. 4 A and B). AME and PME tissue was explanted postgastrulation, DEX treated, and examined after 6 h (Fig. 4 C and D and SI Appendix, Fig. S5). We found that GR-Tbx5 was sufficient to activate wnt2b in the Hh-positive AME but not in the Hh-negative PME (Fig. 4 C and D). We examined whether wnt2b activation by GR-Tbx5 was direct by coadministering DEX with cycloheximide (CHX) to block translation (Fig. 4 A–C). As with DEX administration alone, DEX/CHX coadministration induced wnt2b expression in the AME but not in the PME (Fig. 4B), suggesting that GR-Tbx5 directly activates wnt2b expression in Hh-positive tissue. To validate the requirement for Hh signaling, we coadministered DEX with the Hh antagonist cyclopamine (SI Appendix, Fig. S5) (56). Coadministration of DEX and cyclopamine significantly blunted the activation of wnt2b in AME tissue as compared with DEX alone (97.0-fold decrement, P = 5.48E-3) (Fig. 4D). Last, we examined whether Hh signaling in the AME was unique or whether treatment of PME tissue with the Hh agonist purmorphamine (57) was sufficient for the coinduction of wnt2 expression. PME tissue treated with purmorphamine alone significantly activated gli1 (16.7-fold activation, P = 0.015) but not wnt2b (Fig. 4D and SI Appendix, Fig. S5). However, injection of GR-Tbx5 followed by coadministration of DEX and purmorphamine significantly activated wnt2b expression in the PME (P = 9.29E-3) (Fig. 4D). Together, these data suggested that Tbx5 directly activates wnt2b gene expression in the presence of active Hh signaling.
Fig. 4.
tbx5 directly regulates wnt2b expression for lung development in the presence of Hh signaling. (A) To study the interaction of Tbx5 and Hh signaling, we utilized (1) a DEX-inducible GR-Tbx5 fusion protein; (2) CHX to inhibit protein synthesis; and (3) pharmacological agonists (purmorphamine) and antagonists (cyclopamine) of Smoothened (SMO) to activate or repress Hh signaling. (B) Strategy used in C and D for examining the regulation of wnt2b by Tbx5 in the presence or absence of Shh signaling in Xenopus. The AME (red) corresponds to shh-expressing tissue, and the PME (blue) corresponds to shh-negative tissue. (C) RNA ISH for wnt2b in AME or PME explants treated with CHX, DEX, or both. Note that the ISH signal is black; the brown color is pigment. (D) qRT-PCR of wnt2b in AME or PME explants, with or without GR-Tbx5, and treated with combinations of DEX, Hh agonist, and Hh antagonist. (E) Strategy used in F and G to examine the regulation of wnt2b by Hh signaling in the presence or absence of tbx5. AME explants were treated with DMSO (gray) or the Hh agonist purmorphamine (blue) for 48 h. (F) RNA ISH for wnt2b on DMSO- or Hh agonist-treated explants from embryos injected with mismatched morpholinos (MM-MO) or tbx5-MO. (G) qRT-PCR of wnt2b normalized to DMSO-treated MM-MO–injected explants. (Scale bars: B, 200 µm; C and E, 400 µm.)
We next evaluated whether Tbx5 is required for the Hh pathway to activate expression of wnt2b and other markers of pulmonary development (Fig. 4 E–G and SI Appendix, Fig. S5). We activated Hh signaling using the Hh agonist purmorphamine in control or tbx5-MO–injected AME foregut explants (Fig. 4E). Purmorphamine treatment of control embryos expanded the endogenous wnt2b expression domain, consistent with activation by Hh signaling (Fig. 4 F and G). However, purmorphamine was unable to promote wnt2b in explants from tbx5-MO embryos, which, similar to vehicle-treated tbx5-MO explants (Fig. 4 F and G), had little detectable wnt2b. Similar to Tbx5−/− mice (Figs. 1 and 3), tbx5-MO explants displayed decreased expression of nkx2-1 (P = 1.13E-9), shh (P = 6.81E-3), and dhh (P = 1.11E-5) in the foregut endoderm and decreased expression of wnt2b (P = 1.20E-3) and gli1 (P = 1.09E-3) in the mesoderm (Fig. 4 F and G and SI Appendix, Fig. S5). These explant data demonstrate that tbx5 and Hh signaling are corequired for wnt2b expression and further demonstrate that Tbx5 expression is required for Shh and dhh expression in the PE. Overall, these findings suggest a hierarchical series of signaling loops: TBX5 drives canonical Wnt mesoderm-to-endoderm signaling for pulmonary induction, and Shh signaling from the PE to mesoderm collaborates with mesodermal TBX5 to drive ongoing WNT2B mesoderm-to-endoderm signaling for pulmonary morphogenesis.
Identification of TBX5-Dependent Wnt2 Enhancers.
