Significance
RNA viruses usurp and reprogram host cells using short RNA genomes. RNA viruses encode the information required for their replication in both their primary sequences and higher-order structures formed when the RNA genome strand folds back on itself, but the extent of higher-order structure has remained unclear. We use a new high-throughput RNA structure probing technology to identify RNA regions with tertiary folds and discover that roughly one-third of the dengue virus RNA genome forms higher-order interactions, many in regions functionally important for replication. This work suggests that tertiary structure elements might be common in large RNAs, and that these regions might contain pockets targetable by small molecules in the design of antiviral therapeutics.
Keywords: RNA tertiary structure, RING-MaP, viral packaging, genome circularization
Abstract
RNA virus genomes are efficient and compact carriers of biological information, encoding information required for replication both in their primary sequences and in higher-order RNA structures. However, the ubiquity of RNA elements with higher-order folds—in which helices pack together to form complex 3D structures—and the extent to which these elements affect viral fitness are largely unknown. Here we used single-molecule correlated chemical probing to define secondary and tertiary structures across the RNA genome of dengue virus serotype 2 (DENV2). Higher-order RNA structures are pervasive and involve more than one-third of nucleotides in the DENV2 genomic RNA. These 3D structures promote a compact overall architecture and contribute to viral fitness. Disrupting RNA regions with higher-order structures leads to stable, nonreverting mutants and could guide the development of vaccines based on attenuated RNA viruses. The existence of extensive regions of functional RNA elements with tertiary folds in viral RNAs, and likely many other messenger and noncoding RNAs, means that there are significant regions with pocket-containing surfaces that may serve as novel RNA-directed drug targets.
RNA viruses are ubiquitous and are among the most efficient and compact carriers of biological information in nature. RNA viruses account for many serious ongoing and emerging threats, including HIV and the Ebola, influenza, dengue (DENV), and Zika viruses. The short genomes of these viruses encode information in both primary sequences and in higher-order secondary and tertiary structures that often enable the viruses to efficiently usurp and reprogram host cells. Regions in RNA genomes that contain significant well-defined base pairings (secondary structures) often have functions that enhance viral infectivity and replication (1–3). Tertiary structures in RNA viruses have not yet been defined on an RNA genome-wide basis but might be harbingers of as-yet undiscovered functions, may be regions in which mutations could create live attenuated virus vaccines with a low likelihood of reverting to virulence, or might prove to be high-value targets for antiviral drugs.
DENV is a member of the Flavivirus genus, which includes West Nile, yellow fever, tick-borne encephalitis, and Zika viruses. Roughly one-half of the global population is at risk of infection with DENV; it imposes formidable social, productivity, and healthcare burdens in dengue-endemic countries (4); and the future of a current vaccine (CYD-TDV) is in doubt (5). The DENV RNA genome is 10.7-kb long and encodes a single ORF flanked by highly structured 5′- and 3′-UTRs. The capped, nonpolyadenylated genomic RNA encodes a single polypeptide that is cotranslationally processed to produce viral structural and replicative proteins (6). Conserved RNA structures in the 5′- and 3′-UTRs and in internal regions function as promoters and regulators of translation, RNA replication, mRNA degradation, and production of subgenomic regulatory fragments (7–9). Complementary sequences at the 5′ and 3′ ends interact to circularize the viral genome, which is necessary for RNA synthesis (8). As is the case with essentially all viral and long cellular RNAs, the full extent of structure throughout the DENV2 genome is unknown.
Here we describe comprehensive interrogation of the secondary and tertiary structures of a DENV2 RNA genome. We first used selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) to obtain nucleotide-resolution measurements of structure across the entire DENV2 genome both inside authentic viral particles (in virion) and in the absence of proteins (ex virion). Modeling based on these data enabled accurate identification of every previously identified DENV RNA secondary structure (7–9) and numerous previously unidentified elements, many with well-determined structures and evolutionary conservation comparable to functional elements previously identified in DENV and other RNA viruses (1–3).
We then identified through-space interactions using the RNA interaction groups by mutational profiling (RING-MaP) strategy (10), detecting eight regions with dense internucleotide correlations, consistent with 3D tertiary folds. Mutations designed to destabilize selected RNA structures disrupted the global RNA genome architecture and reduced viral fitness. This work suggests that roughly one-third of the DENV2 RNA genome contains RNA elements that harbor tertiary folds that may also contain clefts and pockets, and implies that tertiary structures may be pervasive in large RNAs.
Results
Nucleotide-Resolution Structural Interrogation of Authentic DENV2 Genomic RNA.
We obtained comprehensive nucleotide-resolution structural data for 99% of the nucleotides in DENV2 strain S16803 (11) both inside intact viral particles and gently extracted from viral particles (ex virion). RNAs were probed using the SHAPE-MaP strategy with 1-methyl-7-nitroisatoic anhydride (1M7), which reports individual nucleotide flexibility, and with differential SHAPE reagents that specifically detect nucleotides that form noncanonical interactions, to allow highly accurate secondary structure modeling (1, 12, 13).
