Significance
IFN-γ is an example of a pleiotropic cytokine that plays critical roles in promoting both protective immune responses and immunopathological processes. In response to this cytokine, cells activate a well-identified and conserved JAK/STAT signaling pathway that can still elicit distinct responses, even in the same cell type. How this outcome specificity is achieved remains largely unknown. We have found that IFN-γ regulates CD8 T cell differentiation through noncanonical pathways that are enabled by integrin costimulation of the STAT1 pathway. This leads to cooperative effects between activating cell types that is dictated by proximity. This study demonstrates the importance of cofactors and microenvironment in eliciting specific cytokine functions.
Keywords: T cell differentiation, cytokines, cell biology, integrins
Abstract
The cytokine IFN-γ is a critical regulator of immune system development and function. Almost all leukocytes express the receptor for IFN-γ, yet each cell type elicits a different response to this cytokine. Cell type-specific effects of IFN-γ make it difficult to predict the outcomes of the systemic IFN-γ blockade and limit its clinical application, despite many years of research. To better understand the cell–cell interactions and cofactors that specify IFN-γ functions, we focused on the function of IFN-γ on CD8 T cell differentiation. We demonstrated that during bacterial infection, IFN-γ is a dominant paracrine trigger that skews CD8 T cell differentiation toward memory. This skewing is preferentially driven by contact-dependent T cell–T cell (T-T) interactions and the localized IFN-γ secretion among activated CD8 T cells in a unique splenic microenvironment, and is less sensitive to concurrent IFN-γ production by other immune cell populations such as natural killer (NK) cells. Modulation of CD8 T cell differentiation by IFN-γ relies on a nonconventional IFN-γ outcome that occurs specifically within 24 hours following infection. This is driven by IFN-γ costimulation by integrins at T-T synapses, and leads to synergistic phosphorylation of the proximal STAT1 molecule and accelerated IL-2 receptor down-regulation. This study provides evidence of the importance of context-dependent cytokine signaling and gives another example of how cell clusters and the microenvironment drive unique biology.
IFN-γ is a pleiotropic cytokine potentially produced by all immune cells. It is crucial for immune responses against tumors (1) and infections (2), and is implicated in many autoimmune diseases (3). IFN-γ uses the same signaling machinery to elicit distinct and diverse responses (4). It consistently binds to a single receptor composed of two chains, IFN-γ receptor 1 (IFN-γR1) and IFN-γR2 (5), and triggers a conserved Janus kinase (JAK)/signal transducer and activator of transcription 1 (STAT1) pathway (6). How specificity of IFN-γ action is achieved is still not understood. Differential expression of receptors and signaling components, leading to the integration of negative and positive feedbacks, has been described as a molecular basis of the cell specificity of cytokine action (7). Overall, however, deciphering the relevant microenvironment, cellular interactions, and cofactors specifying the outcome of IFN-γ stimulation is still an outstanding question.
In CD8 T cells, IFN-γ production is traditionally associated with effector function and cytotoxicity. Interestingly, naive CD8 T cells are able to rapidly, although transiently, produce IFN-γ following T cell receptor (TCR) triggering (8) and Toll-like receptor 2/7 signals (9). This early IFN-γ production by CD8 T cells has been suggested to mediate innate protection against Listeria monocytogenes (LM) (10), although others suggest that such IFN-γ derived from CD8 T cells may have a regulatory role rather than a direct innate function (9). Given the fact that IFN-γ is a key factor modulating the differentiation of CD4 T cells (11), it has been suggested that IFN-γ might also modulate CD8 T cell differentiation.
The balanced differentiation of CD8 T cells in effector and long-term memory subsets is crucial for immunity against intracellular pathogens. Variations in CD8 T cell fate have been extensively described based on their transcriptional profile, phenotype, function, and final anatomical location (12–14). The underlying dynamic interactions that take place during early effector and memory CD8 T cell development are still poorly understood, however (15). The initial process of CD8 T cell activation is dependent on three signals (16): (i) antigenic stimulation, (ii) engagement of costimulatory molecules, and (iii) exposure to cytokines. These signals are largely provided by other cell types and integrated by the responding T cell, a well-understood paradigm of CD8 T cell fate choice (17). However, several lines of evidence suggest that CD8 T cells can themselves tune their own fate. A T cell autonomous program of differentiation is initiated following brief antigenic stimulation and, in an antigen-independent manner, modulates the acquisition of effector and memory functions (18–20). In this context, T cell fate can then be influenced by stochastic levels of protein expression (21), autocrine or paracrine feedback loops (22–24), or direct T cell–T cell (T-T) coregulation through soluble factors or synapse formation (25–29).
In this study, we took a top-down approach to study the direct involvement of IFN-γ in CD8 T cell differentiation in the context of LM infection. We determined that IFN-γ skews CD8 T cell generation toward memory generation during a specific temporal window. IFN-γ–dependent T cell fate occurs through a paracrine “self-sufficient” mechanism, as CD8 T cells have a dominant sensitivity to their own IFN-γ, the first wave of which they produce within hours following antigen exposure. Inhibition of effector generation by IFN-γ involves accelerated down-regulation of the IL-2 receptor chain CD25, a known requirement for CD8 T cell memory commitment (30). By using a combination of in vivo imaging and in vitro experiments, we determined that activated CD8 T cells rely on the integration between IFN-γR and integrin signaling, locally within T-T synapses to increase their responsiveness to IFN-γ through Src kinase signaling from integrins. Our data highlight a model by which the integrin-rich environment of T-T synapses specifically provides context and differentiation cues to newly activated CD8 T cells through specification of the signaling response to local IFN-γ.
Results and Discussion
Regulation of CD8 T Cell Differentiation Through Early Secretion of IFN-γ.
