Significance
The true physical form of the proteins within the silk glands of spiders that permits storage at very high concentrations rather than as precipitated material prior to being transformed into solid silk fibers remains one of the fundamental mysteries that has limited our ability to produce artificial silks of the quality of natural silks. Here we determine that spider silk proteins are stored as complex micellar nanoparticles composed of assembled subdomains. When extruded during the silk spinning process, these subdomains undergo fibrillization while remaining assembled in micelles. Knowledge of the nanostructured protein assemblies in the dope is critical to the basic understanding of the spinning process and to our ability to mimic the natural material properties in synthetic analogues.
Keywords: biomimetic materials, spider silk formation, hierarchical micelles, natural protein nanostructures
Abstract
Many natural silks produced by spiders and insects are unique materials in their exceptional toughness and tensile strength, while being lightweight and biodegradable–properties that are currently unparalleled in synthetic materials. Myriad approaches have been attempted to prepare artificial silks from recombinant spider silk spidroins but have each failed to achieve the advantageous properties of the natural material. This is because of an incomplete understanding of the in vivo spidroin-to-fiber spinning process and, particularly, because of a lack of knowledge of the true morphological nature of spidroin nanostructures in the precursor dope solution and the mechanisms by which these nanostructures transform into micrometer-scale silk fibers. Herein we determine the physical form of the natural spidroin precursor nanostructures stored within spider glands that seed the formation of their silks and reveal the fundamental structural transformations that occur during the initial stages of extrusion en route to fiber formation. Using a combination of solution phase diffusion NMR and cryogenic transmission electron microscopy (cryo-TEM), we reveal direct evidence that the concentrated spidroin proteins are stored in the silk glands of black widow spiders as complex, hierarchical nanoassemblies (∼300 nm diameter) that are composed of micellar subdomains, substructures that themselves are engaged in the initial nanoscale transformations that occur in response to shear. We find that the established micelle theory of silk fiber precursor storage is incomplete and that the first steps toward liquid crystalline organization during silk spinning involve the fibrillization of nanoscale hierarchical micelle subdomains.
The ability to replicate the natural silk spinning process with an aqueous synthetic analog at bulk scale, to truly mimic the properties of natural silks, holds tremendous promise for biomedical materials, architectural design, and civil and mechanical engineering. However, to date, the development of synthetic silks that exhibit the mechanical properties of the natural product has been limited (1–5). Currently, we know the primary sequence of the natural spider silk spidroins, and we have significant information on the biochemical triggers of the spinning process and how to mimic it (6–12). The gap in our knowledge involves the nanoscale processes at work within the silk gland where the highly concentrated protein precursor is stored and then on demand transformed into silk fibers (2, 6–9, 12–16). Knowledge of this central part of the natural process is critical to the development of synthetic analogs and key to preparative methods, including microfluidics, for producing artificial fibers from recombinant proteins that exhibit the mechanical properties of native silks (2–4, 6, 17–22). Beyond spider silks, analogous mechanisms might be involved in the formation of protein fibers generally, such as the detrimental nucleation and growth of amyloid fibers from oligomers (23–25). Critical insights gleaned for one fibrous protein system can potentially be translated more generally to further our understanding of how concentrated proteins are stored and then assembled to yield structurally organized 3D materials.
The dragline [major ampullate (MA)] silk-precursor dope solution of Latrodectus hesperus, or black widow spiders, is predominantly composed of two proteins (spidroins) that are stored in high concentration (26–28) (25–50 wt %) in the glands before extrusion, acidification, and fiber formation in the duct (12). These spidroins, MaSp1 and MaSp2, are large and highly repetitive (250 and 312 kDa, respectively). In the gland environment, the repetitive core regions of the spidroins are predominantly unstructured (26–29) (random coil), whereas the termini exhibit pH-sensitive helical bundles (8, 14, 30). The pH sensitivity of the termini has been implicated as an important characteristic for the assembly of spider silk (8, 14, 30, 31). The hydropathy plots for the spidroins are roughly sinusoidal in form, with rapidly alternating hydrophilic–hydrophobic units (approximately +0.3 to −0.6; SI Appendix, Fig. S1) (10). The C- and N-terminal domains contain the units with the highest hydrophilicity in each monomer, whereas the long, central, repetitive domain is generally neutral and hydrophobic, giving these proteins an amphiphilic character analogous to a complex block copolymer amphiphile (32, 33). This amphiphilicity suggests, according to the micelle theory of silk assembly (2, 6–9, 14, 16), that the concentrated protein dope solution is composed of nanoscale assemblies (34)—the essential starting structures needed for the formation of robust, microscale silk fibers (7, 8, 14). Microscopic characterization of synthetically formed silkworm fibroin/PEO fibers has shown evidence of spherical micelle-like structures on the micrometer scale (7). Similarly, scanning electron microscopy and atomic force microscopy imaging of the fully and partially dried silk gland dope from Nephilia clavata spiders have found the existence of micrometer-sized granule particles (35). These appear to be present within the hierarchical structure of the spiders’ silk fibers themselves (35). However, no direct experimental evidence from the hydrated native protein dope solution has been provided in support of the micelle theory. The true physical form of the protein assemblies when stored in the gland, and the process by which this dope solution is transformed into a hierarchical polycrystalline structure when spun through the duct, remain largely unknown.
Here we analyze the native protein dope from the silk glands of black widow spiders to determine the physical form of the liquid precursor of natural silk fibers and test the validity of the existing micelle theory (2, 6–9, 16). We use a combination of indirect and direct observation methods for nanoscale characterization, applying solution NMR spectroscopy and cryo-TEM tomographic imaging. Measuring the diffusion behavior of the spidroins in the natural silk gland dope by NMR reveals that the spidroin proteins in the silk precursor are predominantly confined within entangled volumes of several-hundred-nanometer diameters, suggesting the existence of tightly packed spherical micelles, as postulated in the existing micelle theory (2, 6–9, 16). However, 3D imaging of these nanostructures by cryo-TEM tomography shows that the spidroin micellar assemblies are far more morphologically complex, existing as hierarchical micellar nanoparticles (several-hundred-nanometer diameters) composed of networks of flake-like subdomains. When the native dope is physically sheared, mimicking the extrusion process occurring through a spider’s spinning duct, the subdomains within the hierarchical micelles transform, becoming narrowed and elongated fibrils that remain assembled as interwoven networks in the parent micelle.