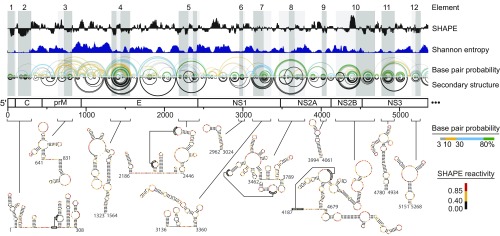
We first used ex virion SHAPE reactivities to model the secondary structure of the entire DENV2 RNA genome (Fig. 1, black arcs and SI Appendix, Fig. S1, panel 1) (13). We assessed the degree of structural heterogeneity at each position by summing base-pairing probabilities for each base pair combination within the predicted ensemble of structures to calculate a Shannon entropy term for each nucleotide (Fig. 1, dark blue and SI Appendix, Fig. S1, panel 1). Regions with well-determined structures (“elements”) were identified by considering both highly structured regions (low SHAPE reactivity) and well-determined structured regions (low experimentally-informed Shannon entropy).
Fig. 1.
Well-determined secondary structure elements in the DENV2 RNA genome. The first half of the genome is shown (the entire genome is shown in SI Appendix, Fig. S1, panel 1). Median ex virion 1M7 SHAPE reactivities (black) and Shannon entropies (dark blue) are plotted over centered 55-nt windows. Regions with both low SHAPE and low Shannon entropy are highlighted by dark-gray shading, with light-gray shading extended to encompass entire intersecting helices. The first 12 elements (out of 24 total elements) with well-determined structures are numbered. Base pair probability arcs are colored by probability (see scale), with green arcs indicating the most probable base pairs; black arcs indicate plausible pseudoknots (PK). The minimum free-energy secondary structure (inverted black arcs) was obtained using both 1M7 and differential SHAPE reactivities as constraints (1, 12, 13). Secondary structures of elements 1–12 are colored by SHAPE reactivity; high-resolution structures are provided in SI Appendix, Fig. S1, panel 2.
Viral and bacterial RNA elements with both low SHAPE reactivity and low entropy are often functionally important (1, 2, 14). Twenty-four elements in the DENV2 genome show low SHAPE reactivity and low Shannon entropy (Fig. 1, numbered gray regions and secondary structures, and SI Appendix, Fig. S1). Only the 5′-UTR, the 5′ portion of the capsid-coding region, and the 3′-UTR have been characterized previously (7, 8, 15), and the SHAPE-directed models agree with these accepted structures (SI Appendix, Fig. S2).
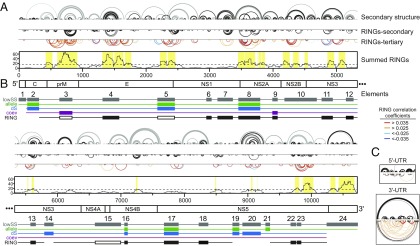
We recently developed a strategy called RING-MaP to detect nucleotides involved in through-space structural communication (10, 16). Here RNA is modified by dimethyl sulfate such that each strand contains multiple structure-specific modifications. Nucleotide pairs that are modified in a correlated fashion belong to an RNA interaction group (RING); for example, two nucleotides in the same helix will be protected when that helix is stable but will be more likely to be modified in a coordinated way under destabilizing conditions. Similar behavior is observed for nucleotides involved in tertiary interactions. We characterized through-space structural communication throughout the DENV2 RNA genome using RING-MaP (Fig. 2) and performed additional experiments on the 5′- and 3′-UTRs to achieve sufficient read depth in these terminal regions (Fig. 2C).
Fig. 2.
Tertiary interactions and structural conservation in the DENV2 RNA genome. (A and B) The entire genome is split into two panels. (A) Characterization of DENV2 RNA secondary and tertiary structures. Base pairs common to both ex virion and refolded RNA are depicted by black arcs; base pairs unique to refolded RNA are in gray. RINGs are depicted by inverted arcs. Secondary structure RINGs are in black. Tertiary RINGs (colored by correlation coefficient; see key) were summed over 101-nt windows, and positions with a value greater than the median plus 1 SD were assigned as having a significant degree of tertiary structure (yellow shading). (B) Evolutionary and RING support for the 24 elements with low SHAPE and low entropy (gray boxes; lowSS). Filled boxes indicate element-supporting data: minor allele frequency tests (allele; green), synonymous substitution rates (dS; blue), coevolution (coev; purple), and RINGs (black). For RING data, open boxes indicate regions that fold differently between the ex virion and refolded RNA, and the lack of a black line indicates regions with very few RINGS. (C) 5′-UTR and 3′-UTR RINGs detected using gene-specific primers. Positions shown: 5′-UTR, 1–275; 3′-UTR, 10,247–10,660.
RINGs were binned into those likely to represent secondary vs. tertiary interactions based on their contact distance. The contact distance of two nucleotides is the shortest path between them according to the SHAPE-directed secondary structure model when nested helices are skipped and pseudoknots are ignored. RINGs with contact distances of <20 nucleotides generally reflect base pairing, whereas RINGs spanning longer contact distances tend to report higher-order structural interactions. RINGs reflecting secondary structure correspond closely to those modeled independently by SHAPE (Fig. 2 A and B).
