Significance
The high-affinity methionine importer MetNI belongs to the ATP Binding Cassette (ABC) family of transporters that carry out the ATP-dependent uptake of substrates into cells. As with other ABC importers, MetNI requires a soluble binding protein (MetQ) that in the canonical mechanistic model delivers substrates to the transporter. We made the unexpected observation that a MetQ variant with significantly impaired ligand-binding properties supports d-selenomethionine uptake at a higher rate than wild-type MetQ. A crystal structure of MetNIQ in the outward-facing conformation reveals access channels through the binding protein to the transmembrane translocation pathway. These studies support a noncanonical role for the binding protein in facilitating the uptake of certain substrates directly through the transporter–binding protein complex.
Keywords: ATP Binding Cassette transporter, alternating access transport mechanism, methionine transporter, transinhibition
Abstract
The Escherichia coli methionine ABC transporter MetNI exhibits both high-affinity transport toward l-methionine and broad specificity toward methionine derivatives, including d-methionine. In this work, we characterize the transport of d-methionine derivatives by the MetNI transporter. Unexpectedly, the N229A substrate-binding deficient variant of the cognate binding protein MetQ was found to support high MetNI transport activity toward d-selenomethionine. We determined the crystal structure at 2.95 Å resolution of the ATPγS-bound MetNIQ complex in the outward-facing conformation with the N229A apo MetQ variant. This structure revealed conformational changes in MetQ providing substrate access through the binding protein to the transmembrane translocation pathway. MetQ likely mediates uptake of methionine derivatives through two mechanisms: in the methionine-bound form delivering substrate from the periplasm to the transporter (the canonical mechanism) and in the apo form by facilitating ligand binding when complexed to the transporter (the noncanonical mechanism). This dual role for substrate-binding proteins is proposed to provide a kinetic strategy for ABC transporters to transport both high- and low-affinity substrates present in a physiological concentration range.
The substrate specificity of a transporter represents one of the most basic functional attributes and often serves as the defining characteristic. Transporters can exhibit both high affinity for a particular substrate and broad specificity in translocating related compounds. An example is the Escherichia coli MetNI methionine importer established by Kadner et al. to mediate the high-affinity uptake of l-methionine, while transporting d-methionine with lower affinity; other methionine derivatives including selenomethionine are also transported (1–4). MetNI is a member of the ATP Binding Cassette (ABC) family of transporters (5–9), consisting of two transmembrane subunits (MetI) that form a translocation pathway and two nucleotide-binding subunits (MetN) that couple ATP binding and hydrolysis to transport (10, 11). As for other ABC importers (5, 12–15), MetNI requires an additional component, MetQ (8, 10, 11, 16–18), which is a substrate-binding protein (SBP), generally viewed to deliver substrates to their cognate transporters.
The role of SBPs has been intriguing since their original identification as osmotic sensitive components that were required for the transport of certain nutrients (19). Investigations of a number of SBPs support a model where binding of substrate by free apo-SBP is accompanied by a “Venus flytrap” conformational change (20, 21), followed by delivery of ligand to the ABC transporter (22–25); in this canonical model, the SBP delivers the substrate to the transporter and is predominantly responsible for the substrate specificity of transport. However, these sequential events have been challenged by biochemical and single-molecule studies (26–30), including an early demonstration by Shuman that binding protein-independent variants of the MalFGK2 maltose transporter could be isolated (31, 32). Different roles of the binding protein were also inferred by Merino and Shuman (33), who reported that the unliganded maltose-binding protein, despite its inability to bind lactose, is required for a MalFGK2 variant to transport lactose efficiently. The properties of these variants suggested substrate specificity was determined in the transmembrane regions of the transporter itself (31, 34). Participation of both the SBP and the transporter in determining substrate specificity was reported in a structural analysis of the maltose transporter with various bound substrates (35). More recently, studies have suggested a role for substrates binding directly to a preformed complex between the SBP and transporter, rather than delivered to the transporter by free SBP (28, 30).
Although these studies have provided evidence for a noncanonical role of SBPs in facilitating substrate uptake by ABC importers, no supporting structural evidence has been reported. In the course of characterizing the transport properties of MetNI, we made an unexpected observation that the N229A MetQ variant with greatly reduced ligand binding affinity supported d-selenomethionine uptake at a higher rate than wild-type MetQ. We solved a crystal structure of the ATPγS-bound MetNI in complex with the ligand free N229A variant of MetQ. This structure reveals unprecedented features of the outward-facing conformation that support a noncanonical transport mode of the MetNI transporter in which MetQ facilitates access of substrate to the translocation pathway through the MetNIQ complex. The ability of intracellular methionine to inhibit the uptake of extracellular methionine (transinhibition), a key component of MetNI regulation described by Kadner (3), is also structurally detailed.
Unexpected High Transport Rates for a Substrate-Binding Deficient MetQ
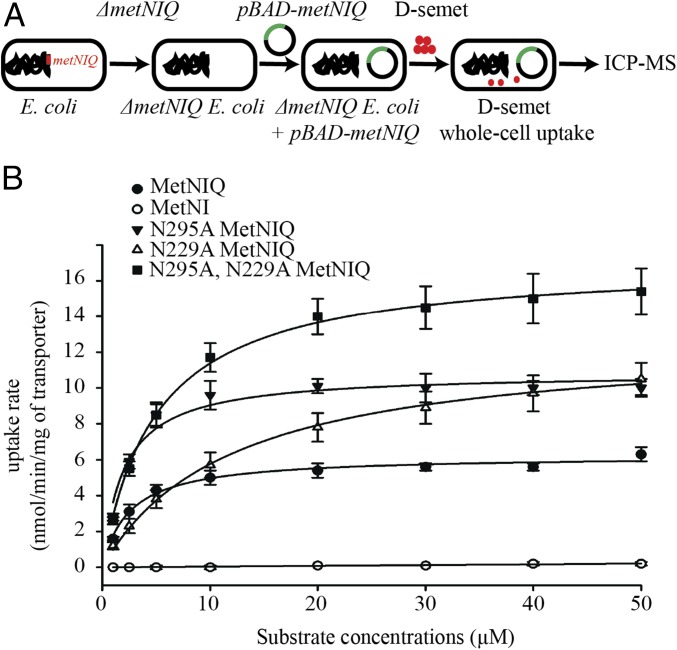
To address the transport function of MetNI, we carried out in vivo transport assays using a ΔmetNIQ knockout E. coli strain. There are two transport systems in E. coli that are responsible for methionine uptake (4): the higher-affinity MetD transport system (MetNIQ operon) and the lower-affinity, and as yet uncharacterized, MetP system (36). The uptake of l-methionine is mediated by both systems, whereas d-methionine is specifically transported by MetD (2, 4, 36). To selectively study MetNI transport, we took advantage of the exclusive role of MetNI in d-methionine transport and used a d-methionine derivative, d-selenomethionine (d-semet), as the transport substrate for quantification by inductively coupled plasma-mass spectrometry (ICP-MS) (Fig. 1A). Kinetic constants for the transport activity of d-semet derived from Michaelis–Menten analysis (Fig. 1B) are shown in Table 1. The maximal uptake rate of the wild-type MetNIQ is 6.3 ± 0.4 nmol⋅min−1⋅mg−1 of transporter, with Km = 1.8 µM (n = 3) (Fig. 1B and Table 1). Because l-methionine is present in the cytoplasm at 0.1–0.3 mM (1, 37), to bypass the potential transinhibition effect, an N295A mutation at the C2 domain of the MetNI was introduced that reduced the binding of l-methionine to undetectable levels as assessed by isothermal titration calorimetry (ITC). This variant results in a higher maximal uptake rate of 10 ± 0.5 nmol⋅min−1⋅mg−1 of transporter (n = 9) with no change in Km (Fig. 1B and Table 1). We infer this represents the uptake rate of d-methionine by MetNI in the absence of transinhibition regulation by the internal pool of l-methionine.
Fig. 1.
