Abstract
Caspase-8 is an apoptotic protease that is activated by a proximity-induced dimerization mechanism within the death-inducing signaling complex (DISC). The death effector domain (DED) of caspase-8 is involved in protein-protein interactions and is essential for the activation. Here, we report two crystal structures of the dimeric DEDs of the F122A mutant of caspase-8, both of which illustrate a novel domain-swapped dimerization, while differ in the relative orientation of the two subunits and the solvent exposure of the conserved hydrophobic patch Phe122/Leu123. We demonstrate that mutations disrupting the dimerization of the DEDs abrogate the formation of cellular death effector filaments (DEFs) and the induced apoptosis by overexpressed DEDs. Furthermore, such dimerization-disrupting mutations also impair the activation of the full-length caspase-8 and the downstream apoptosis cascade. The structures provide new insights into understanding the mechanism underlying the activation of procaspase-8 within the DISC and DEFs.
Introduction
Caspase-8 is a cysteine protease that initiates the extrinsic apoptotic pathway in response to cell surface death receptor activation1,2. Procaspase-8 exists in the cytosol within the cell as an inactive monomer, which is characterized by an N-terminal tandem death effector domains (DEDs) and a C-terminal catalytic protease domain3,4. The activation of procaspase-8 is proposed to occur through an induced proximity mechanism5–7. Upon engagement with the death ligand such as Fas Ligand (FasL), the clustering death receptor Fas recruits the adaptor protein Fas-associated death domain (FADD) to its cytoplasmic tail8. The inactive procaspase-8 is then recruited to FADD via homotypic interactions between the DEDs of procaspase-8 and the adaptor protein FADD, leading to the dimerization and activation of procaspase-89. During this process, dimeric FasL trimers potentially serve as a minimal unit fully capable of activating the Fas pathway10.
The current induced proximity model is proposed based on early studies using drug-induced dimerization of a truncated procaspase-8 without the DEDs11,12, or chimeric caspase-8 that Fpk3 or FKBP is fused to truncated procaspase-8 without the DEDs5,9,13. In these early studies, kosmotrope-induced dimerization of the caspase domain of procaspase-8 leads to its activation in vitro12. Moreover, the cell-permeable ligand-induced dimerization of the FKBP domain also drives the activation of the chimeric procaspase-8 in the intact cell9. However, the function of the DEDs in the activation of procaspase-8 cannot be fully addressed based on these studies using the truncated procaspase-8 that lacks the DEDs. It remains unclear whether the DEDs of procaspase-8 forms dimer, if true, how the DEDs forms the dimeric structure within the DISC.
To address these questions, we tried to determine the structure of the DEDs of caspase-8, but the structural study of the DEDs is markedly hindered by the low solubility and high aggregating propensity for the tandem DEDs of caspase-814. Through mutant screening, we recently solved the structure of the monomeric DEDs of caspase-8 for a soluble F122A/I128D mutant (DEDF122A/I128D)15, which closely resembles the structure of viral FLIP MC15916,17. However, the monomeric DEDF122A/I128D mutant provides limited clues about the homotypic interactions between the DEDs of caspase-8. More recently, the cryo-EM structure of caspase-8 tandem DED filaments was determined18, which provides an elegant structural framework to understand interactions between the DEDs of caspase-8 itself and that between caspase-8 and other proteins such as FADD, cFLIP, and vFLIP. This structure also takes a significant step forward in understanding the mechanism underlying death-inducing signaling complex (DISC) assembly. On the other hand, the filament formation generally occurs under high concentrations of DEDs in vitro18 or intracellularly19–22, it is unclear how the engagement of FasL triggers the downstream filament formation of caspase-8 under physiological condition. Moreover, further studies are still needed to address how the filament structure accommodate the requirement for dimerization in the activation of caspase-8, and how this structure reconciles with the observation that dimeric FasL trimer is a minimal unit fully capable of activating the Fas pathway10.
In this study, we determined the crystal structures of the DEDs of caspase-8 illustrating a novel domain-swapped dimer, which adopts either open or closed conformations in terms of the solvent-exposure of the conserved hydrophobic patch Phe122/Leu123 that is proposed to bind with the DED of FADD. Together with the biochemical data and the computational modeling, the domain-swapped structures of the dimeric DEDs of caspase-8 provide an alternative model to understand the dimerization-driven activation of caspase-8 in the Fas pathway mediated by a minimal dimeric FasL trimer.
Results
Soluble stable dimer of the DEDs mutant
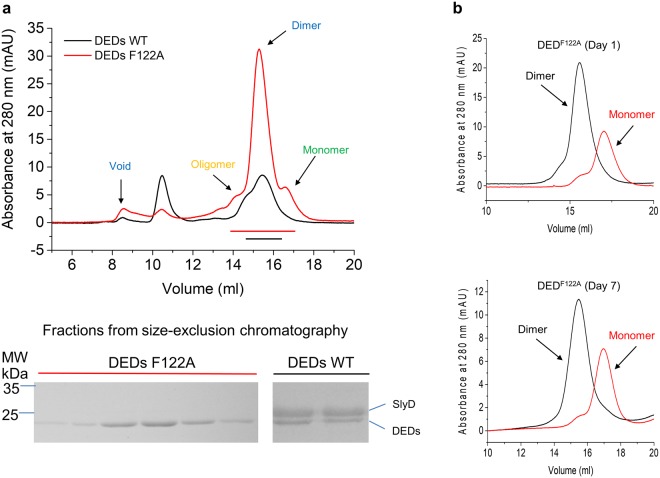
To eliminate the potential impact of the N-terminal fused proteins, such as GFP and MBP, on the homotypic interactions between the DEDs of caspase-8, we tried to express the wild-type DEDs with a C-terminal His6 tag. In consistent with the insoluble and aggregation-prone property of DEDs, we only obtained very low yields of soluble protein (Fig. 1a). Unexpectedly, the protein eluted from the peaks mainly corresponding to dimeric and oligomeric states rather than from the void position of the gel filtration column as shown in the previous study18, suggesting that dimeric DEDs rather than filamentous DEDs could be obtained under low expression level. However, further SDS-PAGE analysis showed that the DEDs dimer mixed with a bacterial folding helper protein slyD23, highlighting that the solubilization of the insoluble wild-type DEDs must be facilitated by slyD (Fig. 1a). In screening for the DED mutants that are soluble and remain self-assembled, we found a F122A mutant of the DEDs of caspase-8 (DEDF122A), which is soluble and less aggregated (Fig. 1a). Interestingly, when expressed with an N-terminal His-SUMO fusion tag, the F122A mutant with tag removal still has dominant dimeric peak during gel filtration albeit having minor peaks corresponding to the monomeric and oligomeric states (Supplementary Fig. 1a). Both the DEDF122A monomer and dimer are highly stable at room temperature without obvious inter-conversion or aggregating in days (Fig. 1b). By contrast, both MBP-DEDWT-SUMO and DEDY8A-SUMO form filaments in the previous study18, highlighting the different properties for DEDs under low and high expression levels.