Conservation of Secondary Structure Elements in Diverse DENV2 Isolates.
If the base-pairing interactions identified by SHAPE and RING experiments are functionally important for viral replication, then evolutionary pressure to maintain these interactions can be detected. We measured allele frequencies, synonymous substitution rates, and the degree of complementary coevolution to determine whether the selective pressures on the base-paired sites in the 24 elements are stronger than those in unpaired sites. Allele frequency distribution tests revealed that mutations at base-paired sites are significantly less well tolerated than those at unpaired sites for 7 of the 24 elements (Fig. 2B, green boxes). Analysis of synonymous substitution rates revealed a statistically significant decrease in mutation rates at base-paired sites compared with unpaired sites in 8 of the 22 elements in the protein-coding region (Fig. 2B, blue boxes). Although allele frequency distribution and synonymous substitution rate analyses measure different evolutionary signals, both indicate that base-paired sites within a similar set of structural elements experience greater degrees of negative selection than unpaired sites. Finally, there is significant support for complementary coevolution of base pairs in two elements (Fig. 2B, purple boxes). Together, evolutionary selection has maintained base pairing in one-half of the 22 elements located in the protein-coding region.
As an independent measure of structure, we examined RING-MaP data for evidence of base-pairing interactions within the 24 well-determined elements. Base pairs in 13 of the 24 elements are supported by RINGs, and no motifs outside of these 24 elements show evidence of extensive through-space interactions (Fig. 2 A and B, black boxes). Finally, structures in the highly conserved 5′- and 3′-UTRs, which correspond to elements 1 and 24, are functionally important (7–9). Collectively, there is strong support, based on evolutionary pressure, secondary structure RINGs, or previous functional studies, for 22 of the 24 elements identified de novo as low-SHAPE/low-entropy elements (Fig. 2 A and B, elements 1–9, 11–14, and 16–24).
The DENV2 RNA genome is thus replete with elements that form well-determined secondary structures, consistent with studies of multiple RNA viruses (1–3, 16–19). The unanswered question now is the extent to which these base-paired elements form higher-order motifs.
Packaged RNA Undergoes a Structural Switch.
Inside a viral particle, the structure of the DENV RNA might be altered by both a requirement to fit into the compact virion interior and by interactions with the protein capsid. As an initial approach to characterize higher-order interactions in the DENV2 RNA genome, we compared the structure of the DENV2 RNA inside intact viral particles (in virion) with that of the ex virion RNA gently extracted from viral particles (examined above). The two 1M7 SHAPE profiles are very similar (SI Appendix, Fig. S3A), but we detected compact regions with statistically significant SHAPE differences between ex virion and in virion RNA states (13, 20) (SI Appendix, Fig. S3B). Of the ∼900 nucleotides with significant SHAPE differences, most (82%) are nucleotides that are single-stranded in the protein-free structural model, and some of the significant differences are clustered in the genome (SI Appendix, Fig. S3C). Nine of these clusters overlap with well-determined structural elements (SI Appendix, Fig. S3D), including in the 5′- and 3′-UTRs. These clusters of SHAPE differences may reflect RNA interactions with the viral capsid, local conformational differences, or global RNA reorganization.
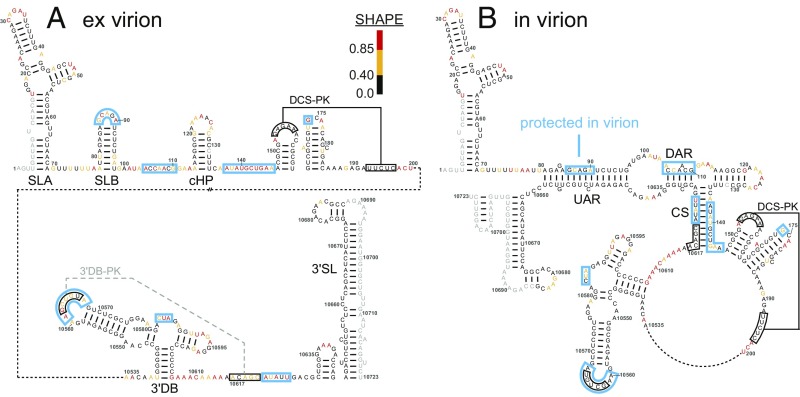
Nucleotides in the 5′- and 3′-UTRs mediate circularization of the RNA genome, which involves a secondary structure rearrangement necessary for replication (8, 15) (Fig. 3 and SI Appendix, Fig. S4). Strikingly, almost every nucleotide predicted to become base-paired on genome circularization is protected from SHAPE modification inside the viral particle (Fig. 3 and SI Appendix, Fig. S4). Moreover, in virion SHAPE reactivities are most consistent with a circularized structure (Fig. 3B), whereas ex virion SHAPE reactivities are consistent with a linear structure (Fig. 3A). Direct structure probing thus suggests that the RNA genome is in the circular form in the native viral particle and relaxes to favor the linear form as a free RNA.
