Significance
The mechanisms of how TRIM5α interferes with the integrity of the HIV-1 capsid to restrict HIV-1 infectivity remain poorly understood. We examined, at atomic resolution, the interactions with TRIM5α in the assembled capsid and the dynamics of capsid’s hexameric and pentameric building blocks. Remarkably, assemblies in the presence of the pentameric subunits are more rigid on microsecond to millisecond timescales at the sites of pentamer incorporation than tubes containing only hexamers. Furthermore, TRIM5α binding to capsid induces global rigidification and perturbs key intermolecular interfaces, essential for higher-order capsid assembly. TRIM5α thus uses several mechanisms to destabilize the capsid lattice, ultimately inducing its disassembly. Our results suggest that dynamic allostery plays a pivotal role in capsid assembly and HIV-1 infectivity.
Keywords: HIV-1 capsid, magic-angle spinning NMR, TRIM5α, CA protein assemblies, HIV-AIDS
Abstract
The host factor protein TRIM5α plays an important role in restricting the host range of HIV-1, interfering with the integrity of the HIV-1 capsid. TRIM5 triggers an antiviral innate immune response by functioning as a capsid pattern recognition receptor, although the precise mechanism by which the restriction is imposed is not completely understood. Here we used an integrated magic-angle spinning nuclear magnetic resonance and molecular dynamics simulations approach to characterize, at atomic resolution, the dynamics of the capsid’s hexameric and pentameric building blocks, and the interactions with TRIM5α in the assembled capsid. Our data indicate that assemblies in the presence of the pentameric subunits are more rigid on the microsecond to millisecond timescales than tubes containing only hexamers. This feature may be of key importance for controlling the capsid’s morphology and stability. In addition, we found that TRIM5α binding to capsid induces global rigidification and perturbs key intermolecular interfaces essential for higher-order capsid assembly, with structural and dynamic changes occurring throughout the entire CA polypeptide chain in the assembly, rather than being limited to a specific protein-protein interface. Taken together, our results suggest that TRIM5α uses several mechanisms to destabilize the capsid lattice, ultimately inducing its disassembly. Our findings add to a growing body of work indicating that dynamic allostery plays a pivotal role in capsid assembly and HIV-1 infectivity.
HIV, the pathogen that causes acquired immunodeficiency syndrome (AIDS), infects ∼37 million people globally. Mature virions contain conical capsids, which are assembled from ∼1,500 copies of the capsid protein (CA) and enclose the two copies of the RNA genome and accessory proteins. The capsid cores are pleomorphic with varied shapes and appearances (1). Cores contain varying numbers of CA hexamer units (∼216) and 12 pentamers (Fig. 1A), with the latter inducing curvature and enabling closure of the ovoid (2–4). CA comprises an N-terminal domain (NTD) and a C-terminal domain (CTD), which are connected by a flexible linker (5–8). Overall, four intermolecular interfaces are essential for the overall structure and the stability of the capsid: specific CTD-CTD contacts are responsible for stable dimer formation, setting up the oligomerization and interhexamer interactions at the pseudotwofold and pseudothreefold axes, while NTD-NTD and NTD-CTD contacts form intrahexamer interactions (8–10). Individual residues crucial in these interfaces have been identified and verified with respect to their functional importance by mutagenesis (1, 2, 11, 12). The NTDs line the outside of the assembly, while the CTDs connect hexameric units on its inside (4, 10). As a result, the NTD harbors the interaction epitopes for restriction factors, such as cyclophilin A (CypA) and tripartite motif isoform 5α (TRIM5α) (13, 14).
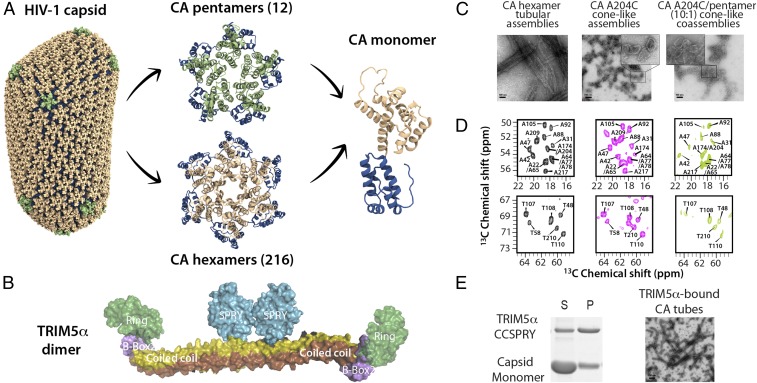
Fig. 1.
(A) Structure of a HIV-1 conical capsid and its building blocks. (B) Domain structure of TRIM5α. (C) TEM images of tubular assemblies of cross-linked hexamer CA (A14C/E45C/W184A/M185A) assemblies (Left), cone-like CA A204C assemblies (Middle), and cone-like coassemblies containing natural abundance CA A204C and 10% cross-linked U-13C,15N-pentamer CA (N21C/A22C/W184A/M185A) (Right). (D) Selected regions of 13C-13C correlation spectra of tubular hexamer CA assemblies (black), CA A204C assemblies (magenta), and coassemblies of A204C/cross-linked pentamer CA (light green). The first contour is set at 3.5 times the noise RMSD. (E) Pelleting assay of cross-linked hexamer CA/TRIM5α CC-SPRY tubular coassemblies and TEM of cross-linked hexamer CA tubular assemblies in the presence of TRIM5α CC-SPRY. P, pellet; S, supernatant. (Scale bars: 100 nm.)