Figure 1.
DEDs dimerization. (a) Gel filtration profiles of DEDWT and DEDF122A expressed without N-terminal fusion protein. The corresponding Coomassie stained SDS gel are shown at the bottom. (b) Gel filtration profile of the monomer and dimer of DEDF122A on the Superdex 200 10/300 GL column in Day 1 and 7, respectively. No obvious inter-conversion occurred during this period. The gels were cropped for clarity, and full-length gels were shown in Supplementary Fig. 9.
Crystal structure of dimeric DEDF122A
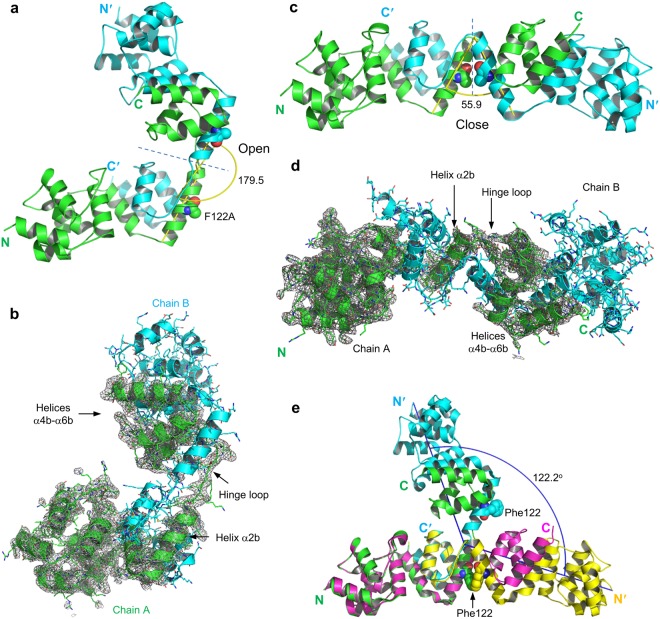
To gain insight into homotypic interactions in dimeric DEDs, we set out to determine the crystal structure of the self-assembled DEDs. The dimeric peak was collected and further concentrated for crystallization (Supplementary Fig. 1a). We obtained crystals of two crystal forms with varied cell dimensions under different crystallization conditions. The crystal structures were determined to resolutions of 3.1 Å and 3.6 Å, respectively, in the P1 space group (Table 1). Both structures reveal a novel domain-swapped conformation involving the exchange of the C-terminal helices α4b-α6b between the two subunits, and the hinge loop for the domain swapping is located at the segment linking helices α2b and α4b (Fig. 2a–d). The overall structure of the dimer displays a dumbbell-like shape with two-fold molecular symmetry. Each globular module, containing helices α1a-α7a and α1b-α2b from one monomer and helices α4b-α6b from the other monomer (Supplementary Fig. 1b,c), closely resembles the crystal structure15 of the monomeric DEDF122A/I128D with a root mean squared deviation of 0.46 Å for 155 equivalent Cα atoms between them (Supplementary Fig. 1d). Such domain-swapped dimerization represents the fourth homotypic interaction type distinct from other three known interaction types in the death-fold superfamily24.
Table 1.
Data collection and refinement statistics.
| PBD ID | 5H31 | 5H33 |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9779 | 1.5418 |
| Space group | P1 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 51.77, 51.71, 171.96 | 56.37, 50.79, 90.31 |
| a, b, g (°) | 90.0, 90.0, 90.0 | 90.0, 105.4, 90.0 |
| Resolution (Å) | 50.0–3.15 (3.26–3.15)* | 54.44–3.60 (3.73–3.60)* |
| Total reflections | 88207 | 19517 |
| R merge | 0.166 (0.945)* | 0.217 (0.560)* |
| I/sI | 12.2 (2.0)* | 4.4 (1.5)* |
| Completeness (%) | 99.7 (99.6)* | 99.6 (99.7)* |
| Redundancy | 11.5 (22.6)* | 3.31 (3.34)* |
| Refinement | ||
| Resolution (Å) | 49.59–3.17 | 43.63–3.60 |
| Unique reflections | 15236 | 5874 |
| Rwork/Rfree | 0.225/0.268 | 0.280/0.313 |
| No. atoms | 6064 | 3014 |
| Protein | 6064 | 3014 |
| Ligand/Ion | — | — |
| Water | — | — |
| B factors | ||
| Protein | 73.0 | 86.0 |
| Water | — | |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.002 | 0.003 |
| Bond angles (°) | 0.475 | 0.644 |
| Ramachandran statistics | ||
| Most Favored (%) | 96.25 | 96.37 |
| Allowed (%) | 3.75 | 3.63 |
| Outlier (%) | 0.0 | 0.0 |
*Values in parentheses are for highest-resolution shell.
Figure 2.