Fig. 3.
Encapsidated DENV2 RNA is in its circular form. Nucleotides protected in virion (blue boxes) are shown on both linear and circular genome structures. (A) Secondary structure of the proposed linear form of the DENV2 genomic RNA 5′-UTR, capsid-coding region, and 3′-UTR with nucleotides colored by ex virion SHAPE reactivity. Pseudoknots DCS-PK and 3′DB-PK are shown. The 3′DB-PK does not form in the ex virion RNA, as indicated by a dashed gray line. Positions with no data are in gray. (B) Secondary structure of the proposed circular form of the DENV2 genomic RNA color-coded by in virion SHAPE reactivity.
Discovery of DENV2 RNA Tertiary Structures by RING Analysis.
We next used RING-MaP data to identify higher-order tertiary structures (10) (Fig. 2, see RINGs-tertiary). To validate the relationship between genome-scale RING analysis and RNA tertiary structure, we first applied the RING-MaP strategy to a previously characterized DENV2 RNA tertiary interaction: the 3′ dumbbell pseudoknot (3′DB-PK) (21). We incubated the DENV2 genomic RNA with an antisense locked nucleic acid (LNA) complementary to one helix in the pseudoknot (LNA-PK1) or with a control LNA that binds outside the 3′-UTR. RINGs consistent with the 3′DB-PK interaction specifically and completely disappeared in the presence of LNA-PK1 but not in the presence of the control LNA (SI Appendix, Fig. S5). New long-range RINGs appeared on binding by LNA-PK1, consistent with a reorganization of the 3′DB-PK region in the absence of pseudoknot formation (SI Appendix, Fig. S5B).
To determine the density of RINGs that corresponds to a tertiary structure motif, we examined the RING pattern of the catalytic domain of RNase P under conditions that promote formation of secondary structures only (absence of Mg2+) or the full tertiary structure (presence of Mg2+) (SI Appendix, Fig. S6). We observed secondary structure RINGs that recapitulate many of the accepted helices in RNase P under both conditions. We only observed significant tertiary structure RINGs (contact distances of ≥20 nucleotides) for the RNA probed in the presence of Mg2+. Summing the number of tertiary RINGs across the RNA revealed a strong peak only in the plus-Mg2+ structure (SI Appendix, Fig. S6).
We next computed the summed number of tertiary RINGs across the DENV2 RNA genome and found eight regions to have RING densities comparable to that of the fully folded RNase P RNA (Fig. 2A, yellow shading). These regions overlap with elements previously identified to have well-determined secondary structures (Fig. 2, elements). There are high densities of RINGs in elements 3, 4, 5, 8, 10, 11, 23, and 24. This is a satisfying result, as a well-determined secondary structure is a prerequisite for formation of a stable tertiary structure (22).
To examine whether these regions with evidence for tertiary structure influence the overall conformation of the DENV2 genomic RNA, we selected three elements that met specific criteria (SI Appendix, Supporting Text) for further analysis: 4 (Env), 8 (NS2A), and 10 (NS2B). As a control, we evaluated an element in a region with a well-determined secondary structure but a low density of tertiary RINGs, which was not expected to have a higher-order fold (element 20; NS5). We introduced synonymous mutations into the DENV2 RNA that disrupted the predicted secondary and tertiary structures of each of these elements while maintaining the amino acid sequence and avoiding rare codons (SI Appendix, Fig. S7).
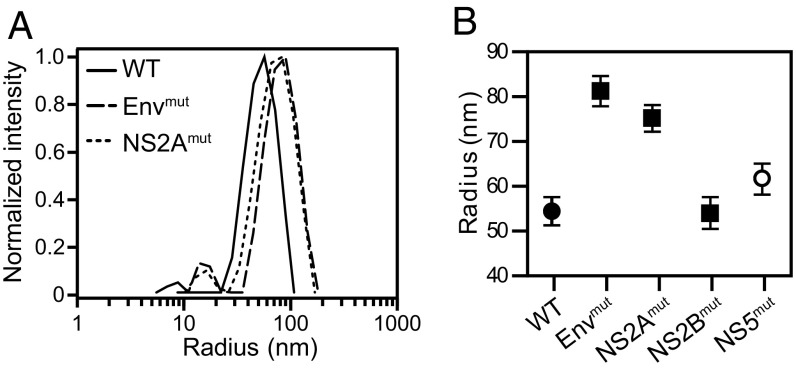
We examined whether these mutations altered the global size and organization of the full-length DENV2 RNA using dynamic light scattering (23). Full-length genomic RNAs were transcribed in vitro and refolded. Mutations in the Env and NS2A elements increased the hydrodynamic radius of the DENV2 RNA relative to wild type (WT) RNA, but mutations in NS2B and the NS5 control have little or no effect on size (Fig. 4). Thus, two of the three examined elements (Env and NS2A) predicted by RING analysis to adopt higher-order structures influence the global architecture of the DENV2 RNA genome.
Fig. 4.
