Abstract
Family, twin and adoption studies demonstrate clearly that alcohol dependence and alcohol use disorders are phenotypically complex and heritable. The heritability of alcohol use disorders is estimated at approximately 50–60% of the total phenotypic variability. Vulnerability to alcohol use disorders can be due to multiple genetic or environmental factors or their interaction which gives rise to extensive and daunting heterogeneity. This heterogeneity makes it a significant challenge in mapping and identifying the specific genes that influence alcohol use disorders. Genetic linkage and (candidate gene) association studies have been used now for decades to map and characterize genomic loci and genes that underlie the genetic vulnerability to alcohol use disorders. These approaches have been moderately successful in identifying several genes that contribute to the complexity of alcohol use disorders. Recently, genome-wide association studies have become one of the major tools for identifying genes for alcohol use disorders by examining correlations between millions of common single-nucleotide polymorphisms with diagnosis status. Genome-wide association studies are just beginning to uncover novel biology; however, the functional significance of results remains a matter of extensive debate and uncertainty. In this review, we present a select group of genome-wide association studies of alcohol dependence, as one example of a way to generate functional hypotheses, within the addiction cycle framework. This analysis may provide novel directions for validating the functional significance of alcohol dependence candidate genes.
This article is part of the Special Issue entitled “Alcoholism”.
Keywords: Genetics, Alcohol use disorder, Alcohol dependence, Genetic mapping, Addiction cycle, Genome-wide association study
1. Introduction
Alcoholism is a chronic relapsing disorder characterized by a loss of volitional control over consumption, impaired decision-making, pathological preoccupation with alcohol seeking at the expense of healthier forms of behavior and a compulsive drive for harmful drinking in the face of serious life consequences (e.g., deteriorating health, job and family loss). The Diagnostic and Statistical Manual of the American Psychiatric Association, fourth edition (DSM-IV), established criteria for both alcohol dependence (alcoholism) and alcohol abuse. Research studies based on diagnosis compare those who meet or do not meet criteria for alcohol dependence. Alcohol dependence is defined by the presence of three or more of a set of seven criteria. Alcohol abuse is defined by the presence of two out of four criteria in the absence of meeting criteria for dependence. The recent publication of DSM-5 in 2013 combined the two separate diagnoses of alcohol abuse and alcohol dependence from the DSM-IV into a single dimensional diagnosis of alcohol use disorder (AUD) with mild, moderate, and severe subclassifications. In DSM–5, anyone meeting any two of eleven criteria during a specified period received the diagnosis of AUD where severity is based on the number of criteria met.
The prevalence of AUD (or alcohol dependence in DSM-IV parlance) has increased in the last decade in the general population of United States adults, 18 years of age or older. Results from the National Epidemiologic Survey on Alcohol and Related Conditions (NESARC) reported that the DSM-IV 12-month and lifetime prevalence rates in 2001–2002 increased from 8.5% to 30.3%, respectively to 12.7% and 43.6% in 2012 through 2013, respectively (Grant et al., 2015). Despite the increase in AUD over the last decade, many individuals never seek or receive evidence-based treatments. The major reasons are due to fears of stigmatization and the surprisingly still commonly held belief that alcohol dependence is not a medical condition that can be effectively treated with medications or behavioral therapies (Cohen et al., 2007). This situation amounts to a clear treatment exigency that is only exacerbated by the limited number of approved and effective pharmacotherapies that currently exist for alcohol dependence. One promising avenue to ameliorate the treatment gap would be to provide many novel pharmacotherapies that have diverse mechanisms of action that are well-tolerated and effective. This strategy has merit because the arrival of many novel pharmacotherapies for depression over the last 30 years, for example, helped catalyze a transformation in the way depression is perceived and treated. In this regard, the power of high-throughput discovery based methods from disciplines such as genomics can be used to identify many new drug targets (Kingsmore et al., 2008; Sanseau et al., 2012).
Phenotypic variability in AUD is influenced by two major components: (1) environment and (2) genes. AUD are significantly polygenic and are highly heterogeneous making it a great challenge to define a specific universal set of genetic and environmental factors that influence risk across every population affected. Therefore, AUD are not unlike other common complex diseases or disorders such as diabetes, asthma or cancer. The ubiquitous problem of pervasive heterogeneity observed in many common diseases including AUD has ushered in the era of personalized medicine which is hoped to replace the largely unsuccessful one-size fits all approach to treatment. In this regard genomic medicine is particularly relevant to fulfilling the goals of tailoring treatments based on an individual's genomic makeup. This is highly relevant to expanding the range of pharmacotherapies for the significantly unmet medical needs of those affected with a AUD.
Although the DSM, now in its 5th edition, has provided a standardized and reliable way of diagnosing AUD and other substance use disorders, there still remains a great amount of heterogeneity that is uncaptured and the DSM in general has not fully integrated the many advances on the neuroscience and genetics of AUD (Litten et al., 2015). Unfortunately, there is burgeoning evidence that the new AUD construct for DSM-5 has further increased the heterogeneity existing in the DSM. A recent study showed that the DSM-5 specific categories of “moderate” to “severe” AUD corresponds to what was previously captured by “alcohol dependence”; the addition of a “mild” AUD category represents a diagnostic label of unknown clinical relevance (Compton et al., 2013). Therefore, attempts have recently been made to move towards a neuroscience-based framework for addictive disorders with the goal of establishing more biological and genetically relevant domains that one day could be incorporated into a diagnostic framework (Kwako et al., 2016). In particular, a more biologically-based diagnostic framework for AUD will undoubtedly aid discovery of more effective pharmacotherapies by reducing patient heterogeneity, for example. To this end, one of the most useful paradigms for conceptualizing AUD and drug addiction is taken from advances in the understanding of the neurocircuitry of addiction and the impact on the brain reward, stress, and executive function systems. (Koob and Volkow, 2016). As such, addiction has been defined as a three stage cycle containing: (1): binge/intoxication, (2): withdrawal/negative affect and (3): preoccupation/anticipation stages that represent distinct neurocircuitry and functional domains. Initial use of alcohol is driven by positive reinforcement mechanisms that involves patterns of binge-and-intoxication. Alcohol or stimuli associated with alcohol act as a positive reinforcer, to strengthen behavior, which engages the mesolimbic dopamine and opioid reward systems in the central nervous system. In addition, engagement of the dorsal striatum during alcohol addiction is thought to help to solidify habitual behaviors associated with drug seeking and taking. Namely, neuroadaptations in the dorsal striatum involve changes in glutamate, GABA and the endocannabinoid system (Koob and Volkow, 2010). After a period of repeated binge/intoxication neuroadaptations occur leading to a negative emotional state (e.g., anxiety, depression, anhedonia) induced by alcohol withdrawal. Negative reinforcement mechanisms predominately drive behavior where motivation to continue excessive consumption is strengthened by the alleviation of negative affect by resumed alcohol drinking. Neuroadaptations in the brain reward and stress systems (e.g., extended amygdala) drive dependent (compulsive) drinking. Neuroadaptations occurring in the circuitry of the mesolimbic dopamine system, particularly the ventral tegmental area (VTA), the nucleus accumbens (NAc), and amygdala, cause enhanced saliency value of the drug and drug stimuli, and the decreased sensitivity to natural reinforcers. For example, decreases in dopamine and GABA system function, and increases in dynorphin-κ opioid activity in the ventral striatum, representing a general decrease in reward system function (within-system opponent processes), are coupled with enhancement of cortioctropin-releasing factor (CRF) in the extended amygdala as well as blunting of the hypothalamic–pituitary–adrenal (HPA) axis (between-system opponent processes). Finally, compulsive dependent consumption involves poor inhibitory control and poor executive functioning, which are mediated by prefrontal cortical regions of the brain. For example, regions of the prefrontal cortex are selectively damaged by chronic intermittent alcohol and result in poor decision making that can engage and perpetuate the addiction cycle. This preoccupation/anticipation stage is associated with a narrowing of behaviors exclusively to those involved with alcohol seeking and consumption. Taken together, neuroadaptations in the brain reward, stress, habit formation and executive functioning centers coalescence to drive compulsive alcohol intake.
There has been a recent explosion in the amount of genomic information now available for common complex diseases including AUD. This is primarily due to the advent of the genome-wide association study (GWAS). However, how new genomic information afforded by GWAS of alcohol dependence as well as information from other gene identification approaches fits within the addiction cycle has not been explored. In this paper, we review a selected series of “first-generation” GWAS of alcohol dependence after briefly tracing the history of the genetics of alcohol dependence that led to the GWAS-era. The first generation of GWAS of alcohol dependence in general have been viewed as yielding disappointing results due to various technical, practical and philosophical issues. Several excellent reviews have highlighted the general findings of these studies and have addressed prevalent GWAS challenges and limitations such as lack of sufficient statistical power due to small sample sizes, small individual gene effects, missing heritability, precision of phenotypes, and the overall lack of replication (Edenberg and Foroud, 2014; Hart and Kranzler, 2015; Tawa et al., 2016). Here we take a more optimistic approach by “mining” the existing GWAS results for logical biological function(s), despite largely non-significant statistical findings predicated (perhaps overly conservative, see (Kanai et al., 2016) by the need to correct for individual (presumably independent) tests of associations of ~1 million single-nucleotide polymorphisms (SNPs) throughout the genome. Specifically, we aim to generate novel biological functional hypotheses for candidate genes identified by GWAS and other approaches within the framework of the addiction cycle.
2. Adoption, family and twin studies: the contribution of genes versus the environment
The genetics of alcoholism (now formally called AUD) was founded on many years of observations that alcohol problems cluster in families (Cotton, 1979). The fact that alcoholism tends to run in families has been documented by almost every civilization dating back to at least the ancient Greeks (Goodwin, 1979). However, a family history of alcoholism, although a critical step in considering whether genetic factors might be important, is not conclusive proof that a familial association is produced by genes, environment, or the interaction of both.
Early studies reported that relatives of alcoholics were approximately at a three to four-fold increased risk for AUD (Goodwin, 1989), which then led to studies that aimed to determine if this increased risk for AUD seen in children of alcoholics was still evident even if the they were not raised by their biological parents in an alcoholic environment. In the early 1970s a well-powered study which included a large sample of half-siblings who were raised by their alcoholic biologic families or raised by foster families who were not alcoholic, concluded that adverse alcohol outcomes in offspring related more closely to alcoholism in a biological parent than compared to alcohol problems in foster families (Schuckit et al., 1972). At about the same time this report was made several large adoption studies from the United States, Denmark, and Sweden reported a threefold or greater increased risk for AUD in sons of alcoholics even when they were adopted as infants and raised by nonalcoholic families (Goodwin et al., 1973, 1977,1974). For example, Goodwin et al. (1973) studied the drinking history of 55 adopted-out sons of alcoholics and 78 adopted-out sons of non-alcoholics. All of the sons were adopted as infants, within the first six weeks of life. Importantly, the sons of alcoholics had no knowledge that their biological parents' suffered from alcoholism. Biological sons of alcoholics who had been adopted by non-related foster families were four times as likely to become alcoholics compared to adopted sons of non-alcoholics. These seminal findings provided convincing support that part of the vulnerability to AUD was mediated by genetic factors.
The second approach used a twin design to estimate the contribution of genes versus the environment in risk for AUD. Identical twins (monozygotic; MZ) share 100% of their genes while fraternal twins (dizygotic; DZ) share only approximately 50% (the same as full siblings). In addition, environmental variability can usually be equated more evenly when twin pairs are used because they are usually raised together in the same homes and experience the same developmental events at the same ages. Therefore, a higher level of similarity for a diagnosis among identical (MZ) compared with fraternal (DZ) twins indicates that genetic factors were likely to have contributed to the development of the disorder, not just childhood environment. Twin studies were carried out in the United States, Finland, Sweden, and the United Kingdom (Gurling et al., 1984; Hrubec and Omenn, 1981; Kendler et al., 1994; Murray et al., 1983; Partanen, 1972), and almost all supported a genetic influence in alcoholism with an estimated proportion of the risk explained by genes of about 60% (Edenberg and Foroud, 2006; Kendler, 2012). Taken together twin, family and adoption studies have convincingly demonstrated that genetic factors account for between 50 and 60% of the vulnerability to alcoholism (McGue, 1999; Prescott and Kendler, 1999). Although these results provide compelling evidence for a genetic influence on alcoholism they do not indicate the specific genes that increase or decrease risk towards developing alcoholism.
3. Genetic mapping studies of AUD: linkage and candidate gene association studies
Linkage and association designs are both used to map Mendelian and complex traits (Table 1). In the 1980s, it became feasible to use DNA markers to map genotype-phenotype correlations and begin to pin-point specific genomic loci that influence alcohol phenotypes. Genetic linkage studies determine inheritance of a trait or disorder (e.g., AUD) among family members in extensive pedigrees by evaluating if one or more genetic markers spaced across the 23 chromosomes segregate with the disorder. Linkage mapping is a locus-driven approach that can usually only roughly locate a broad chromosomal region, which may contain 10s of 100s of genes, that co-segregate with the disorder. Thus, further fine mapping with recombinant genetic markers in informative family members is required to get to the gene. One of the first studies of this kind, the Collaborative Study on the Genetics of Alcoholism (COGA) was formed in 1989 to map and characterize the genetic variants associated with alcoholism (Bierut et al., 2002). COGA is a family-based genetic study with nine research sites throughout the U.S. Nuclear and multigenerational families were ascertained through a proband in treatment for alcohol dependence. The initial sample consisted of 105 families with 987 individuals. Results from the first genome-wide scan provided evidence of linkage for loci on chromosomes 1 and 7 and suggestive linkage on chromosome 2 with alcohol dependence; linkage on chromosome 2, conferring protection against alcohol dependence, near the alcohol dehydrogenase genes was also found (Reich et al., 1998). A follow up study in 157 independent families densely affected with alcoholism was conducted and provided further evidence of linkage to chromosomes 1 and 7 with new loci identified on chromosomes 2 and 3 (Foroud et al., 2000).
Table 1.
General comparison of genetic association and linkage gene mapping approaches.
| Category | Association (candidate gene and genome-wide) |
Linkage |
|---|---|---|
| Subjects | Families or unrelated individuals | Related individuals from a pedigree |
| Mapping principle | Allele-based | Locus- gene based |
| Mapping resolution | High (<100 kb) | Low (<5 Mb) |
| Power | Low-moderate | Moderate-high |
| Best for detection of: | Common-uncommon variants | Rare-very rare variants |
The second approach involves genetic association in population and/or family-based samples. Genetic association is essentially a form of linkage mapping but is allele-based rather than locus-based (Table 1). Association studies identify alleles of a gene that are more or less common in people with a trait or disease versus those without. COGA has conducted several candidate-gene association studies and found association with more than 20 genes affecting both alcohol dependence and endophenotypes associated with alcoholism (Edenberg and Foroud, 2006). Other similar studies by investigators all over the world, in a range of different populations, have identified genetic variants that contribute towards vulnerability to alcoholism (Goldman et al., 2005a; Kalsi et al., 2009). In this group of genes almost all of the major neurotransmitter systems are represented including: GABA, glutamate, serotonin, dopamine, and acetylcholine. Genes involved with alcohol metabolism (alcohol dehydrogenase [ADH] and aldehyde dehydrogenase [ALDH]) as well as neuropeptide signaling (neuropeptide Y [NPY], corticotropin-releasing hormone [CRH]), neuroendocrine signaling, other signaling molecules (multiple PDZ domain protein [MPDZ]), and cellular architecture (ras suppressor protein 1[RSU1]) are also implicated (Edenberg and Foroud, 2006; Kranzler and Edenberg, 2010).
3.1. Functional alcohol dependence variants: ADH and ALDH
The variants in the alcohol metabolizing genes (ADH and ALDH) have clear functional consequences at the molecular and behavioral levels. This is the exception rather than the rule in terms of relating genetic variation to straightforward functional consequences of polymorphisms that have been identified for alcohol dependence. For example, a missense variant in (alcohol dehydrogenase 1B [ADH1B]) leads to the replacement of Arg48 with His48 in the encoded protein and reduces the risk for alcoholism in Asians, Native, European and African Americans. The hypothesized mechanism of action involves an increased aversive reaction when alcohol is consumed by the accumulation of the toxic alcohol metabolic by-product, acetaldehyde in the periphery which is due to a reduction in the function of polymorphic ADH enzyme. This mechanism of action is also the basis for the use of disulfiram, the first FDA-approved medication for alcohol dependence.