Structures of the dimeric DEDF122A. (a) Structure of the domain-swapped dimeric DEDF122A in the open conformation, in which F122A is solvent-exposed. (b) 2Fo − Fc electron density map (contoured at 1.0σ) of chain A of the domain-swapped dimeric DEDF122A in the open conformation. (c) Structure of the domain-swapped dimeric DEDF122A in the closed conformation, in which F122A is buried at the interface of the dimer. (d) 2Fo − Fc electron density map (contoured at 1.0σ) of chain A of the domain-swapped dimeric DEDF122A in the closed conformation. (e) Structural superimposition of the domain-swapped dimeric DEDF122A in the open (PDB ID: 5H33, green and cyan) and closed (PDB ID: 5H31, magenta and yellow) conformations. Phe122 was modeled from F122A and highlighted by sphere representation. The orientation of globule modules was shown by the rotational angle (122.2 degrees).
The two dimeric crystal structures differ in the relative orientation of the two globule modules of the dimer, which represent the closed and open conformations in terms of the inter-helical angle between helices α2b from the monomeric subunits, or in terms of the solvent exposure of the conserved hydrophobic patch Phe122/Leu123 (Fig. 2e). The transition from the open conformation to the closed conformation buries about 500 Å2 solvent-accessible surface area, suggesting that the closed conformation might be more stable in solution. Whereas the open conformation with the solvent-exposed hydrophobic patch Phe122/Leu123 might facilitate the interaction with the partner proteins such as FADD in the DISC (Supplementary Fig. 2)16,25.
We next compared the structure of the monomer subunit of the domain-swapped DEDF122A dimer with that of the monomeric DEDF122A/I128D. Both structures are essentially identical except for helices α4b-α6b, which rotate as a rigid body around the helix α2b from the closed conformation in the monomeric DEDF122A/I128D to the open conformation in the domain-swapped DEDF122A dimer (Supplementary Fig. 1d; Fig. 3a). The opening of helices α4b-α6b exposes about 2,500 Å2 solvent-accessible surface area, suggesting a high activation energy barrier for the transition between the DEDF122A monomer and domain-swapped dimer, as reflected by the observation that both the DEDF122A monomer and domain-swapped dimer are highly stable at room temperature without obvious inter-conversion in days (Fig. 1b).
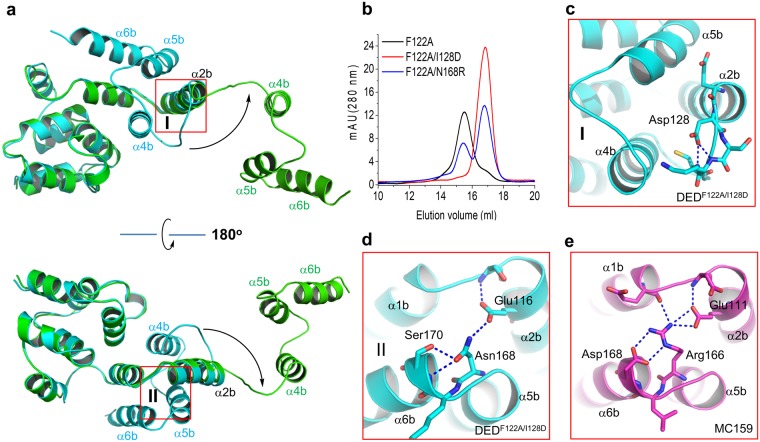
Figure 3.
Conformational changes in domain swapping of DEDF122A. (a) Superposition of structures of the monomeric subunit of the domain-swapped dimer of DEDF122A (green) and the monomeric DEDF122A/I128D (cyan; PDB ID: 4ZBW). Black arrows highlight the flipping of helices α4b-α6b during domain swapping. (b) Gel filtration profiles of DEDF122A, DEDF122A/I128D, DEDF122A/N168R. Both I128D and N168R mutations inhibit the dimerization of DEDF122A. (c) Close-up view of hydrogen bonds formed by I128D with the hinge loop in the monomeric DEDF122A/I128D. Hydrogen bonds are indicated by blue dashed lines in all figures. (d) Close-up view of hydrogen bonds formed by the E-NxSL motif in DEDF122A/I128D. (e) Close-up view of hydrogen bonds formed by the E/D-RxDL motif in vFLIP MC159 (PDB ID: 2BBR).
To further validate that the domain-swapped dimeric structures are not the artifact by the crystal packing, we generated a structure-based Q125C mutation on the helix α2b to facilitate the formation of an intermolecular disulfide bond between the two subunits of the domain-swapped dimer (Supplementary Fig. 3a). The SDS-PAGE analysis indicates that DEDF122A/Q125C migrates as both dimer and monomer in the absence of the reducing agent β-mercaptoethanol (βME), while runs as the monomer in the presence of βME, suggesting the disulfide bond formation between the two subunits in the absence of reducing agent. In contrast, F122A mutant runs as mainly a monomer in the absence or presence of βME (Supplementary Fig. 3b). We tried to perform the similar SDS-PAGE analysis for the wildtype DEDs (DEDWT) and the Q125C (DEDQ125C) mutant. No dimer staining for both WT and Q125C DEDs no matter in the absence or presence of βME, which might be attributed to the low concentration of the proteins and the potential interference of the mixed slyD.