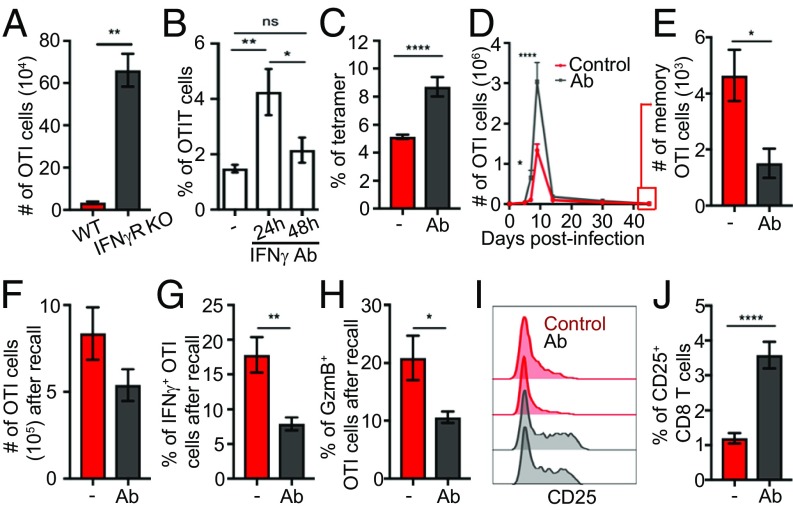
To investigate the direct involvement of IFN-γ in CD8 T cell differentiation, we analyzed the effect of IFN-γR deletion in ovalbumin (OVA)-specific CD8 T (OTI) cells on their expansion following LM-expressing OVA (LMOVA) infection. We found that the number of IFN-γR−/− OTI cells at the peak of the effector response was 20-fold higher than that of their WT counterpart (Fig. 1A), showing that direct IFN-γ signaling restricts CD8 T cell expansion. Because CD8 T cell differentiation is a dynamic process but knocking out the allele is constitutive, we temporally inhibited IFN-γ with blocking Abs and scored OTI cell frequency. An augmentation of OTI numbers was observed when IFN-γ was blocked 24 h postinfection, but not later (Fig. 1B and SI Appendix, Fig. S1 A and B). A similar increase in antigen-specific CD8 T cells following IFN-γ blockade at 24 h was found when the endogenous response was assessed (Fig. 1C), showing that this was not an artifact induced by transfer of transgenic T cells. This critical 24-h window corresponds to a first wave of IFN-γ production following LM infection (SI Appendix, Fig. S1C). Tracking OTI cells over time revealed that blockade of this first wave of IFN-γ not only increased expansion specifically during the effector stage but also restricted memory generation (Fig. 1 D and E). Memory cells generated when early IFN-γ was blocked were able to expand normally upon rechallenge with LMOVA (Fig. 1F), but they displayed decreased IFN-γ and granzyme B production (Fig. 1 G and H). Altogether, we concluded that IFN-γ produced around 24 h after LM infection skews CD8 T cell differentiation toward memory.
Fig. 1.
Regulation of CD8 T cell differentiation through early secretion of IFN-γ. (A) Mice bearing WT and IFN-γR−/− OTI cells (1:1 ratio) were infected with LMOVA. The graph shows the number of effector WT or IFN-γR−/− OTI cells analyzed by flow cytometry at day 10. Data are from two independent experiments (n = 4). WT mice (C) bearing OTI cells (B and D–I) were infected with LMOVA. When indicated, mice were treated with blocking IFN-γ 24 h postinfection (Ab) or as otherwise stated. (B) Phenotype of OTI cells in the spleen was analyzed by flow cytometry. The graph shows the percentage of OTI cells among CD8 T cells 10 d postinfection. Data are from three independent experiments (n = 5). (C) Frequency of OVA-specific cells among endogenous CD8 T cells was analyzed 8 d after infection by flow cytometry. Data are from three independent experiments (n = 15). (D) Graph shows the number of OTI cells over time. Data are from two independent experiments (n = 6). (E) Graph shows the number of memory (KLRG1−CD127+) OTI cells 45 d postinfection. Data are from two independent experiments (n = 6). (F–H) Mice were rechallenged with LMOVA 50–60 d after primary challenge and analyzed after 5 d. Data are from three independent experiments (n = 12). (F) Graph shows the number of OTI cells quantified by flow cytometry. (G and H) Splenocytes were restimulated in vitro with PMA and ionomycin in the presence of brefeldin A. Graphs show the percentage of OTI cells expressing IFN-γ (G) and granzyme B (GzmB; H). (I) Histogram shows CD25 expression on OTI cells 5 d after primary infection. Data are from three independent experiments (n = 7). (J) Frequency of CD25+ endogenous total CD8 T cells was analyzed by flow cytometry 5 d after infection with LMOVA in the presence or absence of IFN-γ Ab blockade at 24 h. Data are from three independent experiments (n = 6). *P < 0.05, **P < 0.001, ****P < 0.0001. ns, not significant.
The fact that early (24–48 h) blocking of IFN-γ increased CD8 T cell numbers starting at day 7 (Fig. 1D) suggested an indirect function of IFN-γ on cell expansion or contraction. Consistent with this, temporal IFN-γ blockade had no direct effect on OTI cell cycle, proliferation, or apoptosis (SI Appendix, Fig. S1 D–F), and it did not affect expression of apoptosis or inhibitory receptors (SI Appendix, Fig. S1 G and H) during early effector expansion. These data suggested that IFN-γ regulated CD8 T cell effector differentiation, per se, as opposed to cell expansion. Surprisingly, IFN-γ did not modulate the ratio between the transcription factors t-bet and eomes (SI Appendix, Fig. S1I), two master regulators of CD8 T cell differentiation. This suggested that IFN-γ impacted CD8 T cell differentiation through a noncanonical mechanism. IFN-γ can synergize or antagonize the effects of growth factors and cytokines (31). Because inflammatory cytokines favor effector differentiation by prolonging the expression of the IL-2 receptor alpha chain CD25 (30, 32), we investigated the function of early IFN-γ secretion on CD25 expression. Regardless of IFN-γ blockade, virtually all primed OTI cells up-regulated CD25 2 d after infection, started to down-regulate CD25 after day 4, and no longer expressed CD25 by day 7 (SI Appendix, Fig. S1J). However, IFN-γ blockade resulted in a higher percentage of OTI cells expressing CD25 during the down-regulation phase, with a higher mean fluorescence intensity (MFI) (Fig. 1I and SI Appendix, Fig. S1J). This resulted in enhanced IL-2–driven signaling in OTI cells at this time, as analyzed by Stat5 phosphorylation (SI Appendix, Fig. S1K). A similar CD25 increase was found on endogenous CD8 T cells following IFN-γ blockade (Fig. 1J). Prolonged CD25 expression at day 5 is in agreement with increased effector and decreased memory formation detected from day 7 and is consistent with IFN-γ antagonizing, or targeting, the same pathway as inflammatory cytokines (33). Altogether, our data show that IFN-γ skews CD8 T cell differentiation toward memory formation by modulating CD25 expression.
Spatiotemporal Behavior of IFN-γ–Producing Cells During LM Infection Suggests Paracrine IFN-γ Signaling in Early Activated CD8 T Cells.
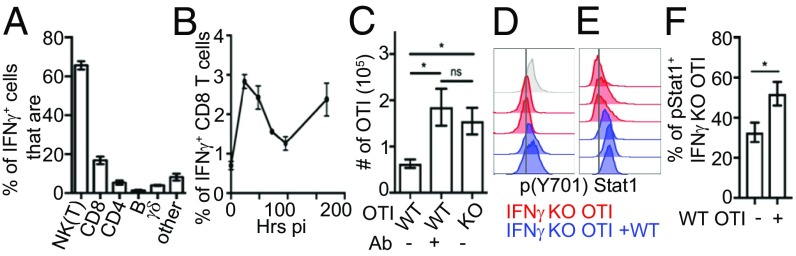
As IFN-γ secretion 24 h postinfection regulated CD8 T cell differentiation, we characterized the cell types producing IFN-γ in situ at this time. Natural killer (NK) cells comprised 60% of IFN-γ–positive cells (Fig. 2A). Additionally, activated CD8 T cells accounted for nearly 20% of the total IFN-γ–positive cells (Fig. 2A and SI Appendix, Fig. S2A), and IFN-γ production by CD8 T cells followed overall the same biphasic pattern (Fig. 2B) seen for global IFN-γ production (SI Appendix, Fig. S1C). Early IFN-γ production by CD8 T cells was not only a feature of memory cells as previously described (34), because activated cells coming from the naive (CD44−) pool also contributed to IFN-γ production (SI Appendix, Fig. S2B), albeit in lower quantity (SI Appendix, Fig. S2C).