3.2. Functional variants: binge-intoxication stage
Examples of other genes for alcohol dependence that have documented evidence of functional consequences at the molecular and/or behavioral levels are shown in Fig. 1 (genes in ovals) and listed in Table 2. For example, alcohol dependence is associated with a variant within neuropeptide Y (NPY) (Mottagui-Tabar et al., 2005). In a landmark study haplotype-driven NPY expression (including the variant associated with alcohol dependence) predicted brain responses to emotional and stress challenges and was inversely correlated with trait anxiety (Zhou et al., 2008). The results are consistent with the function of NPY as an anxiolytic peptide and suggests that the association with alcohol dependence may be functionally mediated by alterations in NPY gene expression that affect individual differences in stress reactivity and anxiety; individuals with NPY variants that result in low brain expression are more likely to have high anxiety that leave them vulnerable to excessive alcohol consumption, particularly in the binge-intoxication phase (Xu et al., 2013).
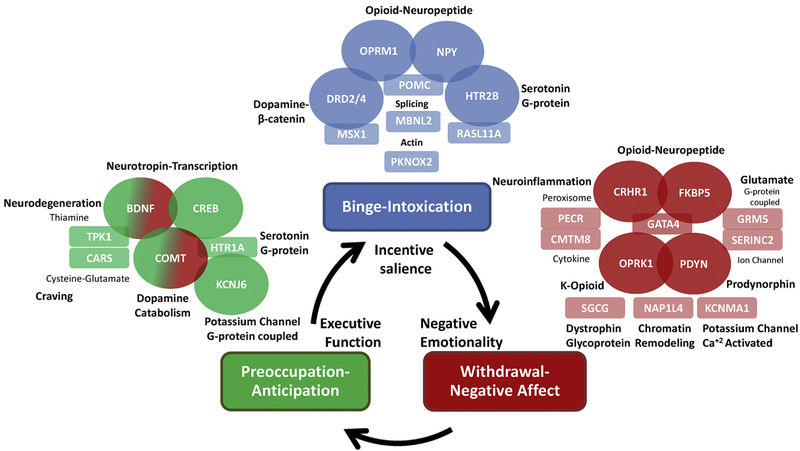
Fig. 1.
Candidate genes for alcohol dependence characterized into stages of the addiction cycle. This schematic shows the three stages of the addiction cycle: (1): binge-intoxication (blue), (2): withdrawal-negative affect (red) and (3): preoccupation-anticipation (green); also shown are behavioral domains linked to each stage of the cycle: incentive salience, negative emotionality, and executive function, respectively. Candidate functional genes identified by non-GWAS approaches are shown in ovals and colored coded according to the relevant addiction stage the gene is hypothesized to function. GWAS candidates are shown in rectangles color coded according to the relevant addiction stage the gene is hypothesized to function. The general biological function of each gene in the figure is highlighted in black. Non-GWAS functional gene relationships are indicated by overlapping ovals. Hypothesized functional relationships between non-GWAS genes and GWAS candidates are indicated by overlapping ovals and rectangles. GWAS candidates in each stage are grouped according to potential biological functional similarities where possible. Finally, one example of pleiotropy is shown in the preoccupation-anticipation stage. BDNF-COMT functional variants have pleiotropic effects in both the preoccupation-anticipation stage and the withdrawal-negative affect stage. This pleiotropic relationship is shown by dual-color shading of the BDNF-COMT ovals reflecting both addiction stages.
Table 2.
Functional genetic variants.
| Gene Symbol |
Description | Discovery Method |
Addiction Cycle |
References |
|---|---|---|---|---|
| NPY | Neuropeptide Y | Candidate Association | Binge-Intoxication | Mottagui-Tabar et al., 2005; Zhou et al 2008; Xu et al., 2013 |
| HTR2B | 5-Hydroxytryptamine Receptor 2B | Candidate Association | Binge-Intoxication | Bevilacqua et al., 2010; Tikkanen et al., 2015 |
| OPRM1 | Opioid Receptor Mu 1 | Candidate Association | Binge-Intoxication | Lovallo et al., 2015; Ray et al., 2012 |
| DRD2 | Dopamine Receptor D2 | Linkage | Binge-Intoxication | Filbey et al., 2012; Savitz et al., 2013 |
| DRD4 | Dopamine Receptor D4 | Linkage | Binge-Intoxication | Filbey et al., 2012; Savitz et al., 2013 |
| CRHR1 | Corticotropin Releasing Hormone Receptor 1 | Candidate Association | Withdrawal-Negative Affect | Chen et al., 2010; Hansson et al., 2006; Hsu et al., 2012; Ray et al., 2013 |
| FKBP5 | FK506 Binding Protein 5 | Candidate Association | Withdrawal-Negative Affect | Bevilacqua et al., 2012; Huang et al., 2014 |
| OPRK1 | Opioid Receptor Kappa 1 | Candidate Association | Withdrawal-Negative Affect | Faisal et al., 2014; Xu et al., 2013; Xuei et al., 2006 |
| PDYN | Prodynorphin | Linkage | Withdrawal-Negative Affect | Karpyak et al., 2013; Preuss et al., 2013 |
| COMT | Catechol-O-Methyltransferase | Linkage | Preoccupation-Anticipation | Tiihonen et al., 1999; Voisey et al., 2011; Dickinson and Elvevag, 2009; Ashare et al., 2013; Goldman et al., 2005b |
| BDNF | Brain Derived Neurotrophic Factor | Candidate Association | Preoccupation-Anticipation | Chen et al., 2015; Benzerouk et al., 2013 |
| CREB1 | CAMP Responsive Element Binding Protein 1 | Candidate Association | Preoccupation-Anticipation | Chen et al., 2015 |
| KCNJ6 | Potassium Voltage-Gated Channel Subfamily J Member 6 | GWAS endophenotype | Preoccupation-Anticipation | Kang et al., 2012; Crabbe et al., 2006; Tipps and Buck, 2015; Aryal et al., 2009; Blednov et al., 2001; Goldman et al., 2005b; Kozell et al., 2009; Lewohl et al., 1999 |
Examples of other candidate genes with experimentally derived functional information that are hypothesized to function within the binge-intoxication domain including (see Table 2): (serotonin type 2B receptor [HTR2B]) implicated in impulsivity, anti-social personality, and aggression (Bevilacqua et al., 2010; Tikkanen et al., 2015); (mu-opioid receptor [OPRM1]) implicated in reward learning, greater subjective reinforcement from alcohol consumption, greater alcohol-cue and stress reactivity; cortisol stress response, and treatment response to naltrexone (Lovallo et al., 2015; Ray et al., 2012). The dopamine receptors, [DRD2 and DRD4] implicated in reward learning, modulation of neural processes underlying response inhibition, and impulsivity (Filbey et al., 2012; Savitz et al., 2013).
3.3. Functional variants: withdrawal-negative affect stage
Candidate functional genes within the withdrawal-negative affect stage include (see Table 2): CRF type 1 receptor (CRHR1) implicated in stress reactivity and negative emotionality. For example, animal studies found an innate up-regulation of the Crhr1 transcript, encoding the corticotropin-releasing hormone receptor 1 (CRH-R1), in several limbic brain areas of msP rats genetically selected for high alcohol preference. Up-regulation of Crhr1 was associated with a genetic polymorphism in the Crhr1 promoter, and was accompanied by increased CRH-R1 density. In addition, a selective CRH-R1 antagonist (antalarmin) significantly reduced oral operant alcohol self-administration and blocked stress-induced reinstatement in the high preference msP rat line. These animal studies support a functional association of variants within CRHR1 and alcohol dependence in human studies(Chen et al., 2010; Hansson et al., 2006; Hsu et al., 2012; Ray et al., 2013). FK506 binding protein 5 (FKBP5), a chaperone protein that regulates HPA axis function, is implicated in severity of withdrawal, childhood trauma, aggression and suicidal behavior (Bevilacqua et al., 2012; Huang et al., 2014). Opioid Receptor Kappa 1 subtype (OPRK1), is one of the major subtypes of opioid receptors in the brain particularly localized to limbic brain regions that regulate emotion and reward. Activation of OPRK1 results in behavioral dysphoria during escalated drug or alcohol consumption and withdrawal leading to a negative affective emotional state characterized by increased stress reactivity, irritability, depression, and anhedonia. Negative reinforcement processes that are initiated alleviate the negative affective state of alcohol abstinence/withdrawal, by resumption of hazardous drinking, which perpetuates the addiction cycle. In support of this, a variant in OPRK1 is associated with stress response and related drug craving, limbic brain activation and cocaine relapse risk (Faisal et al., 2014; Xu et al., 2013; Xuei et al.,2006). Prodynorphin (PDYN) is a preproprotein that is proteolytically processed to form the secreted opioid peptides beta-endorphin, dynorphin, leu-enkephalin, rimorphin, and leumorphin. These endogenous opioids are ligands for several different subtypes of opioid receptors; the proteolytic product, dynorphin, has been studied most extensively, which binds to the kappa opioid receptor (OPRK1). A recent paper reported an association of the PDYN gene with alcohol dependence and the propensity to drink in negative emotional states. Further an association of a PDYN variant with drinking to avoid emotional or somatic discomfort was also reported (Karpyak et al., 2013; Preuss et al., 2013).
3.4. Functional variants: preoccupation-anticipation stage
Finally, candidate functional genes for the preoccupation-anticipation domain (see Table 2) include: genetic variation in a set of known signaling genes of the brain-derived neurotrophic factor (BDNF)-cAMP response element-binding protein 1/5 (CREB1 and CREB5) neuronal pathway that were negatively correlated with fMRI signals during an alcohol-cue task in the precuneus and parietal brain areas; alcohol dependent individuals with these risk variants had greater brain activation during the alcohol-cue task, suggesting that the variants influence alterations in learned drug seeking associations (Chen et al., 2015). In another study, the common functional BDNF Val66Met polymorphism was associated with deficits in executive functions in adult offspring of alcohol-dependent probands (Benzerouk et al., 2013).
The gene encoding the enzyme, catechol-O-methyl transferase (COMT), is involved in the degradation of dopamine and other catecholamines. A previous study reported an association between a functional variant of COMT and type 1 alcoholism, which is a late on-set subtype of the alcohol dependence phenotype characterized by harmful drinking to reduce anxiety, which was replicated in an independent study (Tiihonen et al., 1999; Voisey et al., 2011). However, the association between COMT and alcohol dependence was not observed in another independent sample that consisted of mostly families of European and African American descent (Foroud et al., 2007). The discrepancy may be due to allelic heterogeneity at the COMT locus which has several different polymorphisms within the gene. COMT is one of the major genes that modulates dopamine tone in the prefrontal cortex and has a significant influence on cortical information processing/executive functioning. The common COMT functional variant, Val158Met, bidirectionally regulates the enzymatic activity of the encoded enzyme which impacts the efficiency of prefrontal-mediated cognition, specifically executive functioning, working memory, fluid intelligence and attentional control. (Dickinson and Elvevag, 2009). The same common functional COMT polymorphism was associated with abstinence-related working memory deficits in two independent samples of smokers (Ashare et al., 2013) and differentially modulates stress and anxiety responses as well as brain endogenous opioid function (Goldman et al., 2005b).
Drug and alcohol dependence can be characterized by a dysfunction in learning and memory processes that leads to drug craving and seeking behaviors that involve prefrontal cortical, midbrain (VTA) and hippocampal circuits. G protein-coupled inwardly rectifying potassium (GIRK) channels are known to play a critical role in this process (Crabbe et al., 2006; Tipps and Buck, 2015). GIRK channels are direct targets of alcohol and mediate behaviors associated with alcohol dependence including regulation of VTA dopamine tone during dependence. (Aryal et al., 2009; Blednov et al., 2001; Goldman et al., 2005b; Kozell et al., 2009; Lewohl et al., 1999; Herman et al., 2015). A potassium channel gene variant, KCNJ6 (GIRK2) was associated with alcohol dependence in a study of densely affected families (Kang et al., 2012). The function of this variant mediated alterations in the theta band of event related oscillations, which index the activity of neuronal ensembles while performing a task and are important for processes underlying frontal inhibitory control, conscious awareness, recognition memory and episodic retrieval (Kang et al., 2012). Collectively, potassium/GIRK channels appear to be compelling genetic candidates mediating cognitive and executive function deficits in alcohol dependence.
In summary, it can be reasonably concluded from the preceding discussion of candidate genes identified for alcohol dependence represent bona fide variants that function to alter the phenotype. The majority of these genes were identified in “high-risk” or homogenous populations which resulted in relatively dramatic effects on the phenotype, such as the stop codon found HTR2B which was associated with a very severe early on-set form of alcoholism in individuals that are also characterized as violent criminal offenders and display extreme impulsivity (Bevilacqua and Goldman, 2013). This example highlights the beneficial use of intermediate phenotypes (such as impulsivity) to help guide gene identification, especially in homogenous populations such as in the Finnish population in which this study was conducted. However, the candidate gene approach in particular is useful only to a certain threshold when gene effect sizes are considered. To fully understand the complexity of the genetics of AUD, genes with a range of effect sizes need to be identified because it is almost a certainty that both gene variants with large effects and many variants of small effect will ultimately explain the substantial phenotypic variability in AUD.
Thus, the firsts methodologies used for identifying genes for AUD, namely, genome-wide linkage and candidate gene association studies (Agrawal and Bierut, 2012; Enoch, 2013; Rietschel and Treutlein, 2013) were moderately successful. As a whole these studies explain only a small portion of the genetic variance, because the frequency of the identified variants in these studies are usually rare in the general population and thus cannot explain the entire population variance in AUD. However, the most replicable finding from these initial studies was the identification of genetic variation in the alcohol metabolizing genes and protective effects on risk for AUD in Asian, American Indian, European, and African American populations (Bierut et al., 2012; Chen et al., 1999; Edenberg, 2007; Thomasson et al., 1991). There are several major limitations of linkage and candidate gene association studies that likely contributed to the largely unsuccessful identification of the many genes for AUD. Genetic linkage studies are conducted using large extended families across multiple generations derived from at least one affected family member. Assembling extended pedigrees required for linkage studies is labor intensive and more difficult compared to assembling population-based samples (case-control) required for association studies. In addition, linkage mapping has very poor genomic resolution where large chromosomal regions are identified that contain multiple possible candidate genes within the identified region. Candidate gene association studies require nomination of a biologically relevant candidate gene or pathway to test for association with AUD. Because a priori knowledge is required this limits the potential of finding novel and unexpected genetic associations. The GWAS approach (discussed below) is not limited to nominating a specific candidate gene or pathway because it is a discovery based approach that interrogates a complete panel of common SNPs distrusted across the entire genome without dependence on an a priori hypothesis.
4. Genome-wide association studies of alcohol dependence
The GWAS approach was made possible by completion of large international genomic projects beginning with the Human Genome Project (draft sequence, 2001), the HapMap Project (2003) and the 1000s genomes project (2006) which have provided an enormous amount of information on genomic variation within and between populations as well as providing the foundation for GWAS. For example, the HapMap project provided information on nearly all of the common genetic variation in humans. Approximately 1.3 million single nucleotide polymorphisms (SNPs) were genotyped in Phase I of the project, and published in 2005 (The International HapMap Consortium (2005). Phase II HapMap, characterized over 3.1 million SNPs that were genotyped in 270 individuals from four geographically diverse populations (Frazer et al., 2007). These studies enabled the use of high-throughput genotyping arrays to index millions of common SNPs across the entire genome that are then tested for association with any phenotypic trait or disease. The GWAS approach is theoretically capable of identifying all of the common genetic variants, represented by SNPs, associated with a disease or trait. This is known as the common-disease, common-variant hypothesis which presumes that relatively common genetic variants with low penetrance are the primary genetic factors within a population that underlie common diseases (Chakravarti, 1999; Lander, 1996; Pritchard and Cox, 2002; Reich and Lander, 2001). In addition, initial GWAS assumed that common genetic variants (SNPs) contributing to disease susceptibility would have moderate to large effects and thus would require relatively small sample sizes to detect significant associations. Two of the first GWAS conducted on age-related macular degeneration and exfoliation glaucoma supported this assumption (Haines et al., 2005; Thorleifsson et al., 2007). However, subsequent GWAS indicated that very large sample sizes are required to achieve the statistical power necessary to detect the small effects of risk loci for the majority of complex traits and disease, especially for psychiatric disorders. In addition, very stringent statistical corrections are required to control for multiple testing leading to an increase in false-positives due to >1 million SNPs typically interrogated in a single GWAS. A Bonferroni-corrected genome-wide significance threshold set at p < 10–8 is required.
Another overall finding from the first era of GWAS is that the heritability explained by SNP associations is significantly less than estimates of heritability derived from family studies. More specifically, the variants that reach statistical significance typically explain only a small fraction of the heritability. This is commonly referred to as the “missing heritability” problem (Manolio et al., 2009). There have been several explanations presented to explain the missing heritability, some of which include: undetected rare variants of large effect, epistatic interactions, and the idea that heritability estimates from family studies may actually be overinflated (Golan et al., 2014; Munoz et al., 2016; Zuk et al., 2012, 2014).