Point mutations in DED2 disrupt the dimerization of DEDF122A
The domain swapping in dimeric DEDF122A involves the exchange of helices α4b-α6b in DED2 of DEDF122A. To understand the potential factors underlying the domain swapping, we tried to identify the point mutations disrupting the dimerization of DEDF122A. In the previous study15, Ile128 has been identified as the key residue that confers the insoluble and aggregation-prone property of DEDs of caspase-8. Here I128D mutation do markedly inhibit the dimerization and oligomerization of DEDF122A (Fig. 3b). Ile128 is located at the connection between the helix α2b and the hinge loop. The side chain carboxylate group of I128D form two favorable hydrogen bonds with the backbone N-H of the hinge loop, which would restrain the rotation of the hinge loop and inhibit the domain swapping (Fig. 3c). The furthermore sequence alignment and structural analysis reveals that the corresponding residue of Asp128 in DEDF122A/I128D is Asn123 in the viral DED homologue vFLIP (Supplementary Fig. 4a). Similar to Asp128 in DEDF122A/I128D, Asn123 also forms a hydrogen bond with the backbone N-H of the helix α3b in vFLIP (Supplementary Fig. 4b,c), which might account for the monomeric preference of vFLIP16,17. Moreover, Arg166 in the conserved E/D-RxDL motif on helices α2b and α6b of vFLIP forms extensive hydrogen bonding interactions between helices α2b and α6b, whereas the corresponding residue Asn168 in DEDF122A would disrupts these interactions, and facilitate the opening of helices α4b-α6b and domain swapping of DEDF122A (Fig. 3d,e; Supplementary Fig. 4d). As expected, the N168R mutation significantly inhibits the dimerization of DEDF122A (Fig. 3b).
Dimer formation is important for the filament formation of DEDs
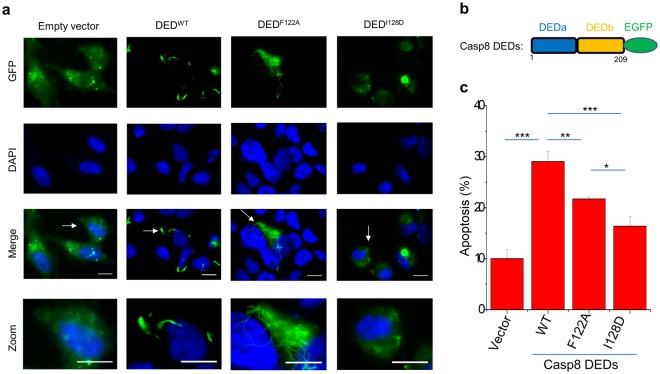
The tandem DEDs of caspase-8 have been shown to form novel cytoplasmic death effector filaments (DEFs) upon transient overexpression19–22. We thus examined whether the dimerization is important for the DEFs formation (Fig. 4a,b). Compared with the wild-type caspase-8 DEDs, the I128D mutation abrogated the DEFs formation for the tandem DEDs of caspase-8, and markedly reduced the apoptosis of HeLa cells induced by the DEFs (Fig. 4c), highlighting the essential role of the domain-swapped dimerization in the DEFs formation. Intriguingly, DEDF122A still formed the distinctive filamentous structure but with a more diffusive pattern (Fig. 4a), which is consistent with the in vitro feature of DEDF122A that still forms dimer but much less aggregates (Fig. 1a). The DEFs formation and induced apoptosis are less prominent for DEDF122A compared with the wild-type DEDWT, suggesting a potential role of Phe122 in the high-order aggregation of DEDs of caspase-8.
Figure 4.
Cellular death effector filament formation. (a) HeLa cells were transfected with either empty vector (EGFP) or the caspase-8 DED variants DEDWT-EGFP or DEDF122A-EGFP or DEDI128D-EGFP for 18 hr before fixing and staining with DAPI. Cells were imaged and a representative field for each transfection is shown. Lower panels show enlargement of those areas arrowed in the merge panels. Scale bar, 20 μm. (b) Domain schematics of the caspase-8 DED variants fused with EGFP. (c) Apoptosis induction of HeLa cells by the caspase-8 DED variants. Apoptosis rates were quantified as the percentage of Annexin V-APC positive versus GFP positive cells. Error bars, s.d. of independent experiments (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 (One-way ANOVA).
The recent cryo-EM structure has revealed the important role of Phe122 in the formation of the filamentous structure by the tandem DEDs of caspase-8 in vitro, in which Phe122 mediates the type I interaction between different DEDs units in the filament (Supplementary Fig. 5a)18. This model might also account for the role of Phe122 in the cytoplasmic DEFs formation. On the other hand, this model does not explicitly explain that the I128D mutation abrogates the DEFs formation, since Ile128 is not directly involved in any types of interactions in the filamentous structure (Supplementary Fig. 5b), and no obvious structural change occurs for the tandem DEDs caused by the I128D mutation (Supplementary Fig. 5c). Alternatively, we tried to reconcile the roles of Phe122 and Ile128 in the DEF formation based on the domain-swapped model. Domain swapping has been recognized as an aggregation mechanism for a number of proteins26,27. Both dimeric DEDF122A exhibit closed-ended domain-swapped structures, which would block the oligomerization of the DEDs and the formation of DEFs. Based on the previous studies28–30, it would be reasonable to propose that the closed-ended domain-swapped structures can switch to the open-ended domain-swapped structures and form the final polymerized filaments due to the flexible hinge loop (Supplementary Fig. 6). The I128D mutation disrupts the domain swapping thus in turn blocks the filament formation, and the burial of the hydrophobic residue Phe122 in the wildtype DEDs would more favor the filament formation compared with F122A in this study and F122E in the previous study18. Moreover, the DEDs of the full-length caspase-8 could also be integrated into the filaments, leading to the activation of caspase-8 and the downstream apoptosis (Supplementary Fig. 7)19.