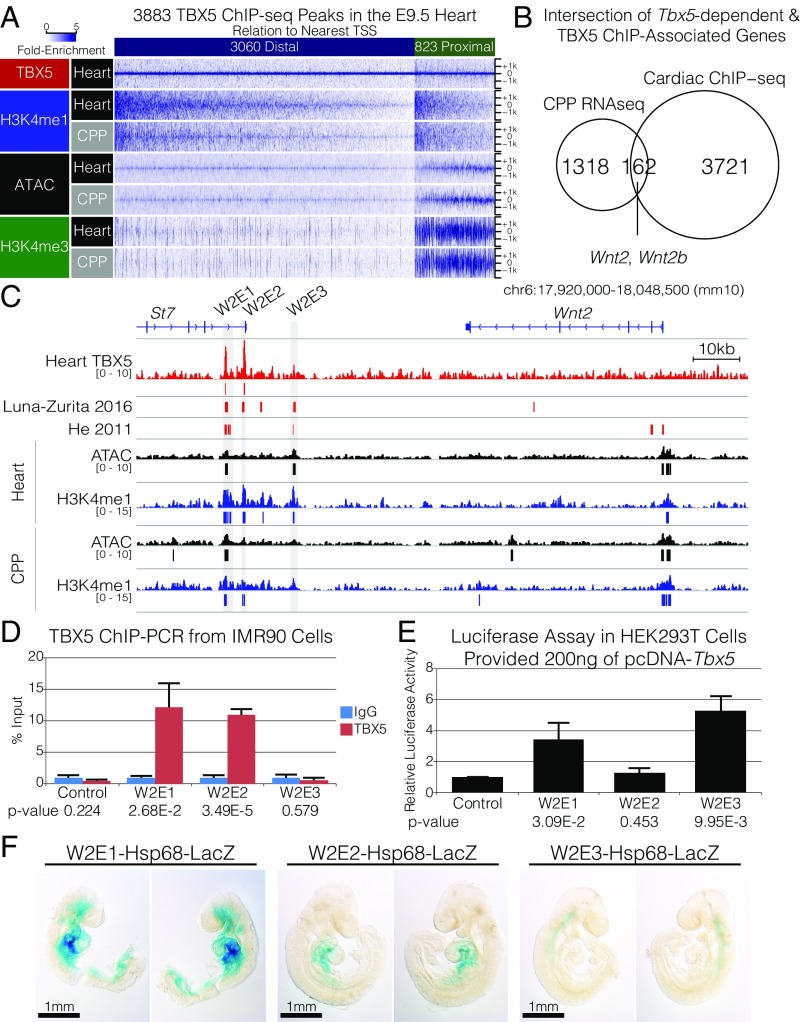
To identify direct targets of TBX5 in coordinating cardiopulmonary specification, we performed TBX5 ChIP-seq on microdissected hearts including the Wnt2-expressing inflow tract from E9.5 mouse embryos (Fig. 1A). We identified 3,883 TBX5-bound regions at E9.5 (Fig. 5A and SI Appendix, Table S2). These locations segregated into 823 promoter-proximal (TBX5 summit ≤2 kbp from an annotated transcription start site) and 3,060 promoter-distal sites. To define TBX5 binding in a genomic context, we identified active promoters and cis-regulatory regions for H3K4me3 and H3K4me1 by ChIP-seq and in microdissected E9.5 heart and CPP tissue by assay for transposase-accessible chromatin sequencing (ATAC-seq) (Figs. 1A and 5A) (58). ATAC-seq, genome-wide and at TBX5-bound regions, showed similar signal (Pearson correlation coefficient = 0.96 and 0.93), H3K4me1 (Pearson correlation coefficient = 0.89 and 0.76), and H3K4me3 (Pearson correlation coefficient = 0.92 and 0.93) in both CPPs and the heart (SI Appendix, Fig. S6). To identify direct TBX5 targets in the CPP tissue, we overlapped the 1,480 Tbx5-dependent genes (Fig. 1C) with the 3,880 ChIP sites (annotated to the nearest gene). This conservative approach identified 162 genes associated with 220 bound sites, including Wnt2 and Wnt2b (Fig. 5 B and C and SI Appendix, Fig. S7 and Table S3). This observation was consistent with the hypothesis that TBX5 directly regulates Wnt2 and Wnt2b transcription (22, 32).
Fig. 5.
Identification of TBX5-bound cis-regulatory elements for Wnt2. (A, Upper) We identified 3,883 peaks by TBX5 ChIP-seq in the E9.5 heart that correspond to 3,060 distal and 823 proximal sites. (Lower) Heatmaps of fold enrichment plotted for TBX5, H3K4me1, and H3K4me3 by ChIP-seq and ATAC-seq from the heart and CPP microdissections at each of the 3,883 summits ± 2,000 bp. (B) Overlap of the 1,318 down-regulated genes in the CPP of Tbx5−/− embryos by RNA-seq and the 3,883 genes nearest to TBX5 ChIP-seq peaks. The 162 genes in the intersection include Wnt2 and Wnt2b. (C) Genome browser view of Wnt2 and St7 (mm10 chr6:17,920,000–18,048,500) with TBX5 ChIP-seq (both from A and published in refs. 59 and 60), H3K4me1 ChIP-seq, and ATAC-seq in the heart and pSHF. Tracks depict fold-enriched signal, and bars below represent significant peak calls. Cloned enhancers W2E1, -2, and -3 are shaded in gray. (D) ChIP-PCR for TBX5 at W2E1, -2, and -3 in the human IMR90 lung fibroblast cell line. Significance is calculated relative to IgG control. (E) Luciferase assay examining activation of W2E1, -2, and -3 in HEK293T cells provided a vector containing Tbx5 relative to a control vector. (F) Transgenic embryos were generated using an Hsp68-LacZ reporter construct upstream of W2E1, -2, or -3 and were stained at E9.5.
While both Wnt2 and Wnt2b are Tbx5 dependent and redundant for lung development (17, 22), only Wnt2 is required for both lung and cardiac morphogenesis in mammals (18). Therefore, we attempted to identify the TBX5-dependent cis-regulatory elements that control Wnt2 expression. We compared our TBX5 ChIP-seq results with previously published TBX5 ChIP-seq from two in vitro systems (59, 60). Using this approach, we identified a cluster of TBX5-bound sites adjacent to the 3′ end of the neighboring gene, St7, which demonstrated the hallmarks of putative regulatory elements including chromatin accessibility and H3K4me1 signal (Fig. 5C). We cloned the regions corresponding to the ChIP-seq signal (mm10 chr6:17938154–17940081, chr6:17941997–17942724, and chr6:17952290–17953703) and named the putative regulatory elements “Wnt2 enhancer 1,” “Wnt2 enhancer 2,” and “Wnt2 enhancer 3” (W2E1–3), respectively (Fig. 5C). We performed ChIP-qPCR in IMR90 human lung fibroblast cells to validate TBX5 localization at these candidate enhancers (Fig. 5D). We observed a significant enrichment of TBX5 at W2E1 and W2E2 over IgG control (12.14 ± 3.81 SD, P = 0.03 and 10.95 ± 0.85 SD, P = 3.5E-5, respectively), while W2E3 did not show enrichment (0.58 ± 0.36 SD, P = 0.44).
We examined the enhancer activity and Tbx5 dependence of W2E1–3 in vitro by luciferase reporter assay using HEK293T cells and exogenous expression of Tbx5, as previously described (7, 8, 61). Tbx5 expression activated W2E1 and W2E3 (3.44 ± 1.07 SEM, P = 0.0309 and 5.29 ± 0.93 SEM, P = 0.0099, respectively) but not W2E2 (1.27 ± 0.33 SEM, P = 0.4532) compared with a control vector (Fig. 5E). As W2E1 was both TBX5 bound and responsive to Tbx5 expression, we examined the dependence of W2E1 activity on the presence of the canonical T-box motif AGGTG (SI Appendix, Fig. S8) (59, 60, 62). Mutation of the minimal canonical T-box motifs in W2E1 resulted in a 3.997-fold decrease compared with wild-type W2E1 (P = 0.0252), whereas mutation of T-box motifs within the control vector had no effect (P = 0.5237).