Results
Solution NMR Indicates Gland-Stored Spidroin Diffusion Confinement and Entanglement Within ∼300-nm-Diameter Volumes.
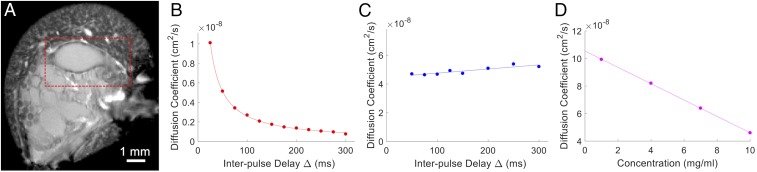
Using pulsed field gradient stimulated-echo (PFG-STE) NMR (Fig. 1 and SI Appendix, Fig. S2), we measured the Einstein self-diffusion coefficient (D) and the observation time (Δ, interpulse delay) dependence of native MA silk gland dope from L. hesperus spiders (Fig. 1A). This NMR technique probes the average temporal displacement of an ensemble of molecules, from which D can be determined (Materials and Methods). The measured displacement and D of a heterogeneous sample, for example, a solution containing micelles, provides information about the physical constraints on motion, distances and metrics, arrangement of molecules, and structural permeability (36, 37). The diffusion of the native gland protein shows a strong Δ dependency, evidenced by the initial decay in Fig. 1B, behavior that indicates restricted diffusion of the silk protein in the gland. Assuming bulk diffusion, the mean squared displacement (MSD) that encloses the volume of restricted diffusion is ∼300 nm (Materials and Methods), suggesting the proteins are confined/entangled within structural elements on this length scale. Once denatured, the diffusion of the protein (48 h in urea) no longer exhibits a Δ dependence (Fig. 1C), indicating that the denatured solution is homogeneous and free of entanglement or diffusion restriction. By sampling multiple spidroin concentrations in 4 M urea and extrapolating to infinite dilution (Fig. 1D), we find a self-diffusion coefficient of ∼10.7 × 10−8 cm2/s corresponding to a hydrodynamic radius (rH) of ∼19 nm for the spidroin monomers (Materials and Methods). This experimental rH is consistent with the theoretical values for the MaSp1 protein (∼22 nm) based on number of residues and chain length (38), indicating that the diffusion restriction was removed following treatment in denaturant for 48 h. These diffusion NMR measurements provide strong evidence that under native conditions in the MA gland, spidroin proteins exist as entangled assemblies several hundred nanometers in diameter. These structures are disrupted and broken down to freely diffusing proteins that remain unstructured (SI Appendix, Fig. S3) when fully denatured in urea for 48 h. We note that for short urea incubation times on the order of 4–10 h (SI Appendix, Fig. S2), restricted diffusion similar to the native dope is found.
Fig. 1.
Native L. hesperus MA silk protein diffusion measured by PFG-STE NMR. (A) Gradient echo magnetic resonance image (GRE-MRI, 18.8 T) of 300-µm-thick cross-section slice of the L. hesperus spider abdomen at the coronal cross-section orientation. Red box indicates one of the two MA glands, which were removed by dissection from L. hesperus spiders for all subsequent NMR, DLS, and cryo-TEM analysis. (B–D) PFG-STE NMR data. (B) Self-diffusion coefficient (D) vs. interpulse delay (Δ) for MA silk protein at native gland conditions (concentration ∼35 wt %, pH ∼7). Strong Δ dependence is observed, and the decreasing D as a function of Δ illustrates that diffusion is restricted at native conditions (MSD ∼300 nm). (C) D vs. Δ for the MA silk protein dope following dilution in 4 M urea (concentration ∼10 wt %, 48 h in urea). The Δ dependence is no longer observed, indicating diffusion is not restricted following solubilizing in urea. (D) MA silk protein D measured by PFG-STE NMR as a function of protein concentration in 4 M urea (48 h). The extracted self-diffusion coefficient at infinite dilution is 10.7 × 10−8 cm2/s (rH ∼ 19 nm).
Direct Morphological Characterization of the Entangled Spidroin Nanostructures by Cryo-TEM Tomography Reveals Hierarchical Micellar Nanoparticles.
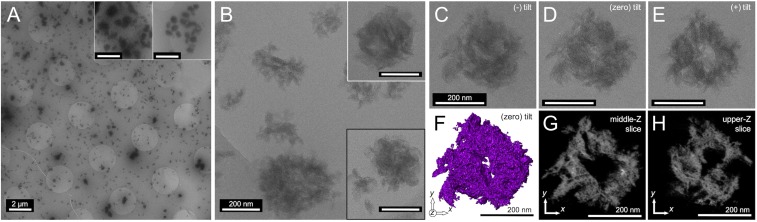
We aimed to directly image and characterize the morphology of the entangled spidroin protein nanostructures in the native silk dope using cryogenic transmission electron microscopy (cryo-TEM; Fig. 2). To prepare cryo-TEM grids (Materials and Methods), the native dope was incubated in urea for 4 h to lower sample viscosity and concentration. At low magnification (Fig. 2A), an abundance of roughly spherical structures was observed, with similar results found for multiple spiders and multiple samples (SI Appendix, Fig. S4). The largest structures were ∼800 nm in diameter (small population), and the predominant population of particles were ∼200–400 nm in diameter, with similar-sized structures implicated by the results from NMR diffusion and dynamic light scattering (DLS) analysis of the MA protein dope following brief urea incubation (SI Appendix, Fig. S5).