In vivo uptake assay of MetNIQ and its variants. (A) Schematic of whole-cell uptake assay for MetNIQ variants. (B) Michaelis–Menten plot of initial velocities for d-semet uptake versus substrate concentrations for MetNIQ and its variants. The corresponding Vmax and Km values are listed in Table 1.
Table 1.
Summary of kinetic constants for transport activity of MetNIQ variants
| MetNIQ variants | Uptake rate Vmax* | Km, µM |
| MetNIQ | 6.3 ± 0.4 | 1.8 ± 0.4 |
| MetNI | 0.2 ± 0.1 | N/A |
| N295A (EcN) MetNIQ | 10 ± 0.5 | 1.7 ± 0.3 |
| N229A (EcQ) MetNIQ | 10.5 ± 0.9 | 7.4 ± 0.5 |
| N295A, N229A MetNIQ | 15.4 ± 1.3 | 4.8 ± 0.7 |
| N295A, F103A (EcI) MetNIQ | — | — |
| N295A, M107A (EcI) MetNIQ | — | — |
| N295A, Y160A (EcI) MetNIQ | 14.0 ± 1.3 | 4.1 ± 0.2 |
| N295A, M163A (EcI) MetNIQ | 4.7 ± 0.5 | 1.4 ± 0.2 |
The maximum uptake rate (Vmax) and Michaelis–Menten constant (Km) were determined from the plot shown in Fig. 1B. N/A, not applicable; dash, not detectable. The molar ratio of the MetQ:MetNI variants in these studies was measured to be ∼4.
nmol⋅min−1⋅mg−1 of transporter.
Deletion of MetQ significantly reduced the uptake rate of d-semet to 0.2 ± 0.1 nmol⋅min−1⋅mg−1 of transporter (n = 3), which indicates that MetQ is required for the transport; however, an unanswered question is whether MetQ must bind substrate and deliver it to the transporter. To address this question, we generated a substrate-binding deficient variant of MetQ, N229A (NA MetQ), that reduces the affinity of MetQ for d-methionine by ∼20-fold and for d-semet by approximately eightfold compared with the wild-type MetQ (SI Appendix, Fig. S1). Unexpectedly, the N229A MetNIQ mutation stimulates the maximal transport rate for d-semet (Vmax = 10.5 ± 0.9 nmol⋅min−1⋅mg−1 of transporter) (n = 6) by ∼1.5-fold compared with the wild-type MetNIQ (Vmax = 6.3 ± 0.4 nmol⋅min−1⋅mg−1 of transporter), with an approximately fourfold increase in Km to 7.4 µM (Fig. 1B and Table 1). A similar stimulation is observed for the MetQ N229A variant in combination with the MetN N295A MetNIQ (Vmax = 15.4 ± 1.3 nmol⋅min−1⋅mg−1 of transporter) (n = 6) compared with the MetN N295A MetNIQ (Vmax = 10 ± 0.5 nmol⋅min−1⋅mg−1 of transporter) (Fig. 1B and Table 1). These results indicate that the binding protein can facilitate substrate uptake through mechanisms in addition to the canonical role where the free SBP delivers the substrate to the transporter.
Conformational Changes in the Outward-Facing State
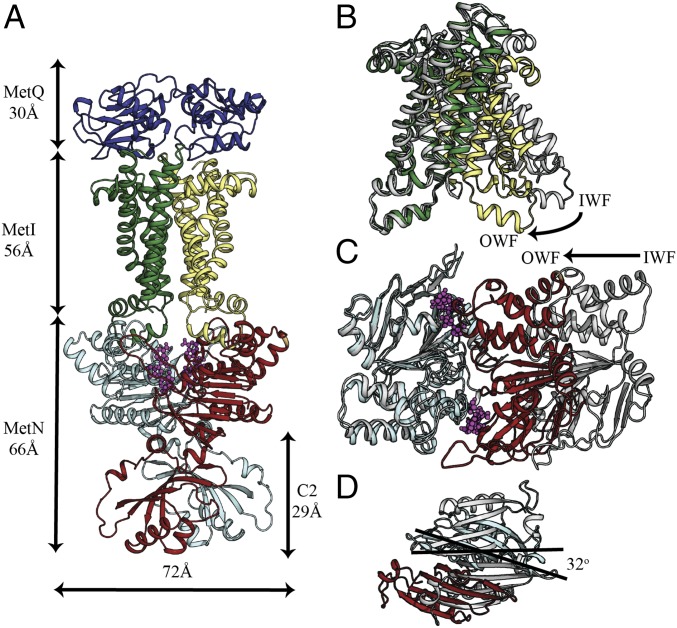
To complement our functional data, we determined the 2.95-Å resolution crystal structure of the ATPγS-bound E166Q N295A MetNI in complex with N229A MetQ (Fig. 2A). As anticipated, MetNI adopts an outward-facing (OWF) conformation exhibiting significant conformational changes relative to the uncomplexed MetNI in the inward-facing (IWF) state (Fig. 2 B–D). A convenient reference frame is the molecular twofold axis that relates the pairs of MetNI subunits in each conformation. Relative to the IWF conformation, the MetI subunits in the MetNIQ complex have rotated ∼31° toward the molecular twofold axis (SI Appendix, Fig. S2) opening the translocation pathway toward the periplasm and closing it at the cytoplasm. Relative to the transmembrane domains, the nucleotide binding domains (NBDs) are rotated ∼18° toward the molecular twofold axis in going from the IWF to OWF conformation, facilitating dimerization of two NBDs in the presence of ATPγS.
Fig. 2.
Crystal structure of the MetNIQ complex. (A) Side-view representation of the MetNIQ. MetQ subunit is colored slate, MetI subunits are forest and pale yellow, and MetN subunits are cyan and firebrick. C-regulatory (C2) domains are at the C termini of MetN subunits. Conformational changes of (B) the transmembrane MetI subunits, (C) the nucleotide-binding MetN subunits, and (D) the C2 domains between their OWF (colored as described in A) and IWF conformations (PDB ID code 3TUJ) (colored gray). One subunit of MetI (colored forest) or MetN (colored cyan) of the OWF conformation was overlaid to that of the IWF conformation to highlight the differences in the relative placement of the opposite subunit (B and C). Superposition of MetNIQ and MetNI reveals the rotation of the C2 domains around the molecular twofold axis between the two structures (D).
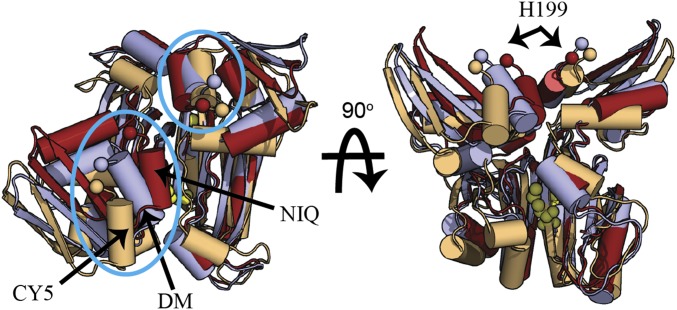
MetNI-catalyzed transport is regulated by the intracellular concentration of methionine, a phenomenon known as transinhibition (1–3), which is mediated by the C2 domains located at the C-terminal end of the MetN subunits (8, 38, 39). Between the IWF [Protein Data Bank (PDB) ID codes 3DHW, 3TUI, and 3TUZ] and OWF conformations, the C2 domains rotate ∼32° around the molecular twofold axis (SI Appendix, Fig. S2); this rearrangement is associated with the binding of l-methionine across the interface between C2 domains in the IWF conformation (SI Appendix, Fig. S3A). The transition to the OWF conformation is accompanied by a repositioning of the C2 domains arising from a register shift (38) in the hydrogen-bonding network across the antiparallel β-sheet stabilizing the C2 dimer (SI Appendix, Fig. S3B). The toggling of the C2 dimer between these two states in the absence and presence of methionine shifts the separation between the two H-motif catalytic residues H199 from 13 to 24 Å (Cα to Cα), respectively. Although H199 and the adjacent 200-helix (residues 200–205) are part of the NBDs, each 200-helix maintains a fixed position with respect to a single C2 subunit (Fig. 3). Consequently, the shift in β-sheet register leads to a significantly increased separation between the H199 residues in the presence of methionine.