4.1. Overview of GWAS of alcohol dependence: populations, sample size, genome-wide significance
An initial group of GWAS of alcohol dependence were published over the last the several years. It is beyond the scope of this review to detail methodological aspects of the published alcohol dependence GWAS. For a detailed description of GWAS study design and experimental details see (Hart and Kranzler, 2015). Briefly, the GWAS we discuss examine the DSM-IV defined phenotype of alcohol dependence using either case-control status of alcohol dependence as the primary phenotype or quantitative phenotypes derived from DSM-IV alcohol dependence (e.g., criterion factor score). In several of these studies secondary or intermediate phenotypes are also examined which include: age of onset, maximum drinks consumed in a 24-h period, and heaviness of drinking. Discovery sample sizes range from approximately 300–16,000 subjects consisting of European American, African American, Australian of European descent, Chinese, Korean, or German populations. Overall, genome-wide significant (p < 10–8) and replicable SNPs were located in the following genes or non-coding RNAs (ncRNAs): ADH1B, ADH1C, ADH7, ALDH2, LOC100507053 (Frank et al., 2012; Gelernter et al., 2014; Park et al., 2013). Not surprisingly, the SNPs in the genes encoding the alcohol metabolizing enzymes are among the common variants with the largest effects on AUD risk. The associations found in the alcohol-metabolizing enzyme genes represent the most consistent overall finding for GWAS of alcohol dependence to date and these results replicate findings from candidate gene and linkage studies which have found similar associations with these enzymes (Edenberg and Foroud, 2013). However, these results were somewhat discouraging because it was widely predicted that the GWAS-era would revolutionize our understanding of the genetics of complex traits including alcohol dependence by uncovering many novel and unexpected common genetic variants for alcohol dependence that have eluded linkage and candidate gene association studies.
4.2. Subset of GWAS and criteria used for generation functional hypotheses
Currently there is great emphasis on increasing sample size, on the order of 50,000–200,000 subjects, for GWAS of alcohol dependence through meta-analyses to provide sufficient power to detect variants at the genome-wide significant threshold of p < 10–8 (Agrawal et al., 2016; Begum et al., 2012; Tawa et al., 2016). Meta-analysis efforts will take a considerable amount of time and involve coordinated efforts among many different laboratories and large consortia such as the Psychiatric Genetics Consortium. However, a plethora of novel biological information can be useful to consider if a plausible functional hypothesis can be generated for candidate genes from the existing GWAS, even when the majority of the identified SNPs do not meet the stringent genome-wide significance threshold (p < 10–8). In particular, attempting to make connections between GWAS candidates, where functional significance is uncertain, with gene variants with strong evidence of functionality identified via candidate gene association or linkage, may provide clues for ultimate validation for GWAS (see Fig. 1). Further, hypothesis-generation is one of the core features of the GWAS design and puts first a primary objective of biological discovery rather than risk prediction as promulgated by Hirschhorn (2009) and others (Cantor et al., 2010; Hall et al., 2016; Hirschhorn, 2009). The GWAS agnostic discovery approach effectively enables the potential to identify hitherto unknown and novel genes displaying a wide range of biological functions, which will be critical for a comprehensive understanding of the etiology of AUD. However, caution should still be taken when functional hypotheses are considered from the existing GWAS of alcohol dependence. Until sufficient sample sizes can be tested and results replicated, any one of the candidate genes we discuss could be a false positive due to the curse of stringent statistical corrections required for reducing false positive results due to multiple testing; however this stringent statistical threshold could very well hide evidence for identification of true positives as novel variants. Below we present the findings of a series of GWAS of alcohol dependence where biological functional information could be derived relevant to the stages of the addiction cycle. We used several criteria for choosing which candidate genes to discuss:
Firstly, because it is commonly acknowledged that the first generation of GWAS of alcohol dependence were grossly underpowered due to small initial sample sizes resulting in many SNPs that do not meet the stringent genome-wide significant threshold (p < 10–8), we focus only on the top-ranking SNPs generally with p values of 10–4 or less.
Second, after considering the set of top-ranking SNPs from each GWAS we selected SNPs for discussion based on whether a logical biological hypothesis could be generated from extant literature and whether this functional hypothesis was relevant to a stage of the addiction cycle.
Third, only GWAS examining the phenotype of alcohol dependence are considered; GWAS examining alcohol consumption in non-dependent subjects are excluded.
Fourth, priority was given to candidate genes with SNPs in non-intergenic regions of the genome The majority of the SNPs we discuss are found in introns (see Table 3)
Fifth, priority was also given to top ranking candidate genes with evidence of multiple different SNPs within the same gene associated with alcohol dependence.
Table 3.
GWAS candidate variants.
| Gene Symbol |
Description | Addiction Cycle |
Function | SNP | p value | Genomic Context | References | Gene/Pathway Tested Animal Models |
|---|---|---|---|---|---|---|---|---|
| MBNL2 | Musdeblind-like protein 2 | Binge-intoxication | Alternative Splicing | rs9556711 | p<10-7 | intron_variant | Heath et al., 2011 | Gene-No; Pathway-Yes |
| RASL11A | RAS Like Family 11 Member A | Binge-intoxication | GTPase Signlaing | rs9512637 | p<10-7 | intergenic_variant | Heath et al., 2011 | Gene-No; Pathway-Yes |
| MSX1 | Msh homeobox 1 | Binge-intoxication | Dopamine-Beta-Catenin Signaling | rs1000579 | p<10-7 | intergenic_variant | Frank et al., 2012 | Gene-No; Pathway-Yes |
| PKNOX2 | PBX/Knotted 1 Homeobox 2 | Binge-intoxication | Actin Signaling | rs750338 | p<10-6 | intron_variant | Wang et al., 2011 | Gene-Yes; Pathway-Yes |
| PKNOX2 | PBX/Knotted 1 Homeobox 2 | Binge-intoxication | Actin Signaling | rs1426153 | P<106 | intron_variant | Zuo et al., 2012 | Gene-Yes; Pathway-Yes |
| PKNOX2 | PBX/Knotted 1 Homeobox 2 | Binge-intoxication | Actin Signaling | rs10893366 | P<10-6 | intron_variant | Zuo et al., 2012 | Gene-Yes; Pathway-Yes |
| PKNOX2 | PBX/Knotted 1 Homeobox 2 | Binge-intoxication | Actin Signaling | rs10893366 | p<10-7 | intron_variant | Bierut et al., 2010 | Gene-Yes; Pathway-Yes |
| POMC | Proopiomelanocortin | Binge-intoxication | Neuroendocrine-Anti-Stress | rs33950052 | p<10-4 | intron_variant | Kendler et al., 2011 | Gene-Yes; Pathway-Yes |
| PECR | Peroxisomal Trans-2-Enoyl-CoA Reductase | Withdrawal-Negative affect | Neuroinflammation-Peroxisome | rs7590720 | p<10-8 | intron_variant | Treutlein et al., 2009 | Gene-No; Pathway-Yes |
| SERINC2 | Serine Incorporator 2 | Withdrawal-Negative affect | Ionotropic Glutamate | rs4478858 | p<10-8 | intron_variant | Zuo et al., 2012 | Gene-No; Pathway-Yes |
| SGCG | Sarcoglycan Gamma | Withdrawal-Negative affect | Dystrophin Glycoprotein Complex | rs4770403 | p<10-6 | 5_prime_UTR_variant | Frank et al., 2012 | Gene-No; Pathway-No |
| CMTM8 | CKLF Like MARVEL Transmembrane Domain Containing 8 | Withdrawal-Negative affect | Neuroinflammation-Cytokine | rs9825310 | p<10-6 | intergenic_variant | Frank et al., 2012 | Gene-No; Pathway-Yes |
| NAP1L4 | Nucleosome Assembly Protein 1 Like 4 | Withdrawal-Negative affect | Chromatin Remodeling | rs8505 | p<10-5 | 3_prime_UTR_variant | Edenberg et al., 2010 | Gene-No; Pathway-Yes |
| KCNMA1 | Potassium Calcium-Activated Channel Subfamily M Alpha 1 | Withdrawal-Negative affect | BK-Potassium Channel, Calcium Activated | rs2077641 | p<10-4 | intron_variant | Kendler et al., 2011 | Gene-Yes; Pathway-Yes |
| GATA4 | GATA Binding Protein 4 | Withdrawal-Negative affect | Neuroendocrine-Stress | rs13273672 | p<10-4 | intron_variant | Treutlein et al., 2009 | Gene-Yes; Pathway-Yes |
| GRM5 | Glutamate Metabotropic Receptor 5 | Withdrawal-Negative affect | Metabotropic Glutamate | rs6483362 | p<10-6 | intron_variant | Bierut et al., 2010 | Gene-Yes; Pathway-Yes |
| HTR1A | 5-Hydroxytryptamine Receptor 1A | Preoccupation-Anticipation | Serotonin-Compulsivity | rs7445832 | p<10-9 | intergenic_variant | Zuo et al., 2012 | Gene-Yes; Pathway-Yes |
| CARS | Cysteinyl-TRNA Synthetase | Preoccupation-Anticipation | Cysteine-Glutamate system | rs4758621 | p<10-4 | 5_prime_UTR_variant | Edenberg et al., 2010 | Gene-No; Pathway-Yes |
| TPK1 | Thiamin Pyrophosphokinase 1 | Preoccupation-Anticipation | Thiamine-Neurodegeneration | rs10224675 | p<10-6 | intron_variant | Bierut et al., 2010 | Gene-No; Pathway-Yes |
4.3. GWAS candidates: PECR and GATA4
Treutlein et al., 2009 conducted a GWAS of alcohol dependence which consisted of a German sample of males with an early age of onset of DSM-IV alcohol dependence (Treutlein et al., 2009). Age of onset was younger than 28 years and each case (n = 487) had alcohol dependence that required hospitalization for the treatment of severe withdrawal symptoms. Control subjects (n = 1358) were derived from three population-based epidemiologic studies conducted in the same geographical location and were ascertained as general population based controls for various biomedical studies. However, it is important to note that Treutlein et al., 2009 do not provide data on whether the population-based control subjects used in their GWAS of alcohol dependence were screened for psychiatric disorders including substance and alcohol use disorders. A total of 524,396 SNPs were tested in the case-control sample for association with alcohol dependence. There were no genomewide significant SNP associations observed (p < 10–8). However, 120 SNPs with a P < 10–4 were selected for genotyping in a follow-up sample of German males with DSM-IV alcohol dependence (age of onset younger than 45 years old). In addition, 19 additional SNPs were selected for genotyping in the follow-up sample which were selected based on differential expression of the gene in the rat brain after long term alcohol consumption. In the follow-up study, 16 SNPs showed association with at least nominal significance (p < 0.05), 15 of which were associated with the same allele as in the GWAS. The authors performed a quasi-meta-analysis by combining data from the GWAS sample and the follow-up sample. Two SNPs, rs7590720 and rs1344694 met a genome-wide significance (p < 10–8) in the combined sample. The two SNPs were in the 3’-flanking region of the gene encoding peroxisomal trans-2-enoyl-coA [coenzyme A] reductase(PECR) (Table 3), located on chromosome region 2q35 (PECR), see Fig. 1. PECR participates in chain elongation of fatty acids and is an integral component of peroxisomes which play a key role in protection against oxidative stress particularly in glial cells (Di Cesare Mannelli et al., 2014). Peroxisomal neurobiology has previously been implicated in alcohol dependence. Peroxisome proliferator-activated receptors (PPARs) are nuclear transcription factors with well-known roles in lipid metabolism and more recently have been associated with neuroinflammatory processes induced by alcohol (Varga et al., 2011). It is logical to assume that PECR and PPAR are part of the same biological pathway and thus likely have cooperative/complementary functions. PPAR agonists, including tesaglitazar and fenofibrate, reduce voluntary alcohol consumption, withdrawal severity and stress-induced relapse in rodents (Blednov et al., 2015, 2016; Stopponi et al., 2013; Stopponi et al., 2011). Re-analysis of GWAS data focusing only on SNPs within PPAR genes found an association of SNPs in PPARA and PPARG with alcohol withdrawal and PPARGC1A with alcohol dependence (Blednov et al., 2015). Reduced alcohol consumption by tesaglitazar and fenofibrate may be associated with signaling at GABAergic interneurons, neuropeptide systems and dopaminergic signaling pathways which ultimately regulate stress-related neurocircuitry (Ferguson et al., 2014). In sum, an association of SNPs in PECR and alcohol dependence fits with recent literature demonstrating a critical role of peroxisome neurobiology, particularly PPARs, in mediating alcohol-related phenotypes including alcohol consumption and withdrawal. A logical hypothesis may then be that genetic variation in PECR functions to alter the alcohol dependence phenotype in a similar fashion (via neuroinflammatory processes) to that of PPARs, particularly in stress-related neurocircuitry including the extended amygdala.
Several additional SNPs at p value of p < 10–4 in the combined sample were noted including one in GATA4 (see Fig. 1 and Table 3). GATA4 encodes the GATA Binding Protein 4, a member of the GATA family of zinc-finger transcription factors. Members of this family recognize the GATA motif which is present in the promoters of many genes and is thus an important cellular transcription regulator. GATA4 binds regulatory genomic regions of the atrial natriuretic peptide (ANP) receptor which are highly expressed in the amygdala, caudate and hypothalamus (Cao and Yang, 2008; Grepin et al., 1994). ANP is produced and released during alcohol withdrawal and may be anxiolytic by inhibition of corticotrophin releasing factor (CRF), adrenocorticotrophic hormone (ACTH) and cortisol (Antoni et al., 1992). A previous study found a significant association between GATA4 with alcohol dependence, relapse risk and treatment response with acamprosate (Kiefer et al., 2011). A follow-up study also reported an association between GATA4 genotype and limbic gray matter volume which was predictive of an increased risk for alcohol relapse (Zois et al., 2016). Thus, GATA4 represents a compelling candidate due to convergent results that suggest GATA4 variation may affect alcohol dependence phenotypes by alterations in transcriptional regulation of brain limbic ANP expression and ANP receptor signaling associated with stress induced by alcohol dependence.
4.4. GWAS candidates: GRM5, TPK1 and PKNOX2
Bierut et al., 2010 conducted a GWAS that examined a well-characterized sample of 1897 DSM-IV alcohol dependent cases and 1932 alcohol-exposed, non-dependent controls from the Study of Addiction: Genetics and Environment (SAGE) (Bierut et al., 2010). The SAGE discovery sample consists of three large, complementary datasets: the Collaborative Study on the Genetics of Alcoholism (COGA), the Family Study of Cocaine Dependence (FSCD), and the Collaborative Genetic Study of Nicotine Dependence (COGEND). Approximately, 950,000 SNPs were analyzed that span the entire genome. About 60% of the cases were male. Roughly 70% of the sample were from European descent and 31% African-American ancestry. A small number of subjects (3%) reported Hispanic ethnicity. Almost half of the alcohol-dependent sample were diagnosed with comorbid marijuana or cocaine dependence. Comorbid substance dependence in half of the cases is an important factor to consider when interpreting the results of this study. In the discovery sample no single SNP met the genome wide significant threshold of p < 10–8. However, the authors selected 15 SNPs with p < 10–5 for further analyses in two replication samples. Of these 15 SNPs several are worth highlighting for functional relevance with the addiction cycle.
A modest association (p < 10–6) was found in a SNP (rs6483362) in GRM5 which encodes the glutamate metabotropic-5 G-protein coupled receptor (mGlu5) (Fig. 1 and Table 3). mGlu1/5 receptor binding is increased in the hippocampus of type 1 alcoholics (Kupila et al., 2013). mGlu5 receptor antagonists reduce alcohol-withdrawal induced anxiety in rats (Kumar et al., 2013). mGlu5 signaling on striatal Dopamine-D1 receptor expressing neurons is necessary for both novelty-seeking behavior and the abstinence-induced escalation of alcohol drinking in mice (Parkitna et al., 2013). mGlu5 receptors in the basolateral amygdala and nucleus accumbens regulate cue-induced reinstatement of ethanol-seeking behavior (Sinclair et al., 2012). Thus, although SNPs in GRM5 did not meet the stringent genome-wide significant threshold in the study by Bierut et al., 2010, results from human alcoholic brain and animal models of alcohol dependence support a key role of GRM5 in alcohol dependence phenotypes and the negative affective state during protracted abstinence.