Hydrodynamic radii of WT and mutant DENV2 RNAs. (A) Dynamic light scattering curves from individual experiments, representative of multiple replicate experiments. The mean radius values for WT, Envmut, and NS2Amut are 54, 81, and 75 nm, respectively. (B) RNA radii of WT and mutant RNAs. Values are mean and SD calculated from four experiments. Control NS5mut is denoted by an open symbol.
Higher-Order Structures in the DENV2 RNA Genome Regulate Infection.
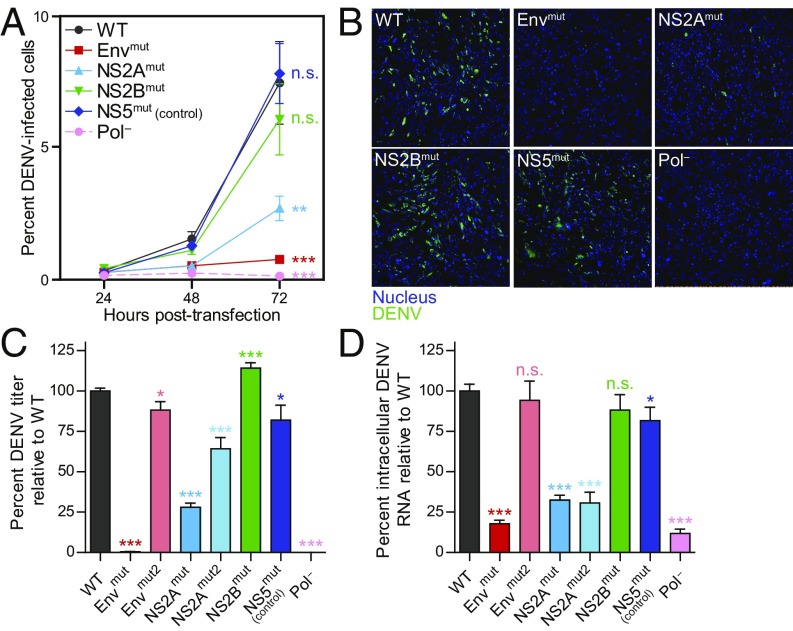
To examine the impact of the higher-order RING-supported structures on DENV2 replication, in vitro transcribed and capped WT and mutant DENV2 RNAs were transfected into BHK-21 cells, and viral replication was assayed by measuring the fraction of infected cells in the cell monolayer (Fig. 5 A and B), infectious viral particles in the supernatant (Fig. 5C), and intracellular viral RNA (Fig. 5D). The Envmut and NS2Amut mutants (described above) severely and moderately attenuated, respectively, all measures of viral replication over the course of infection relative to WT. The percentage of infected cells, viral titers, and viral RNA were reduced by ∼90% for Envmut and by ∼60% for NS2Amut at 72 h posttransfection. In contrast, structure-disrupting mutations in elements 10 (NS2Bmut) and the control (element 20; NS5mut) did not affect these viral replication measurements. Notably, the Envmut and NS2Amut viruses maintained their lower titers compared with the WT even after >60 d of passage (SI Appendix, Fig. S8). Specific disruption of higher-order RNA structures thus created stable attenuated viruses.
Fig. 5.
Replication fitness of DENV2 tertiary structure mutants and control. (A) DENV2-positive BHK-21 cells after transfection of WT or mutant in vitro transcribed DENV2 RNAs. Virus was visualized by immunostaining of DENV2 envelope protein and nuclei (n ≥3, with ≥1,000 cells counted per condition). (B) Representative fields showing DENV2 RNA-transfected cells (green) and nuclei (DAPI, blue) at 72 h posttransfection. (C) Viral titer relative to WT following transfection of DENV2 RNA at 72 h (n ≥3). (D) Percent DENV2 RNA relative to WT at 72 h posttransfection as quantified by RT-qPCR (n ≥3). Values plotted are mean ± SEM of at least three independent experiments. Pol−, a lethal mutation in the DENV2 NS5 RNA-dependent RNA polymerase; n.s., not significant. *P < 0.05; **P < 0.01; ***P < 0.001, two-way ANOVA with Bonferroni correction in A, Student’s t test in C and D.
We also determined the impact of higher-order RNA structures in full replication cycle experiments, an assay that also requires that virus enter cells. The Envmut virus was not tested in this assay, as this mutant is so severely compromised that it was not possible to generate sufficient viral stocks. The NS2Amut virus resulted in a significant (∼85%) reduction in the percentage of infected cells, viral titer, and both intracellular and extracellular RNA (SI Appendix, Fig. S9) compared with WT virus. The NS2Bmut virus and the NS5mut control replicated as efficiently as WT virus. Taken together, these data reveal that higher-order RNA structures in the Env and NS2A-coding regions (elements 4 and 8), identified using the summed RING approach, directly influence DENV2 replication.