Dimer formation of DEDs is essential for the Fas pathway
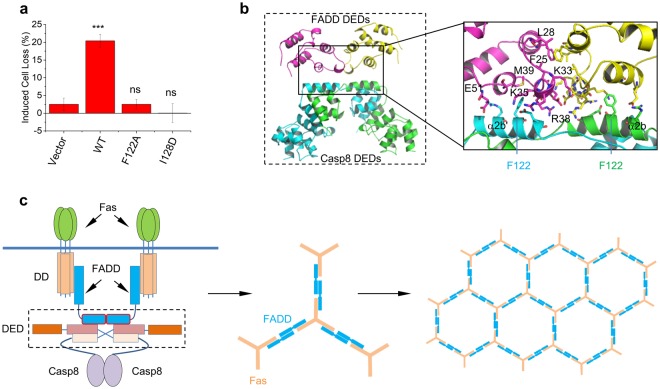
To further assess the physiological relevance of the domain-swapped dimer of DEDs, we determined the ability of anti-Fas agonist antibody to induce apoptosis of caspase-8-deficient Jurkat cells transfected with empty vector (EV) and wild-type (WT), F122A and I128D mutant procaspase 8 (Fig. 5a). Transfection of wild-type caspase-8, but neither F122A nor I128D mutants, rescued the apoptotic response of caspase-8-deficient Jurkat cells towards anti-Fas agonist antibody. The defect of I128D mutant highlights the crucial role of the domain-swapped dimerization of DEDs of caspase-8 in the extrinsic apoptotic pathway. On the other hand, though F122A mutant has the capability to form dimer in solution and in the crystal, and even to form intracellular filaments, it was severely impaired in rescuing the cellular apoptosis of caspase-8-deficient Jurkat cells induced by anti-Fas agonist antibody, suggesting a distinct role of Phe122 rather than promoting dimer and filament formation in the extrinsic apoptotic pathway.
Figure 5.
Model of proximity-induced dimerization of caspase-8. (a) Apoptosis induction of caspase-8-deficient Jurkat T-cells transfected with the full-length caspase-8 variants by agonistic Fas antibody. The percentage cell loss of GFP+ cells that are negative for annexin V was quantitated. Error bars, s.d. of independent experiments (n = 3). ns, not significant, ***P < 0.001 (One-way ANOVA). (b) Modeled structure of the self-associated dimer of FADD DEDs binding with the domain-swapped dimer of caspase-8 DEDs. Inset, details of the interactions at the dimeric interface of FADD DEDs and the binding interface between DEDs of FADDs and caspase-8. Key residues involved in interactions are highlighted by stick representation. (c) Fas receptor clustering array bridged by the self-associated FADDs (middle and right). Fas and FADD are colored in wheat and marine blue, respectively. Minimal DISC model composed of dimeric Fas trimers, dimeric FADDs and caspase-8s is shown in the left panel. The prodomains of caspase-8 exist in a domain-swapped dimer.
Phe122 of caspase-8 has been shown to be important for interaction with FADD DED15,18 and the recruitment of caspase-8 to the DISC31, but it is unclear how Phe122 mediates the interaction between caspase-8 and FADD via their DEDs at the DISC, and how the dimerization of the DEDs of caspase-8 involves in this interaction. The previous study32 has indicated that FADD self-association via the DED is required for stable interaction of FADD with Fas and with caspase-8. The replacement of the DED of FADD by a dimeric leucine-zipper coiled-coil maintains the capability of the death domain (DD) of FADD to bind with the Fas DD, suggesting that the dimerized DEDs of FADD might be the functional unit for the interactions between the DDs of FADD and Fas. In line with this hypothesis, the dimerized DEDs of FADD might serve as the docking site of the domain-swapped dimer of the DEDs of caspase-8 and facilitate the recruitment of caspase-8 to the DISC.
To understand the potential binding mode between FADD and caspase-8, we used molecular modeling to generate a complex model between the DEDs of FADD and caspase-8. Given a small cluster including Phe25, Leu28 and Lys33 critical for the self-assembly of FADD, we first built up the dimeric structures of the DEDs of FADD with this cluster as the interfacial residues. Using Phe122 of the opened domain-swapped dimeric DEDs of caspase-8 as the anchoring points, a potential complex model was generated for the dimeric DEDs of FADD and caspase-8 (Fig. 5b). The solvent exposed Phe122 of caspase-8 fits well into the hydrophobic groove lined by the residues on the helices α3 and α4 of FADD, which is consistent with the proposed interactions between FADD and caspase-8. The dimerization of the DEDs of both FADD and caspase-8 strengthens the interaction between FADD and caspase-8, and vice versa. This model allows the integration of the full-length caspase-8 and provides insight into the proximity-induced dimerization mechanism of caspase-8 activation within the DISC (Supplementary Fig. 8).
Discussion
DISC formation and subsequent caspase-8 activation are critical initial events in the extrinsic apoptotic pathway7. Extensive previous studies have provided significant insights into understanding this process, but the underlying mechanisms still need further investigation and continuing refinement18,21,22,32–34. In this study, we found that the tandem DEDs of caspase-8 form a novel domain-swapped dimer, in which the domain-swapping involves the exchange of the C-terminal helices α4b-α6b between the two subunits. Employing functional studies we demonstrated that dimerization of the DEDs is essential for activation of caspase-8. Furthermore, though the conserved residue Phe122 is not necessary for the dimerization, it is essential for the activation of caspase-8 and the downstream apoptosis cascade.
Our work supports the proximity-induced activation model of caspase-85–7. The domain-swapped dimerization of the DEDs brings the protease domains into proximity and facilitates the dimerization and activation of caspase-8, which closely resembles that occurs in the activation of the chimeric FKBP-caspase-8 induced by the dimerizing agent FK10129. Moreover, upon engaging with the DED of FADD, the domain-swapped dimer of the DEDs of caspase-8 are also easily integrated into the clustered Fas/FADD complexes in which trimeric Fas assemble into arrays bridged by dimeric FADD (Fig. 5c)32,35. This model explicitly explains the observation that dimerized FasL trimers have about 1000-fold enhancement of the signaling efficiency compared with the non-crosslinked FasL trimer10. This model is also consistent with the observation that the membrane-bound CD95 ligand rather than the soluble one is critical for apoptosis36. We noted that it is currently unclear whether the domain-swapped dimerization of the DEDs of caspase-8 occurs before or upon binding with FADD in the DISC, and it is also unclear how the cytosolic monomeric DEDs switches to the domain-swapped dimer upon the upstream death signal in physiological condition.