We examined the sufficiency of W2E1–3 for driving cardiac and SHF gene expression in vivo. Each enhancer was cloned upstream of the Hsp68 minimal promoter driving lacZ expression and was utilized for the generation of transient transgenic mouse embryos, as previously described (7, 63, 64). W2E1 and W2E2 each drove robust activation of lacZ within the CPP and inflow tract domains of Wnt2 expression with W2E1 driving robust activation of lacZ within many domains of Wnt2 at E9.5 (Fig. 5F). W2E3, in contrast, activated lacZ in non–Wnt2-, non–Tbx5-expressing tissues. Taken together, our data suggested that W2E1 and W2E2 represent TBX5-responsive cis-regulatory elements for early cardiopulmonary Wnt2 expression.
Requirement of TBX5-Dependent Regulation of Wnt2.
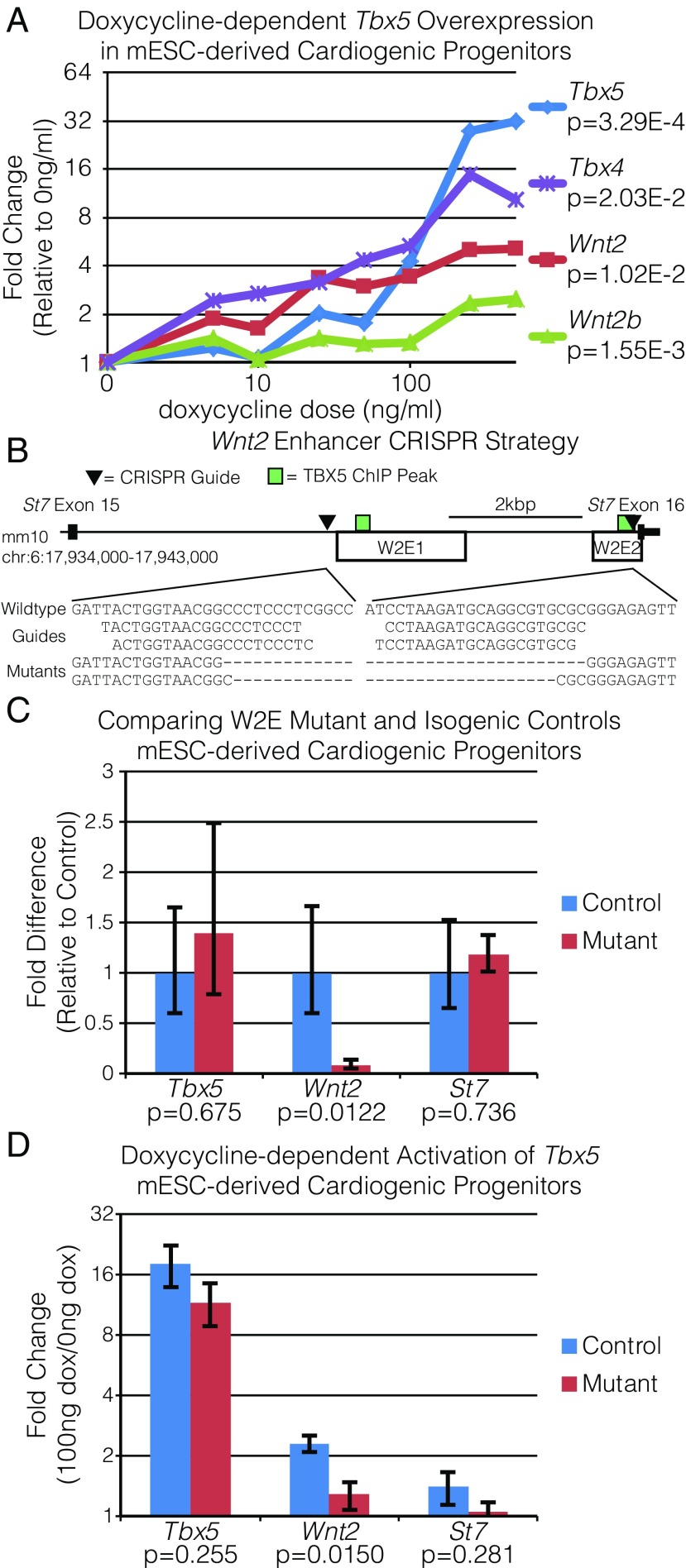
To investigate the direct requirement of TBX5-driven enhancers for pulmonary mesoderm gene expression, we generated a mouse embryonic stem cell (mESC) line with doxycycline (DOX)-inducible Tbx5 expression using the A2Lox.cre mESC line (65). This line (Tbx5OE-mESC) was differentiated along a sequence of ES cells to mesoderm to lateral plate mesoderm to cardiac progenitor as previously described (66). We observed a linear dose-response of Tbx5 in cardiac progenitors (0 ng/μL to 500 ng/μL; 0.07x + 0.60, P = 3.29E-4) after 24 h (Fig. 6A). We observed a significant relationship between DOX dose and expression of Wnt2 (P = 1.02E-2), Wnt2b (P = 1.55E-3), and Tbx4 (P = 2.03E-2), another marker of pulmonary mesoderm. This observation suggested that pulmonary mesodermal markers are directly responsive to Tbx5 expression levels in CPPs in vitro.
Fig. 6.
Cis-regulatory elements are required for Wnt2 expression. (A) Dose-dependent gene-expression changes in mESC-derived cardiogenic progenitors harboring a DOX-dependent Tbx5 construct (Tbx5OE-mESC) measured by qRT-PCR. Cells were treated with DOX for 24 h before analysis. (B, Upper) sgRNAs were designed to induce a 4.5-kb deletion within the last intron of St7, removing a majority of W2E1 and W2E2 in the Tbx5OE-mESC via CRISPR-Cas9. This design maintains the last exon and the predicted splice branch for St7 while removing the TBX5-bound sites in W2E1 and W2E2. (Lower) Amplification and sequencing across the target site demonstrate successful deletion. (C) Expression levels of Tbx5, Wnt2, and St7 were compared in W2E mutants and isogenic controls following differentiation to cardiogenic progenitors by qRT-PCR. (D) Changes in Tbx5, Wnt2, and St7 expression following 24 h of 100 ng/mL DOX relative 0 ng/mL DOX in the W2E mutant and isogenic controls differentiated to cardiogenic precursors.