Fig. 3.
Rotation of the C2 domains facilitates ATP-induced dimerization of MetN subunits. Comparison of the C2 domains following superposition of one of the two C2 domain subunits. C2 domains are presented in the locked transinhibited IWF state (colored wheat; PDB ID code 3TUZ), the unlocked IWF state (colored light blue; PDB ID code 3TUJ), and MetNIQ in the OWF state [colored firebrick, structure 3; PDB ID code 6CVL (this study)]. The superimposed subunit is to the right in each panel. This comparison highlights the significant differences in the two subunits between these states. Of note, the separation between the 200-helices (cyan ellipses, Left) in the unlocked OWF MetNIQ (NIQ) and IWF DM states is significantly shorter than in the IWF locked state (CY5) in the presence of methionine. As a consequence, the catalytic H-motif H199 residues from both NBDs are too far apart in the locked state to form the ATPase active form of the NBDs. l-methionine bound to the regulatory sites on the C2 domains are depicted as yellow spheres.
A Substrate Access Channel in the Outward-Facing State
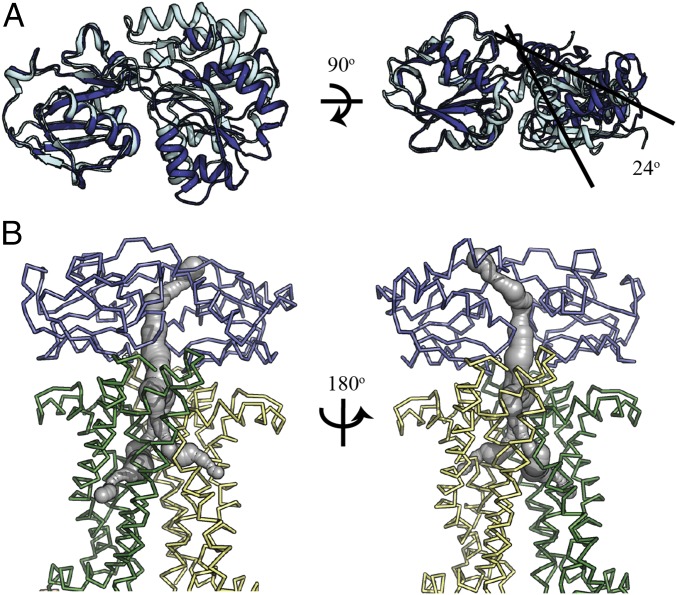
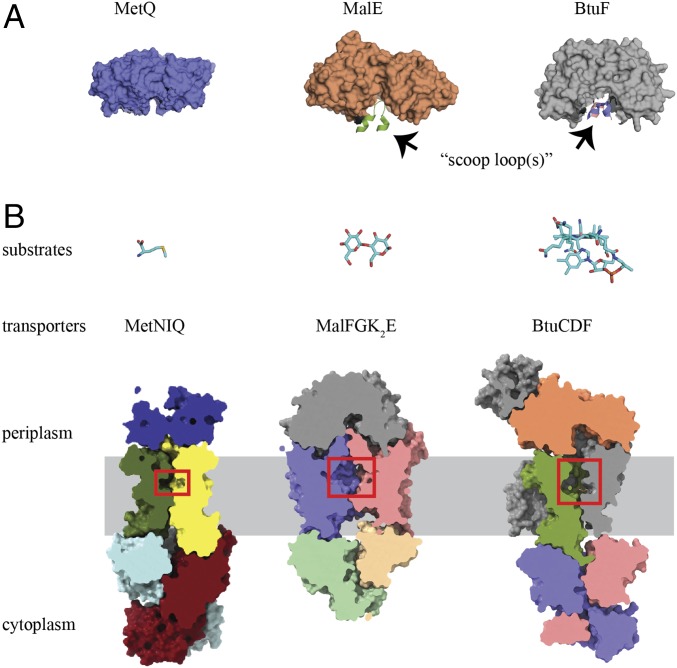
Significant rearrangements are also evident in the conformation of MetQ in the MetNIQ complex relative to the methionine-bound form. In contrast to the Venus flytrap-type of hinge bending motion that separates the two lobes of the MalE and BtuF SBPs in complexes with MalFGK2 and BtuCD, respectively, MetQ in the MetNIQ complex shows a 24° twist around an axis perpendicular to the interface between the two lobes (Fig. 4A). Significantly, this twisting motion creates an opening for substrate access through MetQ that is not present in the MalFGK2E and BtuCDF complexes. The rmsd between the methionine-bound form of MetQ and the conformation of MetQ in complex with MetNI is 4.4 Å, which, despite the smaller ligand size, is larger than observed between the corresponding liganded and transporter-bound forms for MalE (40) and BtuF (41, 42) with rmsds of 3.2 and 1.0 Å, respectively. The outward hinge-type movements of the MalE and BtuF SBP lobes, together with insertions of periplasmic or scoop loops from the transmembrane subunits (Fig. 5A), have been proposed to facilitate substrate delivery from the SBP to the maltose and vitamin B12 transporters (22, 41). In contrast, scoop loops are not present in the MetNIQ complex (Fig. 5A). The combination of the large distortion of MetQ upon complex formation with MetNI and the relatively small size of the methionine substrate may allow exogenous methionine to access the MetNIQ complex through MetQ. To explore this possibility, the CAVER 3.0.1 program (43) was used to identify several possible substrate entry pathways in MetNIQ, using a shell radius of 3 Å. Identification of a continuous channel starting at the cleft between the two lobes of the complexed MetQ suggests that methionine cannot only access the MetQ substrate binding site but can also reach the translocation pathway through MetI in the MetNIQ complex (Fig. 4B). This structure therefore provides evidence for a substrate entry pathway in an ABC importer–SBP complex through the SBP.
Fig. 4.
Conformational changes in MetQ creates substrate entry pathways in MetNIQ. (A) Superposition of the substrate-bound (colored cyan) and MetNI complexed MetQ (colored slate) shows a 24° twist between the two lobes that potentially provides access to potential substrate entry pathways. (B) Possible substrate entry pathways (shown in gray spheres) to the MetNIQ assembly were calculated by CAVER 3.0.2 pymol plugin with shell radii of 3 Å (43).
Fig. 5.
Translocation pathways of different ABC importers. (A) Surface slab views of the outward-facing conformations of three ABC importers, including MetNIQ (this work), MalFGK2 (PDB ID code 2R6G), and BtuCDF (PDB ID code 4FI3), reveal potential substrate-binding pockets (red box in B) with different sizes and shapes in the translocation pathways. Although MetI subunits form a small cavity, larger cavities are observed in the case of maltose and vitamin B12 transporters. Only maltose (shown in ball-and-stick) was structurally observed in the cavity. (B) Although a scoop loop was proposed to facilitate substrate handoff between the maltose- and vitamin B12-binding proteins with their transporters, no scoop loop is present in the methionine transporter.