Fig. 2.
Autonomous regulation of CD8 T cell differentiation through paracrine IFN-γ signaling. (A and B) Mice were infected with LMOVA and treated with brefeldin A 6 h before being killed. (A) Different cell populations among IFN-γ–positive splenocytes 24 h after infection were analyzed by flow cytometry. Splenocytes were first gated on IFN-γ–positive cells, and the different populations within the IFN-γ–positive cells were defined using the Abs NK1.1 [NK(T)], CD8, CD4, CD19 (B), and γδ TCR. (B) Percentage of CD8 T cells producing IFN-γ was analyzed by flow cytometry over time. Data are from at least three independent experiments (n = 6–8). (C) Mice bearing WT or IFN-γ−/− OTI cells were infected with LMOVA. When indicated, mice were treated with blocking IFN-γ 24 h postinfection (Ab). The graph shows the number of effector (KLRG1+CD127−) OTI cells per 1/3 spleen. Data are from two independent experiments (n = 5). (D) WT or IFN-γ−/− OTI cells were stimulated with ionomycin (PI) for 24 h, either separated or in coculture, and analyzed by flow cytometry. Representative histograms show p(Y701)Stat1. WT OTI cells are colored gray, IFN-γ−/− OTI cells are colored red, and IFN-γ−/− OTI cells are colored blue when they are cocultured with WT OTI cells. (E and F) Mice transferred with IFN-γ−/− OTI cells, with or without WT OTI cells (1.1 ratio), were infected with LMOVA. After 24 h, splenocytes were analyzed by flow cytometry. (E) Representative histogram showing p(Y701)Stat1 staining in IFN-γ−/− OTI cells. IFN-γ−/− OTI cells only are colored red, and IFN-γ−/− OTI cells cocultured with WT OTI cells are colored blue. (F) Graph shows the percentage of p(Y701)Stat1-positive IFN-γ−/− OTI cells. Data are from two independent experiments (n = 6). *P < 0.05. ns, not significant.
As IFN-γ derived from CD4 T cells is sufficient to mediate Th1 differentiation in the context of Leishmania major infection (35), we hypothesized that CD8 T cell-derived IFN-γ might likewise be the dominant source regulating OTI cell differentiation. In support of this, genetic ablation of IFN-γ only in OTI cells resulted in a greater number of effector T cells following LMOVA infection, almost to the same extent as seen for total Ab-mediated IFN-γ blockade (Fig. 2C). Although IFN-γ was produced by endogenous CD8 T cells, surprisingly, it did not fully compensate for the absence of IFN-γ secretion by IFN-γ−/− OTI cells (Fig. 2C). This was not due to an overproduction of IFN-γ by OTI cells compared with endogenous T cells, as OTI cells represented approximately only 1/15th of all CD8 T cell IFN-γ producers (SI Appendix, Fig. S2D). It therefore raised the question of whether IFN-γ signaling in T cells was autocrine or paracrine. To investigate this, we asked whether IFN-γ produced by WT OTI cells could induce signaling in IFN-γ−/− OTI cells, which would be indicative of paracrine signaling. In vitro coculture (Fig. 2D) and in vivo coinjection (Fig. 2 E and F) of IFN-γ−/− OTI cells with WT OTI cells enhanced IFN-γ signaling in IFN-γ−/− OTI cells, as assessed by Stat1 phosphorylation on Y701 [p(Y701)Stat1] 24 h after priming, suggesting paracrine IFN-γ signaling. Overall, CD8 T cells autonomously tune their differentiation, at least in part, through early paracrine secretion of IFN-γ from other CD8 T cells.
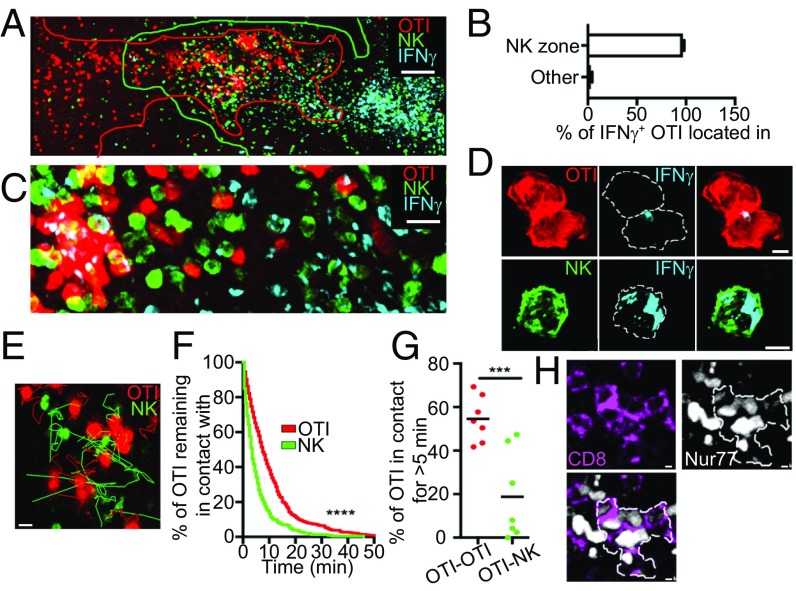
How does this take place in situ? We hypothesized that a specific microenvironment providing other cofactors might be responsible for this alternate scenario. Twenty-four hours after LMOVA infection, IFN-γ–producing CD8 T cells were located in the white pulp, where they colocalize with other IFN-γ–producing cells such as NK cells that invaded the white pulp (Fig. 3 A and B and SI Appendix, Fig. S3 A and B), indicating that CD8 sensitivity to their own IFN-γ was not due to a segregation between CD8 T cells and other IFN-γ–producing cells. Assessment of cytokine distribution at the subcellular level in situ revealed that IFN-γ was vesicular and typically directed toward the interface with other cells in OTI cells, while NK cells exhibited a more ubiquitous cellular localization pattern (Fig. 3 C and D, SI Appendix, Fig. S3C, and Movies S1 and S2). We previously described that T-T interactions occur after priming in the context of vaccination (26), interactions that would be in agreement with the directional IFN-γ secretion observed specifically between OTI cells. Indeed, two-photon microscopy revealed that T-T contacts occurred following LMOVA infection (Movie S3) with a dwell time of interaction of 10 min, while NK cells display shorter contacts with T cells (Fig. 3 E and F). For example, this resulted in 54.5% of OTI-OTI contacts and 18% of OTI-NK contacts when we scored contacts that lasted more than 5 min (Fig. 3G). Using the mouse reporter Nur77-GFP coupled with CD8 in situ staining, we detected and tracked clusters of activated endogenous CD8 T cells 24 h after LMOVA infection (Fig. 3H and Movie S4), showing that clustering events were not due to the high precursor frequency of OTI cells transferred. We noted, however, that OTI clusters rarely contained endogenous activated CD8 T cells (SI Appendix, Fig. S3D), which might be due to antigen specificity and/or the rapid attraction toward chemokine niches in which the higher affinity OTI cell response might dominate over the mixed-affinity endogenous response. This would be consistent with the fact that expression of some chemokine receptors such as CXCR3 is regulated by T cell affinity (36). Importantly, clusters of endogenous CD8 T cells also mostly excluded NK cells, as seen for OTI cell clusters (SI Appendix, Fig. S3E). To conclude, CD8 T cells favored interactions with other CD8 T cells over NK cells in vivo, which could explain the dominance of paracrine IFN-γ signaling and its alternate outcome in early activated CD8 T cells.