Fig. 2.
Native L. hesperus MA silk protein imaged by cryo-TEM (4 h incubation in 4 M urea). (A) Low-magnification image showing abundance of generally spherical (200–400 nm) micellar protein assemblies. Insets highlight the small population of the largest assemblies found (∼500–800 nm). (Scale bar, 2 µm.) (B) High-magnification images of the spidroin hierarchical assemblies, representative of the highest populations; spherical micellar assemblies composed of disordered flake-like subdomains. Black boxed Inset shows similar structures found in a sample prepared from a separate spider following the same procedure (SI Appendix, Fig. S4). (Scale bar for Insets, 200 nm.) (C–E) Cryo-TEM images from a tomography tilt series of one selected hierarchical protein nanoassembly. (F) The 3D isosurface rendering of the assembly in C–E. (G and H) Approximately 0.8-nm-thick z slices extracted at different z heights from the tomography reconstruction in F of the assembly in C–E.
At high magnification (Fig. 2B), we found that the spherical structures are micellar assemblies having hierarchical architectures, composed of flake-like subdomains, ∼50 nm laterally and ∼25 nm in thickness. Many of these protein assemblies appear to contain significant solvent-filled voids within their internal volumes. The 3D intricacy of these architectures, evident from initial 2D transmission observations, led to our acquisition of cryo-TEM tomographic tilt series (Fig. 2 C–E, SI Appendix, Fig. S6, and Movie S1) to allow the reconstruction and rendering of the 3D volume of individual assemblies (Fig. 2 F–H and Movies S2–S4). Using 3° tilt intervals to minimize cumulative dose, a 37-image stack of a selected assembly was acquired, manually aligned, and 3D-reconstructed using a discrete tomography algorithm (Materials and Methods) (39). The 3D isosurface rendering (Fig. 2F and Movie S2) of the hierarchical assembly in Fig. 2 C–E captures the empty internal volume and disordered subdomains of that structure, features that are convoluted in the 2D transmission images. The 1-voxel-thick (∼0.8 nm) z slices (Fig. 2 G and H and Movie S3) of the reconstruction show the interconnected and disordered nature of the flake-like subdomains within each assembly. These cryo-TEM results provide direct evidence for the existence of nanoscale micellar protein assemblies in the native silk dope, assemblies that are hierarchical structures composed of disordered subdomains, more akin to complex compound micelles than the spherical micelles previously postulated (7, 8).
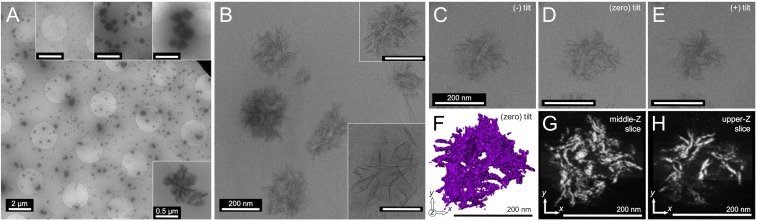
The progressive extrusion (shearing and liquid crystalline alignment) and acidification that occur as the initial native protein dope travels down the spinning duct of the spider are believed to trigger and control the transformation and formation processes that yield the robust, polycrystalline silk fibers (6–8, 13, 14). To probe the initial nanoscale steps of transformation that occur in the native spidroin hierarchical assemblies in response to shear, we prepared a separate set of samples for cryo-TEM analysis where the dope solution was vigorously micropipetted just before deposition on cryo-TEM grids (Fig. 3). Similar to the findings for the unperturbed native dope sample (Fig. 2A), at low magnification, an abundance of spherical, submicron spherical structures are also found for the sheared dope sample (Fig. 3A). However, close inspection of the cryo-TEM data at higher magnification reveals subtle but significant differences in the architecture of the hierarchical assemblies, specifically at the subdomain level (Fig. 3B). The subdomains following micropipette shearing transformed from the initial flake-like morphologies (Fig. 2) into elongated and narrow fibrillar morphologies (∼50–100 nm in length and ∼5–15 nm in width).
Fig. 3.
Native L. hesperus MA silk protein imaged by cryo-TEM (4 h incubation in 4 M urea) after shear (vigorous micropipette pumping). (A) Low-magnification images showing the abundance of generally spherical assemblies (100–300 nm). Insets highlight the small population of the largest assemblies found (∼500–800 nm) and a region found with isolated fibrils (Left inset). (Scale bar, 2 µm.) (B) High-magnification images of the spidroin assemblies, representative of the highest populations; spherical hierarchical assemblies composed of interwoven fibrillar subdomains. Insets show the smaller population of loosely packed fibrillar aggregates. (Scale bar, 200 nm.) (C–E) Cryo-TEM images from a tilt series of one selected hierarchical nanoassembly. (F) The 3D tomography rendering of the assembly in C–E. (G and H) Approximately 0.8-nm-thick z slices extracted at different z heights from the tomography reconstruction in F of the assembly in C–E.
We acquired cryo-TEM tomographic tilt series (Fig. 3 C–E, SI Appendix, Fig. S7, and Movie S5) and created 3D reconstructions and volume renderings (Fig. 3 F–H and Movies S6–S8) of the hierarchical assemblies in the sheared dope sample for comparison with those found in the native dope sample (Fig. 2). The 3D isosurface rendering in Fig. 3F (Movie S6) shows the disordered and interwoven 3D arrangement of the fibril subdomains within each spherical assembly, which also contain significant internal void volume (solvent filled). From the 1-voxel-thick (∼0.8 nm) z slices (Fig. 3 G and H and Movie S7) of the 3D reconstruction, the narrow (∼10 nm) fibrillar structure of the interlaced subdomains is visualized clearly, showing the differences between the sheared subdomain morphologies and flake-like subdomains in the hierarchical assemblies of the native (unperturbed) dope (Fig. 2 G and H). These cryo-TEM results suggest that the initial transformations occurring as the gland-stored spidroin protein dope is extruded along the spinning duct occur at the subdomain level, before transitions at the micellar level or at higher orders (Fig. 4). Immediately following shear, the general morphologies of the micellar assemblies remain roughly spherical, of the same overall size (several-hundred-nanometer diameters) and with disordered arrangements of subdomains. However, the subdomains themselves transition from flake-like morphologies to narrow fibril morphologies. This suggests that the complex, hierarchical architecture of native micellar assemblies (Fig. 2) is integral to the formation of natural-quality spider silk fibers.