Gating Mechanisms
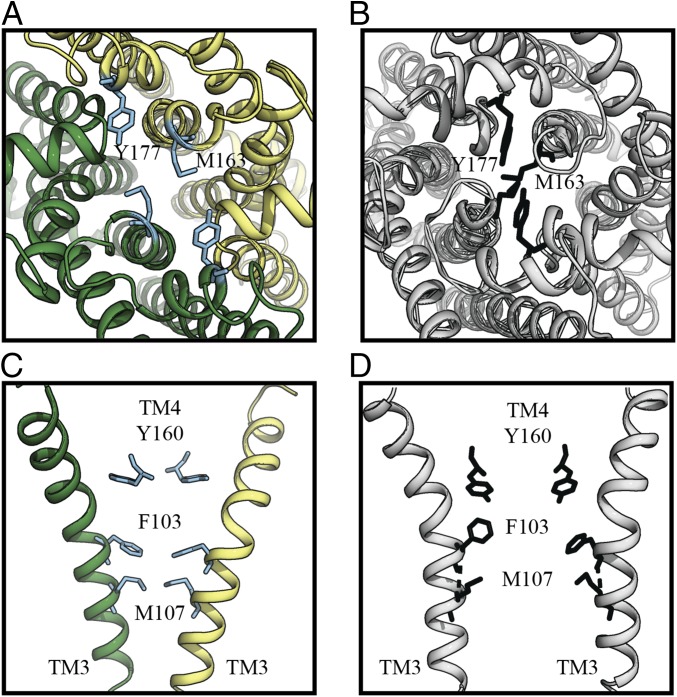
Interconversion of the inward- and outward-facing conformations of MetNI is associated with rearrangements of the cytoplasmic and periplasmic gates (Fig. 6). At the periplasmic gate, residues Y177 and M163 move away from the center of the translocation pathway (Fig. 6A), compared with those in the IWF conformation of MetNI (Fig. 6B). On the other side, gating residue M107 (TM3) from both MetI subunits form a cytoplasmic gate, not seen in the IWF conformation of MetNI (Fig. 6D), restricting the translocation pathway (Fig. 6C). Although substitution of the periplasmic gate M163 to A reduces maximal transport activity (Vm= 4.7 ± 0.5 nmol⋅min−1⋅mg−1 of transporter) (n = 3), the cytoplasmic gate mutation M107A severely disrupts transport activity (Table 1).
Fig. 6.
Conformational changes of the translocation pathway gates of the MetNIQ complex. Structure comparison of the MetI subunits in their outward-facing conformation (A and C) (colored forest and pale yellow) versus inward-facing conformation (B and D) (colored gray) reveals major rearrangements of the periplasmic gate residues, including Y177 and M163 (cyan; shown in ball-and-stick representation) (A and B), and of the cytoplasmic gate residues, including F103, M107, and Y160 (black; shown in ball-and-stick representation) (C and D).
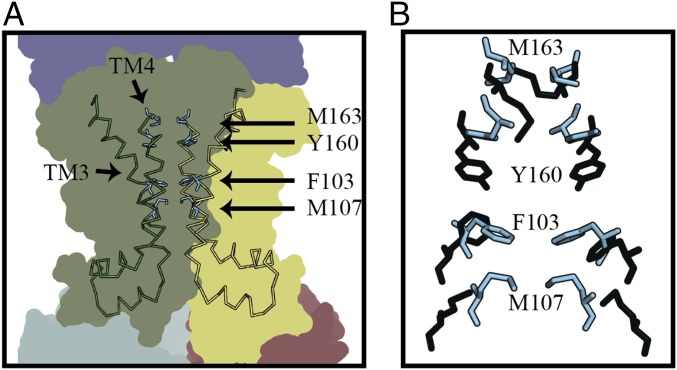
A cavity is present between the MetI subunits in the OWF conformation (Fig. 5B), whose boundary is defined by residues Y160 and F103 at the top and bottom of the cavity, respectively (Fig. 7A), along the translocation pathway. This cavity is smaller than those observed in the maltose and vitamin B12 transporters (40, 42) (Fig. 5B) and could accommodate a methionine. No binding of methionine was observed, however, in the translocation pathway of MetNIQ either in the original structure determination or after soaking crystals in selenomethionine, or as inferred biophysically by ITC measurements using the preformed MetNIQ complex with N229A MetQ to bypass binding of d-methionine to SBP MetQ. This is in contrast to the maltose MalFGK2 and arginine ArtQN transporters where substrates are found in the translocation pathway (40, 44, 45). Together with the gating residues M163 and M107, Y160 and F103 residues from both MetI subunits line the translocation pathway as if to form a selectivity filter, M-Ω-Ω-M (Ω, aromatic ring residue), for methionine selection (Fig. 7B). Aromatic rings from these residues may favor a selection toward the sulfur atom of the methionine residue, as seen in the binding pocket of holo-MetQ (17).
Fig. 7.
Residue rearrangement and potential substrate-binding cavity along the translocation pathway. (A) Movement of TM3 and TM4 of the MetI subunits form a small cavity (shown in Fig. 5A), whose boundary is defined by residues Y160 and F103 along the translocation pathway. (B) Superposition of MetI subunits in the OWF and IWF conformation reveals a rearrangement of a potential selectivity filter, M-Ω-Ω-M (Ω, aromatic ring residues), along the translocation pathway of the OWF MetNIQ. Side chains in the OWF and IWF conformations are colored cyan and black, respectively.
A Noncanonical Transport Mechanism for d-Methionine
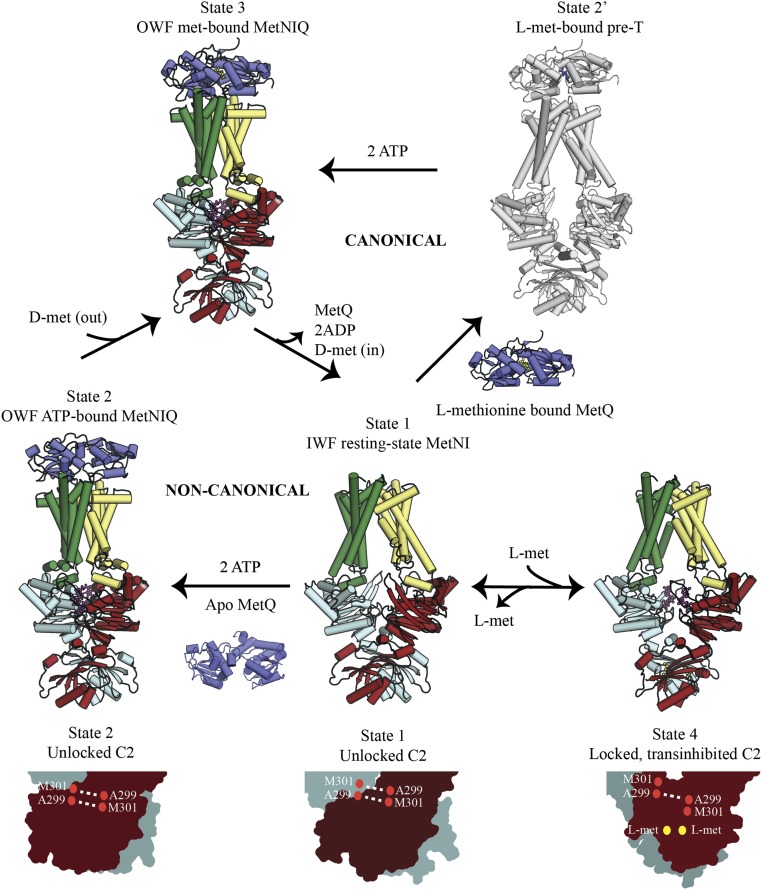
Detailed structural mechanisms have been developed for the transport cycles of MalFGK2E by Chen and coworkers (22) and BtuCDF by Locher and coworkers (23), reflecting a canonical role for the SBP in delivering substrate to the transporter. Our observations on the uptake of d-(seleno)methionine by MetNI support the presence of a distinct (noncanonical) mechanism where the initial substrate binding event is proposed to occur directly to the MetNIQ complex. The noncanonical and canonical models are compared in Fig. 8; a critical parameter distinguishing these pathways will be the affinity of the SBP for substrate, with the noncanonical pathway preferentially utilized by lower-affinity substrates such as d-(seleno)methionine and the canonical pathway favored by higher-affinity substrates such as l-methionine. Two intermediates are common to both pathways in this scheme, the IWF MetNI state 1 and the OWF MetNIQ state 3 containing bound substrate. The transition between state 3 and state 1 is accompanied by the translocation of substrate across the membrane. The distinction between the two pathways is provided by the precise sequence of events for the association of MetQ and substrate with the transporter. In the noncanonical pathway, unliganded MetQ associates with the ATP-bound form of MetNI to form state 2, corresponding to the MetNIQ structure determined in this work. d-(seleno)methionine from outside the cell then binds to generate the transport-competent state 3. In contrast, the canonical pathway proceeds through the association of liganded MetQ to MetNI to generate state 2′, which consequently contains both SBP and substrate. State 2′ has been modeled after the pretranslocation (pre-T) state of the maltose and molybdate transporters where binding of the liganded SBP precedes binding of ATP (22, 25); state 3 is then formed upon binding of ATP.