We examined the requirement of the TBX5-bound cis-regulatory elements W2E1 and W2E2 for Tbx5-dependent Wnt2 expression. Specifically, we utilized CRISPR/cas9 to generate a 4.6-kbp deletion of W2E1 and W2E2 without disrupting the last exon of St7 or its predicted splice acceptor from the mESC line overexpressing Tbx5 (hereafter, the “Tbx5OE-mESC line”) (Fig. 6B). Two homozygous deletion clones (W2E mutants) and two control clones (W2E controls) were generated and evaluated. Following differentiation of clones to cardiac progenitors, the W2E mutants demonstrated a significant reduction in Wnt2 gene expression compared with W2E controls (91.8% reduction, P = 0.0122), while there was no significant difference in Tbx5 or St7 (Fig. 6C). To examine the requirement of W2E1/W2E2 for Tbx5-dependent activation of Wnt2, we induced Tbx5 overexpression and evaluated the response of Tbx5, Wnt2, and St7 expression. We observed that the W2E mutants had significantly reduced Wnt2 expression in response to Tbx5 overexpression compared with the W2E control lines (2.31-fold versus 1.28-fold activation, P = 0.0150); no significant differences between the mutant and control lines were observed for Tbx5 or St7 (Fig. 6D). Taken together, these results demonstrate that W2E1 and W2E2 are required for Wnt2 expression and are necessary for Tbx5-responsive Wnt2 expression in mESC-derived CPPs. These results demonstrate direct molecular control of Wnt2 by TBX5 in an in vitro model of early cardiopulmonary development.
Discussion
TBX5 has been genetically implicated in human cardiac septal defects for over 20 y. Based on its strong expression in the heart, TBX5 was assumed to directly drive a cardiac GRN for cardiac septation. Recently, work by the I.P.M. laboratory has determined that the role of TBX5 resides in the SHF (Fig. 7). We assumed that probing pSHF CPPs for Tbx5-dependent target genes would uncover a direct cardiac progenitor GRN for cardiac morphogenesis. Instead, we observed a primary role for Tbx5 in the initiation of lung development and, secondarily, the establishment of PE-to-mesoderm signaling for cardiac septation.
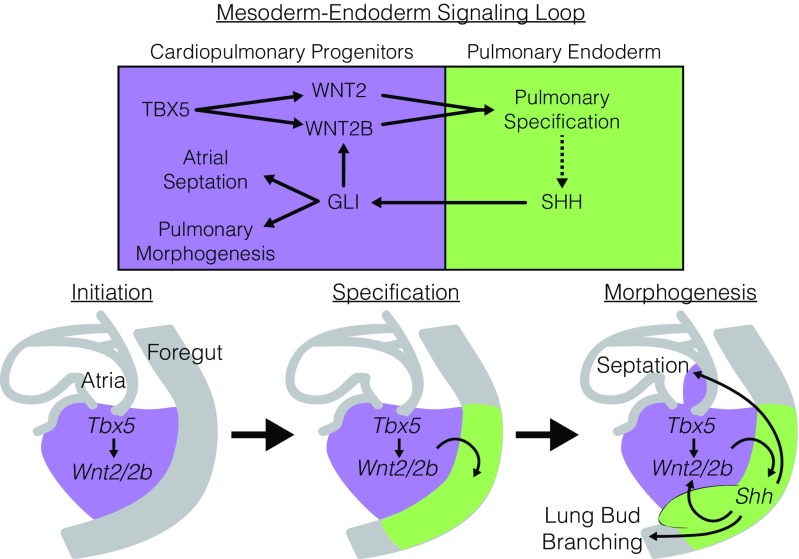
Fig. 7.
Tbx5 is required for a mesoderm–endoderm bidirectional signaling loop for cardiopulmonary development. Model of genetic interaction between Tbx5, canonical Wnt signaling, and Hh signaling for PE specification, pulmonary morphogenesis, and cardiac septation. TBX5, expressed in the CPPs (purple) initiates the bidirectional signaling loop through direct activation of Wnt2 and Wnt2b expression. Canonical Wnt signaling drives pulmonary specification in the foregut endoderm and Nkx2-1 expression. SHH, derived from the PE (green) signals back to the CPPs where it cooperatively activates Wnt2b but not Wnt2. Shh signaling drives both atrial septation and lung bud morphogenesis through previously described downstream targets.
We report that Tbx5 is required for the initiation of lung development through canonical Wnt signaling (Fig. 7). We observed that Tbx5 directly regulates the transcription of both Wnt2 and Wnt2b, wingless-family signaling molecules redundantly required for the earliest aspects of pulmonary development (17). Specifically, we identified cis-regulatory elements for Wnt2 that are required for TBX5-responsive transcription and that drive transcription in the CPPs and inflow tract of the heart. A role for canonical Wnt signaling in inflow tract development is conserved between Drosophila and mammals (67). Drosophila Wingless (wg) is required for the formation of the Drosophila cardiac inflow tract (67), suggesting that the preexisting role of canonical Wnt signaling in inflow tract development may have been coopted for lung development and inflow septation later in vertebrates and early tetrapods. Although the early requirement of Tbx5 for heart and limb development has been well documented across vertebrate species (13–15, 45–47, 68–70), the role of Tbx5 in lung development has not been examined outside of mammals. Overall, our work suggests a fundamental role for Tbx5 in tetrapod lung development and the possibility that the evolutionary origin of lungs may have involved the recruitment of TBX5 from an ancestral cardiac expression domain.
Although a recent model suggested that the lungs and SB are evolutionarily derived from a common structure, we find that tbx5 is not required for the development of both (43, 44). Previous work in zebrafish demonstrates that depletion of wnt2 and wnt2bb causes SB agenesis, similar to their requirement in lung specification (17, 38). However, we find that although Tbx5 is required for lung formation, tbx5a/b is not required for SB formation in the zebrafish. These observations suggest that tbx5a/b-independent regulation of Wnt signaling is required for the initiation of SB development. One question worth future investigation is whether the Tbx5-positive lateral plate mesoderm gives rise to SB components or whether the dorsally derived SB forms from a mesodermal contribution distinct from the ventrally derived lungs. Additionally, zebrafish are part of the derived teleost fish, and further characterization across ray-finned fish is required. We note that the Senegal bichir (Polypterus senegalus), a member of the early-diverging Actinopterygii, has a ventral-sided lung structure for air breathing and was reported to express both tbx5 and tbx4 in the early lung structure (43). Additional work is required to resolve the evolutionary relationship between the SB and lungs.