Fig. 3.
Dynamic localization of IFN-γ–secreting cells suggests a contact-dependent regulation of CD8 T cell differentiation. NCR1-GFP (A–C) or WT (D) mice were transferred with CellTracker Orange (CMTMR)-labeled OTI cells, infected with LMOVA, and killed 24 h after infection. Mice were treated with brefeldin A 6 h before harvest. (A) Photograph is a representative example of the localization of IFN-γ (blue)–producing OTI cells (red) relative to other IFN-γ–positive cells such as NK cells (green). (Scale bar: 100 μm.) (B) Quantification of the localization of IFN-γ–producing OTI cells relative to NK cells. Data are from four independent experiments. (C and D) Photographs are representative examples of IFN-γ–producing NK and OTI cells at different scales. [Scale bars: C, 10 μm; D, 2 μm (OTI) and 3 μm (NK).] Dotted lines in D delimit cell edges. (E–G) NCR1-GFP mice were transferred with 1 × 106 CMTMR-labeled OTI cells and infected with LMOVA. After 24 h, spleens were imaged by two-photon microscopy for a period of 30 min at 30-s intervals. Data are from three independent experiments. (E) Snapshot of OTI cell and NK cell overlapping region. Tracks of OTI (red) and NK (green) cells over a 30-min time lapse are displayed. (Scale bar: 10 μm.) (F) Graph shows the percentage of OTI cells remaining in contact with another OTI cell (red) or an NK cell (green) over time. (G) Graph shows the percentage of OTI cells that are in contact for more than 5 min with another OTI cell (red) or an NK cell (green). Each dot represents an individual field. ***P < 0.0002. (H) Representative images of spleen section of a Nur77-GFP mouse 24 h after infection with LMOVA. Spleen sections were stained for GFP (white) and CD8 (purple). The white edge delineates a cluster of activated CD8 T cells. (Scale bars: 2 μm.)
Cell Contacts Are Required to Maximize IFN-γ Signaling in Activated CD8 T Cells.
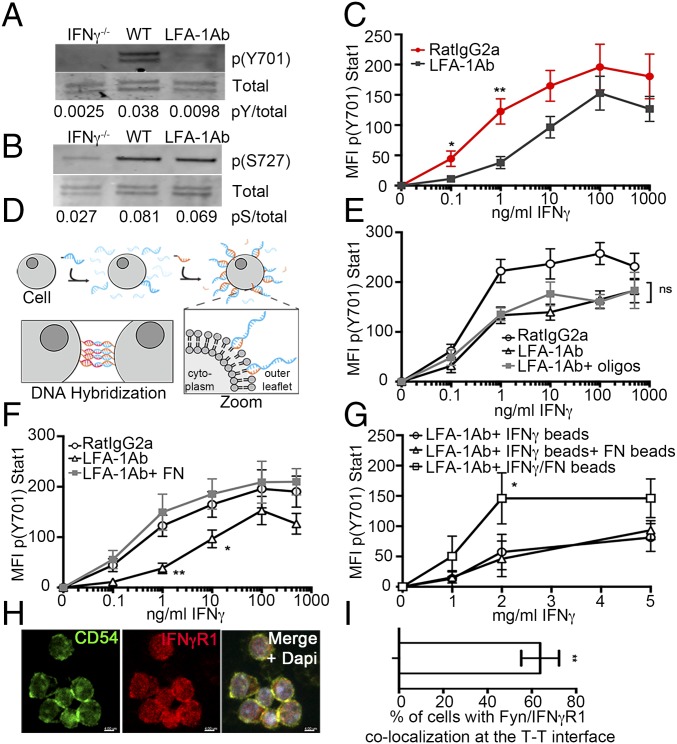
Our findings suggest that T-T contacts dictated the responsiveness of activated CD8 T cells to the IFN-γ they produce. These contacts rely on LFA-1 and ICAM-1 (26, 27, 37) and require LFA-1 activation downstream of T cell priming (38). We tested the effect of blocking these engagements with LFA-1 blocking Abs in cultures where T cells were activated in a system devoid of antigen-presenting cell (APC). Inhibiting T-T contacts in vitro resulted in decreased p(Y701)Stat1 downstream of IFN-γ (Fig. 4A and SI Appendix, Fig. S4A). This was not due to an inability of cells to produce or detect IFN-γ, per se, since phosphorylation of Stat1 at another site, serine 727, was unaffected (Fig. 4B and SI Appendix, Fig. S4B). A contact requirement for optimum IFN-γ signaling was not the consequence of a desensitization mechanism due to chronic IFN-γ exposure, as contacts were also required for increasing both the amplitude and the sensitivity (as defined by EC50: control = 0.5 ± 0.21, LFA-1 Ab = 4.3 ± 0.19) of IFN-γ signaling following acute exposure (Fig. 4C). Requirement of contact for maximum IFN-γ signaling was also not the result of IFN-γR down-regulation (SI Appendix, Fig. S4C), as observed for CD4 T cells differentiating into the Th1 subset (39). We thus concluded that cell–cell interactions potentiated the responsiveness of activated CD8 T cells to IFN-γ.
Fig. 4.