Fig. 8.
Mechanistic model for the MetNI-catalyzed transport of methionine derivatives. The relationship between the canonical and noncanonical pathways in the MetNI transport cycle is illustrated utilizing available structures of MetNI, MetQ, and MetNIQ represented as cylinders. Methionine, ADP, and ATP are shown as ball-and-sticks. Conformational states of the two C2 domains are shown in cyan and firebrick colored surface representations. State 1 represents a resting state of MetNI (PDB ID code 3TUJ). In the noncanonical pathway, state 2 represents an ATP-bound MetNIQ in the absence of methionine (this study; PDB ID code 6CVL). d-methionine can subsequently access the translocation pathway of the MetNIQ assembly through MetQ to form state 3 (also modeled as PDB ID code 6CVL). The canonical pathway proceeds through state 2′ depicted as a hypothetical pre-T conformation of l-methionine–bound IWF MetNIQ, based on structural studies of the maltose and molybdate transporters (22, 25). The transition from state 3 to state 1 is common to both pathways and associated with methionine transport into the cell. During the transport cycle, the C2 domains are in unlocked conformations that can accommodate the changes between IWF and OWF states. State 4 represents the transinhibited state of MetNI transporter (PDB ID code 3TUZ) with the C2 domains in a locked conformation stabilized by methionine binding and rearrangement of the β-sheet hydrogen bonding pattern at the interface between the C2 domains.
The regulation of the MetNI transport cycle through transinhibition arises from the binding of intracellular l-methionine to the C2 domains, resulting in transport termination. As established previously (38), the presence or absence of methionine toggles between two conformational states related by a register shift in the β-sheet hydrogen-bonding pattern at the dimer interface between C2 domains. In the transinhibited state 4, l-methionine bound to one C2 domain hydrogen bonds with N295 of the other (SI Appendix, Fig. S3A), helping stabilize a hydrogen-bonding network including between the peptide NH and CO groups of the two A299 residues on adjacent β strands. In the absence of bound l-methionine (states 1–3), the β-sheet hydrogen-bonding network between C2 domains shifts two residues, so that A299 is now paired with M301 (SI Appendix, Fig. S3B). As discussed above, the two conformations of the C2 domains are coupled to significant changes in the separation between the catalytic H-motif residues H199 and ultimately between the NBDs (Fig. 3). Transinhibition consequently reflects the increased distance between H199 residues in the presence of bound methionine, thereby precluding formation of the catalytically competent NBD dimer for ATP hydrolysis.
A Kinetic Transport Model for both High-Affinity and Low-Affinity Substrates
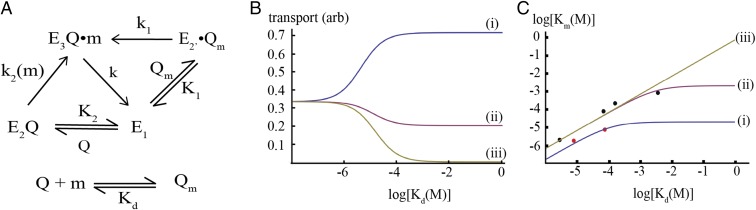
The ability of SBPs to mediate substrate uptake through both free (in the canonical model) and transporter-bound (in the noncanonical model) forms may represent a strategy to transport both high- and low-affinity substrates in physiologically relevant concentration ranges. A basic kinetic model (Fig. 9A) capturing these properties can be devised where the SBP (Q) can bind to the transporter (E1) in either substrate-bound (Qm) or substrate-free (Q) to form E2′Qm and E2Q, respectively. These complexes can then convert into a transport-competent form E3Q•m by isomerization (rate constant k1) and binding of substrate [rate constant k2(m)], which after transport returns to the initial E1 state. An informative comparison can be made between the two cases where the substrate can bind to EQ and when it cannot (the noncanonical and canonical pathways, defined by k2 > 0 and k2 = 0, respectively), using a typical substrate concentration of 10−5 M. For high-affinity substrates (Kd << 10−5 M), there is little difference between these models, but for weakly bound substrates, the ability of E2Q to bind substrates allows transport to occur under condition of high Kd where Qm and E2′Qm would only be present at low concentrations (Fig. 9B). Although the details will depend on the specific parameter values used, evaluation of Km as a function of Kd reveals that Km plateaus above a threshold Kd in this model (Fig. 9C), so that transport can occur at physiological concentrations even for substrates that are weakly bound by the SBP. Together with reports from the maltose and vitamin B12 uptake systems supporting the binding of substrate by the SBP–transporter complex (28, 30), these observations highlight an active role for both the SBP and transporter in the establishing the specificity and affinity of substrates that are imported by ABC transporters (33).
Fig. 9.
Kinetic analysis of canonical and noncanonical substrate uptake by ABC transporters. (A) Kinetic scheme modeling canonical and noncanonical pathways in substrate import by the transporter, E, where E1, E2, E2′, and E3 represent different states of the transporter as shown in Fig. 8. The methionine ligand (m) binds to Q to form the liganded species Qm with a dissociation constant Kd. Qm and Q can bind to E1 to form E2′Qm and E2Q, with effective dissociation constants K1 and K2, respectively. Transport occurs from the state E3Q•m that is generated either by isomerization of E2′Qm with the first-order rate constant k1 or by binding of m to E2Q with a second-order rate constant k2. The steady-state solution to this scheme for the case where E, E2′Qm, E2Q, Q, and Qm are at equilibrium is described in Materials and Methods. (B) Dependence of the transport rate for a substrate at 10 µM concentration on various parameters of the kinetic scheme in A, as a function of the Kd for binding of substrate to the SBP. Curves i, ii, and iii correspond to k2 = 5,000, 500, and 0 M−1⋅s−1, with Qtot = 10−4 M, K1 = K2 = 10−4 M, and k = k1 = 0.01 s−1. For substrates with high affinity to the SBP (Kd < ∼10−6 M) the models are equivalent, whereas for more weakly bound substrates (Kd > ∼10−4 M) the transport rate depends critically on the value of k2. Depending on the value of k2, the rate of transport of a weakly bound substrate can exceed that of a substrate that binds tightly to the SBP (compare curves i and ii). When k2 = 0 (curve iii), transport does not occur under these conditions, which corresponds to the case where substrate is only delivered to E1 by binding to the SBP. (C) Dependence of the Km for transport on the dissociation constant Kd for binding of substrate to the SBP. The red and black circles represent experimental data points for the methionine (this study) and maltose transporter (30), respectively. The curves are generated with the following parameters: i, k = 0.01 s−1, k1 = 0.1 s−1, and k2 = 1,000 M−1⋅s−1; ii, k = 0.1 s−1, k1 = 0.1 s−1, and k2 = 100 M−1⋅s−1; and iii, k = 0.1 s−1, k1 = 0.1 s−1, and k2 = 0 M−1⋅s−1, with Qtot = 10−4 M and K1 = K2 = 10−4 M for all curves. Curves were not explicitly fit to the experimental data but reflect parameter values that approximate the data. For this kinetic scheme, Km reaches a plateau as Kd increases, and depending on the parameter values, this plateau can be tuned to the physiological concentration range. For curve iii, where substrate does not bind to E2Q, a plateau region is not reached, so substrates with poor affinities for Q (high Kd) would have correspondingly high Km values.
Materials and Methods
Cloning, Expression, and Purification.
The metN and metI genes were amplified from E. coli K12 genomic DNA and cloned into a pET-modified dual vector, under two T7 promoters (pTN2) with the N-terminal hexahistidine tag sequence at the 5′ end of metN as described previously (8, 46). The metQ gene encoding for a mature MetQ with no signal sequence was cloned into a separate pET19b (+) plasmid with N-terminal decahistidine tag followed by an enterokinase-cleavage site. All of the mutations, including E166Q, N295A in MetN, and N229A in MetQ, were done by site-directed mutagenesis (Stratagene). The cloned plasmids were expressed separately in E. coli BL21-gold (DE3) cells (EMD) at room temperature in ZY media (47).