Integrating our observations that Tbx5 is required for pulmonary specification with previous work demonstrating a role for Tbx5 in lung morphogenesis (22) suggests sequential roles for Wnt signaling during lung development: an early requirement for initiation and a later requirement for branching morphogenesis. In contrast to the complete loss of lung development in the Tbx5 germline-null mouse, conditional removal of Tbx5 at E8.5 caused malformation of lung bud branching and disruption of canonical Wnt signaling in explant cultures (22). Consistent with a dual-role hypothesis, a partial decrement of canonical Wnt signaling allows lung initiation but causes defects in lung-branching morphogenesis (17, 18), while a complete failure of lung initiation has been observed only by homozygous removal of both Wnt2 and Wnt2b (17). We conclude that Tbx5 initiates a multistep mesoderm–endoderm–mesoderm signaling loop (Fig. 7). TBX5 directly drives WNT2 and WNT2B mesoderm-to-endoderm signaling for pulmonary induction. Secondarily, PE-to-mesoderm Shh signaling collaborates with mesodermal TBX5 for ongoing WNT2B mesoderm-to-endoderm signaling and later pulmonary morphogenesis (Fig. 7).
This study and previously published work suggest that TBX5 mutations may be associated with lung defects. Although rare, lung defects have been described in patients with HOS. Two cases of structural lung disease have been associated with “atypical” HOS: one case of right lung agenesis (71) and one case of horseshoe lungs (72). Additionally, a screen of patients with esophageal atresia and tracheoesophageal fistula identified a patient with HOS (73). Further, rare genetic variants at the TBX5/RBM19 locus have been associated with lung function in smokers by a genome-wide association study (74). These studies suggest the intriguing possibility that TBX5 may play a role in both lung development and adult lung function, similar to its requirement for both cardiac development and adult cardiac function (15, 61, 63, 75).
Cardiac septa are observed in all lunged vertebrates. We have previously demonstrated that GLI-dependent transcription downstream of PE Shh signaling and Tbx5 cooperate in mesodermal SHF CPPs to drive atrial septation (7–9, 49). Here, we demonstrate that Tbx5 is required for the initiation of Shh signaling through the specification of PE. Previous work demonstrated that atrial septal defects caused by removal of Tbx5 from the CPPs were rescued by concomitant activation of Hh signaling in those cells, providing epistatic evidence that Tbx5 acts upstream of Hh signaling for atrial septation (9). In this context, our current results suggest that Shh signaling from the PE to the CPPs is a direct requirement for atrial septation, while Tbx5 may be dispensable following the initiation of lung development and subsequent Shh signaling. Our work demonstrates that Tbx5 haploinsufficiency causes reduced Wnt2 expression and subsequently reduced expression of Shh in the PE, resulting in reduced expression of quantitative markers of Shh reception in the cardiopulmonary mesoderm. Therefore, this quantitative decrement in CPP Hh signaling may contribute to the causation of cardiac septal defects in HOS patients.
The linked mesoderm–endoderm–mesoderm molecular pathways for lung development and cardiac inflow septation are conserved between amphibians and mammals. Remarkably, amphibians with evolutionary loss of lungs exhibit much reduced atrial septation, consistent with necessary instructive cross-talk between these structures (3, 76). We posit that Tbx5–Wnt2/Wnt2b signaling provides a molecular basis for the link between lung formation and the cardiac specializations required for pulmonary blood flow observed in lunged vertebrates.
Materials and Methods
Ethics Statement.
All murine and zebrafish experiments were performed under University of Chicago Institutional Animal Care and Use Committee (IACUC) protocols no. 71737 and no. 71112. X. laevis and X. tropicalis adults were housed according to Cincinnati Children’s Hospital Medical Center or University of North Carolina, Chapel Hill IACUC protocols. Handling of lizards (Anolis sagrei) and harvest of tissues complied with national and institutional guidelines and were approved by the IACUC of the University of Amsterdam (DAE101617).
Mouse Lines.
Tbx5 germline mutant animals (Tbx5+/−) were produced by crossing the Tbx5tm1Jse allele (Tbx5flox) with a germline cre-recombinase and were out-crossed for multiple generations with CD1 animals as previously described (15, 23). Additionally, the Shhtm1Amc (77) and Smotm1Amc (78) germline mutants have been previously described.
Xenopus Experiments and CRISPR-Based Genome Editing.
Ovulation, in vitro fertilization, and dejellying of embryos were performed as described (79). The pCS2+GR Tbx5 plasmid (70) was used to synthesize mRNA for injection using the Ambion mMessage mMachine SP6 RNA synthesis kit. GR-Tbx5 RNA (125 pg) was injected into either the dorsal or ventral marginal zone (targeting the AME or PME, respectively) at the eight-cell stage. Validated Tbx5 translation-blocking MOs (35) were injected at the eight-cell stage (3.5 ng of each MO). See SI Appendix for full details.
A small guide RNA (sgRNA) designed to target exon 5 of the X. tropicalis locus (GGGGTTCTGATATGAAGTGA) was coinjected at 200 pg with 2 ng Cas9 protein (PNA Bio) in 2-nL drops into one-cell-stage wild-type X. tropicalis embryos (80). To screen rapidly for altered loci, an ∼500-bp genomic fragment asymmetrically flanking the sgRNA target sequence was amplified by PCR and subjected to digestion by T7 endonuclease (New England Biolabs).
Zebrafish Lines and Experiments.
Zebrafish were maintained under standard laboratory conditions (81). MO injections were performed as described (82). tbx5a MO (3.7 ng) (47) and 5 ng of tbx5b translation-blocking MO (48) were injected into each embryo. Lines used were *AB and heartstrings (hst) mutants (45).
Transcriptional Profiling by RNA-Seq.
RNA-seq was performed on microdissected CPP tissue at E9.5. Microdissected tissues from four embryos were pooled, and total RNA was extracted from five Tbx5+/+ and two Tbx5−/− pools and was sequenced using the Illumina HiSeq 2500 platform by the Genomics Core Facility at the University of Chicago. Analysis was performed as previously described (23). See SI Appendix for full details.
qRT-PCR.
For mice, RNA was extracted from microdissected tissue as was done for RNA-seq. The reverse-transcription reaction was performed using SuperScript III First-Strand Synthesis SuperMix (Invitrogen). qRT-PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems) and was run on an AB7500 machine (Applied Biosystems). Gene-expression level was normalized by Gapdh. For Xenopus, RNA was collected from three biological replicates containing four explants each. RNA was extracted using the Direct-zol RNA MiniPrep Plus kit (R2070; Zymo Research), and cDNA was generated using SuperScript VILO Master Mix (11755050; Thermo Fisher). Real-time PCR reactions were carried out using PowerUp SYBR Green Master Mix (A25742; Thermo Fisher) on ABI StepOnePlus qPCR machines (Applied Biosystems). Ornithine decarboxylase (odc) was used as a reference gene.