Contact-dependent IFN-γ signaling in early activated CD8 T cells. WT (A, B, and H) or IFN-γ−/− (A–C and E–G) OTI cells were stimulated with PMA and ionomycin for 24 h. Where indicated, cells were treated with LFA-1 blocking Ab (LFA-1 Ab) or control Ab (RatIgG2a) 5 h before harvest. The p(Y701)Stat 1 (pY; A) and p(S727)Stat1 (pS; B) were analyzed by Western blot. Quantification (bottom) corresponds to the intensity ratio between phosphorylated and total Stat1. (C–F) Indicated amount of IFN-γ was added 20 min before harvest. Data are from three independent experiments. (C) Graph shows p(Y701)Stat1 according to the dose of IFN-γ. (D and E) LFA-1 Ab–treated cells were forced to interact in an integrin-independent manner using fatty acid-modified oligonucleotides for 3 h. (D) Cartoon illustrating the strategy of oligonucleotide coating and forced proximity. Fatty acid-linked DNA oligos (blue) were inserted in the outer leaflet of the plasma membrane and stabilized with DNA oligo anchors (orange). Oligo-labeled T cells were forced to stay aggregated by using complementary oligos (blue and red), leading to DNA hybridization. (E) Graph shows p(Y701)Stat1 according to the dose of IFN-γ. The black line with circles represents mock-treated cells, the black line with triangles represents LFA-1 Ab–treated cells, and the gray line with squares represents LFA-1 Ab–treated cells with oligonucleotides (n = 6). (F) When indicated, cells were plated on fibronectin (FN) for 2 h before IFN-γ treatment. The graph shows p(Y701)Stat1 according to the dose of IFN-γ. The black line with circles represents mock-treated cells, the black line with triangles represents LFA-1 Ab–treated cells, and the gray line with squares represents LFA-1 Ab–treated cells deposited on FN-coated plates (n = 6). (G) Cells were incubated with IFN-γ–coated beads (black line with circles), IFN-γ–coated beads plus FN-coated beads (black line with triangles), or beads coated with both IFN-γ and FN (black line with squares) for 20 min. The graph shows p(Y701)Stat1 according to the quantity of IFN-γ coated on the beads. (H and I) At 24 h, activated OTI cells stained for ICAM-1 (green, Left), IFN-γR1 (red, Center), and the nucleus (DAPI; blue in the merge, Right). (H) Representative image of T cell clusters. (Scale bars: 4 μm.) (I) Quantification of ICAM-1 and IFN-γR1 colocalization at T-T contacts. *P < 0.05, **P < 0.001.
LFA-1 promotes cellular adherence and signaling in response to ligation (40), which could both potentially maximize IFN-γ signaling. We first addressed whether adherence and proximity were responsible for enhanced IFN-γ signaling by forcing OTI cells treated with LFA-1 blocking Ab (“LFA-1less”) to cluster in an integrin-independent manner by using a DNA zippering method (modified from refs. 41, 42) (Fig. 4D). Although this method allowed LFA-1less T cells to cluster to the same extent as control T cells (SI Appendix, Fig. S4D), but it did not lead to increased sensitivity (EC50: LFA-1 Ab = 0.52 ± 0.17, LFA-1 Ab + oligos = 0.32 ± 0.18) or amplitude of p(Y701)Stat1 (Fig. 4E). Conversely, addition of plate-bound coated integrin was able to rescue IFN-γ–induced p(Y701)Stat1 of LFA-1less OTI cells (Fig. 4F) (EC50: LFA-1 Ab = 4.1 ± 0.17, LFA-1 Ab + FN = 0.36 ± 0.22), suggesting that integrin engagement was sufficient to potentiate IFN-γ signaling in activated CD8 T cells. To further assess the sufficiency of integrins to mediate this effect, we used a system of beads where IFN-γ and integrins were codelivered or delivered independently. Codelivery was required for maximum amplitude of p(Y701)Stat1 generation (Fig. 4G), suggesting that integrins had to colocalize with IFN-γR to enable signal integration, consistent with IFN-γR and ICAM-1 colocalization at T-T contacts (Fig. 4 H and I).
Altogether, we concluded that activated CD8 T cells responded to polarized engagement of integrin ligands and IFN-γ in a manner that costimulated IFN-γ signaling. We believe the term “costimulation” is warranted in this situation since LFA-1 engagement on its own had little effect on Stat1 phosphorylation.
Integrin-Mediated Activation of Src Kinases Enhances IFN-γ Responsiveness of Activated CD8 T Cells and Restricts Effector Differentiation.
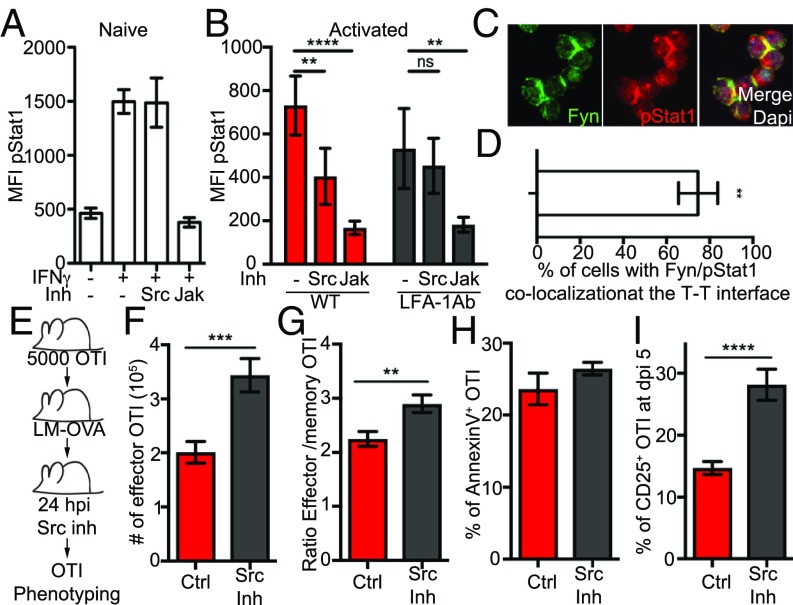
To understand how integrin engagement potentiated IFN-γ signaling, we tested the function of the canonical integrin signaling intermediate Src kinases (Fyn or Lck in T cells) in potentiating IFN-γ signaling. We inhibited Src kinases using the inhibitor PP2 and compared this with inhibiting the JAK1/2 pathway using the selective inhibitor ruxolitinib. While IFN-γ signaling in naive OTI cells, which cannot make T-T contacts, was inhibited by Jak, but not Src, inhibitors (Fig. 5A), maximum p(Y701)Stat1 in activated OTI cells was blocked by inhibitors of both pathways (Fig. 5B). Src kinase inhibitor treatment did not result in decreased cell clustering (SI Appendix, Fig. S5A). Consistent with integrin and IFN-γ signal integration, p(Y701)Stat1 was located at the T-T contact interface and colocalized with the Src kinase Fyn (Fig. 5 C and D).
Fig. 5.