Purification of MetNI mutants was carried out by homogenizing 10 g cell paste in 100 mL of lysis buffer containing 20 mM Tris⋅HCl, pH 8, 100 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol (βMe), 20 μg/mL DNase I, 200 μg/mL lysozyme. The cells were broken by three passes through a cell disruptor, M110L pneumatic microfluidizer (Microfluidics). C12E7 was added at a final concentration of 0.5% (vol/vol) to solubilize membranes at 4 °C for 2 h. Unlysed cells and cell debris were removed by centrifugation at 100,000 × g, 4 °C, and 30 min. The supernatant was collected, 70 mM imidazole, pH 8.0, was added, and the sample was then loaded onto a 5-mL Ni-Sepharose HP column (GE Healthcare) equilibrated with the MetNI purification buffer containing 20 mM Tris⋅HCl, pH 7.5, 100 mM NaCl, 0.1% C12E7, 70 mM imidazole. After sample loading, the column was washed with 12 column volumes of the purification buffer. Two column volumes of elution buffer containing 20 mM Tris⋅HCl, pH 7.5, 100 mM NaCl, 0.1% C12E7, 300 mM imidazole were used to elute protein off the Ni column. The eluate was then subjected to size-exclusion chromatography. The eluted peaks were collected, concentrated, ultracentrifuged (267,000 × g, 20 min, 4 °C) to remove aggregation, and adjusted to 12 mg/mL. Three hundred-µL-per-tube aliquots of MetNI were flash frozen in liquid nitrogen and stored at −80 °C.
Purification of signal sequence cleaved MetQ was done by resuspending 10 g cell paste in 100 mL of lysis buffer as mentioned above. Cell lysis was done by freezing and thawing for three cycles in liquid nitrogen and in a 42 °C water bath, respectively. Clearing of cell lysates was done by centrifugation at 37,500 × g, 30 min, and 4 °C. After the supernatant was collected, 70 mM imidazole, pH 8.0, was added before loading onto a 5-mL Ni-Sepharose HP column (GE Healthcare) equilibrated with purification buffer containing 20 mM Tris⋅HCl, pH 7.5, 100 mM NaCl, 70 mM midazole. After sample loading, the column was washed with 12 column volumes of the same buffer. Two column volumes of elution buffer containing 20 mM Tris⋅HCl, pH 7.5, 100 mM NaCl, 300 mM imidazole were used to elute protein off the Ni column. The eluate was then subjected to size-exclusion chromatography. The eluted peaks were pooled, incubated with enterokinase (NEB) to cleave the His tag, and passed through a hand-packed Ni column again to remove uncut MetQ. The His-tag–cleaved MetQ was then concentrated to 20 mg/mL and flash-frozen in aliquots of 300 μL per tube.
Selenomethionine-Substituted Proteins.
To prepare selenomethionine-substituted MetNI and MetQ proteins, the cloned plasmids were transformed in E. coli auxotroph B834 (DE3) cells (EMD), which were then grown in phosphate-, amino acids-, and selenomethionine (PASM)-containing autoinduction media (47), containing 125 µg/mL selenomethionine for 3–5 d at room temperature.
Crystallization of the E. coli MetNIQ Protein Complex.
The native MetNIQ and selenomethionine-substituted MetNIQ complexes were prepared as described previously (17). Crystallization screens for MetNIQ were conducted at different concentrations, 5–20 mg/mL, by vapor diffusion in hanging drops at 20 °C using three different commercial kits, including MemGold2, MORPHEUS (Molecular Dimension), and PEGRx (Hampton Research). MetNIQ was crystallized in a reservoir containing 0.1 M Mes, pH 6, and 22% PEG 400, using 5 mg/mL protein at a ratio of protein to reservoir of 2:1. Crystals appeared after 2–3 d, fully grew after 5–7 d, and shattered after 10–14 d. Crystals were harvested at day 7 and cryoprotected by increasing PEG 400 concentration to 25, 30, and then 35%, followed by flash-freezing for data collection. Selenomethionine-substituted MetNIQ crystals often diffracted better than the native ones.
Heavy-Metal Derivatives.
MetNIQ crystals were soaked in crystallization buffer (0.1 M Mes, pH 6, and 22% PEG 400) containing 1 mM K2HgI4 for an hour before back-soaking, cryoprotecting, and flash-freezing.
Data Collection and Structure Determination.
All X-ray diffraction datasets were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) beamline 12-2 equipped with a PILATUS 6M pixel array detector. Diffraction images were processed and scaled with X-ray Detector Software (XDS) (SI Appendix, Table S1).
Several datasets diffracting anisotropically to 4–3 Å resolution were collected at wavelengths of 1.0000 Å (native crystals), 0.9794 Å (Semet-substituted crystals), and 1.00582 Å (Hg-soaked crystals). Ellipsoidal truncation and anisotropic scaling were applied to the MetNIQ diffraction datasets using the University of California, Los Angeles, Molecular Biology Institute Diffraction Anisotropy server (services.mbi.ucla.edu/anisoscale) (48) (SI Appendix, Fig. S4). All crystals belonged to space group P3221 and showed partial twinning with twinning fractions ranging from 0.10 to 0.28 determined by Xtriage (Phenix) (49); therefore, a twin law with operator -h, -k, l was applied. Initial phases were obtained by molecular replacement–single anomalous dispersion (SAD) (AutoSol, Phenix) using SAD data from a 3.8-Å selenomethionine derivative crystal and a MalFGK2-E (PDB ID code 2R6H)–derived model of MetNI in an OWF conformation as a partial model. SAD experimental phasing (Phaser-EP; Phenix) using heavy-atom (selenium) sites was carried out to reduce model bias (SI Appendix, Fig. S5). These phases were extended to 3.5 Å resolution and then 2.95 Å resolution using two other higher-resolution datasets, the latter collected from a K2HgI4-soaked crystal. Coupled with density modifications (AutoSol; Phenix), the latter showed clear electron densities for MetQ, which allowed us to manually dock the two individual lobes of MetQ. Validation of the sequence register was obtained from the selenium positions and the location of mercury sites near cysteines in MetN observed from a K2HgI4-soaked MetNIQ crystal. The MetNIQ model was refined using a combination of Phenix.refine (49), phenix.rosetta_refine (50), and Refmac5 (Collaborative Computation Project Number 4) (51). Although the densities of MetN and MetI subunits are well defined, that of MetQ is weaker (SI Appendix, Fig. S6). The absence of crystal contact in the MetQ of the MetNIQ crystal may result in some flexibility or even some orientational disorder. In general, the final model of MetNIQ fit the density calculated from the 2.95-Å resolution dataset well, and the final Rwork/Rfree = 0.21/0.23 (refinement statistics are in SI Appendix, Table S1).
Software used in the project was installed and configured by SBGrid (52).
Structure Comparisons.
The OWF conformation of MetNI determined in this work (PDB ID code 6CVL) was compared with IWF conformations determined previously (8, 38): unlocked C2 domains in the presence of the detergent n-decyl-β-d-maltoside (DM; PDB ID code 3TUJ) and locked C2 domains in the presence of the detergent cyclohexyl-pentyl-β-d-maltoside (CY5; PDB ID code 3TUI) and with bound selenomethionine (PDB ID code 3TUZ). The structure of methionine-bound MetQ (PDB ID code 4YAH) is described in Nguyen et al. (17).
In Vivo Transport Assays.