Bierut et al., 2010 report a modest association (p < 10–6) with a SNP (rs10224675) in TPK1. TPK1 encodes thiamine pyrophosphokinase 1 which catalyzes the conversion of thiamine to thiamine pyrophosphate and plays a central role in thiamine metabolism (Fig. 1 and Table 3). Alcohol dependence is a well-known factor that can lead to a thiamine deficiency (Singleton and Martin, 2001). Thiamine deficiency is a known cause of Wernicke's Encephalopathy and Korsakoff's Syndrome(WKS) that is commonly associated with chronic alcohol dependence (Sutherland et al., 2014). Wernicke’s Encephalopathy is an acute life– threatening neurologic disorder that includes mental confusion, paralysis of the nerves that move the eyes (i.e., oculomotor disturbances), and an impaired ability to coordinate movements, particularly of the lower extremities (i.e., ataxia). Whereas, Korsakoff's Psychosis is a chronic neuropsychiatric syndrome characterized by behavioral abnormalities and memory impairments. Approximately 80–90 percent of alcoholics with Wernicke's Encephalopathy develop Korsakoff's psychosis (Butterworth, 1989). In economically advantaged countries such as the United States, where other forms of malnutrition are uncommon, thiamine deficiency and the resulting WKS occur most commonly among alcoholics. In postmortem studies, brain abnormalities characteristic of WKS were present in approximately 13 percent of alcoholics (Harper, 1988). However, thiamine deficiency resulting from chronic alcohol is also associated with general alcohol neurotoxicity and cerebellar degeneration without WKS (Martin et al., 1994). In addition, a subclinical thiamine deficiency during chronic heavy alcohol consumption can lead to significant cognitive impairments and reduced neuroplasticity (Vedder et al., 2015). The moderate association between a TPK1 SNP and alcohol dependence (Bierut et al., 2010), is supported by previous work showing a link between thiamine deficiency, neuropathology and associated cognitive impairments in cases of chronic alcohol consumption. Therefore, if the association with TPK1 is confirmed by future studies the genetic variation in TPK1 can be hypothesized to function in altering thiamine signaling and metabolism that affects alcohol dependence induced cognitive deficits and neuronal degeneration. This may have relevance to cognitive deficits associated with poor decision making that is characteristic of the preoccupation-anticipation addiction domain.
A SNP (rs10893366), in PKNOX2 (p < 10–7) was also reported by Bierut et al. (2010). While subsequent GWAS identified the same SNP (rs10893366) (p < 10–6) (Zuo et al., 2012), as well as two different SNPs (rs750338 and rs1426153) (p < 10–6) in PKNOX2 associated with alcohol dependence, which strongly supports a critical role of this gene in addiction (Table 3). How might genetic variation in PKNOX2 alter cellular function underlying alcohol dependence phenotypes? PBX/Knotted 1 Homeobox 2 (PKNOX2) belongs to the TALE (3-amino acid loop extension) class of homeodomain proteins characterized by a 3-amino acid extension between alpha helices 1 and 2 within the homeodomain (Imoto et al., 2001) (Fig. 1 and Table 3). PKNOX2 can function as a nuclear transcription factor that regulates gene expression by binding to DNA in a sequence-specific manner to unique DNA recognition motifs (Fognani et al., 2002). There is also concordance between rodents and humans for a role of PKNOX2 in alcohol-related phenotypes. A massive brain transcriptome study of gene expression alterations in rodents that display an innate tendency for a high or low degree of alcohol preference found a correlation between PKNOX2 expression and preference (Mulligan et al., 2006). Of functional interest, multiple isoforms of PKNOX2 exist due to alternative splicing that generates cytoplasmic localized versions of PKNOX2 that bind actin monomers to regulate the actin cytoskeleton. (Haller et al., 2004). In this regard all three PKNOX2 SNPs are located within introns of the gene and thus may potentially facilitate alternative splicing to generate PKNOX2 actin-interacting isoforms. This hypothesis directly relates to an accumulating body of work that implicates regulation of the actin cytoskeleton in binge drinking, the neuroadaptations associated with alcohol dependence and the negative affective state associated with protracted abstinence (Moonat et al., 2011). In addition, a recent analysis in drosophila and humans found that Ras suppressor 1 (Rsu1), a regulator of the actin cytoskeleton, affects alcohol consumption across species (Ojelade et al., 2015). Rsu1 acts downstream of the integrin cell adhesion molecule and upstream of the Ras-related C3 botulinum toxin substrate 1 (Rac1) GTPase to regulate the actin cytoskeleton and is required for normal alcohol preference in Drosophila. Genetic variation in RSU1 is associated with brain activation in the ventral striatum during reward anticipation in adolescents and alcohol consumption in both adolescents and adults (Ojelade et al., 2015). Collectively, PKNOX2 is a novel candidate gene associated with alcohol and substance dependence as well as alcohol consumption and likely mediates these phenotypes by alterations in the actin cytoskeleton that modulates synaptic plasticity particularly at dendritic spines (Mulholland and Chandler, 2007).
4.5. GWAS candidates: CARS and NAP1L4
Using a family-based sample from the Collaborative Study on the Genetics of Alcoholism (COGA), Edenberg et al. (2010) performed a GWAS which interrogated over 850,000 SNPs in European and African Americans for association with DSM-IV alcohol dependence (n = 1399) (Edenberg et al., 2010). Alcohol-dependent probands were ascertained through alcohol treatment programs at seven centers across the United States. Thus cases represent a population with more severe alcohol dependence. Control subjects (age greater than 25 years old) were drawn from the community across the seven centers and were required to have consumed alcohol but not to have a diagnosis of alcohol abuse, dependence or harmful use at any time in their lives. The selected controls also could not meet criteria for DSM-3R or DSM-4 diagnoses of abuse or dependence on cocaine, marijuana, opioids, sedatives, or stimulants. However, study exclusion based on a diagnosis of substance dependence was not a requirement for cases because of the significant co-morbidity between alcohol and substance dependence. Several analyses were performed with the European American case-control GWAS being the primary analysis. Although no genome-wide significant SNPs (p < 10–8) were reported in the primary analysis, combined evidence from the case-control GWAS, a family-based replication study and differential gene expression analysis converged on a cluster of genes on chromosome 11 with the strongest evidence of association. These genes included SNPs within or near: SLC22A18, PHLDA2, NAP1L4, SNORA54, CARS, and OSBPL5. Below we describe the potential functional significance of two of these genes, CARS and NAP1L4, because of possible relevance to one or more addiction cycle domains (Fig. 1 and Table 3).
CARS encodes cysteinyl-tRNA synthetase, an enzyme that catalyzes the addition of cysteine to its cognate tRNA (Davidson et al., 2001) (Fig. 1 and Table 3). There is potential functional evidence that CARS plays a role in the addiction cycle. The thiol group on cysteine is highly nucleophilic and is easily oxidized into cystine. Due to its high reactivity it plays a role in many biological functions, most importantly as a mediator of the cellular redox state which mediates oxidative stress. The mammalian genome encodes approximately 214,000 cysteine residues of which at least 10–20% are redox sensitive under biologic conditions. Protection against oxidative stress by cysteine is partially mediated by glutathione (GSH), a highly abundant peptide formed from cysteine, glutamic acid and glycine. Chronic alcohol is well known to produce reactive oxygen species which plays a major role in neurodegeneration. Neurodegeneration is mediated in part by a reduction in cysteine containing GSH in human alcoholic brain and in binge drinking mice (Marballi et al., 2016; Wrona et al., 1997). In addition, increased glutamatergic excitatory signaling within corticostriatal pathways is associated with craving in humans and is necessary for reinstatement in rodents. The cystine-glutamate antiporter (xCT) exchanges extracellular cysteine for intracellular glutamate and functions as the primary regulator of extracellular glutamate in many brain regions. N-acetylcysteine is a l-cysteine prodrug and is known to act by normalizing dependence-induced extra-synaptic glutamate through the activation of the cystine-glutamate exchanger (Massie et al., 2015). Indeed, repeated N-acetyl cysteine reduces drug seeking in rodents and craving in humans (Amen et al., 2011). Therefore, it is intriguing to speculate that genetic variation in CARS may be related to functional alterations in cysteine containing proteins, alterations in oxidative stress, and dysfunction of the cystine-glutamate exchange system. Collectively, these alterations in cysteine biology could be hypothesized to impose significant dysfunction to corticostriatal circuits associated with drug abstinence and craving relevant to the preoccupation-anticipation addiction domain.
NAP1L4 encodes a nucleosome assembly protein that plays a general and integral role in chromatin remodeling, a fundamental epigenetic mechanism that regulates gene expression (Rodriguez et al., 1997) (Fig. 1 and Table 3). Specifically, nucleosome assembly proteins, such as NAP1L4, can function to recruit histones, chromatin remodeling complexes (i.e., SWI/SNF or ACF), or transcription factors which all work to regulate gene expression (Park and Luger, 2006b). Noteworthy is the observation that the NAP1 gene family (NAP1L1–1L5) appears to be exclusively expressed in brain compared to other families of nucleosome assembly proteins (Park and Luger, 2006a). There is a growing literature demonstrating that addiction to drugs and alcohol alters chromatin remodeling mechanisms, including alterations in nucleosome assembly, in neurons which generates massive gene expression dysregulation leading to altered synaptic plasticity and ultimately behavioral dysfunction (Medrano-Fernandez and Barco, 2016; Robison and Nestler, 2011; Vogel-Ciernia and Wood, 2014). For example, alcohol dependence is mechanistically associated with altered chromatin remodeling that leads to alcohol withdrawal induced anxiety, a phenotype arising in part from altered synaptic plasticity in limbic regions of the brain (Krishnan et al., 2014). This chromatin remodeling causes a reduction in expression of NPY in the amygdala which fits with enhanced anxiety during alcohol withdrawal (Pandey et al., 2008). In addition, the SWI/SWF chromatin remodeling complex is required in adult neurons of C.elegans for acute alcohol tolerance and genetic variation in a member of this complex is associated with alcohol dependence in humans (Mathies et al., 2015). Interestingly, a recent study demonstrated that the sigma-1 receptor mediates cocaine-induced transcriptional regulation by recruiting chromatin-remodeling factors at the nuclear envelope, suppressing the expression of monoamine oxide B (MAOB) in the nucleus accumbens (Tsai et al., 2015). This is a remarkably novel mechanism of action of cocaine mediated by transcriptional mechanisms associated with the sigma-1 receptor. In animal models, sigma-1 receptor antagonists decrease alcohol intake and reinforcement learning (Sabino et al., 2009a) and reduce excessive alcohol consumption in alcohol preferring rat selectively bred lines (Sabino et al., 2009b). Therefore, the sigma-1 receptor may perform a cellular function similar to nucleosome assembly proteins (NAP1) because both the sigma-1 receptor and NAP1 proteins play a key role in recruiting chromatin-remodeling complexes to regulate gene expression. Thus, one could hypothesize that genetic variation in NAP1L4 contributes generally to alterations in chromatin remodeling which could alter expression of neuropeptide genes (NPY) or dopamine-related genes (MAOB) affecting the binge-intoxication and/or withdrawal-negative affect addiction cycle domains.
4.6. GWAS candidates: KCNMA1 and POMC
Kendler et al. (2011) performed a GWAS which used a quantitative measure of symptoms of alcohol dependence in a general population from the United States consisting of 2357 European Americans and 812 African Americans subjects from the Molecular Genetics of Schizophrenia (MGS2) control sample (Kendler et al., 2011). As compared to the dichotomous clinical diagnosis of alcohol dependence a symptom count of alcohol dependence phenotype is presumed to provide greater power for genetic analysis while remaining highly correlated with a clinical diagnosis (Grant et al., 2009; Whitfield et al., 2004). Subjects who reported drinking greater than 4 drinks in one day were retained and administered a 7-item questionnaire which assessed craving for alcohol, DSM-IV criterion for alcohol abuse (dangerous use) and criteria 1 (tolerance), 3 (loss of control), 5 (“great deal of time … ”), 6 (activities given up), and 7 (use despite knowledge of harm) for alcohol dependence. Subjects who reported consuming less than three drinks (excluding liftetime abstainers) in a day were considered social drinkers and served as the control comparison group. Factor analysis of the alcohol dependence symptom counts indicated that cases represented a population with a wide range of vulnerability to alcohol dependence. In the primary GWAS case-control sample there were no genome-wide significant SNPs detected. However, SNPs within KCNMA1 and POMC were among the most significant associations observed in the European American and African American sample respectively (Fig. 1 and Table 3).
KCNMA1 encodes the calcium activated large conductance (BK) potassium channel, alpha1 subunit. BK channels are direct molecular targets of ethanol (Dopico et al., 2016), underlie a molecular mechanism of cellular tolerance to alcohol (Bettinger and Davies, 2014), and play a key role in alcohol withdrawal related phenotypes (Ghezzi et al., 2014; N'Gouemo and Morad, 2014; Kreifeldt et al., 2013; Kreifeldt et al., 2015). Along with evidence from Edenberg et al., 2010 who reported a similar association between SNPs within KCNMA1 and alcohol dependence in both European and African American samples, KCNMA1 represents a compelling candidate gene requiring further analysis. In particular, because there is direct molecular evidence implicating the BK channel as an alcohol-responsive protein, it will be interesting to determine how the specific SNPs identified in the alcohol dependence GWAS change the function of the channel in vitro and in vivo (see Table 3). For example, humanized mice could be generated that express the human variant of the BK channel associated with alcohol dependence and these transgenic mice could be tested for various alcohol-related behaviors such as withdrawal-induced drinking.
POMC encodes proopiomelanocortin which is a preproprotein that undergoes extensive, tissue-specific, post-translational enzymatic processing yielding as many as ten biologically active peptides involved in diverse cellular functions. Adrenocorticotropic hormone (ACTH) and the endogenous opiates Beta-endorphin and Met-enkephalin are the most notable peptides derived from POMC; ACTH controls the secretion of glucocorticoids from the adrenal glands and is an integral signaling peptide along the hypothalamic–pituitary–adrenal axis (HPA) with its well-known roles in regulation of the stress response. Endorphins and enkephalins are the primary endogenous ligands for mu-opioid receptors in the brain. Genetic variation in POMC, which produces ACTH and the endogenous opiate receptor ligands, therefore could have several logical functional roles in mediating the alcohol dependence phenotype because of the documented role of these peptides in stress and reward mediated behaviors (Belda et al., 2015; Charbogne et al., 2014). For example, individuals with a family history of alcoholism have lower plasma concentrations for beta-endorphin and ACTH (Gianoulakis et al., 2005), which suggests a dysfunction of the HPA-axis that may be related to genetic variation in POMC associated with alcohol dependence. In addition, the CRF brain-stress system, which plays a key role in the negative affective state and compulsive drug seeking and intake associated with protracted abstinence, is most likely engaged because mice that lack the CRF-1 receptor show a blunted ACTH response to acute alcohol (Lee et al., 2001), and fail to show dependence-induced drinking (Chu et al., 2007). SNPs within the CRF1 receptor gene (CRHR1) were associated with compulsive alcohol consumption and binge drinking phenotypes in an independent sample (Treutlein et al., 2006), further supporting a possible epistatic effect of the HPA-ACTH stress system and extra-hypothalamic CRF stress system.
4.7. GWAS candidates: RASL11A and MBNL2
Heath et al., 2011 performed a GWAS of alcohol dependence using several quantitative phenotypes: alcohol dependence factor score, heaviness of drinking factor score, and several measures of consumption (e.g., past 12-month frequency and maximum drinks per day) (Heath et al., 2011). Cases (n = 2062) were obtained from an Australian twin community sample and their nuclear families, while the control sample was drawn from the Australian population (n = 3393). This was the largest GWAS to date in terms of the sample size. However, even with the increased sample size and the presumed increased power from the use of quantitative phenotypes, Heath et al., 2011 report no SNPs that meet a genome-wide significance level. SNPs with the most significant p values (p < 10–7) of the study were found in or near these candidate genes: SNP (rs9512637) RASL11A, and a SNP (rs9556711) MBNL2. Several of these top signals may have functional relevance underlying alcohol dependence.
RASL11A encodes RAS Like Family 11 Member A, a member of the small GTPase protein family with a high degree of similarity to RAS proteins (Fig. 1 and Table 3). The GTPase H-ras encoded by HRAS in the nucleus accumbens plays a key role in neuroadaptations that underlie excessive alcohol-drinking behaviors in mice (Ben Hamida et al., 2012), while the GTPase K-ras encoded by KRAS is differentially expressed after repeated alcohol in the anterior cingulate cortex and alcohol differentially affected various pathways in a K-ras dependent manner suggesting that K-ras-regulatory pathways play a key role in excessive alcohol drinking after a history of dependence (Repunte-Canonigo et al., 2010). Thus genetic variation affecting the regulation of RASL11A could likely play a role in behaviors associated with protracted abstinence from alcohol and thus represents a novel and compelling candidate gene based on human GWAS and molecular biology studies in rodents.