Capsid cores are highly dynamic (2, 15–18), and motions in capsid assemblies occur over a wide range of timescales, from picoseconds to milliseconds and slower. These dynamics control capsid morphology and, in its final outcome, retroviral replication. We recently described the critical role of dynamics in the capsid’s escape from CypA (17) and specifically showed that motions on the microsecond-to-nanosecond timescale in the CypA binding loop play a role. Our results imply that by fine-tuning these motions via mutations (e.g., A92E and G94D), the virus can escape from CypA dependence. Similarly, the millisecond timescale motions in the NTD-CTD linker are essential for the CA protein to adopt multiple, slightly different conformations in the assembled capsid (15). In addition, conformational plasticity has been observed in the N-terminal β-hairpin and interhexameric interfaces (19, 20). The conformational plasticity in the NTD-CTD linker and in the interhexameric/pentameric interfaces (2, 4, 21) are important for the varied curvature in the conical capsid.
TRIM5α is a central cellular restriction factor for HIV/simian immunodeficiency virus (22) that functions as a capsid pattern recognition receptor and modulates its disassembly by interacting with the CA-NTD in the assembled capsid lattice (23–26). CA recognition is mediated by the PRY/SPRY domain (27–30), and its four flexible loops are believed to be important for pattern sensing (28, 31). Rhesus TRIM5α interacts with the HIV-1 capsid, while human TRIM5α has only modest affinity (22, 25, 32). TRIM5α recognizes capsids of different shapes and curvatures, and TRIM5α’s macromolecular arrangements and symmetry have been proposed to play roles in recognition and immune signaling (33, 34).
Despite extensive studies, the mechanistic details of capsid assembly and disassembly are not well understood. Furthermore, while biochemical and structural studies have provided insight into TRIM5α–capsid interactions (23, 26, 34, 35), atomic-level characterization has not been possible to date. Magic-angle spinning (MAS) nuclear magnetic resonance (NMR) is uniquely suited to elucidate the molecular details of the TRIM5α–capsid interactions and has proven to be a powerful technique for studying HIV-1 protein assemblies (36). Here we report direct, residue-specific evidence of intermolecular interactions between TRIM5α CC-SPRY (coiled-coil and SPRY domains; Fig. 1B) and tubular assemblies of capsid. Our data show that capsid undergoes conformational changes upon TRIM5α CC-SPRY binding, including structural and dynamic changes at the binding interface, as well as sites distal from the binding site. We postulate that the differences in dynamics may play a key role in controlling capsid’s stability and interactions with TRIM5α.
Our present results represent a significant advancement in understanding TRIM5α engagement of the HIV-1 capsid and support a growing body of work indicating that dynamic allostery is a key property of HIV-1 capsid assembly and capsid–cellular protein interactions.
Results
Hexameric and Pentameric Subunits Have Distinct Dynamic Signatures in Capsid Assemblies.
To evaluate the dynamic signatures of pentameric and hexameric building blocks in the conical capsid, we used covalently cross-linked units of CA hexamers (A14C/E45C/W184A/M185A) and pentamers (N21C/A22C/W184A/M185A) in assemblies used for MAS NMR. Crystal structures of the cross-linked hexamer and pentamer subunits were solved by Pornillos et al. (4, 10), revealing conformational differences at the NTD-NTD intrahexamer and CTD-CTD interhexamer interfaces. Cross-linked hexamers can efficiently assemble into tubes at high salt concentration (16) (Fig. 1C). These tubes yield high-quality MAS NMR spectra (Fig. 1D), similar to those previously observed for wild-type CA assemblies (16, 17, 37). CA A204C forms cone-like assemblies at high salt concentrations (2) (Fig. 1C). Using CA A204C and cross-linked pentamers at a 10:1 ratio, assemblies of cone-like morphology are formed similarly (Fig. 1C).
To selectively study pentamers in the context of these coassemblies, we prepared mixed labeled samples with uniformly 13C,15N-labeled cross-linked pentamers, and natural abundance A204C. This sample allowed us to collect direct, atomic-resolution data on the pentamers. The tubular assemblies of cross-linked CA hexamers and the cone-like assemblies of CA A204C served as control samples, reporting on the hexamer units in the context of these two different morphologies. As illustrated in Fig. 1D, the NMR spectra exhibit excellent resolution, suggesting significant local order. The chemical shifts in the 13C-13C correlation spectra of the A204C/cross-linked pentamer coassembly, A204C assembly, and cross-linked hexamer tubular assembly are similar, indicating no major changes in overall structures, although many residue-specific chemical shift differences are present. Not surprisingly, these differences are associated with residues near the mutation sites, for example, A42, A47, and T48 near the E45C amino acid change involved in the C-C crosslink. Interestingly, in A204C/cross-linked pentamer coassemblies, differences are also observed in resonances of residues distal from the mutation sites, indicating conformational changes in both the NTD and CTD. For instance, the resonance of A217 is different, possibly indicating conformational rearrangements in the helix 7/helix 11 NTD-CTD interface (10). In addition, residue T110 in helix 5 exhibits different chemical shifts with respect to the other two assemblies. These observations are consistent with the local structural differences for these regions detected in the X-ray crystal structures (10).