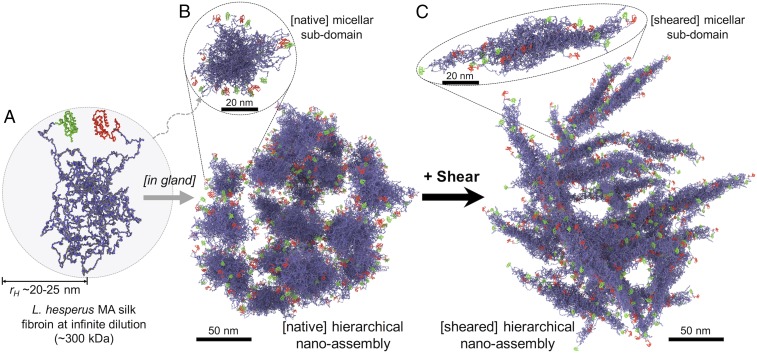
Fig. 4.
Graphical interpretation of the cryo-TEM and diffusion NMR data; illustrative scheme of the modified micelle theory. (A) Spidroin monomer at infinite dilution [the building blocks for the nanoassemblies that form at native gland conditions (B)]. The C- and N-terminal domains (green and red) form helical structures, and the long, central domain (blue and gray) is in the molten-globule state (rH ∼ 20–25 nm) (8, 26–29). (B) At neutral pH and native concentration within the MA gland, the spidroin monomers assemble into small, flake-like subdomains (here shown composed of 12 monomers) that hierarchically assemble into larger, spherical micellar structures (several hundred nanometers in diameter). (C) When sheared, the initial transformations occur at the subdomain level. Individual subdomains transform into narrow, elongated fibrils that remain assembled (disordered and interwoven network) within the spherical hierarchical assemblies.
Discussion
From these NMR and cryo-TEM data, we revise the existing micelle theory (7, 8) and propose instead a hierarchical micelle theory for the native spidroin storage and initial transformation, shown in Fig. 4. At very low concentrations, the free spidroin monomers pack into hydrodynamic radii of ∼20–25 nm, balancing their hydrophobic/hydrophilic interactions with the local solvent (Fig. 4A). At native concentration (when stored in the MA gland), the spidroins assemble into hierarchical, spherical architectures composed of a disordered and interconnected network of flake-like subdomains with interspersed internal voids (Fig. 4B). The physical dimensions of the subdomains from the cryo-TEM data suggest that individual subdomains are composed of ∼10–15 spidroin monomers (SI Appendix). We propose that these hierarchical micellar assemblies are the essential starting structures critical for the formation of natural silk fibers. Further, when sheared, the individual subdomains transform into narrow and elongated fibrils that remain disordered and interwoven within the hierarchical spherical assemblies (Fig. 4C).
These data imply that the structural transformations (fibrillization) at the subdomain level within the hierarchical micelles are critical first steps during silk fiber formation in the natural silk spinning system, before transitions, liquid crystalline organization, and/or assembly/disassembly events at the micellar level. Nonclassical two-step growth processes have been identified for many natural materials, from amyloid fiber formation (24, 40) to CaCO3 biomineralization (41, 42), where the initial formation of specific seed nanostructures is essential for the progression of the second growth/transition step and the ultimate formation of the final material. Synthetically reproducing the quality of fibers that are naturally spun by spiders could be intrinsically dependent on our ability to start the spinning process with the same types of precursors (seeds), not only from a biological and chemical perspective but also in their nanoscale morphologies. Indeed, Rising, Johansson, and coworkers (2, 3) recently developed methods to spin artificial spider silk fibers using recombinant spidroins, starting by first forming small (∼20–50 nm) spherical micelles (nonhierarchical). However, the artificial fibers exhibited significantly inferior mechanical properties compared with natural spider silks, although they are among the best synthetic fibers reported to date (2, 3). In this work we have shown that larger, hierarchical micellar nanoassemblies are the initial protein structures in the native dope—the seeds from which natural silk fibers are spun. Achieving similar synthetically produced hierarchical nanoassemblies (several hundred nanometers in diameter) will be critical for bulk-producing robust synthetic silk fibers from recombinant spidroin proteins.
Materials and Methods
Dissection of L. hesperus (Black Widow) Spider and Extraction of MA Silk Glands.
Large western black widows (Latrodectus hesperus) were fed ∼100 µL of saturated U-13C-15N Alanine solution to label the silk proteins (MaSp1 and MaSp2) with NMR-accessible nuclei for heteronuclear solution NMR experiments. Labeling was done three times a week for 2 wk. While feeding, the spiders were forcibly silked to deplete the native, unlabeled spidroin supply and encourage the synthesis of labeled proteins. At the end of 2 wk the spiders were dissected, and the major ampullate glands were extracted in deionized water. For the studies on native silk solution, major ampullate glands were rinsed with DI water and transferred directly to a 5-mm Shigemi tube filled with 90:10 H2O:D2O. For the urea studies, glands were carefully removed, rinsed, and cleaned of the outer, insoluble membrane. This silk dope was then diluted with a solution of 4 M urea with 1% sodium azide and allowed to incubate at 4 °C for 1 h. The final solution was then either transferred to 5 mm NMR tube for analysis or prepared for cryo-TEM or DLS.
Cryo-TEM Sample Preparation.