Integrin signaling is required for optimum IFN-γ signaling in early activated CD8 T cells and for CD8 T cell differentiation in vivo. (A) Naive OTI cells were incubated with Src or Jak inhibitor (Inh) for 3 h and treated for 20 min with IFN-γ. IFN-γ signaling was analyzed by p(Y701)Stat1 using flow cytometry. (B–D) OTI cells were stimulated at low density with PMA and ionomycin for 24 h. (B) When indicated, cells were treated with LFA-1 blocking Abs (LFA-1 Ab) from the time of activation and Src or Jak Inh 5 h before harvest. IFN-γ signaling was analyzed by p(Y701)Stat1 using flow cytometry. (C and D) At 24 h, activated OTI cells were stained for Fyn (green, Left), p(Y701)Stat1 (red, Center) and the nucleus (DAPI; blue in the merge, Right). (C) Representative image of T cell clusters. (D) Quantification of Fyn and p(Y701)Stat1 colocalization at T-T contacts. (E–I) Mice bearing OTI cells were infected with LMOVA. When indicated, mice (n = 10) were treated with Src Inh 24 h postinfection. The phenotype of OTI cells in the spleen was analyzed by flow cytometry using the Abs CD8, CD45.1, KLRGI, CD127, and CD25. (E) Cartoon illustrating the experimental setup. (F) Graph shows the number of effector (KLRGI+CD127−) OTI cells per 1/3 spleen 10 d after infection (n = 18). Ctrl, control. (G) Graph shows the ratio between effector (KLRGI+CD127−) and precursor memory (KLRGI−CD127+) OTI cells 10 d after infection (n = 15). (H) Graph shows the percentage of annexin V+ OTI cells 5 d postinfection (dpi). (I) Graph shows the percentage of CD25+ OTI cells 5 d after infection. Data are from four (A–D), two (F and H), or three (G and I) independent experiments. **P < 0.001, ***P < 0.0002 and ****P < 0.0001. ns, not significant.
Because integrin signaling was necessary to potentiate IFN-γ signaling in activated OTI cells, we hypothesized that inhibiting Src kinases specifically during the first wave of IFN-γ would mimic the effect of IFN-γ temporal blockade on CD8 T cell differentiation (Fig. 1B). Similar to IFN-γ blockade, injection of Src kinase inhibitor 24 h after LMOVA infection (Fig. 5E) resulted in nearly a doubling of the number of effector OTI cells (Fig. 5F) and an increase in the effector-to-memory ratio (Fig. 5G). Src inhibition did not affect apoptosis (Fig. 5H) but resulted in prolonged CD25 expression (Fig. 5I), phenocopying early IFN-γ blockade. The same effect on expansion (SI Appendix, Fig. S5B) and CD25 expression (SI Appendix, Fig. S5C) could be observed at the endogenous level. Finally, as Src kinases are also downstream of other events relevant to CD8 T cell activation (i.e., TCR triggering), we also controlled that the effect of the Src inhibitor on OTI cell effector expansion we detected in vivo was not due to an interference with TCR triggering. To do so, we interrogated whether the TCR component CD3 was clustered at the T-T interface, which would be indicative of signaling. We did not find any evidence of CD3 localization at T-T synapses in vitro (SI Appendix, Fig. S5D) and in vivo (SI Appendix, Fig. S5E). We then blocked TCR triggering using a blocking Ab against MHC class 1 in vivo. Blocking MHC class 1 resulted in reduced OTI cell expansion when injected at the beginning of the infection as expected, and the same result was observed when blockade happened during clustering events (SI Appendix, Fig. S5F). While these data do not necessarily negate a possible involvement of TCR signaling at T-T contacts, they suggest that this is not the main pathway affected by Src inhibition at this time. Overall, these data support the requirement of integrin signaling during a specific time window to restrict the CD8 T cell commitment to effector.
Discussion
To conclude, we provide evidence of context-dependent IFN-γ signaling resulting in control of the balance between effector and memory CD8 T cell differentiation. We demonstrated that early IFN-γ production by CD8 T cells acts in a paracrine manner to limit effector CD8 T cell differentiation, and relies on coengagement with integrins. The costimulation of IFN-γ by integrins at T-T synapses results in enhanced Stat1 phosphorylation, and is required in vivo for controlling CD8 T cell fate.
Altogether, our data argue that coengagement of IFN-γR and integrins at T-T synapses results in enhanced IFN-γ signaling. Although we do not know whether additional factors are required for signal integration, the fact that IFN-γR must be recruited to lipid nanodomains to elicit signaling (43) raises the possibility that in activated CD8 T cells, integrins recruit IFN-γR to specific lipid nanodomains at T-T synapses, enabling distinct downstream events. Consistent with this, we noted that conditions that could restrict plasma membrane “fluidity,” such as coating IFN-γ on beads or inserting lipid-DNA oligos in the plasma membrane, resulted in inhibition of integrin-mediated sensitivity to IFN-γ, as analyzed by EC50, but did not have a major effect on the increased amplitude of IFN-γ signaling. We speculate that this reflects the fact that reorganization of proteins at the plasma membrane is important for integrin-enhanced sensitivity to IFN-γ. Alternately, this could also reflect a requirement of endocytosis of the IFN-γR/IFN-γ complex.
How IFN-γ regulates CD25 down-regulation is unclear. The fact that we observe a lag of 4 d between IFN-γ secretion and CD25 down-regulation suggests that IFN-γ does not directly regulate CD25 expression. Alternately, it has recently been shown that transient IFN-γ exposure elicits long-lived inflammatory responses in cancer cells due to IFN-γ retention by phosphatidylserine (PS) on the surface of viable cells (44). As TCR activation induces PS exposure at the surface of T cells (45), a similar mechanism could lead to long-term exposure of T cells to IFN-γ.
IFN-γ is typically associated with proinflammatory processes, but it also has regulatory properties (46). Our data provide another example where IFN-γ can be considered as regulating inflammation, as it counteracts prolonged CD25 expression known to be potentiated by inflammatory cytokines (30, 32). The balance between the proinflammatory and antiinflammatory properties of IFN-γ could explain discrepancies between models, where IFN-γ limits effector expansion in some models (e.g., our study, refs. 47, 48), but not in all (e.g., refs. 24, 49). Our data demonstrating that integrin costimulation is required for limiting T cell effector generation suggest that the presence of cofactors represents one of a series of possible modifiers of IFN-γ signaling and function specification.
We also demonstrated that early IFN-γ production by CD8 T cells acts in a paracrine manner to limit effector CD8 T cell differentiation through integrin-mediated T-T contacts. However, it is important to note that blocking IFN-γ (this study) does not phenocopy blocking T-T contacts as a whole (26). Whereas integration of IFN-γ and integrin signaling pathways restricts T cell effector differentiation, T-T contacts promote both effector and central memory differentiation (26). This discrepancy is likely due to the role T-T synapses play as a “platform,” fostering a network of diverse signals shared exclusively between T cells. Some of these may also be temporal and include contributions from other cell-cell contacts in vivo. Our study provides one instance of such a platform function. Importantly, where integrin activation is dictated by the encounter with the antigen early during priming and will reflect the strength of activation, IFN-γ secretion is determined by cell location and environment. Together, a platform of T-T synapses allows for those distinct signals to integrate themselves through costimulation of IFN-γ signaling by integrin coengagement.
Although it is usually accepted that cytokine signals are tightly localized, affecting only cells near the cytokine source, cytokines can permeate a tissue and modify the majority of cells therein (50). The polarized secretion of IFN-γ in CD8 T cells argues for localized cytokine delivery, but our data not do not contradict the existence of IFN-γ permeation, as NK cells do not seem to secrete IFN-γ in a polarized manner. Overall, this might imply that NK cells would be the source of systemic IFN-γ (51), whereas low levels of IFN-γ produced by CD8 T cells would be specifically directed toward its target with signaling amplified or specified further by integrin coengagement. The fact that previous T cell activation is necessary for this (26) means that while IFN-γ might be spread throughout the spleen, only cells in the correct state of activation and engaging integrins will adequately respond to this cue. Overall, having methods to locally boost IFN-γ signaling by integrins at a synapse may optimize efficient T cell-mediated pathogen clearance programs while restricting harmful bystander responses.