To further examine whether differences in dynamics may also be present, 15N-13C REDOR and 1H-13C DIPSHIFT experiments were recorded. Magnetization transfer profiles in these experiments are dependent on 15N-13C (REDOR) and 1H-13C (DIPSHIFT) dipolar couplings—that is, sensitive to motions on millisecond to microsecond and microsecond to nanosecond timescales, respectively (38). We evaluated Cα integrated intensities as a probe for dynamics. Remarkably, the 15N-13C REDOR dephasing curves (Fig. 2A) reveal that cross-linked pentamers in the coassemblies with A204C are more rigid than the hexamers on the millisecond to microsecond timescale. As shown in Fig. 2A, the REDOR dephasing for hexamers in the tubular and cone-like assemblies is essentially identical; however, pentamers in the A204C coassemblies exhibit distinctly different dephasing profiles.
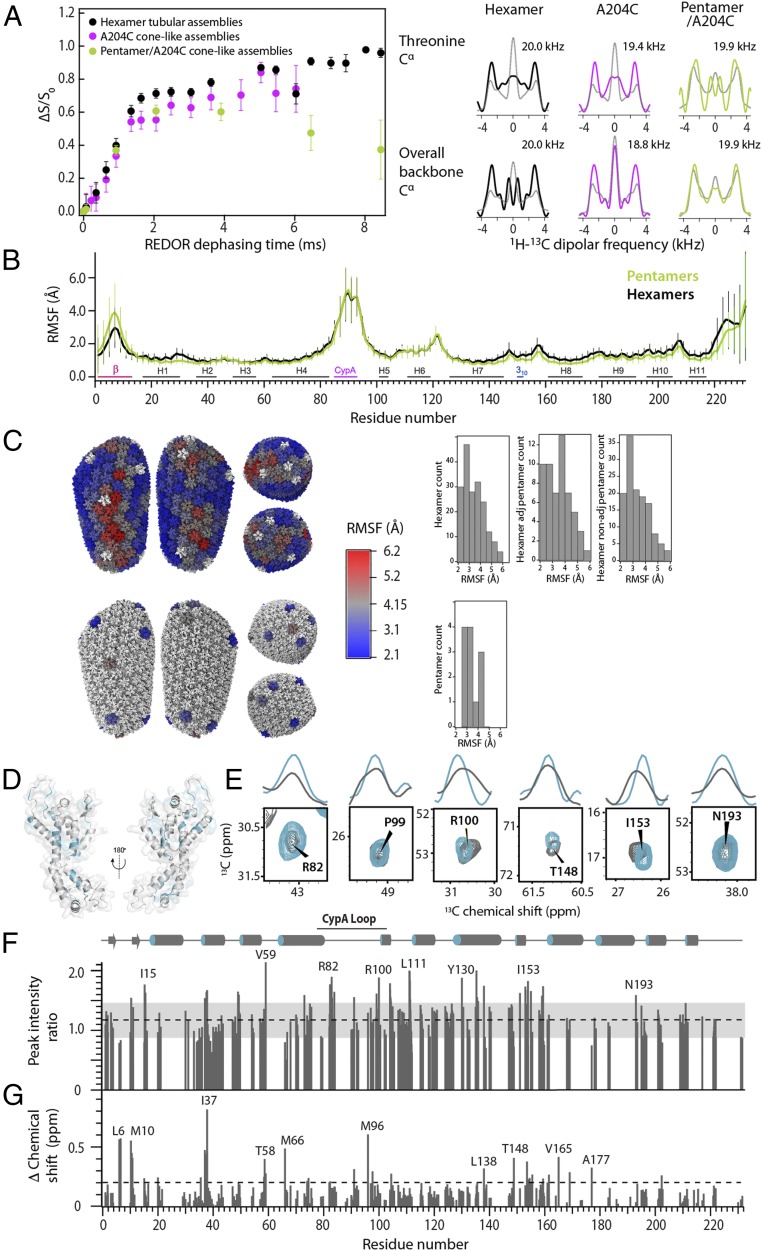
Fig. 2.
(A) 15NH-13Cα REDOR dephasing curves (Left) and 1H-13Cα DIPSHIFT line shapes (Right) for tubular assemblies of U-13C,15N cross-linked hexamers (black), cone-like assemblies of CA A204C (magenta), and cone-like coassemblies of 10:1 natural abundance A204C/U-13C,15N cross-linked pentamers (light green). The best-fitted 1H-13Cα dipolar line shapes are shown in gray, and the dipolar coupling constants are indicated. (B) RMSFs of CA oligomers [Protein Data Bank (PDB) ID code 3J3Q] over the 1.2-μs MD simulations vs. residue number. Hexamer and pentamer atoms are shown in black and light green, respectively. The RMSFs were calculated after minimizing the translational and rotational movements of the CA oligomers. The error bars represent RMSF variations over the entire ensemble of all hexamers and pentamers. (C) RMSF of CA hexamers (Top) and pentamers (Bottom) mapped onto the all-atom CA model and presented in histograms. The translational and rotational degrees of freedom of the whole capsid, rather than individual CA oligomers, were removed before RMSF calculations. (D) Summary of chemical shift and peak intensity differences between the tubular CA assemblies of cross-linked hexamers in the absence and presence of TRIM5α CC-SPRY, mapped onto an isolated CA monomer (PDB ID code 3J34). (E) Superpositions of select regions of the 2D 13C-13C correlation spectra of free (gray) and TRIM5α CC-SPRY–bound (cyan) tubular assembly of cross-linked CA hexamers. The first contour is set at three times the noise level. The 1D traces illustrate peak width and/or intensity changes. (F) Normalized peak intensity ratios extracted from the 13C-13C correlation spectra of TRIM5α CC-SPRY–bound CA tubular assemblies vs. TRIM5α CC-SPRY–free assemblies. Ratios >1 SD above or below the average (dashed line) are considered significant (outside the gray box). (G) 13C chemical shift changes of CA on binding of TRIM5α CC-SPRY, extracted from 13C-13C correlation spectra. The dashed line at 0.2 ppm marks the boundary between 13C chemical shift differences >1 SD above the average.