ISH.
Mouse embryonic ISH was performed as previously described (7, 83, 84). ISH of Xenopus embryos and explants was performed as described (79). ISH was performed on stage-11 lizards as previously described (85). The alligator and chicken sections come from a stained series used in previous publications (86, 87), but the sections shown have not been published before. See SI Appendix for full details.
Histology and 3D Reconstruction.
All mouse and zebrafish embryonic histology was performed by the University of Chicago Human Tissue Resource Center. All tissues were fixed in formalin, embedded in paraffin wax, and sectioned to 10-μM thickness. Tissue was counterstained with H&E. Reconstructions of embryonic lung histology were performed using AMIRA (5.3.2). See SI Appendix for full details.
ChIP and Analysis.
Chromatin extract was prepared from microdissected tissue from E9.5 CD-1 mouse embryos (2× from 50 embryo pools each) obtained from Charles River or from pelleted IMR90 cells (4× from 5 million cells each). For immunoprecipitation, the chromatin extract was incubated with anti-TBX5 antibody (sc-17866; lot no. G1516; Santa Cruz Biotechnology), anti-H3K4me3 (no. 305-34819; lot no. 14004; Wako Chemicals), or anti-H3K4me1 (ab8895; lot no. GR257926-1; Abcam). High-throughput sequencing libraries from ChIP and input DNA were prepared using the NEBNext Ultra DNA Library Prep Kit (E7370S; New England Biolabs) and were sequenced using Illumina HiSeq instruments by the Genomics Core Facility at the University of Chicago. ChIP-seq analysis was performed using a typical pipeline involving Bowtie2 (88) and MACS2 (89, 90). See SI Appendix for full details.
ATAC-Seq and Analysis.
ATAC-seq was performed as previously described (58) on an Illumina HiSeq system by the Genomics Core Facility at the University of Chicago. Analysis was performed in a similar manner to ChIP-seq. See SI Appendix for full details.
Luciferase Assays.
pCDNA3.1 expression vectors for Tbx5 were previously described (63). W2E1–3 were cloned into the pGL4.23 vector (Promega). Expression and reporter vectors were transfected into HEK293T cells using FuGENE (Promega). Cells were cultured for 48 h after transfection and then were lysed and assayed using the Dual-Luciferase Reporter Assay System (Promega).
Transient Transgenics.
Transient transgenic experiments were performed at E9.5 as previously described (7, 63, 64). W2E1–3 were subcloned into the Hsp68-LacZ vector. The resulting construct was digested with NotI enzyme to remove the backbone, gel-purified, and injected into fertilized mouse eggs at the University of Chicago Transgenics Core Facility.
Tbx5OE-mESC Generation, CRISPR, and in Vitro Differentiation.
The inducible Tbx5OE-mESC line was generated as previously published (65). To generate the W2E mutants, we transfected the mESC with pSpCas9(BB)-2-Puro (PX459) plasmid vectors containing guides designed to generate an ∼4.6-kbp deletion of W2E1 and W2E2. Following clone selection and expansion, two homozygous deletion clones (W2E mutants) and two wild-type clones (W2E controls) were evaluated. Cardiac stem cell differentiation was based on the original protocol from the laboratory of Keller and coworkers (66) with some modifications. For all mESC experiments utilizing overexpression, cells were treated with DOX (Sigma D9891) at the cardiac progenitor-like stage (day 6) and were harvested for RNA 24 h later. For CRISPR cell line evaluation, a dose of 100 ng/mL was used. See SI Appendix for full details.
Supplementary Material
Acknowledgments
We thank Lorenzo Pesce for use of the Beagle2 super computer partly supported by NIH Grant 1S10OD018495-01. This work was funded by NIH Grants R01 HL092153 and R01 HL124836 (to I.P.M.), R01 HD089275 (to F.L.C.), R01 HL126509 (to F.L.C. and I.P.M.), R01 DK070858, R01 HL114898, and P01 HD093363 (to A.M.Z.), R01 HD072598 (to R.K.H.), R01 HD084409 and P40 OD010997 (to M.E.H.), U01 HL100407 (to M.K.), R21 AG054770 (to K.I.), and R21 LM012619 (to X.H.Y.); support was also provided by NIH Grants T32 GM007183 (J.D.S.), T32 HL007381 (J.D.S., A.B.R., R.D.N., M.R., and A.D.H.), T32 HD055164 (S.L.), and T32 GM007197 (A.D.H. and E.A.T.B.A.) and by Regenerative Medicine Minnesota Grant RMM 102516 001 (to S.S.-K.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-sequencing, ChIP-sequencing, and ATAC-sequencing data have been deposited in the Gene Expression Omnibus databank (accession nos. GSE75077 and GSE119885).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811624115/-/DCSupplemental.