Whole-cell uptake assays were conducted following an established protocol by the Poolman laboratory (24). In brief, different arabinose-inducible (pBAD) plasmids that carry variants of metN, metI, and metQ genes were transformed into ΔmetNIQ E. coli strain BW2115. Cells were grown in LB media to an OD600 of 1.5, induced for 2 h with 0.2% arabinose, and harvested by centrifuging for 10 min at 5,000 × g, 4 °C. The cell pellets were then washed twice in 50 mM sodium phosphate buffer, pH 6.5, and resuspended to OD600 ∼ 10 in the same buffer. Before transport assays, cells were energized with 0.5% glucose and 5 mM MgCl2 for 10 min at 37 °C. The transport assays were started by adding d-selenomethionine (d-semet) at different concentrations at 37 °C, shaking at 300 rpm. The reactions were then stopped by centrifugation at 18,000 × g, 4 °C, for 1 min, followed by discarding the supernatant. The pellet was washed three more times in the same buffer before homogenizing in 20% HNO3. The samples were incubated at 85 °C overnight and diluted 10-fold for quantifying selenium content by ICP-MS (8800 QqQ; Agilent) [Environmental Analysis Center, California Institute of Technology (Caltech)]. The amount of MetNI and MetQ in each reaction was quantified by Western blot using an anti-His antibody against His-tagged MetN and MetQ, respectively. A correlation between different substrate concentrations and uptake rate was shown in a final Michaelis–Menten plot (Fig. 1B) and a summary table (Table 1). Data were processed using SigmaPlot 10.0 (SigmaPlot).
Isothermal Titration Calorimetry.
MetQ variants were dialyzed overnight in their purification buffers using a Slide-a-lyser mini dialysis device (Thermo Scientific). The samples were then ultracentrifuged at 267,000 × g, 4 °C, for 20 min to remove aggregates and the protein concentration adjusted to 100 µM. Substrate d-methionine was prepared in the last dialysis buffer of each sample. Titrations of d-methionine to MetQ variants were done in triplicate on a MicroCal iTC-200 calorimeter at 25 °C. Titration curves were shown in SI Appendix, Fig. S1. Data were processed using Origin v7.0 (OriginLab).
Steady-State Rate Solution to Kinetic Scheme in Fig. 9.
E1, E2Q, E2′Qm, Q, and Qm are assumed to be in equilibrium with dissociation constants defined in Fig. 9A. The total SBP concentration, Qtot = (Q) + (Qm), is assumed to be much larger than the total transporter concentration, Etot = (E1) + (E2Q) + (E2′Qm) + (E3Q•m), and so the contributions of (E2Q), (E2′Qm), and (E3Q•m) to Qtot are neglected.
The concentrations of (E3Q•m) and (X) = (E1) + (E2Q) + (E2′Qm) may be determined using the steady-state treatment detailed in Segel (53) and Cha (54), yielding
The rate of transport [v = k(E3Q•m)] relative to the maximum possible rate (V° = kEtot) is
Putting this into standard form allows the evaluation of Vmax and Km (Vmax in this context represents the fraction of the maximum possible rate V° ):
In the limits as Kd → 0 and Kd → ∞, the values of Km approach
so that for high-affinity substrates (Kd → 0), Km ∝ Kd, and for low-affinity substrates, Km plateaus to a constant value.
This scheme focuses on the interactions of the transporter, SBP, and ligand m and omits many features of the transport cycle, including the binding of ATP, ATP hydrolysis, and dissociation of the products ADP and Pi, and the interconversion of inward- and outward-facing conformations. Because there is a strong nucleotide state dependence to SBP binding, the dissociation constants in scheme Fig. 9A have been approximated as effective dissociation constants described by the ratio of the dissociation rate constants for E2Q or E2′Qm and the association rate constant between E1 and Q or between E1 and Qm. For MetNIQ, the association rate constant is ∼103 M−1⋅s−1, independent of whether MetQ has bound methionine or not and independent of whether MetNI is in the ATP or nucleotide free states (29, 55). Under turnover conditions, dissociation of the complex will be set by the rate of ATP hydrolysis, which has been measured to be ∼25 min−1 or ∼0.3 s−1 (39) so that as an order of magnitude estimate, K1 ∼ K2 ∼ 10−4 M−1. For comparison, the dissociation constants for the binding of unliganded and liganded MetQ to the ATP-bound form of MetNI are 27 nM and 1 µM (17), respectively, when ATP hydrolysis is prevented through either mutation or absence of Mg+2. The calculations for Fig. 9 were performed with Mathematica version 9.0.0.0.
The transport rate determined for the N295A MetNI transporter with d-selenomethionine [∼10 nmol⋅min−1⋅mg−1 (Table 1)] is equivalent to a turnover time of ∼0.02 s−1, which provides an estimate for the rate constant k; this rate is consistent with measurements made on other ABC transporters, with turnover times of ∼0.002–0.03 s−1 (24, 26, 56–58).
Data and Materials Availability.
Coordinates and structure factors have been deposited in the Protein Data Bank of the Research Collaboratory for Structural Bioinformatics, with PDB ID code 6CVL. The MetNI and MetQ constructs have been deposited with Addgene, with IDs 118269 (N295A E166Q MetNI) and 118268 (N229A MetQ) for the structural work, and IDs 118253, 118254, 118256–118261 and 118581 for the transport assay (Table 1).
Supplementary Material
Acknowledgments
We thank Dr. Raymond Liu for critical reading of the manuscript and Dr. Nathan Dalleska and Stefan Petrovic for assistance with ICP-MS and ITC, respectively. A Vietnam International Education Development scholarship from the Vietnam Ministry of Education and Training (to P.T.N.) is gratefully acknowledged. We gratefully acknowledge the Gordon and Betty Moore Foundation and the Beckman Institute at Caltech for their generous support of the Molecular Observatory at Caltech and the staff at Beamline 12-2, SSRL, for their assistance with data collection. SSRL is operated for the US Department of Energy and supported by its Office of Biological and Environmental Research and by the NIH, National Institute of General Medical Sciences (P41GM103393), and the National Center for Research Resources (P41RR001209). We thank the Center for Environmental Microbial Interactions for their support of microbiology research at Caltech. D.C.R. is an investigator at Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6CVL). The MetNI and MetQ constructs generated in this work have been deposited in Addgene [ID codes 118268 (N229A MetQ) and 118269 (N295A E166Q MetNI) for the structural work, and ID codes 118253, 118254, 118256–118261, and 118581 for the transport assay].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811003115/-/DCSupplemental.