L. hesperus spiders were asphyxiated with N2 gas and quickly killed with a sharp blade. Spiders were then dissected to remove the major ampullate gland (see dissection methods for details) and to manually remove the outer membrane and clean the gland (done at ambient conditions, ∼27 °C). This process of dissection and gland removal and cleaning took ∼35 min for the gland sample used in Figs. 2 and 3 (these two are from the same gland sample prepared on the same day) but only took ∼15 min for the gland sample in SI Appendix, Fig. S4 (for this gland, dissection and cryo-TEM sample preparation were done using a different spider than for Figs. 2 and 3 and on a different day). For both glands, the surface of the gland was cleaned very thoroughly of membrane without perturbing or rupturing the dope. The cleaned gland was immediately added to a vial (100 µL) of urea denaturant solution, and 10 min later, the gland–urea solution was transferred to a 4 °C refrigerator. During this time, the mass of the solution with the gland was measured to determine approximate solution concentration (∼1 wt %). For the gland used in Figs. 2 and 3, the gland–urea solution was incubated at 4 °C for ∼240 min. For the gland used in SI Appendix, Fig. S4, the gland–urea solution was incubated at 4 °C for ∼175 min. After this incubation time at 4 °C, gland–urea solution was immediately transferred to a 4 °C (cold room) cryo-TEM grid preparation laboratory.
For all samples (Figs. 2 and 3 and SI Appendix, Fig. S4), graphene-oxide coated holey-carbon (Quantifoil R2/2, 400 mesh Cu support) TEM grids (Structure Probe, Inc.) were used. The graphene-oxide coating was applied according to refs. 43 and 44 using a 0.15 mg/mL aqueous solution (drop-cast 4 µL onto plasma-cleaned grid), and the coating was prepared several days before the grids were used for cryo-TEM sample prep.
After incubation, cryo-TEM sample preparation and plunging were done in a zero-humidity controlled cold room (4 °C). For the preparation of the Native MA protein samples (Fig. 2 and SI Appendix, Fig. S4), no micropipette was used, as would be done for conventional cryo-TEM sample prep. Instead, a borosilicate Pasteur pipette (13-678-20B; Fischer Scientific International, Inc.) was used to transfer a small volume of the sample solution from the vial to the surface of the GO-coated TEM grid. By eye, the volume of the droplet applied was ∼4–5 µL of solution, applied to the coated surface of the grid while the grid was held aloft in self-clamping tweezers. The tweezers were then quickly, but carefully, attached into a homemade manual plunger, with a small liquid-ethane bath set below the plunging apparatus. Using filter paper, the grid with the droplet on its surface was manually blotted for a 4-s count, applying the filter paper to the coated side of the grid (the side with the droplet). The grid was immediately plunged into the L-ethane bath after the 4-s blot, and the grid was then transferred into an L-N2 bath where it was placed into a storage box. The storage box was transferred into a large L-N2 Dewar storage container and stored.
For the preparation of the native MA protein after vigorous micropipette pumping of the sample (Fig. 3), conventional preparation was done using a micropipette. However, before drop-casting the droplet of solution onto the GO-coated surface of the grid, the micropipette (20-µL-volume micropipette, set to 4-µL fill) was used to vigorously pump in and out on the sample solution 40 times. A 4-µL droplet was then applied to the coated surface of the grid while the grid was held aloft in self-clamping tweezers. The grid was left to sit undisturbed for ∼5 min, and then the tweezers were carefully attached into the homemade manual plunger, with a small liquid-ethane bath set below the plunging apparatus. Using filter paper, the grid with the droplet on its surface was manually blotted for a 4-s count, applying the filter paper to the coated side of the grid (the side with the droplet). The grid was immediately plunged into the L-ethane bath after the 4-s blot, and the grid was then transferred into an L-N2 bath where it was placed into a storage box. The storage box was transferred into a large L-N2 Dewar storage container and stored.
Cryo-TEM Imaging and Tomography Tilt Series Acquisition.
All cryo-TEM imaging (Figs. 2 and 3 and SI Appendix, Figs. S4, S6, and S7) was done using an FEI (FEI Company) Polara cryo-TEM operating at 200 keV equipped with a Gatan (Roper Technologies) K2 direct electron detector (FEG extraction, 4350 V; FEG emission, 110 µA; spot size 5, 70-µm objective lens aperture, 70-µm condenser lens aperture). The Polara cryo-TEM is a cartridge-loaded microscope, where the column is continually maintained at L-N2 temperature (less than −175 °C). The grids that were prepared previously and stored in L-N2 were loaded into the microscope’s cartridge while submerged in L-N2, and the cartridge with grids was inserted into the microscope. The Polara was aligned for low-dose imaging, measuring the dose rate on the K2 detector through vacuum (no grid inserted). The dose rate used was 1.25 e−/Å2 s for high-magnification [50,000× magnification (50 kx)] images (4.096-Å pixel size, 3,708 × 3,838 pixels; Figs. 2 B–E and 3 B–E and SI Appendix, Fig. S4 B–D). The dose rate was not measured for the low-magnification images (Figs. 2A and 3A and SI Appendix, Fig. S4A), because the dose rate was orders of magnitude lower for those images (1-s exposure time, 10.1034 nm pixel size, 1,852 × 1,918 pixels, binning 2). The high-magnification images associated with tomography tilt series (Figs. 2 C–E and 3 C–E) were acquired using a 2-s exposure time (cumulative dose, 2.5 e−/Å2 per image), and all other high-magnification images (not part of tilt series; Figs. 2B and 3B and SI Appendix, Fig. S4 B–D) were acquired using a 3-s exposure time (cumulative dose, 3.75 e−/Å2 per image). All imaging and tomography tilt series acquisition was done using SerialEM software (bio3d.colorado.edu/SerialEM/), which applies autofocusing on adjacent regions of the grid to minimize dose on the sample and uses an automated cross-correlation drift/shift correction during acquisition of tomography tilt series. Raw cryo-TEM images were saved as .MRC stacks.
