Abstract
Semantic features are common radiological traits used to characterize a lesion by a trained radiologist. These features have been recently formulated, quantified on a point scale in the context of lung nodules by our group. Certain radiological semantic traits have been shown to extremely predictive of malignancy [26]. Semantic traits observed by a radiologist at examination describe the nodules and the morphology of the lung nodule shape, size, border, attachment to vessel or pleural wall, location and texture etc. Deep features are numeric descriptors often obtained from a convolutional neural network (CNN) which are widely used for classification and recognition. Deep features may contain information about texture and shape, primarily. Lately, with the advancement of deep learning, convolutional neural networks (CNN) are also being used to analyze lung nodules. In this study, we relate deep features to semantic features by looking for similarity in ability to classify. Deep features were obtained using a transfer learning approach from both an ImageNet pre-trained CNN and our trained CNN architecture. We found that some of the semantic features can be represented by one or more deep features. In this process, we can infer that some deep feature(s) have similar discriminatory ability as semantic features.
Keywords: Convolutional neural network, semantic features, deep features
I. Introduction
Lung cancer is the leading cause of cancer related deaths globally [1]. For early detection and diagnosis of lung cancers, Low Dose Computed Tomography (LDCT) is the most extensively used imaging approach. Using LDCT scans, a radiologist can provide important individual information about one’s lung tumor or nodule, which can provide useful guidance for prognosis and diagnosis. These unique characteristics are termed ‘Semantic features’ which can help to predict prognosis in lung tumors. Semantic features can be subdivided into various categories: location, size, shape, margin, attenuation etc. The existence of cavitation (cavity within the nodule), a semantic feature we used, corresponds to a worse prognosis for lung cancers [2]. Nodule Emphysema, another semantic feature we checked, will increase the lung cancer risk for a lung nodule for a patient with emphysema [3]. Depending on a nodule being solid or non-solid the cancer treatment varies. According to Fleischner criteria [4] solid nodules of size >8mm, require follow up scans every 3 months, and if the solid nodule size is between 6 to 8 mm, a follow up scan at 6-12 months is required followed by another scan at 18-24 months when no change occurred in the earlier one. For non-solid nodules if the nodule size is >6 mm, a follow up scan at 6-12 months is recommended and then every 2 years until 5 years.
Deep learning, is an emerging technique widely used for classification, segmentation and recognition tasks. Deep learning algorithms enable classification of complex data via multiple hidden layers in a neural network. In recent years, for image data, the data are typically first processed through convolution layers and then via multiple hidden layers. This type of neural network is called a convolutional neural network (CNN). Convolutional neural networks gained popularity though LeCun’s LeNet [5]. A breakthrough for CNNs in the classification task came during the 2012 ILSVRC challenge, when Krizhevsky [6] proposed ‘ALEXNET’. Since then CNNs have been used extensively in computer vision, medical image analysis etc. In the medical field, due to the availability of less data, pre-trained CNNs can be used effectively [7] with a transfer learning approach [8]. Deep features contain various low level image feature information such as textures, shape etc. There is no specific naming approach for deep featurs other than representing them using the extracted feature column number (position in a hidden layer treated as a row vector).
For extracting deep features, we used two pre-trained CNNs: a Vgg-s [9] architecture, which was trained on the ImageNet dataset and our designed CNN [7] which was trained on lung nodule images. We also obtained 20 semantic features from the radiologists of the H. Lee Moffitt Cancer Center. This study is focused on showing the similarity of deep feature(s) with a semantic feature. We showed that by replacing one or more deep feature columns by a semantic feature, equivalent classification performance can be achieved. That means, those replaced deep features provided the same information as the semantic features and we can equate those deep feature columns with the name of the corresponding semantic feature.
We found that location based semantic features are hard to interpret and replace, but size, shape, and texture based semantic features can be interpreted with respect to deep feature column(s). We successfully interpreted or explained 9 out of 20 semantic features in our study.