Interestingly, the REDOR curves also differ from those arising from pentamers in the coassemblies with cross-linked hexamers (SI Appendix, Fig. S1). While it is difficult to quantify dynamics from these curves, given the complexity of the system, the dephasing profiles of the pentamers clearly diverge from the hexamer profiles beyond 4 ms of dephasing time. It is possible that two different environments exist in the hexamers, a nearly rigid one and a more dynamic one, while for the pentamers, the profiles are consistent with a single nearly rigid-limit environment (SI Appendix). Further qualitative support for this interpretation is provided by the cross-polarization and double cross-polarization buildup curves (SI Appendix, Fig. S2). The 1H-13Cα DIPSHIFT experiments, in contrast, suggest that the backbone dynamics on the nanosecond to microsecond timescale is similar for hexamers and pentamers (Fig. 2A). Taken together, the NMR data suggest that the pentamers are more rigid than hexamers on the millisecond to microsecond timescale, while on the faster microsecond to nanosecond timescale, similar local backbone motions are present.
The motions in wild-type hexamer and pentamer units were further examined using all-atom molecular dynamics (MD) simulations. In particular, a 1.2-μs MD trajectory of the entire conical, fully solvated empty HIV-1 capsid (64,423,983 atoms) suggests the presence of motions (39). Root-mean-square fluctuations (RMSF) extracted from the MD trajectory for the pentamers exhibit lower values, and thus pentamers appear more rigid than hexamers on average (Fig. 2 B and C). The dynamics in the N-terminal domain, especially in the CypA binding loop and helices 2, 3, 5, and 6, are similar for hexamers and pentamers. The NTDs in hexamers and pentamers are structurally very similar; however, pentamers appear more condensed than hexamers. In the structures of the CA hexamers and pentamers (SI Appendix, Fig. S3), the average distances from the center of mass of the oligomer to every helix 4 are 36.6 Å and 33.9 Å, respectively. Similarly, the average distance from the center of mass of the oligomer to every helix 10 in the pentamer is 35.16 Å, significantly smaller than 48.18 Å, the value for the hexamer. The more compact pentamer results in closer contacts at the pseudothreefold interfaces in the CA lattice.
Thus, based on the NMR data shown in Fig. 2A, we determined that the cross-linked pentamers in the context of the cone-like A204C/pentamer coassembly are more rigid than cross-linked hexamers in tubular or cone-like assemblies, on the microsecond to millisecond timescale. Remarkably, the MD simulations also suggest that the pentamers are more rigid than the hexamers in the context of the conical capsid. Structurally, closer contacts at the pseudothreefold interface (2, 21) are present in the pentameric subunits, and it may well be possible that the differing dynamics of hexamers and pentamers play a role in capsid’s morphology and stability.
Conformational Changes in CA on TRIM5α CC-SPRY Binding: Mapping the Interaction Interface.
To investigate structural and dynamic changes induced by TRIM5α binding, CA assemblies of cross-linked hexamers were incubated with the CC-SPRY domain of TRIM5α. Cosedimentation studies confirmed complex formation (Fig. 1E, Left). Using cross-linked CA hexamers allowed us to assess the interaction with TRIM5α without disrupting the tubes seen with wild-type CA assemblies (40), as shown in the TEM image in Fig. 1E, Right. The resulting sample yielded well-resolved MAS NMR spectra comparable in quality to those of assembled CA tubes, shown above and previously (16, 17, 37). Using these 13C-13C and 15N-13C correlation datasets, chemical shift perturbations were detailed. Many changes were observed throughout the spectrum, including chemical shift and peak intensity changes (Fig. 2 D–G). Interestingly, the 13C chemical shift perturbations were relatively small (0.2–0.8 ppm; Fig. 2G), indicating that the secondary structure was not perturbed. A large number of changes in peak intensities was also observed (Fig. 2F), likely associated with changes in dynamics and/or the presence of multiple conformations. A significant number of resonances appear to sharpen upon binding (Fig. 2E), suggesting a global rigidification. Notable conformational changes occur in the CypA loop and at the NTD-NTD intrahexamer interface, as well as in the major homology region.