All cryo-TEM tomography tilt series were acquired using 3° tilt intervals. For the tilt series in Fig. 2 C–H, the range covered was +49° to −59 (37 images in the series), and the total cumulative dose experienced was ∼92.5 e−/Å2. For the tilt series in Fig. 3 C–H, the range covered was +38° to −58 (33 images in the series), and the total cumulative dose experienced was ∼82.5 e−/Å2.
Tomography Reconstruction of Cryo-TEM Tilt Series and 3D Rendering/Visualization.
The raw cryo-TEM tilt series images (.MRC stack) were manually aligned (Movies S1 and S5) using IMOD software (bio3d.colorado.edu/imod/) (45). The negative logs of the TEM intensities were used to form a pseudo–dark-field image and convert the intensity of the object into a positive value, with the background intensity close to zero. The total variation regularized discrete algebraic reconstruction technique (TVR-DRAFT) tomography reconstruction algorithm (39, 46) was downloaded from GitHub (https://github.com/astra-toolbox/ContributedTools) and was implemented to generate the reconstructions of the protein nanostructures in Fig. 2 F–H (Movie S4) and Fig. 3 F–H (Movie S8). A value of 50 was used for the lambda parameter to reduce noise contributions, and 100 iterations of the simultaneous iterative reconstruction technique algorithm were used with three gray levels (object, background, and vacuum). After reconstruction, a 3D Gaussian filter was applied to further reduce noise and remove isolated voxels, and the objects were globally thresholded and visualized using FEI Avizo 9.0 software (FEI Company). Fig. 2 uses sigma relative (rel.) of 1, kernel size of 7, and threshold value of 80 after the Gaussian filter was applied. Fig. 3 uses sigma rel. of 1, kernel size of 5, and threshold value of 100 after the Gaussian filter was applied. Threshold values were applied using the magic wand tool, which only selects connected pixels. Fig. 2F (Movie S2) and Fig. 3F (Movie S6) are isosurface renderings of the reconstructions, and Fig. 2 G and H (Movie S3) and Fig. 3 G and H (Movie S7) are 1-voxel-thick orthoslice images (0.8 nm thick) from the reconstruction at various z heights. Movies S2 and S6 show first the isosurface rendering as it is rotated around the y axis (360°), and Movies S3 and S7 show the rendering of 1-voxel-thick orthoslices sequentially, going down the z height of the rendered volume from the top to the bottom.
Magnetic Resonance Imaging of Black Widow Spider.
Gradient echo (GRE)-MRI images of a black widow spider (Fig. 1A) were collected using an 18.8T magnet (800 MHz) with a Varian VNMRS console and Doty Scientific 12 mm ID triple axis MRI gradient probe capable of producing up to 300 Gs/cm PFG along each axis. GRE-MRI images with 40 µm × 40 µm in plane resolution and 300-µm slice thickness were collected using a 4-ms echo time and 50-ms repetition time. Localized spectra utilized the STEAM2, a stimulated echo acquisition mode localization scheme, with an echo time of 2.3 ms, a mixing time of 4.5 ms, and a repetition time of 2 s. Additionally, outer voxel suppression pulses and variable power radiofrequency pulses with optimized relaxation delays (VAPOR) water suppression was employed (47). A 1.6-μL volume of interest was selected in the interior of the MA gland using pilot gradient echo images.
PFG-STE NMR Acquisition and Analysis.
NMR diffusion experiments (Fig. 2 and SI Appendix, Figs. S2 and S8) were conducted using the same magnet as used for GRE-MRI imaging (18.8T), and with an 800 MHz Agilent spectrometer and a Doty diffusion probe using an STE pulse sequence with bipolar pulse-field gradient pulses lasting 2 ms (δ) with a 50-ms Δ diffusion delay and a 10-s d1 recycle delay (48, 49). To reduce the effects of eddy currents in the probe, a 2-ms delay was added at the end of each phase of the bipolar gradient pulses. The diffusion experiments were collected by incrementing the gradient magnetic field strength in 10 steps. The probe gradients were calibrated using anhydrous glycerol sample. The calibrated temperature was 26 °C, and the corresponding diffusion coefficient for glycerol is 2.50 10−12 m2/s (48). The gradient pulse (δ) was set to 2 ms, and the delay (Δ) between two bipolar gradient pulses was 50 ms. Twenty experiments were collected with an array of field strengths from 6.4 to 1,273 Gs/cm.
For 1H diffusion measurements on silk proteins, the experiments were carried out at 23 °C. The general experimental parameters used were spectrum width of 9,615 Hz, 32 transients, 10 s recycle delay, 2.45 s of acquisition time, δ = 1 ms, = 50 ms, and gradient field strengths arrayed from 7 to 600 Gs/cm. When arraying the diffusion delay, Δ, all other parameters were kept constant with Δ ranging from 25 to 500 ms with 25-ms increments. The diffusion coefficient change with time in 4 M urea was tracked for 36 h with 2-h sampling intervals and following 1 wk time.
The 2D 1H/15N heteronuclear single-quantum correlation (HSQC) NMR experiments (SI Appendix, Figs. S3 and S9) were carried out with a 600-MHz spectrometer using a Bruker TXI probe controlled by a Bruker Avance III console at room temperature. 2k points were collected in F2, zero-filling up to 4k, and 128 points up to 512 used for F1. Thirty-two scans were collected per slice. A line broadening function of 0.3 Hz was applied to the spectrum in the F2 dimension. Phasing and data processing were carried out in Topspin 3.5. All chemical shifts are referenced to 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS). (See SI Appendix for data analysis methodology.)
Dynamic Light Scattering.
DLS (SI Appendix, Fig. S5) was done using a Malvern Instruments Zetasizer Nano-ZS. All measurements were conducted at 25 °C with a backscatter angle of 173°. The manufacturer’s suggested refractive index of protein, 1.450, was used. The viscosity of 4 M urea was previously determined to be 1.0854cP with a refractive index of 1.364 determined with a refractometer. The data were fitted using the protein analysis model for data processing.