References
- 1.Hsia CC, Schmitz A, Lambertz M, Perry SF, Maina JN. Evolution of air breathing: Oxygen homeostasis and the transitions from water to land and sky. Compr Physiol. 2013;3:849–915. doi: 10.1002/cphy.c120003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farmer CG. Evolution of the vertebrate cardio-pulmonary system. Annu Rev Physiol. 1999;61:573–592. doi: 10.1146/annurev.physiol.61.1.573. [DOI] [PubMed] [Google Scholar]
- 3.Jensen B, Spicer DE, Sheppard MN, Anderson RH. Development of the atrial septum in relation to postnatal anatomy and interatrial communications. Heart. 2017;103:456–462. doi: 10.1136/heartjnl-2016-310660. [DOI] [PubMed] [Google Scholar]
- 4.Jensen B, Wang T, Christoffels VM, Moorman AF. Evolution and development of the building plan of the vertebrate heart. Biochim Biophys Acta. 2013;1833:783–794. doi: 10.1016/j.bbamcr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Peng T, et al. Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature. 2013;500:589–592. doi: 10.1038/nature12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herriges M, Morrisey EE. Lung development: Orchestrating the generation and regeneration of a complex organ. Development. 2014;141:502–513. doi: 10.1242/dev.098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann AD, et al. Foxf genes integrate tbx5 and hedgehog pathways in the second heart field for cardiac septation. PLoS Genet. 2014;10:e1004604. doi: 10.1371/journal.pgen.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann AD, Peterson MA, Friedland-Little JM, Anderson SA, Moskowitz IP. Sonic hedgehog is required in pulmonary endoderm for atrial septation. Development. 2009;136:1761–1770. doi: 10.1242/dev.034157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie L, et al. Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Dev Cell. 2012;23:280–291. doi: 10.1016/j.devcel.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basson CT, et al. The clinical and genetic spectrum of the Holt-Oram syndrome (heart-hand syndrome) N Engl J Med. 1994;330:885–891. doi: 10.1056/NEJM199403313301302. [DOI] [PubMed] [Google Scholar]
- 11.Newbury-Ecob RA, Leanage R, Raeburn JA, Young ID. Holt-Oram syndrome: A clinical genetic study. J Med Genet. 1996;33:300–307. doi: 10.1136/jmg.33.4.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt M, Oram S. Familial heart disease with skeletal malformations. Br Heart J. 1960;22:236–242. doi: 10.1136/hrt.22.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li QY, et al. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 14.Basson CT, et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 15.Bruneau BG, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 16.Goddeeris MM, Schwartz R, Klingensmith J, Meyers EN. Independent requirements for hedgehog signaling by both the anterior heart field and neural crest cells for outflow tract development. Development. 2007;134:1593–1604. doi: 10.1242/dev.02824. [DOI] [PubMed] [Google Scholar]
- 17.Goss AM, et al. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev Cell. 2009;17:290–298. doi: 10.1016/j.devcel.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, et al. A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat Genet. 2008;40:862–870. doi: 10.1038/ng.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rankin SA, et al. A retinoic acid-hedgehog cascade coordinates mesoderm-inducing signals and endoderm competence during lung specification. Cell Rep. 2016;16:66–78. doi: 10.1016/j.celrep.2016.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris-Johnson KS, Domyan ET, Vezina CM, Sun X. Beta-catenin promotes respiratory progenitor identity in mouse foregut. Proc Natl Acad Sci USA. 2009;106:16287–16292. doi: 10.1073/pnas.0902274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domyan ET, et al. Signaling through BMP receptors promotes respiratory identity in the foregut via repression of Sox2. Development. 2011;138:971–981. doi: 10.1242/dev.053694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arora R, Metzger RJ, Papaioannou VE. Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS Genet. 2012;8:e1002866. doi: 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waldron L, et al. The cardiac TBX5 interactome reveals a chromatin remodeling network essential for cardiac septation. Dev Cell. 2016;36:262–275. doi: 10.1016/j.devcel.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burnicka-Turek O, et al. Cilia gene mutations cause atrioventricular septal defects by multiple mechanisms. Hum Mol Genet. 2016;25:3011–3028. doi: 10.1093/hmg/ddw155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minoo P, et al. TTF-1 regulates lung epithelial morphogenesis. Dev Biol. 1995;172:694–698. doi: 10.1006/dbio.1995.8080. [DOI] [PubMed] [Google Scholar]
- 26.Minoo P, Li C, Liu HB, Hamdan H, deLemos R. TTF-1 is an epithelial morphoregulatory transcriptional factor. Chest. 1997;111(Suppl):135S–137S. doi: 10.1378/chest.111.6_supplement.135s. [DOI] [PubMed] [Google Scholar]
- 27.Herriges MJ, et al. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 2014;28:1363–1379. doi: 10.1101/gad.238782.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazzaro D, Price M, de Felice M, Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- 29.Cardoso WV, Lü J. Regulation of early lung morphogenesis: Questions, facts and controversies. Development. 2006;133:1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- 30.Warburton D, et al. Lung organogenesis. Curr Top Dev Biol. 2010;90:73–158. doi: 10.1016/S0070-2153(10)90003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCulley D, Wienhold M, Sun X. The pulmonary mesenchyme directs lung development. Curr Opin Genet Dev. 2015;32:98–105. doi: 10.1016/j.gde.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rankin SA, Zorn AM. Gene regulatory networks governing lung specification. J Cell Biochem. 2014;115:1343–1350. doi: 10.1002/jcb.24810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koshiba-Takeuchi K, et al. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature. 2009;461:95–98, and erratum (2009) 461:550. doi: 10.1038/nature08324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rankin SA, et al. A molecular atlas of Xenopus respiratory system development. Dev Dyn. 2015;244:69–85. doi: 10.1002/dvdy.24180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown DD, et al. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132:553–563. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goetz SC, Brown DD, Conlon FL. TBX5 is required for embryonic cardiac cell cycle progression. Development. 2006;133:2575–2584. doi: 10.1242/dev.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura S, et al. The T/ebp null mouse: Thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 38.Poulain M, Ober EA. Interplay between Wnt2 and Wnt2bb controls multiple steps of early foregut-derived organ development. Development. 2011;138:3557–3568. doi: 10.1242/dev.055921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winata CL, et al. Development of zebrafish swimbladder: The requirement of hedgehog signaling in specification and organization of the three tissue layers. Dev Biol. 2009;331:222–236. doi: 10.1016/j.ydbio.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 40.Yin A, Korzh S, Winata CL, Korzh V, Gong Z. Wnt signaling is required for early development of zebrafish swimbladder. PLoS One. 2011;6:e18431. doi: 10.1371/journal.pone.0018431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cass AN, Servetnick MD, McCune AR. Expression of a lung developmental cassette in the adult and developing zebrafish swimbladder. Evol Dev. 2013;15:119–132. doi: 10.1111/ede.12022. [DOI] [PubMed] [Google Scholar]