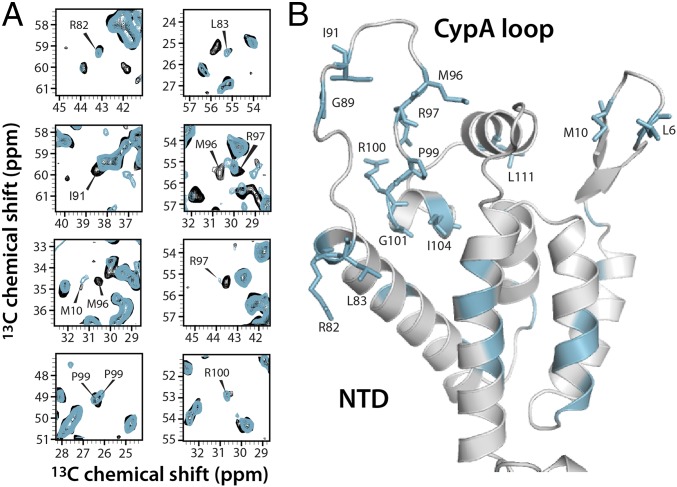
The CypA loop of CA is known to play an important role in binding of retroviral cofactors (41) and has been hypothesized to constitute an intermolecular interface with TRIM5α and other restriction factors in previous biochemical (35, 42), cryo-EM (26, 43), and NMR studies (17, 23). Here we observed a significant number of conformational and dynamic differences in the CypA loop and the connecting residues on binding of TRIM5α (Fig. 3). Specifically, R82, L83, and R100 peaks sharpen up in the CC-SPRY/CA spectrum, suggesting less motion of the associated hinge residues. Furthermore, resonances of residues residing in the loop, such as I91, M96, and R97, exhibit reduced peak intensities or missing intensities in the spectrum of the complex, indicative of slowed dynamics (i.e., motions on the nanosecond timescales are slowed down to microseconds to milliseconds). This observation is similar to the results of previous studies, where significant attenuation of dynamics in the CypA loop was observed on CypA binding (17). Moreover, a small number of residues, such as P99, exhibit two resonances, indicating the presence of two conformations in the CA-TRIM5α complex, possibly associated with binding of CC-SPRY to cis and trans proline conformers of CA in the CypA loop, as seen in solution (44). The data reported here provide atomic-level experimental evidence that motions of residues of the CypA loop in assembled capsid are directly affected by TRIM5α CC-SPRY binding. We note that the relevance of the structural perturbations observed here is borne out by previous biochemical and biophysical studies (23, 26, 35).
Fig. 3.
Structural changes in the CA NTD on TRIM5α CC-SPRY binding. (A) Superposition of select regions of the 13C-13C combined -driven (CORD) correlation spectra of free (gray) and TRIM5α CC-SPRY–bound (cyan) tubular assembly of cross-linked CA hexamers. Affected resonances are labeled by residue name and number. (B) Structural mapping of residues that experience chemical shift changes (cyan) onto the CA structure (PDB ID code 3J34).
In addition, chemical shift changes are also observed for L6 and M10 resonances, which are associated with residues in the N-terminal β1/β2 hairpin (residues 1–13). The presence of the β-hairpin has been proposed to be an important feature in the assembly of the mature viral core (45, 46) and may be exploited by TRIM5α as a molecular recognition motif (47), although this hypothesis is currently debated. A recent study suggested that the TRIM5α B30.2 domain recognizes the capsid through the capsid’s trimeric interface, conferring species-specific HIV-1 restriction activity (48).
Conformational Changes Remote from the Binding Interface.
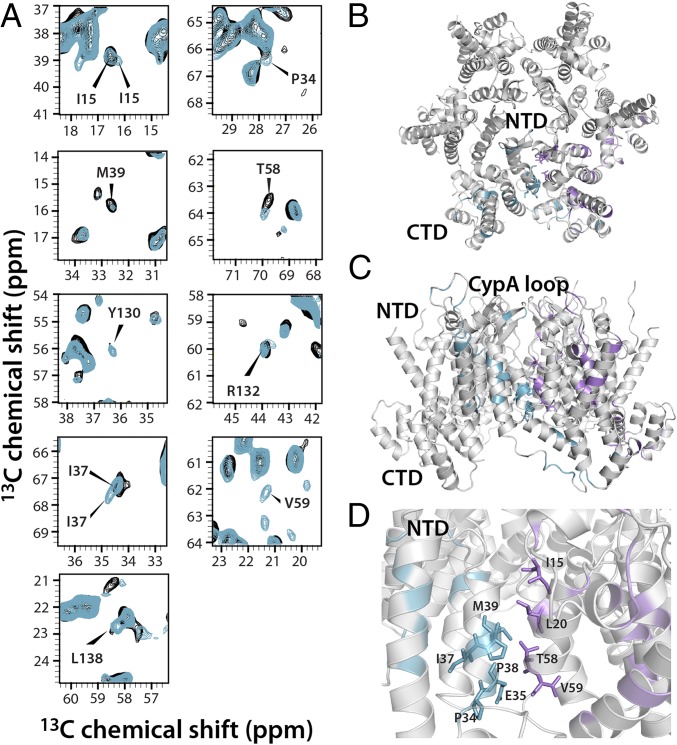
Interactions of TRIM5α CC-SPRY with the CA assemblies also induce conformational changes distal to the binding interface (Figs. 4 and 5). Numerous chemical shift changes are observed for residues at the NTD-NTD intrahexameric interface (Fig. 4). In the crystal structure of the cross-linked CA hexamer M39 and T58 engage in intermolecular contacts (10), which appear to be perturbed upon CC-SPRY binding. Many additional resonances associated with interface residues, such as L20, P34, E35, I37, P38, and V59, are affected as well. Mutations of these interface residues interfere with viral infectivity (49) or result in noninfectious viruses (50). Therefore, the observation of such allosteric perturbations upon TRIM5α CC-SPRY binding may suggest unfavorable consequences with respect to the integrity of the mature capsid core. Other changes are seen for helix 7 residues, close to helix 2, which may affect the relative positioning of these two helices (Fig. 4D). Changes noted for the I15 resonance may reflect a reorientation of helices 1 and 3, in line with previous reports on the importance of residue I15 for accurate core assembly (45, 46).
Fig. 4.