It is not uncommon to observe the domain swapping of functional proteins in the death domain superfamily. For example, the N-terminal caspase-recruitment domain (CARD) of Nod1 forms a domain-swapped homodimer by swapping the H6 helices at the carboxy termini37–39. Moreover, the N-terminal pyrin domain (PYD) of NLRP14 also forms a homodimer by a rearrangement of the α5/6 stem-helix of the C-termini40. In addition, domain swapping has been well described for the dimerization/oligomerization of both the pro-apoptotic proteins Bax/Bak41,42 and the pro-survival protein BCL-XL43 in the intrinsic apoptosis pathway, in which the transition from the monomer to the domain-swapped dimer is facilitated by the detergents or membrane lipid. We speculated that the domain-swapped dimerization of the DEDs of caspase-8 might also be facilitated by the membrane lipid when recruited to the membranous DISC, or possibly assisted by yet identified chaperones44–46.
DEDs can form death effector filaments (DEFs) intracellularly or in vitro at the high concentration of DEDs18–22. The recent cryo-EM structure of caspase-8 tandem DED filaments has revealed that the DEFs formation likely occurs through repeated self-assembly by type I, II, and III interactions of the DEDs, in which both DED1 and DED2 are critical for the self-assembly18. This structure does not provide a clear understanding for the previous study that only DED2 but not DED1 still forms fine cytoplasmic filaments19, while this discrepancy can be explained by the domain-swapped filament model in which only DED2 is involved in oligomerization (Supplementary Fig. 6c). On the other hand, the filamentous structure generally forms under high concentrations of DEDs with the length of about 100 nm or even longer18–22, raising a question about its physiological relevance. Given a limited number of caspase-8 in cell under physiological condition, the probability of such long filamentous structure for caspase-8 is very low during the apoptotic pathway triggered by the death ligand. Consistent with this analysis, the DEDs of caspase-8 forms spot-like rather than filamentous structure under a low overexpression level upon FasL stimulation22, more likely reflecting the arrayed FADD in signaling protein oligomerization transduction structures (SPOTS)32,35,47. This observation can be easily explained by our model in which the domain-swapped dimer of the DEDs of caspase-8 bind with the dimeric DEDs of FADD in the membranous DISC, the fluorescent signal of caspase-8 therefore reflects the array of FADD. In contrast, upon a high level of overexpression of the DEDs of caspase-8, the cytoplasmic DEFs were detected even without FasL stimulation18–22.
In summary, we reported the crystal structures of a domain-swapped dimer of the DEDs of caspase-8. Together with functional analysis, we demonstrated that the domain-swapped dimerization is critical to the activation of caspase-8. Furthermore, we generated a potential complex model for the homotypic interactions between the DEDs of FADD and caspase-8, which would shed new light on the DISC in the extrinsic apoptotic pathway. Meanwhile, the domain-swapped dimerization represents the fourth homotypic interaction type, together with other three known interaction types, for understanding the functions of proteins in the death-fold superfamily.
Methods
Reagents and cell lines
The monoclonal agonist antibody against CD95 (Anti-Fas) was purchased from Millipore (Bedford, MA). Caspase-8 deficient Jurkat cells were kind gift from Dr. Junying Yuan at Harvard University. HeLa cells were obtained from Cellbank (Shanghai, China). Jurkat cells were cultured in RPMI1640 and HeLa cells were cultured in DMEM (Corning, Manassas, VA), supplemented with 10% fetal bovine serum (PAN-Biotech, Aidenbach, Germany). All the other biochemical reagents were from Sigma Aldrich.
Protein expression and purification
The DNA encoding the death effector domain (DED) of human caspase-8 (residues 1–188, Uniprot accession number Q14790-1) were subcloned into a pET-21b vector with a C-terminal His6 tag or subcloned into a pET-28a vector with an N-terminal His-SUMO tag by a standard PCR method. The relevant mutants of Caspase-8 DED (residues1-188) were generated by Fast Mutagenesis System (TransGen Biotech, China). All the Caspase-8 DED constructs mentioned above were transformed into Rosetta (DE3) pLysS cells (Novagen). All of the recombinant protein expression were induced with 0.1 mM IPTG at 20 °C overnight. His-tagged proteins were purified with Qiagen Ni-NTA agarose according to the manufacturer’s instructions. After the first step purification, His-SUMO tag was removed by ULP-1 enzyme, and the proteins were re-chromatographed on a Ni-NTA column. The His-SUMO-tag-cleaved protein was further purified by gel-filtration chromatography (Superdex 200 10/300 GL, GE Healthcare).
Crystallization, data collection and structure determination
The dimeric peak of DEDF122A was collected and concentrated to 6–8 mg/ml in buffer A (20 mM Tris, pH 8.0, 1 mM DTT and 150 mM sodium chloride) for crystallization. Using hanging drop vapor diffusion method, crystals of DEDF122A in two crystal forms grew in a few days in condition A (50 mM sodium chloride, 100 mM Tris pH 8.5, 22.5% PEG3350, at 20 °C, for the crystal form A with short cell dimension and the crystal structure in the open conformation) and condition B (100 mM sodium chloride, 100 mM Tris, pH 8.5, 21% PEG3350, 10 mM Sarcosine, at 20 °C, for the crystal form B with long cell dimension and the crystal structure in the closed conformation), respectively. For cryo-freezing of the crystals, 20% glycerol in mother liquor was used as Cryo-protectant by multi-steps transferring. The crystals were flash frozen in liquid nitrogen for a further X-ray diffraction data collection.
The diffraction data for the crystal form A were collected in homesource Rigaku micromax-002+. The diffraction data for the crystal form B were collected at beamline BL17U of Shanghai Synchrotron Radiation facility. Data were integrated, scaled and processed by HKL200048. The detailed statistics are shown in Extended Data Table 1.
The crystal structure was determined by molecular replacement carried out by Phaser-PHENIX program suite49,50, the monomer of DEDF122A/I128D (PDB accession code 4ZBW15) was used as the initial search model. The final models were manually built in Coot51 and refined by Refmac52,53 in CCP4. The refinement statistics were listed in Supplementary Table 1. All crystallographic figures were drawn in PyMOL54.