- 42.Zheng W, et al. Comparative transcriptome analyses indicate molecular homology of zebrafish swimbladder and mammalian lung. PLoS One. 2011;6:e24019. doi: 10.1371/journal.pone.0024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatsumi N, et al. Molecular developmental mechanism in polypterid fish provides insight into the origin of vertebrate lungs. Sci Rep. 2016;6:30580. doi: 10.1038/srep30580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longo S, Riccio M, McCune AR. Homology of lungs and gas bladders: Insights from arterial vasculature. J Morphol. 2013;274:687–703. doi: 10.1002/jmor.20128. [DOI] [PubMed] [Google Scholar]
- 45.Garrity DM, Childs S, Fishman MC. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 2002;129:4635–4645. doi: 10.1242/dev.129.19.4635. [DOI] [PubMed] [Google Scholar]
- 46.Ng JK, et al. The limb identity gene Tbx5 promotes limb initiation by interacting with Wnt2b and Fgf10. Development. 2002;129:5161–5170. doi: 10.1242/dev.129.22.5161. [DOI] [PubMed] [Google Scholar]
- 47.Ahn DG, Kourakis MJ, Rohde LA, Silver LM, Ho RK. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature. 2002;417:754–758. doi: 10.1038/nature00814. [DOI] [PubMed] [Google Scholar]
- 48.Parrie LE, Renfrew EM, Wal AV, Mueller RL, Garrity DM. Zebrafish tbx5 paralogs demonstrate independent essential requirements in cardiac and pectoral fin development. Dev Dyn. 2013;242:485–502. doi: 10.1002/dvdy.23953. [DOI] [PubMed] [Google Scholar]
- 49.Zhou L, et al. Tbx5 and Osr1 interact to regulate posterior second heart field cell cycle progression for cardiac septation. J Mol Cell Cardiol. 2015;85:1–12. doi: 10.1016/j.yjmcc.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bellusci S, et al. Involvement of sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development. 1997;124:53–63. doi: 10.1242/dev.124.1.53. [DOI] [PubMed] [Google Scholar]
- 51.Pepicelli CV, Lewis PM, McMahon AP. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr Biol. 1998;8:1083–1086. doi: 10.1016/s0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- 52.Peyrot SM, Wallingford JB, Harland RM. A revised model of Xenopus dorsal midline development: Differential and separable requirements for notch and Shh signaling. Dev Biol. 2011;352:254–266. doi: 10.1016/j.ydbio.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowes JB, et al. Xenbase: Gene expression and improved integration. Nucleic Acids Res. 2010;38:D607–D612. doi: 10.1093/nar/gkp953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.James-Zorn C, et al. Navigating Xenbase: An integrated Xenopus genomics and gene expression database. Methods Mol Biol. 2018;1757:251–305. doi: 10.1007/978-1-4939-7737-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karpinka JB, et al. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res. 2015;43:D756–D763. doi: 10.1093/nar/gku956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of hedgehog signaling by direct binding of cyclopamine to smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu X, Ding S, Ding Q, Gray NS, Schultz PG. A small molecule with osteogenesis-inducing activity in multipotent mesenchymal progenitor cells. J Am Chem Soc. 2002;124:14520–14521. doi: 10.1021/ja0283908. [DOI] [PubMed] [Google Scholar]
- 58.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci USA. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luna-Zurita L, et al. Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell. 2016;164:999–1014. doi: 10.1016/j.cell.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nadadur RD, et al. Pitx2 modulates a Tbx5-dependent gene regulatory network to maintain atrial rhythm. Sci Transl Med. 2016;8:354ra115. doi: 10.1126/scitranslmed.aaf4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jolma A, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 63.Arnolds DE, et al. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J Clin Invest. 2012;122:2509–2518. doi: 10.1172/JCI62617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smemo S, et al. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum Mol Genet. 2012;21:3255–3263. doi: 10.1093/hmg/dds165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iacovino M, et al. Inducible cassette exchange: A rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells. 2011;29:1580–1588. doi: 10.1002/stem.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 67.Trujillo GV, et al. The canonical Wingless signaling pathway is required but not sufficient for inflow tract formation in the Drosophila melanogaster heart. Dev Biol. 2016;413:16–25. doi: 10.1016/j.ydbio.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Agarwal P, et al. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 2003;130:623–633. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- 69.Rallis C, et al. Tbx5 is required for forelimb bud formation and continued outgrowth. Development. 2003;130:2741–2751. doi: 10.1242/dev.00473. [DOI] [PubMed] [Google Scholar]
- 70.Horb ME, Thomsen GH. Tbx5 is essential for heart development. Development. 1999;126:1739–1751. doi: 10.1242/dev.126.8.1739. [DOI] [PubMed] [Google Scholar]
- 71.Tseng YR, et al. Holt-Oram syndrome with right lung agenesis caused by a de novo mutation in the TBX5 gene. Am J Med Genet A. 2007;143A:1012–1014. doi: 10.1002/ajmg.a.31672. [DOI] [PubMed] [Google Scholar]
- 72.Qin X, Wei W, Fangqi G. Horseshoe lung associated with Holt-Oram syndrome. Iran J Pediatr. 2015;25:e251. doi: 10.5812/ijp.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Jong EM, Felix JF, de Klein A, Tibboel D. Etiology of esophageal atresia and tracheoesophageal fistula: “Mind the gap”. Curr Gastroenterol Rep. 2010;12:215–222. doi: 10.1007/s11894-010-0108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wain LV, et al. UK Brain Expression Consortium (UKBEC); OxGSK Consortium Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): A genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu Y, et al. Tbx5-dependent pathway regulating diastolic function in congenital heart disease. Proc Natl Acad Sci USA. 2008;105:5519–5524. doi: 10.1073/pnas.0801779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lewis ZR, Hanken J. Convergent evolutionary reduction of atrial septation in lungless salamanders. J Anat. 2017;230:16–29. doi: 10.1111/joa.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.St-Jacques B, et al. Sonic hedgehog signaling is essential for hair development. Curr Biol. 1998;8:1058–1068. doi: 10.1016/s0960-9822(98)70443-9. [DOI] [PubMed] [Google Scholar]
- 78.Zhang XM, Ramalho-Santos M, McMahon AP. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell. 2001;106:781–792. [PubMed] [Google Scholar]
- 79.Sive HL, Grainger RM, Harland RM. 2000. Early Development of Xenopus laevis: A Laboratory Manual (Cold Spring Harbor Lab Press, Cold Spring Harbor, NY), p ix, 338 p.
- 80.Tandon P, Showell C, Christine K, Conlon FL. Morpholino injection in Xenopus. Methods Mol Biol. 2012;843:29–46. doi: 10.1007/978-1-61779-523-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Westerfield M. The Zebrafish Book. 5th Ed Univ Oregon Press; Eugene, OR: 2007. [Google Scholar]
- 82.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 83.Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- 84.Takada S, et al. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 1994;8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 85.Jensen B, et al. Identifying the evolutionary building blocks of the cardiac conduction system. PLoS One. 2012;7:e44231. doi: 10.1371/journal.pone.0044231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jensen B, et al. Specialized impulse conduction pathway in the alligator heart. eLife. 2018;7:e32120. doi: 10.7554/eLife.32120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Poelmann RE, et al. Evolution and development of ventricular septation in the amniote heart. PLoS One. 2014;9:e106569. doi: 10.1371/journal.pone.0106569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Y, et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.