Materials and Methods
Mice.
All experiments involving mice were approved by the Institutional Animal Care and Use Committee of the University of California and in agreement with the United Kingdom Scientific Procedures Act of 1986.
Cell Isolation, Activation, and in Vitro Treatment.
OTI cells were isolated from lymph nodes of 6- to 12-wk-old mice. Selection was carried out with a negative CD8 isolation kit (Stemcell Technologies). For in vitro experiments, cells were activated in vitro at low density with phorbol 12-myristate 13-acetate (PMA) (2 ng/mL) and ionomycin (20 ng/mL), and treated, where indicated, with 10 μg/mL anti–LFA-1 (M17.4; BioXCell), 10 μM Src inhibitor PP2 (Sigma), or 1 μM Jak inhibitor ruxolitinib (Santa Cruz Biotechnology) for 3–6 h at 37 °C in 5% CO2. Then, cells were treated with the indicated dose of IFN-γ (Peprotech) for 20 min at 37 °C in 5% CO2. In some experiments, cells were plated on fibronectin (EMD Millipore)-coated plates.
Infection and Treatments.
OTI cells (5 × 103 cells for effector/memory assessment, 5 × 104 cells for phenotyping at day 5, and 5 × 105 cells for Hoechst and proliferation experiments), isolated as described above, were transferred into recipient C57Bl6 mice by retroorbital or i.v. injection. Mice were infected 16 h later. Mice were given an i.v. injection of 103 colony-forming units of LM expressing a secreted form of OVA (LMOVA) (52).
In some experiments, mice received one or two i.p. injections (separated by 12 h and centered around the indicated time point) of 75 μg of isotype-matched control Ab (rat IgG1; BioXCell), anti–IFN-γ (XMG1.2; BioXCell), one single injection of 125–250 μg of the Src inhibitor PP2 24 h after infection, or one single injection of 250 μg anti-MHC class 1 (AF6-88.5.5.3; BioXCell) at 0 or 24 h after infection.
For recall experiments, mice were rechallenged 50–60 d after the primary infection with i.v. injection of 103 colony-forming units of LMOVA and analyzed after 5 d.
Supplementary Material
Acknowledgments
We thank E. Roberts, M. Headley, J. Bezbradica-Mirkovic, and E. Thompson for critical reading of the manuscript. We thank the Kennedy Institute Imaging Facility and the Biological Imaging Development Center personnel for technical assistance with imaging, and the NIH tetramer facility for the SIINFEKL tetramer. J.M.M. was supported by the Laboratory of Excellence Development Cancer and Targeted Therapies from the University of Lyon, France. This work was supported by grants from the NIH (R01AI052116 and R01AI114787 to M.F.K. and R03AI119220 to A.G.), Kennedy Trust (to A.G.), and Biotechnology and Biological Sciences Research Council (BB/R015651/1 to A.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804556115/-/DCSupplemental.
References
- 1.Ikeda H, Old LJ, Schreiber RD. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002;13:95–109. doi: 10.1016/s1359-6101(01)00038-7. [DOI] [PubMed] [Google Scholar]
- 2.Billiau A, Matthys P. Interferon-gamma: A historical perspective. Cytokine Growth Factor Rev. 2009;20:97–113. doi: 10.1016/j.cytogfr.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Lees JR. Interferon gamma in autoimmunity: A complicated player on a complex stage. Cytokine. 2015;74:18–26. doi: 10.1016/j.cyto.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: An overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 5.Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: A paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 6.Murray PJ. The JAK-STAT signaling pathway: Input and output integration. J Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 7.Ishihara K, Hirano T. Molecular basis of the cell specificity of cytokine action. Biochim Biophys Acta. 2002;1592:281–296. doi: 10.1016/s0167-4889(02)00321-x. [DOI] [PubMed] [Google Scholar]
- 8.Hosking MP, Flynn CT, Whitton JL. Antigen-specific naive CD8+ T cells produce a single pulse of IFN-γ in vivo within hours of infection, but without antiviral effect. J Immunol. 2014;193:1873–1885. doi: 10.4049/jimmunol.1400348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salerno F, Guislain A, Cansever D, Wolkers MC. TLR-mediated innate production of IFN-γ by CD8+ T cells is independent of glycolysis. J Immunol. 2016;196:3695–3705. doi: 10.4049/jimmunol.1501997. [DOI] [PubMed] [Google Scholar]
- 10.Berg RE, Crossley E, Murray S, Forman J. Relative contributions of NK and CD8 T cells to IFN-gamma mediated innate immune protection against Listeria monocytogenes. J Immunol. 2005;175:1751–1757. doi: 10.4049/jimmunol.175.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradley LM, Dalton DK, Croft M. A direct role for IFN-gamma in regulation of Th1 cell development. J Immunol. 1996;157:1350–1358. [PubMed] [Google Scholar]
- 12.Buchholz VR, Gräf P, Busch DH. The smallest unit: Effector and memory CD8(+) T cell differentiation on the single cell level. Front Immunol. 2013;4:31. doi: 10.3389/fimmu.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi NS, Kaech SM. Effector CD8 T cell development: A balancing act between memory cell potential and terminal differentiation. J Immunol. 2008;180:1309–1315. doi: 10.4049/jimmunol.180.3.1309. [DOI] [PubMed] [Google Scholar]
- 15.Krummel MF, Bartumeus F, Gérard A. T cell migration, search strategies and mechanisms. Nat Rev Immunol. 2016;16:193–201. doi: 10.1038/nri.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pennock ND, et al. T cell responses: Naive to memory and everything in between. Adv Physiol Educ. 2013;37:273–283. doi: 10.1152/advan.00066.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obar JJ, Lefrançois L. Early events governing memory CD8+ T-cell differentiation. Int Immunol. 2010;22:619–625. doi: 10.1093/intimm/dxq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Stipdonk MJ, Lemmens EE, Schoenberger SP. Naïve CTLs require a single brief period of antigenic stimulation for clonal expansion and differentiation. Nat Immunol. 2001;2:423–429. doi: 10.1038/87730. [DOI] [PubMed] [Google Scholar]
- 19.Kaech SM, Ahmed R. Memory CD8+ T cell differentiation: Initial antigen encounter triggers a developmental program in naïve cells. Nat Immunol. 2001;2:415–422. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercado R, et al. Early programming of T cell populations responding to bacterial infection. J Immunol. 2000;165:6833–6839. doi: 10.4049/jimmunol.165.12.6833. [DOI] [PubMed] [Google Scholar]