II. DATASET
The NLST study was conducted for three years: a baseline scan (T0) in the first year, followed by two subsequent scans (T1 and T2) in the following two years with a gap of one year [10]. In this study, from the baseline scans (T0) we chose a subset of participants of control positive (sizable nodule that does not become cancer) and screen detected lung cancer (SDLC) from the CT arm of the NLST study. The subset was divided into Cohort 1 and Cohort 2. In Cohort 1, there was a baseline scan (T0) followed by another scan after 1 year (T1) and some of the positive screened nodules become cancerous. In Cohort 2, some of the positive screened nodule become cancerous after two years (T2 scan) of baseline scan (T0). There were no statistically significant differences between the SDLC and control positive cases for age, sex, race, ethnicity, and smoking [11]. For our study, we used only Cohort 2 (85 SDLC and 176 control positive cases) which had available semantic features. The Definiens software suite [12] was used for nodule segmentation. From our initial cases, 76 cases were excluded due to one or more of the following reasons: multiple malignant nodules, not identifying the nodule, or unknown location of the tumor. For this study, we finally used 185 cases (58 SDLC and 127 control positive cases).
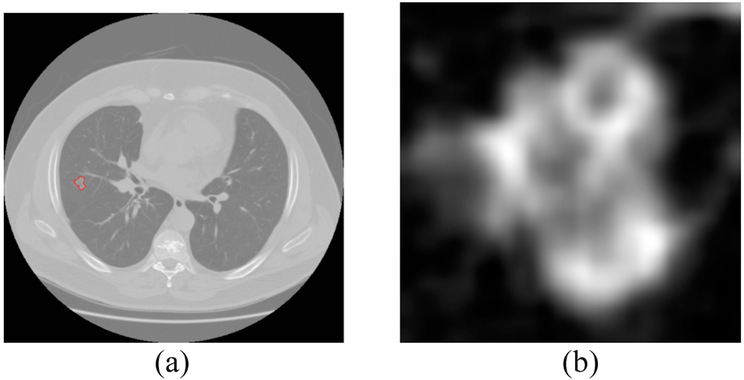
Semantic features [13] from Cohort 2 were created by radiologists (Y.L.) from H. Lee Moffitt Cancer Center. For deep feature extraction, we chose a CT slice for every case which had the largest nodule area, and extracted only the nodule region via a rectangular patch which incorporated the whole nodule. In Fig. 1 we show an extracted nodule along with the slice from the CT scan. Using bi-cubic interpolation, the extracted nodule region was resized as required for input to the pre-trained CNNs. The CT nodule images were grayscale (no color component and we changed the voxel intensities of the CT images to 0-255), but the Vgg-s network was trained on RGB images (24-bit natural camera images). So, we used the same grayscale image three times to simulate an image with three color channels and did normalization using the appropriate color channel image.
Fig. 1.
(a) lung image with nodule inside outlined by red (nodule pixel size= 0.74 mm) (b) extracted nodule
III. Semantic features
A radiologist can provide important individual information about one’s lung tumor, which can provide useful guidance for prognosis and diagnosis. These unique characteristics are termed ‘Semantic features’. Twenty semantic features [14,15] were described by an experienced radiologist with 7 years of experience (Y.L). These features have been recently formulated, quantified on a point scale in the context of lung nodules by our group [25]. These features can be subdivided into the following categories: location, size, shape, margin, attenuation, external and associated findings. In Table 1 feature description details are given.
TABLE 1.