This initial set of virus mutants was designed to comprehensively disrupt RNA structure in each element, such that multiple synonymous mutations (that did not create rare codons) were introduced (SI Appendix, Fig. S7). To more selectively test the functional role of nucleotides specifically predicted to be involved in tertiary contacts without disrupting their overall secondary structure, we examined a second set of Env and NS2A mutants (Envmut2 and NS2Amut2), focusing on nucleotides involved in or adjacent to RING correlations (SI Appendix, Fig. S10). Although available site-specific synonymous substitutions were severely limited, these “focused” Envmut2 and NS2Amut2 mutants yielded viruses that were modestly to moderately attenuated, respectively (Fig. 5 C and D), consistent with the overall importance of higher-order structure in these RNA elements.
3D Models for Functional DENV2 RNA Structures.
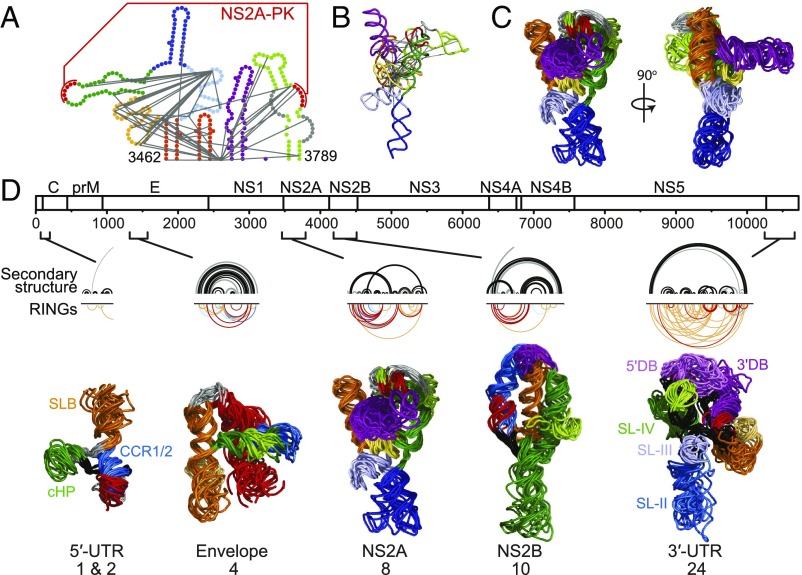
RINGs report through-space secondary and tertiary interactions and can be used as restraints to generate 3D models that recapitulate an overall RNA fold with good accuracy (SI Appendix, Fig. S6C) (10). We modeled well-determined elements within the DENV2 RNA genome that contain dense networks of tertiary RINGs. We illustrate our approach with the structure in element 8, located within the DENV2 NS2A protein-coding region (Fig. 6A). RING-directed refinement yielded a compact structure that minimizes the distances between nucleotides with RING interactions (Fig. 6B). One helix (yellow) forms the core of the structure and is closely surrounded by several other helices (Fig. 6 B and C; helices in light blue, orange, and green). There are only a few RINGs in the remaining (dark blue and purple) regions and, correspondingly, these helices sample a wider range of conformations relative to the well-defined core.
Fig. 6.
Higher-order structures throughout the DENV2 genome. (A) Secondary structure of the NS2A element. RINGs used as tertiary structure constraints in the DMD simulation are depicted by gray lines. (B) Medoid of DMD models of the NS2A element. (C) The 10 lowest aligned free-energy DMD models of the NS2A element. (D) From top to bottom, the DENV2 RNA genome, the secondary structure (black arcs) and tertiary RINGS (inverted colored arcs) of elements with tertiary structures, and tertiary structure models. RINGs reporting tertiary interactions are color-coded by correlation coefficient as in Fig. 2, and 3D folds are color-coded by secondary structure (SI Appendix, Figs. S11–S14). Previously identified elements in the 5′- and 3′-UTRs (7–9) are labeled. For each element, the 10 lowest free-energy models from the largest cluster are shown.
At the 5′ end of the genome (elements 1 and 2), a small number of RINGs connect stem-loop B (SLB) and capsid region hairpin (cHP) to the downstream DCS-PK pseudoknotted structure (SI Appendix, Fig. S11) (7, 8, 15). The RING-constrained DMD simulation supports a model in which the CCR1 and CCR2 helices form a continuous helix, resulting in a compact structure in the pseudoknotted region (Fig. 6D, 5′-UTR). Consistent with the relatively small number of RING constraints, the CCR1/CCR2 domain samples a range of orientations relative to SLB and cHP (SI Appendix, Fig. S11). Incorporation of RING constraints into the DMD simulation of structural element 4 in the envelope-coding region resulted in a highly collapsed state compared with the no-constraint control (Fig. 6D, envelope). DMD simulations yielded two clusters of models for this element, which differ in the orientation of one helix, suggesting that two conformations may be populated (SI Appendix, Fig. S12). The dense RINGs in element 10 yield a single well-defined structure (Fig. 6D, NS2B and SI Appendix, Fig. S13).
Finally, the 3′-UTR (element 24) contains a large number of RINGs connecting multiple secondary structural features (SI Appendix, Fig. S14). Consistent with the extensive RING network, the structural model is highly compact, in part because sequences at the beginning and end of this motif are predicted to form a long-range helix (between nucleotides 10,250 and 10,650) that constrains the topologically allowed structural space of the enclosed domains (Fig. 6D, 3′-UTR). The RING data emphasize that the DENV2 3′-UTR forms a complex 3D fold.