MBNL2 encodes Muscleblind Like Splicing Regulator 2 which is C3H-type zinc finger protein that modulates alternative splicing of pre-mRNAs (Fig. 1 and Table 3). There is compelling evidence that alcohol and alcohol dependence affect alternative splicing mechanisms of various genes (Sasabe and Ishiura, 2010). For example, alcohol affects pre-mRNA alternative splicing at DRD2 which encodes the dopamine type 2 receptor (Wernicke et al., 2010), NMDA receptor NR1 subunit encoded by GRIN1 that can produce as many as eight spliced variants (Hardy et al., 1999; Honse et al., 2003; Kumari, 2001), and BK channel alpha1 subunit (Pietrzykowski et al., 2008). In addition, altered GABA-B receptor subunit 1 splicing is observed in human alcoholic post-mortem brain (Lee et al., 2014). Thus it can be reasonably hypothesized that genetic variation in MBNL2 may affect alcohol dependence phenotypes by altering alternative splicing mechanisms that are known to contribute to splicing of alcohol-responsive proteins.
4.8. GWAS candidates: SGCG, MSX1, CMTM8
Frank et al., 2012, conducted a GWAS of alcohol dependence in order to enlarge the sample size from the study by (Treutlein et al., 2009) in an attempt to increase statistical power (Frank et al., 2012). An additional 900 German cases and 862 controls were added to the GWAS by Treutlein et al., 2009 resulting in a combined sample of 1333 male DSM-IV alcohol dependent cases receiving treatment and 2168 control subjects. Frank et al., 2012 reported a genomewide significant SNP (rs1789891) association located between ADH1B and ADH1C, alcohol-metabolizing enzyme genes, that is in complete linkage disequilibrium with a nonsynomous SNP in ADH1C (rs1693482). The significant association with the ADH1B/1C genes is encouraging and likely has a clear functional implication related to the well described role of genetic variation in the alcohol metabolizing genes and risk for alcohol dependence. However, considering this significant finding alone does not take advantage of potentially informative and novel functional information from other top-ranking candidate genes. Several other of the top ranked SNPs (p < 10E-6) within or near SGCG, MSX1, and CMTM8 are described below with potential functional implications.
SGCG encodes gamma-sarcoglycan, one of several sarcolemmal transmembrane glycoproteins that interact with dystrophin (Fig. 1 and Table 3). The dystrophin-glycoprotein complex (DGC) spans the sarcolemma and is comprised of dystrophin, syntrophin, alpha- and beta-dystroglycans and sarcoglycans (alpha, beta, gamma, epsilon) (Blake et al., 2002). Several forms of muscular dystrophy are caused by defects in the function and assembly of the DGC (Blake et al., 2002). Dystrophin and members of the DGC including sarcoglycans are found in the brain where they participate in macromolecular assemblies that anchor receptors to specialized sites within the membrane. In neurons, dystrophin and the DGC participate in the postsynaptic clustering and stabilization of some inhibitory GABAergic synapses (Waite et al., 2009). Polymorphisms located within epsilon-sarcoglycan results in a neurogenic movement disorder, myoclonus-dystonia syndrome, which suggests that members of the sarcoglycan family of glycoproteins (including gamma-sarcoglycan) play a key role in dopaminergic neurotransmission (Peall et al., 2013). In addition, polymorphisms within epsilon-sarcoglycan are associated with a psychiatric phenotype consisting of compulsivity, anxiety and alcohol dependence in addition to the characteristic motor phenotype (Hess et al., 2007; Peall et al., 2011). Collectively, these results provide strong functional support for the association of a SNP (rs4770403 p < 10-6) in a regulatory genomic region near SGCG and alcohol dependence (Frank et al., 2012). A functional mechanism is hypothesized whereby genetic variation in SGCG affects GABAergic synaptic transmission within the ventral tegmental area that leads to a hypodopaminergic tone that manifests during protracted abstinence and contributes to craving and compulsive drinking.
MSX1 encodes Msh homeobox 1, a regulator of early central nervous system and craniofacial development that is also expressed in the adult brain (Fig. 1 and Table 3). Indeed, MSX1 is expressed at higher levels in the adult hippocampus than in the fetal hippocampus (Ramos et al., 2004). In a genome-wide DNA methylation study examining hippocampal expression of GABA-related genes, MSX1 contained the greatest number of differentially methylated positions associated with schizophrenia and bipolar disorder (Ruzicka et al., 2015). In addition, MSX1 is differentially expressed in the striatum of rats that display a conditioned place preference for psychostimulants (Dela Pena et al., 2013). Functionally, MSX1 is essential for maintaining the neurogenic potential of midbrain dopaminergic neurons through regulation of β-catenin signaling (Joksimovic and Awatramani, 2014). The cross-talk between MSX1 and β-catenin signaling may mediate stress-related phenotypes because intact β-catenin signaling is required for stress-resilience (Dias et al., 2014). Taken together, it can be speculated that genetic variation in MSX1 may lead to altered gene expression of the transcript in the hippocampus and/or striatum associated with compulsive drug and alcohol consumption; whereas a functional disruption of MSX1-β-catenin interactions in the ventral tegmental area may give rise to reduced dopamine signaling characterized by acute and protracted abstinence.
CMTM8 encodes CKLF Like MARVEL Transmembrane Domain Containing 8, a chemokine-like factor gene involved in cytokine signaling (Fig. 1 and Table 3). Compulsive alcohol consumption is known to cause increases in the expression of neuroimmune signaling genes that alter cognitive processes and promote alcohol dependence (Blednov et al., 2005; Crews and Vetreno, 2016). Stress and chronic alcohol withdrawal selectively alters the brain expression of a number of neuroimmune-related genes, including chemokines and cytokines (Knapp et al., 2016). Thus, a feasible hypothesis is that polymorphism in CMTM8 alters neuroimmune-signaling associated with alcohol dependence and is related to heightened stress and negative emotional states during alcohol abstinence.
4.9. GWAS candidates: SERINC2 and HTR1A
Zuo et al., 2012 performed an analysis on samples from two previous studies, SAGE and COGA (Bierut et al., 2010; Edenberg et al., 2010; Zuo et al., 2012). The combined European American sample consisted of 1409 DSM-IV alcohol dependent cases and 1518 non-dependent control subjects which were used as the discovery sample in the GWAS. The combined African American sample consisted of 681 DSM-IV alcohol dependent cases and 608 non-dependent control subjects which was used for replication. The authors reported several SNPs meeting a p < 10–7 including 5 in SERINC2, and 1 in HTR1A. Functional hypotheses relevant to the addiction cycle could be generated for SNPs within or near SERINC2 and HTR1A and are described below.
SERINC2 encodes the serine incorporator 2 which functions as a l-serine transmembrane transporter (Fig. 1 and Table 3). l-serine plays an essential role in neuronal development and function. l-serine serves as a precursor in the synthesis of l-cysteine, sphingolipids, and the neuromodulators d-serine and glycine, which modulate excitatory NMDA receptors and inhibitory glycine receptors in the brain (Snell, 1984). l-serine is a non-essential amino acid; however, its de novo synthesis within the brain is essential because of the limited permeability of the blood–brain barrier for l-serine (Smith, 2000). l-serine is generated in astrocytes and requires a dedicated amino-acid transporter for entry into neuronal cells by ASCT1 (alanine-serine-cysteine transporter) which is highly similar in structure and function to SERINC2 (Yamasaki et al., 2001). Mutations in the gene encoding ASCT1 results in a brain disorder characterized by developmental delay, microcephaly and hypomyelination (Damseh et al., 2015). In addition, knockout of Asct1 in mice results in a significant decrease in brain glycine levels, deficits in glycine neurotransmission and a hyperekplexia-like phenotype. The clinical and pre-clinical phenotypes resulting from genetic variation in the cellular mechanisms that mediate l-serine transport in the brain fit with the finding that SERINC2 SNPs are associated with alcohol dependence. In particular, variation in SERINC2 may likely alter inhibitory glycine neurotransmission as well as excitatory glutamate neurotransmission that would be consistent with hyperexcitability and negative affect during alcohol abstinence or drug seeking and craving during protracted abstinence.
HTR1A encodes the 5-hydroxytryptamine (Serotonin) receptor 1A subunit, a G protein-coupled receptor that mediates the actions of serotonin in the brain (Fig. 1 and Table 3). Rodent animal models and human clinical studies suggest dysfunction in serotonin (5-HT) innervation and transmission in early-onset genetically determined or type 2 alcoholism. A candidate gene association study found an association with variants of HTR1A with suicide attempts in alcohol dependent patients (Wrzosek et al., 2011). A 5HT1A/B antagonist (WAY100635) reduces drug seeking during initial abstinence in rats (Kohtz and Aston-Jones, 2017). In addition, serotonin specific neuronal reduction in expression of Htr1A in mice contributes to cocaine seeking behavior and inhibition of dorsal raphe 5-HT1A autoreceptors attenuates cocaine self-administration in rats with prolonged extended access, but not 1 h access to the drug (You et al., 2016). Finally, a quantitative trait gene, the multiple PDZ domain protein (MPDZ), mediates drug withdrawal responses in mice and humans (Shirley et al., 2004; Fehr et al., 2002; Reilly et al., 2008; Milner et al., 2015; Chen and Buck, 2010; Chen et al., 2011). MPDZ is critical for serotonin type 2 receptor signaling and likely mediates variation in alcohol withdrawal severity via alterations in serotonin signaling fidelity (Reilly et al., 2008; Chen et al., 2009; Buck et al., 2004). Thus, SNPs within HTR1A associated with alcohol dependence (Zuo et al., 2012) could alter compulsive drug seeking behavior which predominates during the preoccupation-anticipation stage of addiction.
5. Conclusions
The GWAS approach is beginning to uncover novel biology but will require further testing and confirmation, especially functional validation. We have attempted to present a new heuristic framework for GWAS of alcohol dependence, as one example of an approach for generating plausible functional hypotheses for candidate genes that may function within one or more stages of the addiction cycle. This heuristic framework could also be viewed as a reverse translational approach: information from a human genetic study used to guide functional genetic studies in animal models (see Table 3). For example, any one of the many well-validated animal models of alcohol-related phenotypes could be used to test the hypothesis that a candidate GWAS gene plays a specific role in a particular stage of the addiction cycle.
The ultimate goal of genomic discovery-based approaches is to identify and characterize the biological function of gene networks that are causal to the disorder rather than individual gene functions (Mamdani et al., 2015; Ponomarev et al., 2012; Repunte-Canonigo et al., 2015; Zhou et al., 2014). In support of this hypothesis a recent study found that functional brain networks measured with resting-state fMRI are directly linked to correlated gene expression in post mortem human brain, suggesting that the emergent properties of gene networks endow function to neurocircuits (Richiardi et al., 2015). Although we largely discussed potential functions of candidate genes in isolation, it is almost certain that there exists an unknown biological connection between genes that function as a network that is yet to be discovered (Morozova et al., 2012; Spanagel et al., 2013; Wolen and Miles, 2012). However, our heuristic framework, parsing genes into an addiction cycle domain, is one way that begins to hint at gene network connectivity and function (see Fig. 1). Big data generating efforts, especially in genomics and the NIH Big Data to Knowledge (BD2K) program, have become an attractive approach for advancing biological discoveries. However, pure big data approaches have been criticized for failing to provide conceptual insights into biological processes (Coveney et al., 2016). It is therefore suggested that big data approaches should rely on theoretical frameworks to avoid circumventing the proven modern scientific method of inquiry and digressing to pre-Baconism methods of radical empiricism (i.e., data generation without reason). The addiction cycle theoretical framework used here, is one theoretical framework, to anchor future genomic studies, rather than fall susceptible to ‘pure blind data gathering,’ which seems to be on the rise. Theoretical frameworks applied to big data (e.g., GWAS) will maximize the production of reliable predictive models, the understanding of the structure and stochastic properties of complex systems, which will ultimately advance actionable conceptual knowledge.
A general observation from comparing across studies is that each GWAS results in largely a different set of candidate genes. This observation is not surprising and underscores the presence of extensive genetic heterogeneity that is well documented for alcohol dependence (Litten et al., 2015). A parsimonious explanation can be derived when genetic heterogeneity is viewed in terms of self-organizing, chaotic, and complex genetic networks, which some have described as a new biology for medicine. (Coffey, 1998; Hawrylycz et al., 2015). Thus, key biological networks may be disrupted via many different genetic perturbations. Even if the phenotype in every affected individual arises from a different specific genetic polymorphism, each will nonetheless share disruption of related key biological processes. This notion is supported by the finding that brain gene expression networks from human alcoholic brain were enriched for signals from GWAS of alcohol dependence (Farris et al., 2015). Defining the ways in which biological networks for alcohol dependence are impacted by genetic variation remains the challenge for the future and will contribute substantially to the understanding of the etiology and provide important targets for pharmacological intervention.
A major goal of the current analysis was to provide one example of how genetic information from GWAS and other approaches can be sorted based on potential function into a heuristic neurobiologic-functional framework (i.e., addiction cycle domains) that may enable improvements in prioritization of novel drug targets, improvements in the predictive value of genetic bio-markers and ultimately the integration of genomic information into a clinical diagnostic system. This approach is supported by the finding that the DSM-IV syndrome of alcohol dependence, and the current DSM-5 syndrome of AUD, do not reflect a single dimension of genetic liability, rather, dependence criteria from DSM-IV reflect at least three underlying dimensions that index genetic risk which is consistent with the genetic animal model literature (Kendler et al., 2012). This phenomenon is also seen for other common genetically complex psychiatric disorders such as depression (Kendler et al., 2013). These results underscore that the DSM disorders are primarily evolving theoretical constructs of unknown etiology rather than discrete neuro-pathologically defined diseases; while on the other hand, an accumulation of direct evidence supports an etiological basis of the disease arising from diverse genetic and environmental risk factors. Therefore, approaches that incorporate biological information such as genetic variants into a neurobiologic-functional framework will undoubtedly result in substantial improvements in treatment and risk prediction for AUD in the future.
Acknowledgements
The authors thank Dr. Justin Rhodes for suggestions on a draft of this manuscript.
Abbreviations
- BDNF-CREB
brain derived neurotropic factor, cAMP response element-binding protein
- CARS
Cysteinyl-TRNA Synthetase
- COMT
Catechol-O-methyltransferase
- CRHR1
CRF receptor 1
- CMTM8
CKLF Like MARVEL Transmembrane Domain Containing 8
- DRD2/DRD4
dopamine receptors type 2 and 4
- FKBP5
FK506 binding protein 5
- GATA4
GATA Binding Protein 4
- GRM5
Glutamate Metabotropic Receptor 5
- HTR1A
5-Hydroxytryptamine Receptor 1A
- HTR2B
serotonin type 2B receptor
- KCNMA1
Potassium Calcium-Activated Channel Subfamily M Alpha 1
- KCNJ6
G protein-activated inward rectifier potassium channel 2
- MBNL2
Muscle-blind Like Splicing Regulator 2
- MSX1
Msh Homeobox 1
- NAP1L4
Nucleosome Assembly Protein 1 Like 4
- OPRM1
mu-opioid receptor
- OPRK1
kappa-opioid receptor
- PDYN
prodynorphin
- PECR
Peroxisomal Trans-2-Enoyl-CoA Reductase
- PKNOX2
PBX/Knotted 1 Homeobox 2
- POMC
Proopiomelanocortin
- NPY
neuropeptide Y
- RASL11A
RAS Like Family 11 Member A
- SERINC2
Serine Incorporator 2
- SGCG
Sarcoglycan Gamma
- SLC10A2
Solute Carrier Family 10 Member 2
- TPK1
Thiamin Pyrophosphokinase 1.