Immunofluorescence assay
HeLa cells were cultured on coverslips in 12 well plates for 24 hr before transfected with 1.4 μg/well of vectors using Lipofectamine® 2000 Reagent (Invitrogen) according to the manufacturer’s instructions. 18 hr later, cells were fixed in Formalin solution for 30 min at room temperature and the nuclei was stained with DAPI for 20 min. Cells were washed in TBS buffer (20 mM Tris, 200 mM NaCl, pH = 7.6) 3 times. Images were taken with AxioImager A1 microscope and AxioCam digital camera (Zeiss, Oberkochen, Germany).
Apoptosis assays
For cell-based apoptosis assay, the wild type (WT) and F122A, I128D mutants of full length Caspase-8 (residues 1–479) and DEDs (residues1–209) were subcloned into pEGFP-N1 vector. HeLa cells (3 × 105) were transfected with plasmids (4 μg) as indicated in figure legends using TransIn EL Transfection Reagent (TransGen Biotech, China). For Jurkat cells, 2 μg plasmids were electroporated into 1 × 106 cells with Amaxa SE Cell Line 4D-Nucleofector X Kit (Lonza Group Ltd, Basel, Switzerland). 18 hr post transfection/electroporation, cells were further stimulated with anti-Fas (1 μg/ml) for 20 hr before harvest. The cell sample were then analyzed for apoptosis via Annexin V staining (eBioscience) using FACSCalibur flow cytometer (Becton Dickinson, NJ, USA). Apoptosis rates were quantified as the percentage of Annexin V-APC positive versus GFP positive cells.
Electronic supplementary material
Acknowledgements
This work was supported by funds from the NSFC (21521003, 81373326, and 21290183 to J.Q.; 81400826 to Q.L.), program of the Science and Technology committee of Guangdong (2014A030312004 and 2015A030313887 to Q.L.) and Shenzhen Science and Technology innovation program (JCYJ20150529095420031, JCYJ20170412150827191 to J.Q.; JCYJ20170818090609800 to Q.L.). We thank Dr. Junying Yuan (Harvard Medical School, Cambridge, MA) for providing caspase-8-deficient Jurkat T cells. We thank the staff of beamline BL17U at Shanghai Synchrotron Radiation Facility for the technical assistance during data collection.
Author Contributions
C.S. and J.Q. designed all experiments. C.S., J.P. and X.G. performed the experiments. L.Z. contributes to the X-ray diffraction data collection and processing. Q.L. assisted J.P. in cellular experiments. All authors discussed the results and commented on the manuscript. J.Q. supervised all aspects of the project and wrote the manuscript.
Data Availability
The atomic coordinates and structure factors have been deposited at the Protein Data Bank under accession numbers 5H31 and 5H33. The data that support the findings of this study are available within the article and its Supplementary Information files, or available from the corresponding author on request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chen Shen, Jianwen Pei and Xiaomin Guo contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-35153-5.
References
- 1.Oberst A, Green DR. It cuts both ways: reconciling the dual roles of caspase 8 in cell death and survival. Nat. Rev. Mol. Cell Biol. 2011;12:757–763. doi: 10.1038/nrm3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salvesen GS, Walsh CM. Functions of caspase-8: the identified and the mysterious. Seminars in Immunol. 2014;26:246–252. doi: 10.1016/j.smim.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muzio M, et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/S0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 4.Boatright KM, et al. A Unified Model for Apical Caspase Activation. Mol. cell. 2003;11:529–541. doi: 10.1016/S1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 5.Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An Induced Proximity Model for Caspase-8 Activation. J. Biol. Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 6.Salvesen GS, Dixit VM. Caspase activation: The induced-proximity model. Proc. Natl Acad. Sci. USA. 1999;96:10964–10967. doi: 10.1073/pnas.96.20.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr. Opin. Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Mace PD, Riedl SJ. Molecular cell death platforms and assemblies. Curr. Opin. Cell Biol. 2010;22:828–836. doi: 10.1016/j.ceb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oberst A, et al. Inducible dimerization and inducible cleavage reveal a requirement for both processes in caspase-8 activation. J. Biol. Chem. 2010;285:16632–16642. doi: 10.1074/jbc.M109.095083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holler N, et al. Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol. Cell. Biol. 2003;23:1428–1440. doi: 10.1128/MCB.23.4.1428-1440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang DW, Xing Z, Capacio VL, Peter ME, Yang X. Interdimer processing mechanism of procaspase-8 activation. EMBO J. 2003;22:4132–4142. doi: 10.1093/emboj/cdg414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pop C, Fitzgerald P, Green DR, Salvesen GS. Role of proteolysis in caspase-8 activation and stabilization. Biochemistry. 2007;46:4398–4407. doi: 10.1021/bi602623b. [DOI] [PubMed] [Google Scholar]
- 13.Fan L, Freeman KW, Khan T, Pham E, Spencer DM. Improved artificial death switches based on caspases and FADD. Hum. Gene Ther. 1999;10:2273–2285. doi: 10.1089/10430349950016924. [DOI] [PubMed] [Google Scholar]
- 14.Yu JW, Shi Y. FLIP and the death effector domain family. Oncogene. 2008;27:6216–6227. doi: 10.1038/onc.2008.299. [DOI] [PubMed] [Google Scholar]
- 15.Shen C, et al. Crystal structure of the death effector domains of caspase-8. Biochem. Biophys. Res. Commun. 2015;463:297–302. doi: 10.1016/j.bbrc.2015.05.054. [DOI] [PubMed] [Google Scholar]
- 16.Yang JK, et al. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol. cell. 2005;20:939–949. doi: 10.1016/j.molcel.2005.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li FY, Jeffery PT, Yu JW, Shi Y. Crystal structure of a viral FLIP: insights into flip-mediated inhibition of death receptor signaling. J. Biol. Chem. 2006;281:2960–2968. doi: 10.1074/jbc.M511074200. [DOI] [PubMed] [Google Scholar]