Discussion
RNA viruses make efficient use of primary sequences, RNA modifications (24), and all levels of RNA structure as they usurp host cell machinery to regulate their replication, protein synthesis, packaging, and evasion of host immune factors. Regions with significant well-determined secondary structures are common across positive-sense RNA virus genomes, many of which are critical for viral fitness (1–3, 17–19). It remains a challenge to identify functionally important RNA structural elements in the context of extensive secondary structure folding. We posited that functional RNA structural elements might be overrepresented among motifs that form higher-order structures, analogous to tertiary structure elements in viral UTRs (25–28). To examine this hypothesis, we characterized the DENV2 RNA genome structure at the levels of secondary (Fig. 1 and SI Appendix, Fig. S1), tertiary (Figs. 2 and 4–6), and quaternary (Fig. 3) genome organization.
We identified 24 elements with well-determined secondary structures. At least one-half of those in the protein-coding region are detectably evolving under evolutionary pressures reflective of biological functionality (Figs. 1 and 2 and SI Appendix, Fig. S1). These results are consistent with studies of other positive-sense RNA virus genomes (1–3, 17–19). We then probed through-space structural communication across the DENV RNA genome and found tertiary structure to be pervasive, spanning at least eight distinct regions (Fig. 2). We modeled 3D folds for a subset of these regions and identified numerous RNA motifs with potential higher-order folds (Fig. 6). Structure-disrupting mutants revealed several of these higher-order RNA structures to be important for global genome organization and viral replication (Figs. 4 and 5). Finally, the genomic RNA is linear in the absence of the protein capsid quaternary organization, whereas encapsidated RNA is likely in a circular form (Fig. 3). Circularization is essential for virus replication (8, 15), and here we show that it occurs during encapsidation and viral packaging. Interactions with host proteins or release from the viral capsid may trigger conversion of the RNA genome to its linear form, as required for translation.
In summary, direct experimental interrogation of RNA structures across the DENV2 RNA genome reveals that motifs with complex, higher-order architectures are pervasive features of this large viral RNA and are functionally integrated with viral replication and packaging. Complex tertiary structure elements are widely used by viruses to mediate interactions with protein, RNA, and small-molecule partners (8, 25, 27, 29, 30). The precise mechanisms by which the higher-order RNA structures identified here affect viral fitness remain to be resolved, but it is clear that these structures modulate overall genome architecture, which might affect packaging efficiency or tune interactions with viral replication components and cellular factors.
Viruses compress a full suite of biological functions into a small replicating package. Discoveries about the organization of biological information in viruses are likely to be broadly important in other systems as well. RING-MaP can detect RNA regions with higher-order tertiary folds across the length of large RNAs and will make tertiary structure discovery tractable for RNA viruses, messenger RNAs, and long noncoding RNAs, facilitating a deeper understanding of the regulatory roles of RNA structure in gene expression. The ability to rapidly and accurately discover regions with higher-order structure that directly modulate viral fitness (Fig. 5 and SI Appendix, Figs. S8 and S9) has immediate applications for the development of live attenuated virus vaccines against dengue, Zika, and other RNA viruses. Finally, any large RNA harboring elements with tertiary folds (Fig. 6) may also contain high information content pockets (31) that comprise novel RNA-directed drug targets.
Materials and Methods
DENV2 experiments were based on strains S16803 and 16681. SHAPE-MaP experiments, structure modeling, and bioinformatics analyses were performed (1, 2, 13) on RNA gently extracted from virions (ex virion) or on intact virions (in virion). SHAPE reactivity differences were evaluated using the deltaSHAPE framework (20). RING-MaP experiments, structure modeling, and bioinformatics analyses were performed (1, 10, 13) on refolded DENV2 RNA. Tertiary structure models were generated using RINGs as constraints in DMD simulations (10). Detailed descriptions of these methods, sequence conservation analysis, and the construction, physical size evaluation, and phenotypic testing of DENV2 mutants are provided in SI Appendix, Methods.
Supplementary Material
Acknowledgments
This work was supported by the NIH (Grants R35 GM122532, to K.M.W.; R01 AI125416 and R43 AI129095, to S.M.H.; U54 AI057157, to W.B.M.; and T32 CA009111, to M.T.S. and M.J.M.), the Burroughs Wellcome Fund (S.M.H.), and the Sunlin and Priscilla Chou Foundation (W.B.M.). E.A.D. was a Lineberger Postdoctoral Fellow in the Basic Sciences (T32 CA009156) and a Ruth L. Kirschstein Postdoctoral Fellow (F32 GM103180). M.A.B. is a Ruth L. Kirschstein Postdoctoral Fellow (F32 GM128330). N.S.G. is supported by an American Heart Association Predoctoral Award (17PRE33670017).
Footnotes
Conflict of interest statement: K.M.W. is an advisor to and holds equity in Ribometrix.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716689115/-/DCSupplemental.