References
- A haplotype map of the human genome. Nature 437, 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Bierut LJ, 2012. Identifying genetic variation for alcohol dependence. Alcohol Res. 34, 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Edenberg HJ, Gelernter J, 2016. Meta-analyses of genome-wide association data hold new promise for addiction genetics. J. Stud. Alcohol Drugs 77, 676–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amen SL, Piacentine LB, Ahmad ME, Li SJ, Mantsch JR, Risinger RC, Baker DA, 2011. Repeated N-acetyl cysteine reduces cocaine seeking in rodents and craving in cocaine-dependent humans. Neuropsychopharmacology 36, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni FA, Hunter EF, Lowry PJ, Noble JM, Seckl JR, 1992. Atriopeptin: an endogenous corticotropin-release inhibiting hormone. Endocrinology 130, 1753–1755. [DOI] [PubMed] [Google Scholar]
- Aryal P, Dvir H, Choe S, Slesinger PA, 2009. A discrete alcohol pocket involved in GIRK channel activation. Nat. Neurosci 12, 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashare RL, Valdez JN, Ruparel K, Albelda B, Hopson RD, Keefe JR, Loughead J, Lerman C, 2013. Association of abstinence-induced alterations in working memory function and COMT genotype in smokers. Psychopharmacol. (Berl) 230, 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum F, Ghosh D, Tseng GC, Feingold E, 2012. Comprehensive literature review and statistical considerations for GWAS meta-analysis. Nucleic Acids Res. 40, 3777–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda X, Fuentes S, Daviu N, Nadal R, Armario A, 2015. Stress-induced sensitization: the hypothalamic-pituitary-adrenal axis and beyond. Stress 18, 269–279. [DOI] [PubMed] [Google Scholar]
- Ben Hamida S, Neasta J, Lasek AW, Kharazia V, Zou M, Carnicella S, Janak PH, Ron D, 2012. The small G protein H-Ras in the mesolimbic system is a molecular gateway to alcohol-seeking and excessive drinking behaviors. J. Neurosci 32, 15849–15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzerouk F, Gierski F, Gorwood P, Ramoz N, Stefaniak N, Hubsch B, Kaladjian A, Limosin F, 2013. Brain-derived neurotrophic factor (BDNF) Val66Met polymorphism and its implication in executive functions in adult offspring of alcohol-dependent probands. Alcohol 47, 271–274. [DOI] [PubMed] [Google Scholar]
- Bettinger JC, Davies AG, 2014. The role of the BK channel in ethanol response behaviors: evidence from model organism and human studies. Front. Physiol 5, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua L, Carli V, Sarchiapone M, George DK, Goldman D, Roy A, Enoch MA, 2012. Interaction between FKBP5 and childhood trauma and risk of aggressive behavior. Arch. Gen. Psychiatry 69, 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua L, Goldman D, 2013. Genetics of impulsive behaviour. Philos Trans. R Soc. Lond. B Biol. Sci 368 (1615), 20120380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua L, Doly S, Kaprio J, Yuan Q, Tikkanen R, Paunio T, Zhou Z, Wedenoja J, Maroteaux L, Diaz S, Belmer A, Hodgkinson CA, Dell'osso L, Suvisaari J, Coccaro E, Rose RJ, Peltonen L, Virkkunen M, Goldman D, 2010. A population-specific HTR2B stop codon predisposes to severe impulsivity. Nature 468, 1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI Jr., Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP, 2010. A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. U. S. A. 107, 5082–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Porjesz B, Saccone NL, Schuckit M, Tischfield J, Wang JC, Foroud T, Rice JP, Edenberg HJ, 2012. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol. Psychiatry 17, 445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Saccone NL, Rice JP, Goate A, Foroud T, Edenberg H, Almasy L, Conneally PM, Crowe R, Hesselbrock V, Li TK, Nurnberger J Jr., Porjesz B, Schuckit MA, Tischfield J, Begleiter H, Reich T, 2002. Defining alcohol-related phenotypes in humans. The collaborative study on the genetics of alcoholism. Alcohol Res. Health 26, 208–213. [PMC free article] [PubMed] [Google Scholar]
- Blake DJ, Weir A, Newey SE, Davies KE, 2002. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol. Rev 82, 291–329. [DOI] [PubMed] [Google Scholar]
- Blednov YA, Benavidez JM, Black M, Ferguson LB, Schoenhard GL, Goate AM, Edenberg HJ, Wetherill L, Hesselbrock V, Foroud T, Harris RA, 2015. Peroxisome proliferator-activated receptors alpha and gamma are linked with alcohol consumption in mice and withdrawal and dependence in humans. Alcohol Clin. Exp. Res 39, 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Bergeson SE, Walker D, Ferreira VM, Kuziel WA, Harris RA, 2005. Perturbation of chemokine networks by gene deletion alters the reinforcing actions of ethanol. Behav. Brain Res 165, 110–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Black M, Benavidez JM, Stamatakis EE, Harris RA, 2016. PPAR agonists: II. Fenofibrate and tesaglitazar alter behaviors related to voluntary alcohol consumption. Alcohol Clin. Exp. Res 40, 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Stoffel M, Chang SR, Harris RA, 2001. Potassium channels as targets for ethanol: studies of G-protein-coupled inwardly rectifying potassium channel 2 (GIRK2) null mutant mice. J. Pharmacol. Exp. Ther 298, 521–530. [PubMed] [Google Scholar]
- Buck KJ, Reilly MT, Rogers LM, Szeliga K, Grant K, Brodie MS, 2004. Serotonin 5-HT2 receptors and alcohol: reward, withdrawal and discrimination. Alcohol Clin. Exp. Res 28 (2), 211–216. [DOI] [PubMed] [Google Scholar]
- Butterworth RF, 1989. Effects of thiamine deficiency on brain metabolism: implications for the pathogenesis of the Wernicke-Korsakoff syndrome. Alcohol Alcohol 24, 271–279. [DOI] [PubMed] [Google Scholar]
- Cantor RM, Lange K, Sinsheimer JS, 2010. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. Am. J. Hum. Genet. 86, 6–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao LH, Yang XL, 2008. Natriuretic peptides and their receptors in the central nervous system. Prog. Neurobiol 84, 234–248. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, 1999. Population genetics–making sense out of sequence. Nat. Genet 21, 56–60. [DOI] [PubMed] [Google Scholar]
- Charbogne P, Kieffer BL, Befort K, 2014. 15 years of genetic approaches in vivo for addiction research: opioid receptor and peptide gene knockout in mouse models of drug abuse. Neuropharmacology 76 (Pt B), 204–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC, Manz N, Tang Y, Rangaswamy M, Almasy L, Kuperman S, Nurnberger J Jr., O'Connor SJ, Edenberg HJ, Schuckit MA, Tischfield J, Foroud T, Bierut LJ, Rohrbaugh J, Rice JP, Goate A, Hesselbrock V, Porjesz B, 2010. Single-nucleotide polymorphisms in corticotropin releasing hormone receptor 1 gene (CRHR1) are associated with quantitative trait of event-related potential and alcohol dependence. Alcohol Clin. Exp. Res 34, 988–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Buck KJ, 2010. Rostroventral caudate putamen involvement in ethanol withdrawal is influenced by a chromosome 4 locus. Genes Brain Behav 9 (7), 768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Cuzon Carlson VC, Wang J, Beck A, Heinz A, Ron D, et al. , 2011. Striatal involvement in human alcoholism and alcohol consumption, and withdrawal in animal models. Alcohol Clin. Exp. Res 35 (10), 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Reilly MT, Kozell LB, Hitzemann R, Buck KJ, 2009. Differential activation of limbic circuitry associated with chronic ethanol withdrawal in DBA/2J and C57BL/6J mice. Alcohol 43 (6), 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Hutchison KE, Calhoun VD, Claus ED, Turner JA, Sui J, Liu J, 2015. CREB-BDNF pathway influences alcohol cue-elicited activation in drinkers. Hum. Brain Mapp 36, 3007–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Lu RB, Peng GS, Wang MF, Wang HK, Ko HC, Chang YC, Lu JJ, Li TK, Yin SJ, 1999. Alcohol metabolism and cardiovascular response in an alcoholic patient homozygous for the ALDH2*2 variant gene allele. Alcohol Clin. Exp. Res 23, 1853–1860. [PubMed] [Google Scholar]
- Chu K, Koob GF, Cole M, Zorrilla EP, Roberts AJ, 2007. Dependence-induced increases in ethanol self-administration in mice are blocked by the CRF1 receptor antagonist antalarmin and by CRF1 receptor knockout. Pharmacol. Biochem. Behav 86 (4), 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey DS, 1998. Self-organization, complexity and chaos: the new biology for medicine. Nat. Med 4, 882–885. [DOI] [PubMed] [Google Scholar]
- Cohen E, Feinn R, Arias A, Kranzler HR, 2007. Alcohol treatment utilization: findings from the national epidemiologic Survey on alcohol and related conditions. Drug Alcohol Depend. 86, 214–221. [DOI] [PubMed] [Google Scholar]
- Compton WM, Dawson DA, Goldstein RB, Grant BF, 2013. Crosswalk between DSM-IV dependence and DSM-5 substance use disorders for opioids, cannabis, cocaine and alcohol. Drug Alcohol Depend. 132, 387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton NS, 1979. The familial incidence of alcoholism: a review. J. Stud. Alcohol 40, 89–116. [DOI] [PubMed] [Google Scholar]
- Coveney PV, Dougherty ER, Highfield RR, 2016. Big data need big theory too. Philos. Trans. A Math. Phys. Eng. Sci 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF, 2006. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict. Biol 11, 195–269. [DOI] [PubMed] [Google Scholar]
- Crews FT, Vetreno RP, 2016. Mechanisms of neuroimmune gene induction in alcoholism. Psychopharmacol. (Berl) 233, 1543–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damseh N, Simonin A, Jalas C, Picoraro JA, Shaag A, Cho MT, Yaacov B, Neidich J, Al-Ashhab M, Juusola J, Bale S, Telegrafi A, Retterer K, Pappas JG, Moran E, Cappell J, Anyane Yeboa K, Abu-Libdeh B, Hediger MA, Chung WK, Elpeleg O, Edvardson S, 2015. Mutations in SLC1A4, encoding the brain serine transporter, are associated with developmental delay, microcephaly and hypomyelination. J. Med. Genet. 52, 541–547. [DOI] [PubMed] [Google Scholar]
- Davidson E, Caffarella J, Vitseva O, Hou YM, King MP, 2001. Isolation of two cDNAs encoding functional human cytoplasmic cysteinyl-tRNA synthetase. Biol. Chem 382, 399–406. [DOI] [PubMed] [Google Scholar]
- Dela Pena I, Jeon SJ, Lee E, Ryu JH, Shin CY, Noh M, Cheong JH, 2013. Neuronal development genes are key elements mediating the reinforcing effects of methamphetamine, amphetamine, and methylphenidate. Psychopharmacol. (Berl) 230, 399–413. [DOI] [PubMed] [Google Scholar]
- Di Cesare Mannelli L, Zanardelli M, Micheli L, Ghelardini C, 2014. PPAR- gamma impairment alters peroxisome functionality in primary astrocyte cell cultures. Biomed. Res. Int 2014, 546453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias C, Feng J, Sun H, Shao NY, Mazei-Robison MS, Damez-Werno D, Scobie K, Bagot R, LaBonte B, Ribeiro E, Liu X, Kennedy P, Vialou V, Ferguson D, Pena C, Calipari ES, Koo JW, Mouzon E, Ghose S, Tamminga C, Neve R, Shen L, Nestler EJ, 2014. beta-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature 516, 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D, Elvevag B, 2009. Genes, cognition and brain through a COMT lens. Neuroscience 164, 72–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopico AM, Bukiya AN, Kuntamallappanavar G, Liu J, 2016. Modulation of BK channels by ethanol. Int. Rev. Neurobiol 128, 239–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, 2007. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res. Health 30, 5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T, 2006. The genetics of alcoholism: identifying specific genes through family studies. Addict. Biol 11, 386–396. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T, 2013. Genetics and alcoholism. Nat Rev Gastroenterol Hepatol 10, 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T, 2014. Genetics of alcoholism. Handb. Clin. Neurol. 125, 561–571. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI Jr., Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T, 2010. Genomewide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin. Exp. Res 34, 840–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, 2013. Genetic influences on the development of alcoholism. Curr. Psychiatry Rep 15, 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisal M, Waseem D, Ismatullah H, Taqi MM, 2014. A molecular prospective provides new insights into implication of PDYN and OPRK1 genes in alcohol dependence. Comput. Biol. Med 53, 250–257. [DOI] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Belknap JK, Crabbe JC, Buck KJ, 2002. Congenic mapping of alcohol and pentobarbital withdrawal liability loci to a <1 centimorgan interval of murine chromosome 4: identification of Mpdz as a candidate gene. J. Neurosci 22 (9), 3730–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris SP, Arasappan D, Hunicke-Smith S, Harris RA, Mayfield RD, 2015. Transcriptome organization for chronic alcohol abuse in human brain. Mol. Psychiatry 20, 1438–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson LB, Most D, Blednov YA, Harris RA, 2014. PPAR agonists regulate brain gene expression: relationship to their effects on ethanol consumption. Neuropharmacology 86, 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbey FM, Claus ED, Morgan M, Forester GR, Hutchison K, 2012. Dopaminergic genes modulate response inhibition in alcohol abusing adults. Addict. Biol 17, 1046–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fognani C, Kilstrup-Nielsen C, Berthelsen J, Ferretti E, Zappavigna V, Blasi F, 2002. Characterization of PREP2, a paralog of PREP1, which defines a novel subfamily of the MEINOX TALE homeodomain transcription factors. Nucleic Acids Res 30, 2043–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, Crowe R, Schuckit M, Porjesz B, Begleiter H, Reich T, 2000. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin. Exp. Res 24, 933–945. [PubMed] [Google Scholar]
- Foroud T, Wetherill LF, Dick DM, Hesselbrock V, Nurnberger JI Jr., Kramer J, Tischfield J, Schuckit M, Bierut LJ, Xuei X, Edenberg HJ, 2007. Lack of association of alcohol dependence and habitual smoking with catechol-O-methyltransferase. Alcohol Clin. Exp. Res. 31, 1773–1779. [DOI] [PubMed] [Google Scholar]
- Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P, Herms S, Wodarz N, Soyka M, Zill P, Maier W, Mossner R, Gaebel W, Dahmen N, Scherbaum N, Schmal C, Steffens M, Lucae S, Ising M, Muller-Myhsok B, Nothen MM, Mann K, Kiefer F, Rietschel M, 2012. Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict. Biol 17, 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, KwokP Y, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe'er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L'Archeveque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J, 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA, 2014. Genome-wide association study of alcohol dependence: significant findings in African- and European-Americans including novel risk loci. Mol. Psychiatry 19, 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi A, Krishnan HR, Atkinson NS, 2014. Susceptibility to ethanol withdrawal seizures is produced by BK channel gene expression. Addict. Biol 19, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianoulakis C, Dai X, Thavundayil J, Brown T, 2005. Levels and circadian rhythmicity of plasma ACTH, cortisol, and beta-endorphin as a function of family history of alcoholism. Psychopharmacol. (Berl) 181, 437–444. [DOI] [PubMed] [Google Scholar]
- Golan D, Lander ES, Rosset S, 2014. Measuring missing heritability: inferring the contribution of common variants. Proc. Natl. Acad. Sci. U. S. A. 111, E5272–E5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F, 2005a. The genetics of addictions: uncovering the genes. Nat. Rev. Genet 6, 521–532. [DOI] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, O'Malley S, Anton R, 2005b. COMBINE genetics study: the pharmacogenetics of alcoholism treatment response: genes and mechanisms. J. Stud. Alcohol Suppl 56–64 discussion 33. [DOI] [PubMed] [Google Scholar]
- Goodwin DW, 1979. The cause of alcoholism and why it runs in families. Br. J. Addict. Alcohol Other Drugs 74, 161–164. [DOI] [PubMed] [Google Scholar]
- Goodwin DW, 1989. The gene for alcoholism. J. Stud. Alcohol 50, 397–398. [DOI] [PubMed] [Google Scholar]
- Goodwin DW, Schulsinger F, Hermansen L, Guze SB, Winokur G, 1973. Alcohol problems in adoptees raised apart from alcoholic biological parents. Arch. Gen. Psychiatry 28, 238–243. [DOI] [PubMed] [Google Scholar]
- Goodwin DW, Schulsinger F, Knop J, Mednick S, Guze SB, 1977. Psychopathology in adopted and nonadopted daughters of alcoholics. Arch. Gen. Psychiatry 34, 1005–1009. [DOI] [PubMed] [Google Scholar]