Description of semantic features
| Characteristic | Definition | Scoring |
|---|---|---|
| Location | ||
| 1. Lobe Location | Lobe location of the nodule | left lower lung (5), left upper lung (4), right lower lung (3), right top lung (2), right upper lung (1) |
| Size | ||
| 2. Long-axis Diameter | Longest diameter of the nodule | NA |
| 3. Short-axis Diameter | Longest perpendicular diameter of nodule in the same section | NA |
| Shape | ||
| 4. Contour | Roundness of the nodule | 1, round; 2, oval; 3, irregular |
| 5. Lobulation | Wavy nodule’s surface | 1, none; 2, yes |
| 6. Concavity | Concave cut on nodule surface | 1, none; 2, slight concavity; 3, deep concavity |
| Margin | ||
| 7. Border Definition | Edge appearance of the nodule | 1, well defined; 2, slight poorly; 3, poorly defined |
| 8. Spiculation | Spike edge in the nodule | 1, none; 2 yes |
| Attenuation | ||
| 9. Texture | Solid, non-solid, part solid | 1, non-solid; 2, part solid; 3, solid |
| External | ||
| 10. Fissure Attachment | Nodule attaches to the fissure | 0, no; 1, yes |
| 11. Pleural Attachment | Nodules attaches to the pleura | 0, no; 1, yes |
| 12. Vascular Convergence | Convergence of vessels to nodule | 0, no significant convergence; 1, significant |
| 13. Pleural Retraction | Retraction of the pleura towards nodule | 0, absence of pleural retraction; 1, present |
| 14. Peripheral emphysema | Peripheral emphysema caused by nodule | 1, absence of emphysema; 2, slight present; 3 severely present |
| 15. Peripheral Fibrosis | Peripheral fibrosis caused by nodule | 1, absence of fibrosis; 2, slight present; 3 severely present |
| 16. Vessel Attachment | Nodule attachment to blood vessel | 0, no; 1, yes |
| 17. Cavitation | Cavity within nodule due to fibrosis | 0, no; 1, yes |
| Associated Findings | ||
| 18. Nodules in primary lobe | Any nodules suspected to be malignant | 0, no; 1, yes |
| 19. Nodules in non-tumor lobe | Any nodules suspected to be malignant | 0, no; 1, yes |
| 20. Lymphadenopathy | Lymph nodes with short- axis diameter greater than 1 cm | 0, no; 1, yes |
IV. Deep features
Convolutional Neural Networks (CNN) [16] have recently been used widely in image classification and object recognition tasks. They are often called deep CNNs because of the number of layers (depth) of the designed architecture and features extracted from these architectures are called deep features. A CNN can be created using a few convolution layers, often followed by a max pooling layer and then fully connected layers and activation layers. Due to the lack of training images, transfer learning has been explored. Transfer learning [8] is an approach, where previously learned knowledge can be applied to another task and task domains can be the same or different. The transfer learning approach was used in our study to extract deep features from a pre-existing CNN when presented with nodule images. Vgg-s, a CNN pre-trained on natural camera images obtained from the ImageNet dataset, and our designed CNN architecture trained using lung nodule images had deep features extracted from them. These two models were architecturally different, as well as created using different training images. Our designed CNN was trained on augmented lung nodule images from Cohort 1. Image augmentation was done by rotating each image 15 degrees and then applying horizontal and vertical flipping. Our architecture was inspired by a cascaded CNN architecture in [21], where the input image was fed through convolution and pooling layers for the primary branch and a re-sized input image was added to the primary branch in the fully connected layer. We trained our CNN in Keras [17] with a Tensorflow [18] backend. The largest Cohort 1 cancer cases and control cases were 104×104 pixels, and 82×68 pixels respectively. So, we chose 100×100 as the input image size for the designed CNN. One hundred epochs were used to train the CNN. A constant learning rate of 0.0001 was used with RMSprop [22] as the gradient descent optimization algorithm. A batch size of 16 was used for both training and validation. Binary crossentropy was used as the loss function.
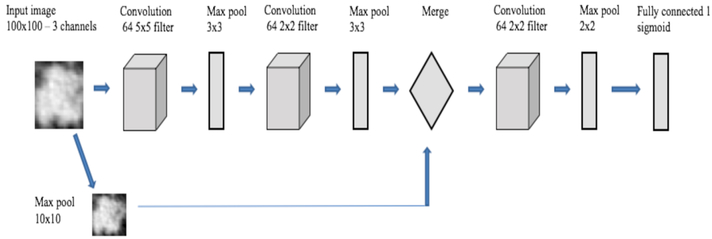
Since our designed CNN was shallow and small, we used L2 regularization [23] along with dropout [24] to reduce overfitting. Our designed CNN is described in more detail in [7] and summarized in Table 2 and Fig. 2. In convolutional layers 1 and 2, leaky ReLU with an alpha value 0.01 was applied. Because of this, some negative values will propagate through the convolution layer and provide non-linearity on the convolution layer output. Our CNN has a cascaded architecture where images are fed to both the “left” branch of the network, followed by a max pooling layer and more complex “right” branch. The right branch consisted of convolution and max pool layers. The cascading happened after getting the same size output (10×10 vector) from both the left and right branches. Features in the convolution layer are more generic (e.g. blobs, textures, edges etc.). So, adding image information directly will create more specific information for each case. After merging, another convolution and max pooling layer before the final classification layer maintains the generic information about the image and can provide more features about the image enabling a better classification result.