References
- 1.Siegfried NA, Busan S, Rice GM, Nelson JAE, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mauger DM, et al. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc Natl Acad Sci USA. 2015;112:3692–3697. doi: 10.1073/pnas.1416266112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pirakitikulr N, Kohlway A, Lindenbach BD, Pyle AM. The coding region of the HCV genome contains a network of regulatory RNA structures. Mol Cell. 2016;62:111–120. doi: 10.1016/j.molcel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shepard DS, Undurraga EA, Halasa YA, Stanaway JD. The global economic burden of dengue: A systematic analysis. Lancet Infect Dis. 2016;16:935–941. doi: 10.1016/S1473-3099(16)00146-8. [DOI] [PubMed] [Google Scholar]
- 5.Flipse J, Smit JM. The complexity of a dengue vaccine: A review of the human antibody response. PLoS Negl Trop Dis. 2015;9:e0003749. doi: 10.1371/journal.pntd.0003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcaraz-Estrada SL, Yocupicio-Monroy M, del Angel RM. Insights into dengue virus genome replication. Future Virol. 2010;5:575–592. [Google Scholar]
- 7.Selisko B, Wang C, Harris E, Canard B. Regulation of flavivirus RNA synthesis and replication. Curr Opin Virol. 2014;9:74–83. doi: 10.1016/j.coviro.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gebhard LG, Filomatori CV, Gamarnik AV. Functional RNA elements in the dengue virus genome. Viruses. 2011;3:1739–1756. doi: 10.3390/v3091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villordo SM, Carballeda JM, Filomatori CV, Gamarnik AV. RNA structure duplications and flavivirus host adaptation. Trends Microbiol. 2016;24:270–283. doi: 10.1016/j.tim.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homan PJ, et al. Single-molecule correlated chemical probing of RNA. Proc Natl Acad Sci USA. 2014;111:13858–13863. doi: 10.1073/pnas.1407306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly EP, Polo S, Sun W, Falgout B. Evolution of attenuating mutations in dengue-2 strain S16803 PDK50 vaccine and comparison of growth kinetics with parent virus. Virus Genes. 2011;43:18–26. doi: 10.1007/s11262-011-0602-z. [DOI] [PubMed] [Google Scholar]
- 12.Rice GM, Leonard CW, Weeks KM. RNA secondary structure modeling at consistent high accuracy using differential SHAPE. RNA. 2014;20:846–854. doi: 10.1261/rna.043323.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc. 2015;10:1643–1669. doi: 10.1038/nprot.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mustoe AM, et al. Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell. 2018;173:181–195.e18. doi: 10.1016/j.cell.2018.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Borba L, et al. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J Virol. 2015;89:3430–3437. doi: 10.1128/JVI.02677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larman BC, Dethoff EA, Weeks KM. Packaged and free satellite tobacco mosaic virus (STMV) RNA genomes adopt distinct conformational states. Biochemistry. 2017;56:2175–2183. doi: 10.1021/acs.biochem.6b01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burrill CP, et al. Global RNA structure analysis of poliovirus identifies a conserved RNA structure involved in viral replication and infectivity. J Virol. 2013;87:11670–11683. doi: 10.1128/JVI.01560-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutchko KM, et al. Structural divergence creates new functional features in alphavirus genomes. Nucleic Acids Res. 2018;46:3657–3670. doi: 10.1093/nar/gky012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson BL, White KA. Exploring the architecture of viral RNA genomes. Curr Opin Virol. 2015;12:66–74. doi: 10.1016/j.coviro.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Smola MJ, Calabrese JM, Weeks KM. Detection of RNA-protein interactions in living cells with SHAPE. Biochemistry. 2015;54:6867–6875. doi: 10.1021/acs.biochem.5b00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsthoorn RC, Bol JF. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA. 2001;7:1370–1377. [PMC free article] [PubMed] [Google Scholar]
- 22.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stetefeld J, McKenna SA, Patel TR. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys Rev. 2016;8:409–427. doi: 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gokhale NS, et al. N6-methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe. 2016;20:654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto H, et al. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat Struct Mol Biol. 2014;21:721–727. doi: 10.1038/nsmb.2859. [DOI] [PubMed] [Google Scholar]
- 26.Jones CP, Cantara WA, Olson ED, Musier-Forsyth K. Small-angle X-ray scattering-derived structure of the HIV-1 5′ UTR reveals 3D tRNA mimicry. Proc Natl Acad Sci USA. 2014;111:3395–3400. doi: 10.1073/pnas.1319658111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapman EG, et al. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science. 2014;344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keane SC, et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science. 2015;348:917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang X, et al. An unusual topological structure of the HIV-1 Rev response element. Cell. 2013;155:594–605. doi: 10.1016/j.cell.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boerneke MA, Dibrov SM, Gu J, Wyles DL, Hermann T. Functional conservation despite structural divergence in ligand-responsive RNA switches. Proc Natl Acad Sci USA. 2014;111:15952–15957. doi: 10.1073/pnas.1414678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warner KD, Hajdin CE, Weeks KM. Principles for targeting RNA with drug-like small molecules. Nat Rev Drug Discov. 2018;17:547–558. doi: 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.