- Goodwin DW, Schulsinger F, Moller N, Hermansen L, Winokur G, Guze SB, 1974. Drinking problems in adopted and nonadopted sons of alcoholics. Arch. Gen. Psychiatry 31, 164–169. [DOI] [PubMed] [Google Scholar]
- Grant BF, Goldstein RB, Saha TD, Chou SP, Jung J, Zhang H, Pickering RP, Ruan WJ, Smith SM, Huang B, Hasin DS, 2015. Epidemiology of DSM-5 alcohol use disorder: results from the national epidemiologic Survey on alcohol and related conditions III. JAMA Psychiatry 72, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JD, Agrawal A, Bucholz KK, Madden PA, Pergadia ML, Nelson EC, Lynskey MT, Todd RD, Todorov AA, Hansell NK, Whitfield JB, Martin NG, Heath AC, 2009. Alcohol consumption indices of genetic risk for alcohol dependence. Biol. Psychiatry 66, 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grepin C, Dagnino L, Robitaille L, Haberstroh L, Antakly T, Nemer M, 1994. A hormone-encoding gene identifies a pathway for cardiac but not skeletal muscle gene transcription. Mol. Cell Biol 14, 3115–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurling HM, Reveley MA, Murray RM, 1984. Increased cerebral ventricular volume in monozygotic twins discordant for alcoholism. Lancet 1, 986–988. [DOI] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA, 2005. Complement factor H variant increases the risk of age-related macular degeneration. Science 308, 419–421. [DOI] [PubMed] [Google Scholar]
- Hall MA, Moore JH, Ritchie MD, 2016. Embracing complex associations in common traits: critical considerations for precision medicine. Trends Genet. 32, 470–484. [DOI] [PubMed] [Google Scholar]
- Haller K, Rambaldi I, Daniels E, Featherstone M, 2004. Subcellular localization of multiple PREP2 isoforms is regulated by actin, tubulin, and nuclear export. J. Biol. Chem 279, 49384–49394. [DOI] [PubMed] [Google Scholar]
- Hansson AC, Cippitelli A, Sommer WH, Fedeli A, Bjork K, Soverchia L, Terasmaa A, Massi M, Heilig M, Ciccocioppo R, 2006. Variation at the rat Crhr1 locus and sensitivity to relapse into alcohol seeking induced by environmental stress. Proc. Natl. Acad. Sci. U. S. A. 103, 15236–15241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy PA, Chen W, Wilce PA, 1999. Chronic ethanol exposure and withdrawal influence NMDA receptor subunit and splice variant mRNA expression in the rat cerebral cortex. Brain Res. 819, 33–39. [DOI] [PubMed] [Google Scholar]
- Harper CG, 1988. Brain damage and alcohol abuse: where do we go from here? Br. J. Addict 83, 613–624. [DOI] [PubMed] [Google Scholar]
- Hart AB, Kranzler HR, 2015. Alcohol dependence genetics: Lessons learned from Genome-Wide Association Studies (GWAS) and post-GWAS analyses. Alcohol Clin. Exp. Res 39, 1312–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T, Guillozet-Bongaarts AL, Jegga AG, Aronow BJ, Lee CK, Bernard A, Glasser MF, Dierker DL, Menche J, Szafer A, Collman F, Grange P, Berman KA, Mihalas S, Yao Z, Stewart L, Barabasi AL, Schulkin J, Phillips J, Ng L, Dang C, Haynor DR, Jones A, Van Essen DC, Koch C, Lein E, 2015. Canonical genetic signatures of the adult human brain. Nat. Neurosci 18, 1832–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW, 2011. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol. Psychiatry 70, 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, Sidhu H, Stouffer DG, Kreifeldt M, Le D, Cates-Gatto C, et al. , 2015. GIRK3 gates activation of the mesolimbic dopaminergic pathway by ethanol. Proc. Natl. Acad. Sci. U. S. A. 112 (22), 7091–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess CW, Raymond D, Aguiar Pde C, Frucht S, Shriberg J, Heiman GA, Kurlan R, Klein C, Bressman SB, Ozelius LJ, Saunders-Pullman R, 2007. Myoclonus-dystonia, obsessive-compulsive disorder, and alcohol dependence in SGCE mutation carriers. Neurology 68, 522–524. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, 2009. Genomewide association studies–illuminating biologic pathways. N. Engl. J. Med 360, 1699–1701. [DOI] [PubMed] [Google Scholar]
- Honse Y, Nixon KM, Browning MD, Leslie SW, 2003. Cell surface expression of NR1 splice variants and NR2 subunits is modified by prenatal ethanol exposure. Neuroscience 122, 689–698. [DOI] [PubMed] [Google Scholar]
- Hrubec Z, Omenn GS, 1981. Evidence of genetic predisposition to alcoholic cirrhosis and psychosis: twin concordances for alcoholism and its biological end points by zygosity among male veterans. Alcohol Clin. Exp. Res 5, 207–215. [DOI] [PubMed] [Google Scholar]
- Hsu DT, Mickey BJ, Langenecker SA, Heitzeg MM, Love TM, Wang H, Kennedy SE, Pecina M, Shafir T, Hodgkinson CA, Enoch MA, Goldman D, Zubieta JK, 2012. Variation in the corticotropin-releasing hormone receptor 1 (CRHR1) gene influences fMRI signal responses during emotional stimulus processing. J. Neurosci. 32, 3253–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MC, Schwandt ML, Chester JA, Kirchhoff AM, Kao CF, Liang T, Tapocik JD, Ramchandani VA, George DT, Hodgkinson CA, Goldman D, Heilig M, 2014. FKBP5 moderates alcohol withdrawal severity: human genetic association and functional validation in knockout mice. Neuropsychopharmacology 39, 2029–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto I, Sonoda I, Yuki Y, Inazawa J, 2001. Identification and characterization of human PKNOX2, a novel homeobox-containing gene. Biochem. Biophys. Res. Commun 287, 270–276. [DOI] [PubMed] [Google Scholar]
- Joksimovic M, Awatramani R, 2014. Wnt/beta-catenin signaling in midbrain dopaminergic neuron specification and neurogenesis. J. Mol. Cell Biol 6, 27–33. [DOI] [PubMed] [Google Scholar]
- Kalsi G, Prescott CA, Kendler KS, Riley BP, 2009. Unraveling the molecular mechanisms of alcohol dependence. Trends Genet. 25, 49–55. [DOI] [PubMed] [Google Scholar]
- Kanai M, Tanaka T, Okada Y, 2016. Empirical estimation of genome-wide significance thresholds based on the 1000 Genomes Project data set. J. Hum. Genet 61 (10), 861–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Rangaswamy M, Manz N, Wang JC, Wetherill L, Hinrichs T, Almasy L, Brooks A, Chorlian DB, Dick D, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J Jr., Rice J, Schuckit M, Tischfield J, Bierut LJ, Edenberg HJ, Goate A, Foroud T, Porjesz B, 2012. Family-based genomewide association study of frontal theta oscillations identifies potassium channel gene KCNJ6. Genes Brain Behav. 11, 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpyak VM, Winham SJ, Preuss UW, Zill P, Cunningham JM, Walker DL, Lewis KA, Geske JR, Colby CL, Abulseoud OA, Hall-Flavin DK, Loukianova LL, Schneekloth TD, Frye MA, Bazov I, Heit JA, Bakalkin G, Mrazek DA, Biernacka JM, 2013. Association of the PDYN gene with alcohol dependence and the propensity to drink in negative emotional states. Int. J. Neuropsychopharmacol 16, 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, 2012. Levels of explanation in psychiatric and substance use disorders: implications for the development of an etiologically based nosology. Mol. Psychiatry 17, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Aggen SH, Neale MC, 2013. Evidence for multiple genetic factors underlying DSM-IV criteria for major depression. JAMA Psychiatry 70, 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Aggen SH, Prescott CA, Crabbe J, Neale MC, 2012. Evidence for multiple genetic factors underlying the DSM-IV criteria for alcohol dependence. Mol. Psychiatry 17, 1306–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV, 2011. Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcohol Clin. Exp. Res 35, 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ, 1994. A twin-family study of alcoholism in women. Am. J. Psychiatry 151, 707–715. [DOI] [PubMed] [Google Scholar]
- Kiefer F, Witt SH, Frank J, Richter A, Treutlein J, Lemenager T, Nothen MM, Cichon S, Batra A, Berner M, Wodarz N, Zimmermann US, Spanagel R, Wiedemann K, Smolka MN, Heinz A, Rietschel M, Mann K, 2011. Involvement of the atrial natriuretic peptide transcription factor GATA4 in alcohol dependence, relapse risk and treatment response to acamprosate. Pharmacogenom. J 11, 368–374. [DOI] [PubMed] [Google Scholar]
- Kingsmore SF, Lindquist IE, Mudge J, Gessler DD, Beavis WD, 2008. Genomewide association studies: progress and potential for drug discovery and development. Nat. Rev. Drug Discov 7, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp DJ, Harper KM, Whitman BA, Zimomra Z, Breese GR, 2016. Stress and withdrawal from chronic ethanol induce selective changes in neuroimmune mRNAs in differing brain sites. Brain Sci. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz AS, Aston-Jones G, 2017. Cocaine seeking during initial abstinence is driven by noradrenergic and serotonergic signaling in Hippocampus in a sex-dependent manner. Neuropsychopharmacology 42 (2), 408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND, 2010. Neurocircuitry of addiction. Neuro psychopharmacology 35, 217–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND, 2016. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry 3, 760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozell LB, Walter NA, Milner LC, Wickman K, Buck KJ, 2009. Mapping a barbiturate withdrawal locus to a 0.44 Mb interval and analysis of a novel null mutant identify a role for Kcnj9 (GIRK3) in withdrawal from pentobarbital, zolpidem, and ethanol. J. Neurosci 29, 11662–11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, Edenberg HJ, 2010. Pharmacogenetics of alcohol and alcohol dependence treatment. Curr. Pharm. Des 16, 2141–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreifeldt M, Cates-Gatto C, Roberts AJ, Contet C, 2015. BK Channel beta1 Subunit Contributes to Behavioral Adaptations Elicited by Chronic Intermittent Ethanol Exposure. Alcohol Clin. Exp. Res 39 (12), 2394–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreifeldt M, Le D, Treistman SN, Koob GF, Contet C, 2013. BK channel beta1 and beta4 auxiliary subunits exert opposite influences on escalated ethanol drinking in dependent mice. Front. Integr. Neurosci 7, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HR, Sakharkar AJ, Teppen TL, Berkel TD, Pandey SC, 2014. The epigenetic landscape of alcoholism. Int. Rev. Neurobiol 115, 75–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Hapidin H, Bee YT, Ismail Z, 2013. Effects of the mGluR5 antagonist MPEP on ethanol withdrawal induced anxiety-like syndrome in rats. Behav. Brain Funct 9, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M, 2001. Differential effects of chronic ethanol treatment on N-methyl-D-aspartate R1 splice variants in fetal cortical neurons. J. Biol. Chem 276, 29764–29771. [DOI] [PubMed] [Google Scholar]
- Kupila J, Karkkainen O, Laukkanen V, Tupala E, Tiihonen J, Storvik M, 2013. mGluR1/5 receptor densities in the brains of alcoholic subjects: a whole-hemisphere autoradiography study. Psychiatry Res 212, 245–250. [DOI] [PubMed] [Google Scholar]
- Kwako LE, Momenan R, Litten RZ, Koob GF, Goldman D, 2016. Addictions neuroclinical assessment: a neuroscience-based framework for addictive disorders. Biol. Psychiatry 80, 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, 1996. The new genomics: global views of biology. Science 274, 536–539. [DOI] [PubMed] [Google Scholar]
- Lee C, Mayfield RD, Harris RA, 2014. Altered gamma-aminobutyric acid type B receptor subunit 1 splicing in alcoholics. Biol. Psychiatry 75 (10), 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Smith GW, Vale W, Lee KF, Rivier C, 2001. Mice that lack corticotropin-releasing factor (CRF) receptors type 1 show a blunted ACTH response to acute alcohol despite up-regulated constitutive hypothalamic CRF gene expression. Alcohol Clin. Exp. Res 25, 427–433. [PubMed] [Google Scholar]
- Lewohl JM, Wilson WR, Mayfield RD, Brozowski SJ, Morrisett RA, Harris RA, 1999. G- protein-coupled inwardly rectifying potassium channels are targets of alcohol action. Nat. Neurosci 2, 1084–1090. [DOI] [PubMed] [Google Scholar]
- Litten RZ, Ryan ML, Falk DE, Reilly M, Fertig JB, Koob GF, 2015. Heterogeneity of alcohol use disorder: understanding mechanisms to advance personalized treatment. Alcohol Clin. Exp. Res 39, 579–584. [DOI] [PubMed] [Google Scholar]
- Lovallo WR, Enoch MA, Acheson A, Cohoon AJ, Sorocco KH, Hodgkinson CA, Vincent AS, Glahn DC, Goldman D, 2015. Cortisol stress response in men and women modulated differentially by the Mu-opioid receptor gene polymorphism OPRM1 A118G. Neuropsychopharmacology 40, 2546–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamdani M, Williamson V, McMichael GO, Blevins T, Aliev F, Adkins A, Hack L, Bigdeli T, van der Vaart AD, Web BT, Bacanu SA, Kalsi G, Kendler KS, Miles MF, Dick D, Riley BP, Dumur C, Vladimirov VI, 2015. Integrating mRNA and miRNA weighted gene Co-Expression networks with eQTLs in the nucleus accumbens of subjects with alcohol dependence. PLoS One 10, e0137671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM, 2009. Finding the missing heritability of complex diseases. Nature 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marballi K, Genabai NK, Blednov YA, Harris RA, Ponomarev I, 2016. Alcohol consumption induces global gene expression changes in VTA dopaminergic neurons. Genes Brain Behav 15, 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin PR, Pekovich SR, McCool BA, Whetsell WO, Singleton CK, 1994. Thiamine utilization in the pathogenesis of alcohol-induced brain damage. Alcohol Alcohol Suppl 2, 273–279. [PubMed] [Google Scholar]
- Massie A, Boillee S, Hewett S, Knackstedt L, Lewerenz J, 2015. Main path and byways: non- vesicular glutamate release by system xc(−) as an important modifier of glutamatergic neurotransmission. J. Neurochem 135, 1062–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathies LD, Blackwell GG, Austin MK, Edwards AC, Riley BP, Davies AG, Bettinger JC, 2015. SWI/SNF chromatin remodeling regulates alcohol response behaviors in Caenorhabditis elegans and is associated with alcohol dependence in humans. Proc. Natl. Acad. Sci. U. S. A. 112, 3032–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, 1999. Phenotyping alcoholism. Alcohol Clin. Exp. Res 23, 757–758. [DOI] [PubMed] [Google Scholar]
- Medrano-Fernandez A, Barco A, 2016. Nuclear organization and 3D chromatin architecture in cognition and neuropsychiatric disorders. Mol. Brain 9, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Pandey SC, 2011. The role of amygdaloid brain-derived neurotrophic factor, activity-regulated cytoskeleton-associated protein and dendritic spines in anxiety and alcoholism. Addict. Biol 16, 238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova TV, Goldman D, Mackay TF, Anholt RR, 2012. The genetic basis of alcoholism: multiple phenotypes, many genes, complex networks. Genome Biol 13, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottagui-Tabar S, Prince JA, Wahlestedt C, Zhu G, Goldman D, Heilig M, 2005. A novel single nucleotide polymorphism of the neuropeptide Y (NPY) gene associated with alcohol dependence. Alcohol Clin. Exp. Res 29, 702–707. [DOI] [PubMed] [Google Scholar]
- Mulholland PJ, Chandler LJ, 2007. The thorny side of addiction: adaptive plasticity and dendritic spines. ScientificWorldJournal 7, 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE, 2006. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc. Natl. Acad. Sci. U. S. A. 103, 6368–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner LC, Shirley RL, Kozell LB, Walter NA, Kruse LC, Komiyama NH, et al. , 2015. Novel MPDZ/MUPP1 transgenic and knockdown models confirm Mpdz's role in ethanol withdrawal and support its role in voluntary ethanol consumption. Addict. Biol 20 (1), 143–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz M, Pong-Wong R, Canela-Xandri O, Rawlik K, Haley CS, Tenesa A, 2016. Evaluating the contribution of genetics and familial shared environment to common disease using the UK Biobank. Nat. Genet 48, 980–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray RM, Clifford C, Gurling HM, Topham A, Clow A, Bernadt M, 1983. Current genetic and biological approaches to alcoholism. Psychiatr. Dev 1, 179–192. [PubMed] [Google Scholar]
- N'Gouemo P, Morad M, 2014. Alcohol withdrawal is associated with a down-regulation of large- conductance Ca(2)(+)-activated K(+) channels in rat inferior colliculus neurons. Psychopharmacol. (Berl) 231, 2009–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojelade SA, Jia T, Rodan AR, Chenyang T, Kadrmas JL, Cattrell A, Ruggeri B, Charoen P, Lemaitre H, Banaschewski T, Buchel C, Bokde AL, Carvalho F, Conrod PJ, Flor H, Frouin V, Gallinat J, Garavan H, Gowland PA, Heinz A, Ittermann B, Lathrop M, Lubbe S, Martinot JL, Paus T, Smolka MN, Spanagel R, O'Reilly PF, Laitinen J, Veijola JM, Feng J, Desrivieres S, Jarvelin MR, Schumann G, Rothenfluh A, 2015. Rsu1 regulates ethanol consumption in Drosophila and humans. Proc. Natl. Acad. Sci. U. S. A. 112, E4085–E4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A, 2008. Brain chromatin remodeling: a novel mechanism of alcoholism. J. Neurosci 28, 3729–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BL, Kim JW, Cheong HS, Kim LH, Lee BC, Seo CH, Kang TC, Nam YW, Kim GB, Shin HD, Choi IG, 2013. Extended genetic effects of ADH cluster genes on the risk of alcohol dependence: from GWAS to replication. Hum. Genet 132, 657–668. [DOI] [PubMed] [Google Scholar]