Chemical shift perturbations on TRIM5α CC-SPRY binding and structural mapping onto the CA hexameric building block at the NTD-NTD interface. (A) Superposition of select regions in the 13C-13C CORD correlation spectra of free (gray) and TRIM5α CC-SPRY–bound (cyan) tubular assemblies of cross-linked CA hexamers. Affected resonances are labeled with residue name and number. (B and C) Top (B) and side (C) views of the CA hexamer (PDB ID code 3J34) in ribbon representation. Residues whose Cα or Cβ resonances are affected are colored cyan in one monomer and in magenta in a neighboring monomer. (D) Expansion of the NTD-NTD interface, highlighting those residues that experience chemical shift changes. Affected residues are shown in stick representation and are labeled with residue name and number.
Fig. 5.
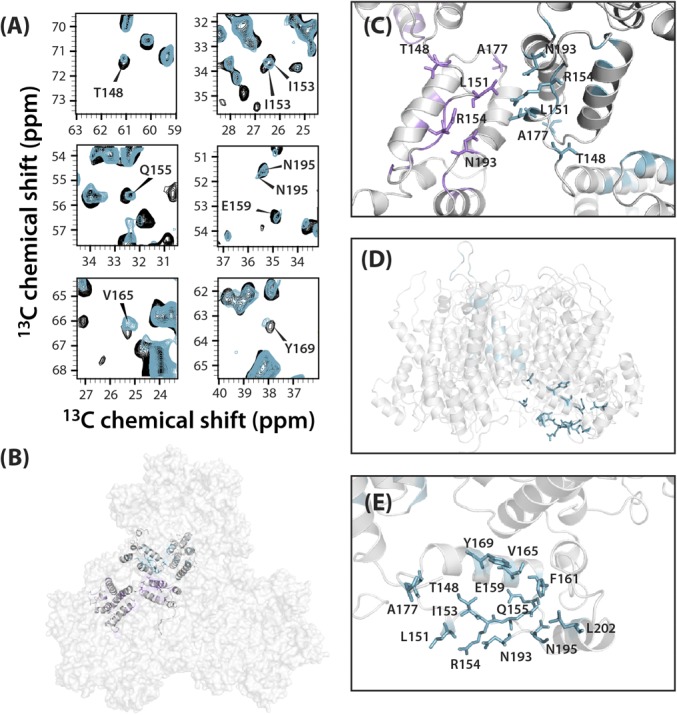
Chemical shift changes observed for resonances associated with the major homology region. (A) Superposition of select regions of the 13C-13C CORD correlation spectra of free (gray) and TRIM5α CC-SPRY–bound (cyan) tubular assemblies of cross-linked CA hexamers. Affected resonances are labeled with residue name and number. (B) Space-filling representation of a trimer of hexamers in tubular CA assemblies (PDB ID code 3J34) with two CA monomer chains at the interhexameric interface in ribbon representation. Residues near the interface whose resonances are affected by TRIM5α CC-SPRY binding are shown in magenta in one monomer and in cyan in the other monomer. (C) CTD-CTD interface in the tubular CA assembly (PDB ID code 3J34). (D) Side view of the hexameric subunit in the tubular CA assembly (PDB ID code 3J34), highlighting affected residues in the major homology region (MHR) and neighboring residues. (E) Expansion of MHR with affected residues shown in cyan stick representation.
Discussion
Using an integrated experimental MAS NMR and computational MD approach, we observed distinct dynamic signatures of cross-linked pentamers in cone-like coassemblies with CA A204C. The experimental NMR results indicate that cross-linked pentamers are more rigid than hexamers in tubular and cone-like assemblies on the microsecond to millisecond timescale. This finding agrees well with the results from MD simulation showing that pentamers fluctuate less than hexamers in the conical capsid. Furthermore, our previous MD study revealed the presence of “surface waves” on the capsid, dynamically connecting large regions of the capsid (39). Such correlated motions may provide a path for allosteric communication across the entire capsid and suggest that dynamic properties may play a role in the HIV-1 capsid core structure and stability. The reduced dynamics in pentamers may be a consequence of the closer contacts at pentamer/hexamer vs. hexamer/hexamer interfaces in the assembled capsid (2, 21). Indeed, a recent cryo-electron tomography study suggested that pentameric and hexameric capsid subunits expose different regions of CA, allowing host cell factors to interact differentially with the hexameric and pentameric subunits (21).
Based on our findings presented in this work, we hypothesize that different dynamics may also play roles in molecular recognition. Our results show that on binding of the restriction factor TRIM5α to CA tubular assemblies, numerous effects on structure and dynamics can be observed. In particular, we present direct evidence that TRIM5α CC-SPRY induces global attenuation of capsid motions, and that site-specific structural and dynamics changes are not limited to the protein-binding interface but occur throughout CA. The latter finding suggests that TRIM5α possibly uses several mechanisms to influence the structure and stability of the capsid core and ultimately its premature disassembly. Furthermore, our results show that TRIM5α CC-SPRY interacts with the CypA loop, attenuating its motions, which are known to be correlated with HIV-1 infectivity (17). TRIM5α binding also affects residues in critical assembly interfaces, including the NTD-NTD and NTD-CTD intrahexameric interfaces and the CTD-CTD interhexameric dimer interface, in accordance with the observation that TRIM5α does not bind appreciably to CA monomers (51). Indeed, the mechanisms of capsid recognition by TRIM5α involve higher-order capsid assembly (48, 26). While emerging evidence suggests that dynamics may play a role in capsid function (2, 15–18), in-depth atomic-level investigations of the relevant timescales are needed to understand how dynamic allostery may fine-tune infectivity. Thus, knowledge of the details of structural changes induced by TRIM5α to destabilize the capsid is essential and may guide the design of novel therapeutics.