- 21.Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321:1081–1084. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long M, Adler AJ. Cutting edge: Paracrine, but not autocrine, IL-2 signaling is sustained during early antiviral CD4 T cell response. J Immunol. 2006;177:4257–4261. doi: 10.4049/jimmunol.177.7.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Brien S, et al. Ikaros imposes a barrier to CD8+ T cell differentiation by restricting autocrine IL-2 production. J Immunol. 2014;192:5118–5129. doi: 10.4049/jimmunol.1301992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curtsinger JM, Agarwal P, Lins DC, Mescher MF. Autocrine IFN-γ promotes naive CD8 T cell differentiation and synergizes with IFN-α to stimulate strong function. J Immunol. 2012;189:659–668. doi: 10.4049/jimmunol.1102727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thaventhiran JE, et al. Activation of the Hippo pathway by CTLA-4 regulates the expression of Blimp-1 in the CD8+ T cell. Proc Natl Acad Sci USA. 2012;109:E2223–E2229. doi: 10.1073/pnas.1209115109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gérard A, et al. Secondary T cell-T cell synaptic interactions drive the differentiation of protective CD8+ T cells. Nat Immunol. 2013;14:356–363. doi: 10.1038/ni.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zumwalde NA, Domae E, Mescher MF, Shimizu Y. ICAM-1-dependent homotypic aggregates regulate CD8 T cell effector function and differentiation during T cell activation. J Immunol. 2013;191:3681–3693. doi: 10.4049/jimmunol.1201954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klebanoff CA, et al. Memory T cell-driven differentiation of naive cells impairs adoptive immunotherapy. J Clin Invest. 2016;126:318–334. doi: 10.1172/JCI81217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polonsky M, et al. Induction of CD4 T cell memory by local cellular collectivity. Science. 2018;360:eaaj1853. doi: 10.1126/science.aaj1853. [DOI] [PubMed] [Google Scholar]
- 30.Kalia V, et al. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Hu X, Ivashkiv LB. Cross-regulation of signaling pathways by interferon-gamma: Implications for immune responses and autoimmune diseases. Immunity. 2009;31:539–550. doi: 10.1016/j.immuni.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pipkin ME, et al. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belz GT, Masson F. Interleukin-2 tickles T cell memory. Immunity. 2010;32:7–9. doi: 10.1016/j.immuni.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Kambayashi T, Assarsson E, Lukacher AE, Ljunggren HG, Jensen PE. Memory CD8+ T cells provide an early source of IFN-gamma. J Immunol. 2003;170:2399–2408. doi: 10.4049/jimmunol.170.5.2399. [DOI] [PubMed] [Google Scholar]
- 35.Wakil AE, Wang ZE, Ryan JC, Fowell DJ, Locksley RM. Interferon gamma derived from CD4(+) T cells is sufficient to mediate T helper cell type 1 development. J Exp Med. 1998;188:1651–1656. doi: 10.1084/jem.188.9.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozga AJ, et al. pMHC affinity controls duration of CD8+ T cell-DC interactions and imprints timing of effector differentiation versus expansion. J Exp Med. 2016;213:2811–2829. doi: 10.1084/jem.20160206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabatos CA, et al. A synaptic basis for paracrine interleukin-2 signaling during homotypic T cell interaction. Immunity. 2008;29:238–248. doi: 10.1016/j.immuni.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dustin ML, Springer TA. T-cell receptor cross-linking transiently stimulates adhesiveness through LFA-1. Nature. 1989;341:619–624. doi: 10.1038/341619a0. [DOI] [PubMed] [Google Scholar]
- 39.Bach EA, et al. Ligand-induced autoregulation of IFN-gamma receptor beta chain expression in T helper cell subsets. Science. 1995;270:1215–1218. doi: 10.1126/science.270.5239.1215. [DOI] [PubMed] [Google Scholar]
- 40.Hogg N, Patzak I, Willenbrock F. The insider’s guide to leukocyte integrin signalling and function. Nat Rev Immunol. 2011;11:416–426. doi: 10.1038/nri2986. [DOI] [PubMed] [Google Scholar]
- 41.Weber RJ, Liang SI, Selden NS, Desai TA, Gartner ZJ. Efficient targeting of fatty-acid modified oligonucleotides to live cell membranes through stepwise assembly. Biomacromolecules. 2014;15:4621–4626. doi: 10.1021/bm501467h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor MJ, Husain K, Gartner ZJ, Mayor S, Vale RD. A DNA-based T cell receptor reveals a role for receptor clustering in ligand discrimination. Cell. 2017;169:108–119.e20. doi: 10.1016/j.cell.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blouin CM, et al. Glycosylation-dependent IFN-γR partitioning in lipid and actin nanodomains is critical for JAK activation. Cell. 2016;166:920–934. doi: 10.1016/j.cell.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 44.Oyler-Yaniv A, et al. A tunable diffusion-consumption mechanism of cytokine propagation enables plasticity in cell-to-cell communication in the immune system. Immunity. 2017;46:609–620. doi: 10.1016/j.immuni.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer K, et al. Antigen recognition induces phosphatidylserine exposure on the cell surface of human CD8+ T cells. Blood. 2006;108:4094–4101. doi: 10.1182/blood-2006-03-011742. [DOI] [PubMed] [Google Scholar]
- 46.Mühl H, Pfeilschifter J. Anti-inflammatory properties of pro-inflammatory interferon-gamma. Int Immunopharmacol. 2003;3:1247–1255. doi: 10.1016/S1567-5769(03)00131-0. [DOI] [PubMed] [Google Scholar]
- 47.Badovinac VP, Tvinnereim AR, Harty JT. Regulation of antigen-specific CD8+ T cell homeostasis by perforin and interferon-gamma. Science. 2000;290:1354–1358. doi: 10.1126/science.290.5495.1354. [DOI] [PubMed] [Google Scholar]
- 48.Sercan O, Stoycheva D, Hämmerling GJ, Arnold B, Schüler T. IFN-gamma receptor signaling regulates memory CD8+ T cell differentiation. J Immunol. 2010;184:2855–2862. doi: 10.4049/jimmunol.0902708. [DOI] [PubMed] [Google Scholar]
- 49.Whitmire JK, Tan JT, Whitton JL. Interferon-gamma acts directly on CD8+ T cells to increase their abundance during virus infection. J Exp Med. 2005;201:1053–1059. doi: 10.1084/jem.20041463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perona-Wright G, Mohrs K, Mohrs M. Sustained signaling by canonical helper T cell cytokines throughout the reactive lymph node. Nat Immunol. 2010;11:520–526. doi: 10.1038/ni.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D, Reilly A, Lawrence DA. Relationships between IFNgamma, IL-6, corticosterone, and Listeria monocytogenes pathogenesis in BALB/c mice. Cell Immunol. 2001;207:13–18. doi: 10.1006/cimm.2000.1749. [DOI] [PubMed] [Google Scholar]
- 52.Pope C, et al. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J Immunol. 2001;166:3402–3409. doi: 10.4049/jimmunol.166.5.3402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.