- Park YJ, Luger K, 2006a. Structure and function of nucleosome assembly proteins. Biochem. Cell Biol 84, 549–558. [DOI] [PubMed] [Google Scholar]
- Park YJ, Luger K, 2006b. The structure of nucleosome assembly protein 1. Proc. Natl. Acad. Sci. U. S. A. 103, 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkitna JR, Sikora M, Golda S, Golembiowska K, Bystrowska B, Engblom D, Bilbao A, Przewlocki R, 2013. Novelty-seeking behaviors and the escalation of alcohol drinking after abstinence in mice are controlled by metabotropic glutamate receptor 5 on neurons expressing dopamine d1 receptors. Biol. Psychiatry 73, 263–270. [DOI] [PubMed] [Google Scholar]
- Partanen J, 1972. On the relevance of twin studies. Ann. N. Y. Acad. Sci 197, 114–116. [DOI] [PubMed] [Google Scholar]
- Peall KJ, Smith DJ, Kurian MA, Wardle M, Waite AJ, Hedderly T, Lin JP, Smith M, Whone A, Pall H, White C, Lux A, Jardine P, Bajaj N, Lynch B, Kirov G, O'Riordan S, Samuel M, Lynch T, King MD, Chinnery PF, Warner TT, Blake DJ, Owen MJ, Morris HR, 2013. SGCE mutations cause psychiatric disorders: clinical and genetic characterization. Brain 136, 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peall KJ, Waite AJ, Blake DJ, Owen MJ, Morris HR, 2011. Psychiatric disorders, myoclonus dystonia, and the epsilon-sarcoglycan gene: a systematic review. Mov. Disord 26, 1939–1942. [DOI] [PubMed] [Google Scholar]
- Pietrzykowski AZ, Friesen RM, Martin GE, Puig SI, Nowak CL, Wynne PM, Siegelmann HT, Treistman SN, 2008. Posttranscriptional regulation of BK channel splice variant stability by miR-9 underlies neuroadaptation to alcohol. Neuron 59, 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD, 2012. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J. Neurosci 32, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS, 1999. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am. J. Psychiatry 156, 34–40. [DOI] [PubMed] [Google Scholar]
- Preuss UW, Winham SJ, Biernacka JM, Geske JR, Bakalkin G, Koller G, Zill P, Soyka M, Karpyak VM, 2013. PDYN rs2281285 variant association with drinking to avoid emotional or somatic discomfort. PLoS One 8, e78688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Cox NJ, 2002. The allelic architecture of human disease genes: common disease- common variant…or not? Hum. Mol. Genet 11, 2417–2423. [DOI] [PubMed] [Google Scholar]
- Ramos C, Martinez A, Robert B, Soriano E, 2004. Msx1 expression in the adult mouse brain: characterization of populations of beta-galactosidase-positive cells in the hippocampus and fimbria. Neuroscience 127, 893–900. [DOI] [PubMed] [Google Scholar]
- Ray LA, Barr CS, Blendy JA, Oslin D, Goldman D, Anton RF, 2012. The role of the Asn40Asp polymorphism of the mu opioid receptor gene (OPRM1) on alcoholism etiology and treatment: a critical review. Alcohol Clin. Exp. Res 36, 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Sehl M, Bujarski S, Hutchison K, Blaine S, Enoch MA, 2013. The CRHR1 gene, trauma exposure, and alcoholism risk: a test of G × E effects. Genes Brain Behav 12, 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Lander ES, 2001. On the allelic spectrum of human disease. Trends Genet 17, 502–510. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI Jr., Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H, 1998. Genome-wide search for genes affecting the risk for alcohol dependence. Am. J. Med. Genet 81, 207–215. [PubMed] [Google Scholar]
- Reilly MT, Milner LC, Shirley RL, Crabbe JC, Buck KJ, 2008. 5-HT2C and GABAB receptors influence handling-induced convulsion severity in chromosome 4 congenic and DBA/2J background strain mice. Brain Res 1198, 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repunte-Canonigo V, Shin W, Vendruscolo LF, Lefebvre C, van der Stap L, Kawamura T, Schlosburg JE, Alvarez M, Koob GF, Califano A, Sanna PP, 2015. Identifying candidate drivers of alcohol dependence-induced excessive drinking by assembly and interrogation of brain-specific regulatory networks. Genome Biol 16, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repunte-Canonigo V, van der Stap LD, Chen J, Sabino V, Wagner U, Zorrilla EP, Schumann G, Roberts AJ, Sanna PP, 2010. Genome-wide gene expression analysis identifies K-ras as a regulator of alcohol intake. Brain Res 1339, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richiardi J, Altmann A, Milazzo AC, Chang C, Chakravarty MM, Banaschewski T, Barker GJ, Bokde AL, Bromberg U, Buchel C, Conrod P, Fauth-Buhler M, Flor H, Frouin V, Gallinat J, Garavan H, Gowland P, Heinz A, Lemaitre H, Mann KF, Martinot JL, Nees F, Paus T, Pausova Z, Rietschel M, Robbins TW, Smolka MN, Spanagel R, Strohle A, Schumann G, Hawrylycz M, Poline JB, Greicius MD, 2015. BRAIN NETWORKS. Correlated gene expression supports synchronous activity in brain networks. Science 348, 1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel M, Treutlein J, 2013. The genetics of alcohol dependence. Ann. N. Y. Acad. Sci 1282, 39–70. [DOI] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ, 2011. Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci 12, 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P, Munroe D, Prawitt D, Chu LL, Bric E, Kim J, Reid LH, Davies C, Nakagama H, Loebbert R, Winterpacht A, Petruzzi MJ, Higgins MJ, Nowak N, Evans G, Shows T, Weissman BE, Zabel B, Housman DE, Pelletier J, 1997. Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics 44, 253–265. [DOI] [PubMed] [Google Scholar]
- Ruzicka WB, Subburaju S, Benes FM, 2015. Circuit- and diagnosis-specific DNA methylation changes at gamma-aminobutyric acid-related genes in postmortem human Hippocampus in schizophrenia and bipolar disorder. JAMA Psychiatry 72, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino V, Cottone P, Zhao Y, Iyer MR, Steardo L Jr., Steardo L, Rice KC, Conti B, Koob GF, Zorrilla EP, 2009a. The sigma-receptor antagonist BD-1063 decreases ethanol intake and reinforcement in animal models of excessive drinking. Neuropsychopharmacology 34, 1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino V, Cottone P, Zhao Y, Steardo L, Koob GF, Zorrilla EP, 2009b. Selective reduction of alcohol drinking in Sardinian alcohol-preferring rats by a sigma-1 receptor antagonist. Psychopharmacology 205, 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanseau P, Agarwal P, Barnes MR, Pastinen T, Richards JB, Cardon LR, Mooser V, 2012. Use of genome-wide association studies for drug repositioning. Nat. Biotechnol 30, 317–320. [DOI] [PubMed] [Google Scholar]
- Sasabe T, Ishiura S, 2010. Alcoholism and alternative splicing of candidate genes. Int. J. Environ. Res. Public Health 7, 1448–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitz J, Hodgkinson CA, Martin-Soelch C, Shen PH, Szczepanik J, Nugent A, Herscovitch P, Grace AA, Goldman D, Drevets WC, 2013. The functional DRD3 Ser9Gly polymorphism (rs6280) is pleiotropic, affecting reward as well as movement. PLoS One 8, e54108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Goodwin DA, Winokur G, 1972. A study of alcoholism in half siblings. Am. J. Psychiatry 128, 1132–1136. [DOI] [PubMed] [Google Scholar]
- Shirley RL, Walter NA, Reilly MT, Fehr C, Buck KJ, 2004. Mpdz is a quantitative trait gene for drug withdrawal seizures. Nat. Neurosci 7 (7), 699–700. [DOI] [PubMed] [Google Scholar]
- Sinclair CM, Cleva RM, Hood LE, Olive MF, Gass JT, 2012. mGluR5 receptors in the basolateral amygdala and nucleus accumbens regulate cue-induced reinstatement of ethanol-seeking behavior. Pharmacol. Biochem. Behav 101, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton CK, Martin PR, 2001. Molecular mechanisms of thiamine utilization. Curr. Mol. Med 1,197–207. [DOI] [PubMed] [Google Scholar]
- Smith QR, 2000. Transport of glutamate and other amino acids at the blood-brain barrier. J. Nutr 130, 1016s–1022s. [DOI] [PubMed] [Google Scholar]
- Snell K, 1984. Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Adv. Enzyme Regul 22, 325–400. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Durstewitz D, Hansson A, Heinz A, Kiefer F, Kohr G, Matthaus F, Nothen MM, Noori HR, Obermayer K, Rietschel M, Schloss P, Scholz H, Schumann G, Smolka M, Sommer W, Vengeliene V, Walter H, Wurst W, Zimmermann US, Stringer S, Smits Y, Derks EM, 2013. A systems medicine research approach for studying alcohol addiction. Addict. Biol 18, 883–896. [DOI] [PubMed] [Google Scholar]
- Stopponi S, de Guglielmo G, Somaini L, Cippitelli A, Cannella N, Kallupi M, Ubaldi M, Heilig M, Demopulos G, Gaitanaris G, Ciccocioppo R, 2013. Activation of PPARgamma by pioglitazone potentiates the effects of naltrexone on alcohol drinking and relapse in msP rats. Alcohol Clin. Exp. Res 37, 1351–1360. [DOI] [PubMed] [Google Scholar]
- Stopponi S, Somaini L, Cippitelli A, Cannella N, Braconi S, Kallupi M, Ruggeri B, Heilig M, Demopulos G, Gaitanaris G, Massi M, Ciccocioppo R, 2011. Activation of nuclear PPARgamma receptors by the antidiabetic agent pioglitazone suppresses alcohol drinking and relapse to alcohol seeking. Biol. Psychiatry 69, 642–649. [DOI] [PubMed] [Google Scholar]
- Sutherland GT, Sheedy D, Kril JJ, 2014. Neuropathology of alcoholism. Handb. Clin. Neurol 125, 603–615. [DOI] [PubMed] [Google Scholar]
- Tawa EA, Hall SD, Lohoff FW, 2016. Overview of the genetics of alcohol use disorder. Alcohol Alcohol 51, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ, 1991. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am. J. Hum. Genet 48, 677–681. [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G, Magnusson KP, Sulem P, Walters GB, Gudbjartsson DF, Stefansson H, Jonsson T, Jonasdottir A, Jonasdottir A, Stefansdottir G, Masson G, Hardarson GA, Petursson H, Arnarsson A, Motallebipour M, Wallerman O, Wadelius C, Gulcher JR, Thorsteinsdottir U, Kong A, Jonasson F, Stefansson K, 2007. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science 317, 1397–1400. [DOI] [PubMed] [Google Scholar]
- Tiihonen J, Hallikainen T, Lachman H, Saito T, Volavka J, Kauhanen J, Salonen JT, Ryynanen OP, Koulu M, Karvonen MK, Pohjalainen T, Syvalahti E, Hietala J, 1999. Association between the functional variant of the catechol-O-methyltransferase (COMT) gene and type 1 alcoholism. Mol. Psychiatry 4, 286–289. [DOI] [PubMed] [Google Scholar]
- Tikkanen R, Tiihonen J, Rautiainen MR, Paunio T, Bevilacqua L, Panarsky R, Goldman D, Virkkunen M, 2015. Impulsive alcohol-related risk-behavior and emotional dysregulation among individuals with a serotonin 2B receptor stop codon. Transl. Psychiatry 5, e681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipps ME, Buck KJ, 2015. GIRK channels: a potential link between learning and addiction. Int. Rev. Neurobiol 123, 239–277. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M, 2009. Genome-wide association study of alcohol dependence. Arch. Gen. Psychiatry 66, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Kissling C, Frank J, Wiemann S, Dong L, Depner M, Saam C, Lascorz J, Soyka M, Preuss UW, Rujescu D, Skowronek MH, Rietschel M, Spanagel R, Heinz A, Laucht M, Mann K, Schumann G, 2006. Genetic association of the human corticotropin releasing hormone receptor 1 (CRHR1) with binge drinking and alcohol intake patterns in two independent samples. Mol. Psychiatry 11, 594–602. [DOI] [PubMed] [Google Scholar]
- Tsai SY, Chuang JY, Tsai MS, Wang XF, Xi ZX, Hung JJ, Chang WC, Bonci A, Su TP, 2015. Sigma-1 receptor mediates cocaine-induced transcriptional regulation by recruiting chromatin- remodeling factors at the nuclear envelope. Proc. Natl. Acad. Sci. U. S. A. 112, E6562–E6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga T, Czimmerer Z, Nagy L, 2011. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta 1812, 1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedder LC, Hall JM, Jabrouin KR, Savage LM, 2015. Interactions between chronic ethanol consumption and thiamine deficiency on neural plasticity, spatial memory, and cognitive flexibility. Alcohol Clin. Exp. Res 39, 2143–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel-Ciernia A, Wood MA, 2014. Neuron-specific chromatin remodeling: a missing link in epigenetic mechanisms underlying synaptic plasticity, memory, and intellectual disability disorders. Neuropharmacology 80, 18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisey J, Swagell CD, Hughes IP, Lawford BR, Young RM, Morris CP, 2011. A novel SNP in COMT is associated with alcohol dependence but not opiate or nicotine dependence: a case control study. Behav. Brain Funct 7, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite A, Tinsley CL, Locke M, Blake DJ, 2009. The neurobiology of the dystrophin-associated glycoprotein complex. Ann. Med 41, 344–359. [DOI] [PubMed] [Google Scholar]
- Wernicke C, Hellmann J, Finckh U, Rommelspacher H, 2010. Chronic ethanol exposure changes dopamine D2 receptor splicing during retinoic acid-induced differentiation of human SH-SY5Y cells. Pharmacol. Rep 62, 649–663. [DOI] [PubMed] [Google Scholar]
- Whitfield JB, Zhu G, Madden PA, Neale MC, Heath AC, Martin NG, 2004. The genetics of alcohol intake and of alcohol dependence. Alcohol Clin. Exp. Res 28, 1153–1160. [DOI] [PubMed] [Google Scholar]
- Wolen AR, Miles MF, 2012. Identifying gene networks underlying the neurobiology of ethanol and alcoholism. Alcohol Res 34, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrona MZ, Waskiewicz J, Han QP, Han J, Li H, Dryhurst G, 1997. Putative oxidative metabolites of 1-methyl-6-hydroxy-1,2,3,4-tetrahydro-beta-carboline of potential relevance to the addictive and neurodegenerative consequences of ethanol abuse. Alcohol 14, 213–223. [DOI] [PubMed] [Google Scholar]
- Wrzosek M, Lukaszkiewicz J, Wrzosek M, Serafin P, Jakubczyk A, Klimkiewicz A, Matsumoto H, Brower KJ, Wojnar M, 2011. Association of polymorphisms in HTR2A, HTR1A and TPH2 genes with suicide attempts in alcohol dependence: a preliminary report. Psychiatry Res. 190, 149–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Seo D, Hodgkinson C, Hu Y, Goldman D, Sinha R, 2013. A variant on the kappa opioid receptor gene (OPRK1) is associated with stress response and related drug craving, limbic brain activation and cocaine relapse risk. Transl. Psychiatry 3, e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, Agrawal A, Bierut L, Goate A, Bucholz K, Schuckit M, Nurnberger J Jr., Tischfield J, Kuperman S, Porjesz B, Begleiter H, Foroud T, Edenberg HJ, 2006. Association of the kappa-opioid system with alcohol dependence. Mol. Psychiatry 11, 1016–1024. [DOI] [PubMed] [Google Scholar]
- Yamasaki M, Yamada K, Furuya S, Mitoma J, Hirabayashi Y, Watanabe M, 2001. 3- Phosphoglycerate dehydrogenase, a key enzyme for l-serine biosynthesis, is preferentially expressed in the radial glia/astrocyte lineage and olfactory ensheathing glia in the mouse brain. J. Neurosci 21, 7691–7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You IJ, Wright SR, Garcia-Garcia AL, Tapper AR, Gardner PD, Koob GF, David Leonardo E, Bohn LM, Wee S, 2016. 5-HT1A autoreceptors in the dorsal raphe nucleus convey vulnerability to compulsive cocaine seeking. Neuropsychopharmacology 41, 1210–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Zhu G, Hariri AR, Enoch MA, Scott D, Sinha R, et al. , 2008. Genetic variation in human NPY expression affects stress response and emotion. Nature 452 (7190), 997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Enoch MA, Goldman D, 2014. Gene expression in the addicted brain. Int. Rev. Neurobiol 116, 251–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zois E, Vollstadt-Klein S, Hoffmann S, Reinhard I, Bach P, Charlet K, Beck A, Treutlein J, Frank J, Jorde A, Kirsch M, Degenhardt F, Walter H, Heinz A, Kiefer F, 2016. GATA4 variant interaction with brain limbic structure and relapse risk: a voxel-based morphometry study. Eur. Neuropsychopharmacol 26, 1431–1437. [DOI] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES, 2012. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. U. S. A. 109, 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, Lander ES, 2014. Searching for missing heritability: designing rare variant association studies. Proc. Natl. Acad. Sci. U. S. A. 111, E455–E464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CS, Wang F, Zhang XY, Deng HW, Krystal JH, Zhang F, Luo X, 2012. Genomewide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology 37, 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]