Materials and Methods
Sample Preparation.
Detailed protocols for expression and purification of HIV-1 (A204C) CA, cross-linked (A14C/E45C/M184A/W185A) hexamers and (N21C/A22C/W184A/M185A) pentamers, and TRIM5α CC-SPRY and the preparation of the assemblies under study are given in SI Appendix.
TEM.
TEM analysis was performed with a Zeiss Libra 120 transmission electron microscope operating at 120 kV. Assemblies were stained with uranyl acetate (2% wt/vol), deposited onto 400-mesh formval/carbon-coated copper grids, and air-dried for 45 min. The copper grids were glow-discharged before staining, so that the assemblies can spread uniformly on the grid surface and adhere to it.
NMR Spectroscopy.
All samples were packed into Bruker 3.2-mm rotors. All 2D 13C-13C and 15N-13C spectra were acquired on a Bruker 19.9 T spectrometer operating at Larmor frequencies of 850.4 MHz (1H), 213.8 MHz (13C) and 86.2 MHz (15N), with a Bruker 3.2 mm E-free HCN probe at a sample temperature of 4 ± 1 °C. Detailed of experimental conditions, data processing, analysis, and numerical simulations are given in the SI Appendix.
MD Simulations.
The HIV-1 capsid, composed of 186 CA hexamers and 12 CA pentamers, was embedded in a water box with 150 mM sodium chloride, resulting in a simulation box of dimensions 70 nm × 76 nm × 121 nm, a total of 64,423,983 atoms, as described previously (2). HIV-1 subtype B, NL4-3 with the A92E mutation, was used for all CA monomers. The CHARMM36 force-field was used with the TIP3P water model at 310 K and 1 atm. Simulations carried out in the present study used the r-RESPA integrator available in NAMD 2.10. Long range electrostatic force calculations used the particle-mesh Ewald method, using a grid spacing of 2.1 Å and eighth-order interpolation, with a 1.2-nm cutoff. The simulations were done using an integration time-step of 2 fs, with nonbonded interactions evaluated every 2 fs and electrostatics updated every 4 fs; all hydrogen bonds were constrained with the SHAKE algorithm.
Supplementary Material
Acknowledgments
The authors thank Shannon Modla (Delaware Biotechnology Institute) for assistance with TEM and Sucharita Sarkar for help with the analysis of cross-polarization and double cross-polarization data. This work was supported by the National Institutes of Health (National Institute of General Medical Sciences and National Institute of Allergy and Infectious Diseases Grant 5P50GM082251). We acknowledge the National Science Foundation (Grant CHE0959496) for acquisition of the 850-MHz NMR spectrometer at the University of Delaware and the National Institutes of Health (Grants P30GM103519 and P30GM110758) for supporting the core instrumentation infrastructure at the University of Delaware. This work used the Extreme Science and Engineering Discovery Environment, which is supported by the National Science Foundation (Grant OCI-1053575). Specifically, it used the Bridges system, which is supported by the National Science Foundation (Grant ACI-1445606) at the Pittsburgh Supercomputing Center. C.M.Q. received support from the National Institutes of Health (Postdoctoral Fellowship Grant F32GM113452).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800796115/-/DCSupplemental.
References
- 1.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol. 2004;78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao G, et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 4.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gamble TR, et al. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 6.Gitti RK, et al. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 7.Gres AT, et al. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science. 2015;349:99–103. doi: 10.1126/science.aaa5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byeon IJL, et al. Structural convergence between cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 10.Pornillos O, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rihn SJ, et al. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog. 2013;9:e1003461. doi: 10.1371/journal.ppat.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wacharapornin P, Lauhakirti D, Auewarakul P. The effect of capsid mutations on HIV-1 uncoating. Virology. 2007;358:48–54. doi: 10.1016/j.virol.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 13.Sanz-Ramos M, Stoye JP. Capsid-binding retrovirus restriction factors: Discovery, restriction specificity and implications for the development of novel therapeutics. J Gen Virol. 2013;94:2587–2598. doi: 10.1099/vir.0.058180-0. [DOI] [PubMed] [Google Scholar]
- 14.Gamble TR, et al. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 15.Byeon IJL, et al. Motions on the millisecond time scale and multiple conformations of HIV-1 capsid protein: Implications for structural polymorphism of CA assemblies. J Am Chem Soc. 2012;134:6455–6466. doi: 10.1021/ja300937v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han Y, et al. Magic-angle spinning NMR reveals sequence-dependent structural plasticity, dynamics, and the spacer peptide 1 conformation in HIV-1 capsid protein assemblies. J Am Chem Soc. 2013;135:17793–17803. doi: 10.1021/ja406907h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu M, et al. Dynamic allostery governs cyclophilin A-HIV capsid interplay. Proc Natl Acad Sci USA. 2015;112:14617–14622. doi: 10.1073/pnas.1516920112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, et al. HIV-1 capsid function is regulated by dynamics: Quantitative atomic-resolution insights by integrating magic-angle-spinning NMR, QM/MM, and MD. J Am Chem Soc. 2016;138:14066–14075. doi: 10.1021/jacs.6b08744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayro MJ, Chen B, Yau WM, Tycko R. Site-specific structural variations accompanying tubular assembly of the HIV-1 capsid protein. J Mol Biol. 2014;426:1109–1127. doi: 10.1016/j.jmb.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deshmukh L, et al. Structure and dynamics of full-length HIV-1 capsid protein in solution. J Am Chem Soc. 2013;135:16133–16147. doi: 10.1021/ja406246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattei S, Glass B, Hagen WJ, Kräusslich HG, Briggs JA. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science. 2016;354:1434–1437. doi: 10.1126/science.aah4972. [DOI] [PubMed] [Google Scholar]