Hydropathy Plot Generation.
The hydropathy plots for the MaSp1 (SI Appendix, Fig. S1A) and MaSp2 (SI Appendix, Fig. S1B) spidroins were created using the respective protein sequences in ref. 10. The hydropathy index was plotted for each individual amino acid using the ExPASy web resource (https://www.expasy.org/).
Supplementary Material
Acknowledgments
We thank Prof. Timothy Baker for access to the instrument, and we thank Dr. James Bower for his assistance with the cryo-TEM experiments. The DLS work was performed with the assistance of David Pullman at SDSU. L.R.P. was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award F32EB021859. D.O. is supported by an SDSU Graduate Research Fellowship. J.D.R.’s work was performed under the auspices of the US Department of Energy, by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344. Grants that supported this work include: Department of Defense - Air Force Office of Scientific Research (DOD-AFOSR) FA9550-17-1-0282 (to G.P.H.), DOD Defense University Research Instrumentation Program (DURIP) FA9550-17-1-0409 (to G.P.H.), National Science Foundation - Division of Materials Research - Biomaterials (NSF-DMR-BMAT) 1809645 (to J.L.Y.), and Army Research Office (ARO) Multidisciplinary University Research Initiative (MURI) W911NF-15-1-0568 (to N.C.G.). The cryo-TEM work was performed at the cryo-electron microscopy facility at the University of California, San Diego, which is supported by our colleague Prof. Timothy Baker (University of California, San Diego) and funded by the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810203115/-/DCSupplemental.
References
- 1.Hardy JG, Romer LM, Scheibel TR. Polymeric materials based on silk proteins. Polymer. 2008;49:4309–4327. [Google Scholar]
- 2.Andersson M, et al. Biomimetic spinning of artificial spider silk from a chimeric minispidroin. Nat Chem Biol. 2017;13:262–264. doi: 10.1038/nchembio.2269. [DOI] [PubMed] [Google Scholar]
- 3.Kronqvist N, et al. Efficient protein production inspired by how spiders make silk. Nat Commun. 2017;8:15504–15515. doi: 10.1038/ncomms15504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heidebrecht A, et al. Biomimetic fibers made of recombinant spidroins with the same toughness as natural spider silk. Adv Mater. 2015;27:2189–2194. doi: 10.1002/adma.201404234. [DOI] [PubMed] [Google Scholar]
- 5.Bowen CH, et al. Recombinant spidroins fully replicate primary mechanical properties of natural spider silk. Biomacromolecules. 2018;19:3853–3860. doi: 10.1021/acs.biomac.8b00980. [DOI] [PubMed] [Google Scholar]
- 6.Tokareva O, Jacobsen M, Buehler M, Wong J, Kaplan DL. Structure-function-property-design interplay in biopolymers: Spider silk. Acta Biomater. 2014;10:1612–1626. doi: 10.1016/j.actbio.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin H-J, Kaplan DL. Mechanism of silk processing in insects and spiders. Nature. 2003;424:1057–1061. doi: 10.1038/nature01809. [DOI] [PubMed] [Google Scholar]
- 8.Hagn F, et al. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature. 2010;465:239–242. doi: 10.1038/nature08936. [DOI] [PubMed] [Google Scholar]
- 9.Jenkins JE, et al. Characterizing the secondary protein structure of black widow dragline silk using solid-state NMR and X-ray diffraction. Biomacromolecules. 2013;14:3472–3483. doi: 10.1021/bm400791u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayoub NA, Garb JE, Tinghitella RM, Collin MA, Hayashi CY. Blueprint for a high-performance biomaterial: Full-length spider dragline silk genes. PLoS One. 2007;2:e514. doi: 10.1371/journal.pone.0000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Beek JD, Hess S, Vollrath F, Meier BH. The molecular structure of spider dragline silk: Folding and orientation of the protein backbone. Proc Natl Acad Sci USA. 2002;99:10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yarger JL, Cherry BR, van der Vaart A. Uncovering the structure–function relationship in spider silk. Nat Rev Mater. 2018;3:18008–18011. [Google Scholar]
- 13.Römer L, Scheibel T. The elaborate structure of spider silk: Structure and function of a natural high performance fiber. Prion. 2008;2:154–161. doi: 10.4161/pri.2.4.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Askarieh G, et al. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature. 2010;465:236–238. doi: 10.1038/nature08962. [DOI] [PubMed] [Google Scholar]
- 15.Eisoldt L, Hardy JG, Heim M, Scheibel TR. The role of salt and shear on the storage and assembly of spider silk proteins. J Struct Biol. 2010;170:413–419. doi: 10.1016/j.jsb.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 16.Hagn F, Thamm C, Scheibel T, Kessler H. pH-dependent dimerization and salt-dependent stabilization of the N-terminal domain of spider dragline silk—Implications for fiber formation. Angew Chem Int Ed Engl. 2011;50:310–313. doi: 10.1002/anie.201003795. [DOI] [PubMed] [Google Scholar]
- 17.Lazaris A, et al. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science. 2002;295:472–476. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- 18.Rising A, Widhe M, Johansson J, Hedhammar M. Spider silk proteins: Recent advances in recombinant production, structure-function relationships and biomedical applications. Cell Mol Life Sci. 2011;68:169–184. doi: 10.1007/s00018-010-0462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rammensee S, Slotta U, Scheibel T, Bausch AR. Assembly mechanism of recombinant spider silk proteins. Proc Natl Acad Sci USA. 2008;105:6590–6595. doi: 10.1073/pnas.0709246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konwarh R, Gupta P, Mandal BB. Silk-microfluidics for advanced biotechnological applications: A progressive review. Biotechnol Adv. 2016;34:845–858. doi: 10.1016/j.biotechadv.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 21.Peng Q, et al. Recombinant spider silk from aqueous solutions via a bio-inspired microfluidic chip. Sci Rep. 2016;6:36473. doi: 10.1038/srep36473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdalla S, Obaid A, Al-Marzouki F, Bahabri F. Preparation and characterization of artificial spider silk produced through microchannel techniques. J Marter Sci Eng. 2017;6:383. [Google Scholar]