References
- 1.Kadner RJ. Transport systems for L-methionine in Escherichia coli. J Bacteriol. 1974;117:232–241. doi: 10.1128/jb.117.1.232-241.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadner RJ. Transport and utilization of D-methionine and other methionine sources in Escherichia coli. J Bacteriol. 1977;129:207–216. doi: 10.1128/jb.129.1.207-216.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadner RJ. Regulation of methionine transport activity in Escherichia coli. J Bacteriol. 1975;122:110–119. doi: 10.1128/jb.122.1.110-119.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadner RJ, Watson WJ. Methionine transport in Escherichia coli: Physiological and genetic evidence for two uptake systems. J Bacteriol. 1974;119:401–409. doi: 10.1128/jb.119.2.401-409.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgins CF. ABC transporters: From microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 6.Locher KP. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23:487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 7.Rees DC, Johnson E, Lewinson O. ABC transporters: The power to change. Nat Rev Mol Cell Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC. The high-affinity E. coli methionine ABC transporter: Structure and allosteric regulation. Science. 2008;321:250–253. doi: 10.1126/science.1157987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson AL, Chen J. ATP-binding cassette transporters in bacteria. Annu Rev Biochem. 2004;73:241–268. doi: 10.1146/annurev.biochem.73.011303.073626. [DOI] [PubMed] [Google Scholar]
- 10.Merlin C, Gardiner G, Durand S, Masters M. The Escherichia coli metD locus encodes an ABC transporter which includes Abc (MetN), YaeE (MetI), and YaeC (MetQ) J Bacteriol. 2002;184:5513–5517. doi: 10.1128/JB.184.19.5513-5517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gál J, Szvetnik A, Schnell R, Kálmán M. The metD D-methionine transporter locus of Escherichia coli is an ABC transporter gene cluster. J Bacteriol. 2002;184:4930–4932. doi: 10.1128/JB.184.17.4930-4932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins CF. ABC transporters: Physiology, structure and mechanism–an overview. Res Microbiol. 2001;152:205–210. doi: 10.1016/s0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- 13.van der Heide T, Poolman B. ABC transporters: One, two or four extracytoplasmic substrate-binding sites? EMBO Rep. 2002;3:938–943. doi: 10.1093/embo-reports/kvf201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berntsson RPA, Smits SHJ, Schmitt L, Slotboom DJ, Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010;584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 15.Maqbool A, et al. The substrate-binding protein in bacterial ABC transporters: Dissecting roles in the evolution of substrate specificity. Biochem Soc Trans. 2015;43:1011–1017. doi: 10.1042/BST20150135. [DOI] [PubMed] [Google Scholar]
- 16.Trötschel C, et al. Methionine uptake in Corynebacterium glutamicum by MetQNI and by MetPS, a novel methionine and alanine importer of the NSS neurotransmitter transporter family. Biochemistry. 2008;47:12698–12709. doi: 10.1021/bi801206t. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen PT, et al. The contribution of methionine to the stability of the Escherichia coli MetNIQ ABC transporter-substrate binding protein complex. Biol Chem. 2015;396:1127–1134. doi: 10.1515/hsz-2015-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams WA, et al. The membrane-associated lipoprotein-9 GmpC from Staphylococcus aureus binds the dipeptide GlyMet via side chain interactions. Biochemistry. 2004;43:16193–16202. doi: 10.1021/bi048877o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heppel LA. The effect of osmotic shock on release of bacterial proteins and on active transport. J Gen Physiol. 1969;54:95–113. doi: 10.1085/jgp.54.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao B, Pear MR, McCammon JA, Quiocho FA. Hinge-bending in L-arabinose-binding protein. The “Venus’s-flytrap” model. J Biol Chem. 1982;257:1131–1133. [PubMed] [Google Scholar]
- 21.Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: Variation of common themes. Mol Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 22.Oldham ML, Chen J. Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science. 2011;332:1202–1205. doi: 10.1126/science.1200767. [DOI] [PubMed] [Google Scholar]
- 23.Korkhov VM, Mireku SA, Veprintsev DB, Locher KP. Structure of AMP-PNP-bound BtuCD and mechanism of ATP-powered vitamin B12 transport by BtuCD-F. Nat Struct Mol Biol. 2014;21:1097–1099. doi: 10.1038/nsmb.2918. [DOI] [PubMed] [Google Scholar]
- 24.Gouridis G, et al. Conformational dynamics in substrate-binding domains influences transport in the ABC importer GlnPQ. Nat Struct Mol Biol. 2015;22:57–64. doi: 10.1038/nsmb.2929. [DOI] [PubMed] [Google Scholar]
- 25.Hollenstein K, Frei DC, Locher KP. Structure of an ABC transporter in complex with its binding protein. Nature. 2007;446:213–216. doi: 10.1038/nature05626. [DOI] [PubMed] [Google Scholar]
- 26.Borths EL, Poolman B, Hvorup RN, Locher KP, Rees DC. In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry. 2005;44:16301–16309. doi: 10.1021/bi0513103. [DOI] [PubMed] [Google Scholar]
- 27.Borths EL, Locher KP, Lee AT, Rees DC. The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter. Proc Natl Acad Sci USA. 2002;99:16642–16647. doi: 10.1073/pnas.262659699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goudsmits JMH, Slotboom DJ, van Oijen AM. Single-molecule visualization of conformational changes and substrate transport in the vitamin B12 ABC importer BtuCD-F. Nat Commun. 2017;8:1652. doi: 10.1038/s41467-017-01815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewinson O, Lee AT, Locher KP, Rees DC. A distinct mechanism for the ABC transporter BtuCD-BtuF revealed by the dynamics of complex formation. Nat Struct Mol Biol. 2010;17:332–338. doi: 10.1038/nsmb.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao H, Duong F. Discovery of an auto-regulation mechanism for the maltose ABC transporter MalFGK2. PLoS One. 2012;7:e34836. doi: 10.1371/journal.pone.0034836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Covitz KM, et al. Mutations that alter the transmembrane signalling pathway in an ATP binding cassette (ABC) transporter. EMBO J. 1994;13:1752–1759. doi: 10.1002/j.1460-2075.1994.tb06439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Treptow NA, Shuman HA. Genetic evidence for substrate and periplasmic-binding-protein recognition by the MalF and MalG proteins, cytoplasmic membrane components of the Escherichia coli maltose transport system. J Bacteriol. 1985;163:654–660. doi: 10.1128/jb.163.2.654-660.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merino G, Shuman HA. Unliganded maltose-binding protein triggers lactose transport in an Escherichia coli mutant with an alteration in the maltose transport system. J Bacteriol. 1997;179:7687–7694. doi: 10.1128/jb.179.24.7687-7694.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merino G, Shuman HA. Truncation of MalF results in lactose transport via the maltose transport system of Escherichia coli. J Biol Chem. 1998;273:2435–2444. doi: 10.1074/jbc.273.4.2435. [DOI] [PubMed] [Google Scholar]
- 35.Oldham ML, Chen S, Chen J. Structural basis for substrate specificity in the Escherichia coli maltose transport system. Proc Natl Acad Sci USA. 2013;110:18132–18137. doi: 10.1073/pnas.1311407110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, et al. A transporter of Escherichia coli specific for L- and D-methionine is the prototype for a new family within the ABC superfamily. Arch Microbiol. 2003;180:88–100. doi: 10.1007/s00203-003-0561-4. [DOI] [PubMed] [Google Scholar]
- 37.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson E, Nguyen PT, Yeates TO, Rees DC. Inward facing conformations of the MetNI methionine ABC transporter: Implications for the mechanism of transinhibition. Protein Sci. 2012;21:84–96. doi: 10.1002/pro.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang JG, Rees DC. The allosteric regulatory mechanism of the Escherichia coli MetNI methionine ATP binding cassette (ABC) transporter. J Biol Chem. 2015;290:9135–9140. doi: 10.1074/jbc.M114.603365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–521. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 41.Hvorup RN, et al. Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF. Science. 2007;317:1387–1390. doi: 10.1126/science.1145950. [DOI] [PubMed] [Google Scholar]
- 42.Korkhov VM, Mireku SA, Locher KP. Structure of AMP-PNP-bound vitamin B12 transporter BtuCD-F. Nature. 2012;490:367–372. doi: 10.1038/nature11442. [DOI] [PubMed] [Google Scholar]
- 43.Chovancova E, et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLOS Comput Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu J, Ge J, Heuveling J, Schneider E, Yang M. Structural basis for substrate specificity of an amino acid ABC transporter. Proc Natl Acad Sci USA. 2015;112:5243–5248. doi: 10.1073/pnas.1415037112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khare D, Oldham ML, Orelle C, Davidson AL, Chen J. Alternating access in maltose transporter mediated by rigid-body rotations. Mol Cell. 2009;33:528–536. doi: 10.1016/j.molcel.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 47.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 48.Strong M, et al. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 50.DiMaio F, et al. Improved low-resolution crystallographic refinement with Phenix and Rosetta. Nat Methods. 2013;10:1102–1104. doi: 10.1038/nmeth.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morin A, et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. Wiley; New York: 1993. [Google Scholar]
- 54.Cha S. A simple method for derivation of rate equations for enzyme-catalyzed reactions under the rapid equilibrium assumption or combined assumptions of equilibrium and steady state. J Biol Chem. 1968;243:820–825. [PubMed] [Google Scholar]
- 55.Li QW. 2017. Biophysical characterization of an ABC L-methionine transporter. PhD dissertation (Calif Inst Technol, Pasadena, CA)
- 56.Chen J, Sharma S, Quiocho FA, Davidson AL. Trapping the transition state of an ATP-binding cassette transporter: Evidence for a concerted mechanism of maltose transport. Proc Natl Acad Sci USA. 2001;98:1525–1530. doi: 10.1073/pnas.041542498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davidson AL, Nikaido H. Purification and characterization of the membrane-associated components of the maltose transport system from Escherichia coli. J Biol Chem. 1991;266:8946–8951. [PubMed] [Google Scholar]
- 58.Liu CE, Ames GFL. Characterization of transport through the periplasmic histidine permease using proteoliposomes reconstituted by dialysis. J Biol Chem. 1997;272:859–866. doi: 10.1074/jbc.272.2.859. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.