- 22.Stremlau M, et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 23.Biris N, Tomashevski A, Bhattacharya A, Diaz-Griffero F, Ivanov DN. Rhesus monkey TRIM5α SPRY domain recognizes multiple epitopes that span several capsid monomers on the surface of the HIV-1 mature viral core. J Mol Biol. 2013;425:5032–5044. doi: 10.1016/j.jmb.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sebastian S, Luban J. TRIM5α selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, et al. Structural insight into HIV-1 capsid recognition by rhesus TRIM5α. Proc Natl Acad Sci USA. 2012;109:18372–18377. doi: 10.1073/pnas.1210903109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roganowicz MD, et al. TRIM5α SPRY/coiled-coil interactions optimize avid retroviral capsid recognition. PLoS Pathog. 2017;13:e1006686. doi: 10.1371/journal.ppat.1006686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Li X, Stremlau M, Lee M, Sodroski J. Removal of arginine 332 allows human TRIM5α to bind human immunodeficiency virus capsids and to restrict infection. J Virol. 2006;80:6738–6744. doi: 10.1128/JVI.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 31.Biris N, et al. Structure of the rhesus monkey TRIM5α PRYSPRY domain, the HIV capsid recognition module. Proc Natl Acad Sci USA. 2012;109:13278–13283. doi: 10.1073/pnas.1203536109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sastri J, Campbell EM. Recent insights into the mechanism and consequences of TRIM5α retroviral restriction. AIDS Res Hum Retroviruses. 2011;27:231–238. doi: 10.1089/aid.2010.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganser-Pornillos BK, et al. Hexagonal assembly of a restricting TRIM5α protein. Proc Natl Acad Sci USA. 2011;108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li YL, et al. Primate TRIM5 proteins form hexagonal nets on HIV-1 capsids. eLife. 2016;5:e16269. doi: 10.7554/eLife.16269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohkura S, et al. Novel escape mutants suggest an extensive TRIM5α binding site spanning the entire outer surface of the murine leukemia virus capsid protein. PLoS Pathog. 2011;7:e1002011. doi: 10.1371/journal.ppat.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinn CM, et al. Magic-angle spinning NMR of viruses. Prog Nucl Magn Reson Spectrosc. 2015;86-87:21–40. doi: 10.1016/j.pnmrs.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han Y, et al. Solid-state NMR studies of HIV-1 capsid protein assemblies. J Am Chem Soc. 2010;132:1976–1987. doi: 10.1021/ja908687k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolodziejski W, Klinowski J. Kinetics of cross-polarization in solid-state NMR: A guide for chemists. Chem Rev. 2002;102:613–628. doi: 10.1021/cr000060n. [DOI] [PubMed] [Google Scholar]
- 39.Perilla JR, Schulten K. Physical properties of the HIV-1 capsid from all-atom molecular dynamics simulations. Nat Commun. 2017;8:15959. doi: 10.1038/ncomms15959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao G, Zhang P. CryoEM analysis of capsid assembly and structural changes upon interactions with a host restriction factor, TRIM5α. Methods Mol Biol. 2014;1087:13–28. doi: 10.1007/978-1-62703-670-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatziioannou T, Perez-Caballero D, Cowan S, Bieniasz PD. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J Virol. 2005;79:176–183. doi: 10.1128/JVI.79.1.176-183.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikeda Y, Ylinen LM, Kahar-Bador M, Towers GJ. Influence of gag on human immunodeficiency virus type 1 species-specific tropism. J Virol. 2004;78:11816–11822. doi: 10.1128/JVI.78.21.11816-11822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C, et al. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat Commun. 2016;7:10714. doi: 10.1038/ncomms10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bosco DA, Eisenmesser EZ, Pochapsky S, Sundquist WI, Kern D. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc Natl Acad Sci USA. 2002;99:5247–5252. doi: 10.1073/pnas.082100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.López CS, et al. Determinants of the HIV-1 core assembly pathway. Virology. 2011;417:137–146. doi: 10.1016/j.virol.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.López CS, Tsagli SM, Sloan R, Eccles J, Barklis E. Second site reversion of a mutation near the amino terminus of the HIV-1 capsid protein. Virology. 2013;447:95–103. doi: 10.1016/j.virol.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCarthy KR, et al. Gain-of-sensitivity mutations in a Trim5-resistant primary isolate of pathogenic SIV identify two independent conserved determinants of Trim5α specificity. PLoS Pathog. 2013;9:e1003352. doi: 10.1371/journal.ppat.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morger D, et al. The three-fold axis of the HIV-1 capsid lattice is the species-specific binding interface for TRIM5α. J Virol. 2017;92:e01541–17. doi: 10.1128/JVI.01541-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manocheewa S, Swain JV, Lanxon-Cookson E, Rolland M, Mullins JI. Fitness costs of mutations at the HIV-1 capsid hexamerization interface. PLoS One. 2013;8:e66065. doi: 10.1371/journal.pone.0066065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luban J. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. J Virol. 2007;81:1054–1061. doi: 10.1128/JVI.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.