- 23.Vollrath F, Knight DP. Liquid crystalline spinning of spider silk. Nature. 2001;410:541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- 24.Šarić A, Chebaro YC, Knowles TPJ, Frenkel D. Crucial role of nonspecific interactions in amyloid nucleation. Proc Natl Acad Sci USA. 2014;111:17869–17874. doi: 10.1073/pnas.1410159111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenney JM, Knight D, Wise MJ, Vollrath F. Amyloidogenic nature of spider silk. Eur J Biochem. 2002;269:4159–4163. doi: 10.1046/j.1432-1033.2002.03112.x. [DOI] [PubMed] [Google Scholar]
- 26.Hijirida DH, et al. 13C NMR of Nephila clavipes major ampullate silk gland. Biophys J. 1996;71:3442–3447. doi: 10.1016/S0006-3495(96)79539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hronska M, van Beek JD, Williamson PTF, Vollrath F, Meier BH. NMR characterization of native liquid spider dragline silk from Nephila edulis. Biomacromolecules. 2004;5:834–839. doi: 10.1021/bm0343904. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins JE, Holland GP, Yarger JL. High resolution magic angle spinning NMR investigation of silk protein structure within major ampullate glands of orb weaving spiders. Soft Matter. 2012;8:1947–1954. [Google Scholar]
- 29.Xu D, Yarger JL, Holland GP. Exploring the backbone dynamics of native spider silk proteins in black widow silk glands with solution-state NMR spectroscopy. Polymer. 2014;55:3879–3885. [Google Scholar]
- 30.Andersson M, et al. Carbonic anhydrase generates CO2 and H+ that drive spider silk formation via opposite effects on the terminal domains. PLoS Biol. 2014;12:e1001921. doi: 10.1371/journal.pbio.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu D, Guo C, Holland GP. Probing the impact of acidification on spider silk assembly kinetics. Biomacromolecules. 2015;16:2072–2079. doi: 10.1021/acs.biomac.5b00487. [DOI] [PubMed] [Google Scholar]
- 32.Alexandridis P, Lindman B, editors. Amphiphilic Block Copolymers: Self-Assembly and Applications. 1st Ed Elsevier Science B.V.; Amsterdam: 2000. [Google Scholar]
- 33.Alexandridis P. Amphiphilic copolymers and their applications. Curr Opin Colloid Interface Sci. 1996;1:490–501. [Google Scholar]
- 34.Israelachvili JN, Mitchell DJ, Ninham BW. Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayers. J Chem Soc Faraday Trans 2. 1975;72:1525–1568. [Google Scholar]
- 35.Lin T-Y, et al. Liquid crystalline granules align in a hierarchical structure to produce spider dragline microfibrils. Biomacromolecules. 2017;18:1350–1355. doi: 10.1021/acs.biomac.7b00086. [DOI] [PubMed] [Google Scholar]
- 36.van Dusschoten D, de Jager PA, Van As H. Extracting diffusion constants from echo-time-dependent PFG NMR data using relaxation-time information. J Magn Reson A. 1995;116:22–28. [Google Scholar]
- 37.Sorland GH. Dynamic Pulsed-Field-Gradient NMR. Springer; Berlin: 2014. [Google Scholar]
- 38.Wilkins DK, et al. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 39.Xiaodong Zhuge, Palenstijn WJ, Batenburg KJ. TVR-DART: A more robust algorithm for discrete tomography from limited projection data with automated gray value estimation. IEEE Trans Image Process. 2016;25:455–468. doi: 10.1109/TIP.2015.2504869. [DOI] [PubMed] [Google Scholar]
- 40.Castello F, Casares S, Ruedas-Rama MJ, Orte A. The first step of amyloidogenic aggregation. J Phys Chem B. 2015;119:8260–8267. doi: 10.1021/acs.jpcb.5b01957. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen MH, Aloni S, De Yoreo JJ. In situ TEM imaging of CaCO3 nucleation reveals coexistence of direct and indirect pathways. Science. 2014;345:1158–1162. doi: 10.1126/science.1254051. [DOI] [PubMed] [Google Scholar]
- 42.De Yoreo JJ, et al. CRYSTAL GROWTH. Crystallization by particle attachment in synthetic, biogenic, and geologic environments. Science. 2015;349:aaa6760. doi: 10.1126/science.aaa6760. [DOI] [PubMed] [Google Scholar]
- 43.Pantelic RS, Meyer JC, Kaiser U, Baumeister W, Plitzko JM. Graphene oxide: A substrate for optimizing preparations of frozen-hydrated samples. J Struct Biol. 2010;170:152–156. doi: 10.1016/j.jsb.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 44.Patterson JP, et al. A simple approach to characterizing block copolymer assemblies: Graphene oxide supports for high contrast multi-technique imaging. Soft Matter. 2012;8:3322–3328. doi: 10.1039/C2SM07040E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 46.Zhuge X, et al. Automated discrete electron tomography—Towards routine high-fidelity reconstruction of nanomaterials. Ultramicroscopy. 2017;175:87–96. doi: 10.1016/j.ultramic.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Tkáč I, Gruetter R. Methodology of H NMR spectroscopy of the human brain at very high magnetic fields. Appl Magn Reson. 2005;29:139–157. doi: 10.1007/BF03166960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stejskal EO, Tanner JE. Spin diffusion measurements: Spin echoes in the presence of a time-dependent field gradient. J Chem Phys. 1965;42:288–292. [Google Scholar]
- 49.Wu D, Chen A, Johnson CS. An improved diffusion-ordered spectroscopy experiment incorporating bipolar-gradient pulses. J Magn Reson A. 1995;115:260–264. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.