- 18.Fu TM, et al. Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex. Mol. Cell. 2016;64:236–250. doi: 10.1016/j.molcel.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegel RM, et al. Death-effector Filaments: novel cytoplasmic structures that recruit caspases and trigger apoptosis. J. Cell Biol. 1998;141:1243–1253. doi: 10.1083/jcb.141.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbero S, et al. Identification of a critical tyrosine residue in caspase 8 that promotes cell migration. J. Biol. Chem. 2008;283:13031–13034. doi: 10.1074/jbc.M800549200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickens LS, et al. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Mol. Cell. 2012;47:291–305. doi: 10.1016/j.molcel.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schleich K, et al. Stoichiometry of the CD95 death-inducing signaling complex: experimental and modeling evidence for a death effector domain chain model. Mol. Cell. 2012;47:306–319. doi: 10.1016/j.molcel.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Scholz C, et al. SlyD proteins from different species exhibit high prolyl isomerase and chaperone activities. Biochemistry. 2006;45:20–33. doi: 10.1021/bi051922n. [DOI] [PubMed] [Google Scholar]
- 24.Kersse K, Verspurten J, Vanden Berghe T, Vandenabeele P. The death-fold superfamily of homotypic interaction motifs. Trends Biochem. Sci. 2011;36:541–552. doi: 10.1016/j.tibs.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Carrington PE, et al. The structure of FADD and its mode of interaction with procaspase-8. Mol. cell. 2006;22:599–610. doi: 10.1016/j.molcel.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 26.Knaus KJ, et al. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 2001;8:770–774. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 27.Eakin CM, Attenello FJ, Morgan CJ, Miranker AD. Oligomeric assembly of native-like precursors precedes amyloid formation by beta-2 microglobulin. Biochemistry. 2004;43:7808–7815. doi: 10.1021/bi049792q. [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki M, Li W, Johnson DJ, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455:1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- 29.Das P, King JA, Zhou R. Aggregation of γ-crystallins associated with human cataracts via domain swapping at the C-terminal β-strands. Proc. Natl. Acad. Sci. USA. 2011;108:10514–10519. doi: 10.1073/pnas.1019152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gronenborn AM. Protein acrobatics in pairs–dimerization via domain swapping. Curr. Opin. Struct. Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes MA, et al. Co-operative and Hierarchical Binding of c-FLIP and Caspase-8: A Unified Model Defines How c-FLIP Isoforms Differentially Control Cell Fate. Mol. Cell. 2016;61:834–849. doi: 10.1016/j.molcel.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandu C, et al. FADD self-association is required for stable interaction with an activated death receptor. Cell Death Differ. 2006;13:2052–2061. doi: 10.1038/sj.cdd.4401966. [DOI] [PubMed] [Google Scholar]
- 33.Scott FL, et al. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature. 2009;457:1019–1022. doi: 10.1038/nature07606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, et al. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat. Struct. Mol. Biol. 2010;17:1324–1329. doi: 10.1038/nsmb.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Werner MH, Wu C, Walsh CM. Emerging roles for the death adaptor FADD in death receptor avidity and cell cycle regulation. Cell Cycle. 2006;5:2332–2338. doi: 10.4161/cc.5.20.3385. [DOI] [PubMed] [Google Scholar]
- 36.O’Reilly LA, et al. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature. 2009;461:659–663. doi: 10.1038/nature08402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coussens NP, Mowers JC, McDonald C, Nun˜ez G, Ramaswamy S. Crystal structure of the Nod1 caspase activation and recruitment domain. Biochem. Biophys. Res. Commun. 2007;353:1–5. doi: 10.1016/j.bbrc.2006.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srimathi T, et al. Monomer/dimer transition of the caspase-recruitment domain of human Nod1. Biochemistry. 2008;47:1319–1325. doi: 10.1021/bi7016602. [DOI] [PubMed] [Google Scholar]
- 39.Ver Heul AM, Gakhar L, Piper RC, Subramanian R. Crystal structure of a complex of NOD1 CARD and ubiquitin. PLoS One. 2014;9:e104017. doi: 10.1371/journal.pone.0104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eibl C, Hessenberger M, Wenger J, Brandstetter H. Structures of the NLRP14 pyrin domain reveal a conformational switch mechanism regulating its molecular interactions. Acta Crystallogr D Biol Crystallogr. 2014;70:2007–2018. doi: 10.1107/S1399004714010311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Czabotar PE, et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 42.Brouwer JM, et al. Bak core and latch domains separate during activation, and freed core domains form symmetric homodimers. Mol. Cell. 2014;55:938–946. doi: 10.1016/j.molcel.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 43.O’Neill JW, Manion MK, Maguire B, Hockenbery DM. BCL-XL dimerization by three-dimensional domain swapping. J. Mol. Biol. 2006;356:367–381. doi: 10.1016/j.jmb.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 44.Takayama S, John C, Reed JC, Homma S. Heat-shock proteins as regulators of apoptosis. Oncogene. 2003;22:9041–9047. doi: 10.1038/sj.onc.1207114. [DOI] [PubMed] [Google Scholar]
- 45.Marianayagam NJ, Sunde M, Matthews JM. The power of two: protein dimerization in biology. Trends Biochem. Sci. 2004;29:618–625. doi: 10.1016/j.tibs.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Ali MH, Imperiali B. Protein oligomerization: how and why. Bioorg. Med. Chem. 2005;13:5013–5020. doi: 10.1016/j.bmc.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 47.Siegel RM, et al. SPOTS: SPOTS: signaling protein oligomeric transduction structures are early mediators of death receptor-induced apoptosis at the plasma membrane. J. Cell Biol. 2004;167:735–744. doi: 10.1083/jcb.200406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 53.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delano, W. L. The PyMol Molecular Graphics System, Delano Scientific LLC, San Carlos, CA, USA, http://www.pymol.org/ (2008).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited at the Protein Data Bank under accession numbers 5H31 and 5H33. The data that support the findings of this study are available within the article and its Supplementary Information files, or available from the corresponding author on request.