TABLE 2.
Our Designed CNN architecture
| Layers | Parameter | Total Parameters |
|---|---|---|
| Left branch: | ||
| Input Image | 100×100 | |
| Max Pool 1 | 10×10 | |
| Dropout | 0.1 | |
| Right branch: | ||
| Input Image | 100×100 | |
| Conv 1 | 64 X 5 X 5, pad 0, stride 1 | |
| Leaky ReLU | alpha = 0.01 | |
| Max Pool 2a | 3×3, pad 0, stride 3 | |
| Conv 2 | 64 X 2 X 2, pad 0, stride 1 | |
| Leaky ReLU | alpha = 0.01 | |
| Max Pool 2b | 3×3, pad 0, stride 3 | |
| Dropout | 0.1 | |
| Concatenate Left Branch + Right Branch | 39,553 | |
| Conv 3+ReLU | 64 X 2 X 2, pad 0, stride 1 | |
| Max Pool 3 | 2×2, pad 0, stride 2 | |
| L2 regularizer | 0.01 | |
| Dropout | 0.1 | |
| Fully connected 1 | 1 sigmoid |
Fig. 2.
Overview of CNN architecture
Using these two models, we extracted deep features from the last layer before the classification layer after applying the ReLU activation function. In Table 3, 64×11×11 means 64 convolutions of window size 11×11 and st= stride, LRN is local response normalization and x3 pool means 3×3 max pooling. The deep features from the Vgg-s architecture were the output of the last fully connected layer (the Full 2 layer as shown in Table 3). In our designed CNN architecture, we obtained deep features after applying max-pooling (Max Pool 3 layer as shown in Table 2).
TABLE 3.
Pre-trained Vgg-s CNN architecture
| Arch. | Conv 1 | Conv 2 | Conv 3 | Conv 4 | Conv 5 | Full 1 | Full 2 | Full 3 |
|---|---|---|---|---|---|---|---|---|
| Vgg-S | 64×11×11 | 256×5×5 | 512×3×3 | 512×3×3 | 512×3×3 | 4096 | 4096 | 1000 |
| st. 4, pad 0 | st. 1, pad 1 | st. 1, pad 1 | st. 1, pad 1 | st. 1, pad 1 | dropout | dropout | softmax | |
| LRN, ×3 pool | ×2 pool | ×3 pool |
Utilizing the Vgg-s pre-trained CNN and our designed CNN architecture we obtained 4096 and 1024 features respectively. After applying the ReLU function, some feature columns turned to all zeros, because, the ReLU function will convert negative feature values to zero. So, for our experiments, we removed those all zero columns from both the pre-trained CNN feature vectors and our trained CNN feature vectors. After removing those columns, the final feature vector size of the Vgg-s pre-trained CNN and our trained CNN became 3844 and 560 respectively. The architectures and parameters for the pre-trained CNNs used are shown in Tables 2 and 3. These deep features don’t have any specific naming approach other than identifying them using the extracted feature column number (position in a hidden layer treated as a row vector).
V. Experiments and results
In this section, we analyze the approach taken to represent one semantic feature using one or several deep feature columns.
Wrapper feature selection [19] was applied to the semantic features of Cohort 2 using the best first strategy to choose the subset of features which generated the best accuracy possible. Backward selection with a random forests classifier [20] with 200 trees was done. A 10-fold cross validation was used to choose the best subset from the training data. A subset of 13 features were chosen using the wrapper approach resulting in an accuracy of 83.78% (AUC 0.837). This is, of course, an optimistic accuracy given the features were chosen on training data. However, our purpose is to use good semantic features to explain deep features.
The chosen features (13) were: Location, Long axis diameter, Short axis diameter, Lobulation, Concavity, Border definition, Spiculation, Texture, Cavitation, Vascular convergence, Attachment to vessel, Peri-nodule fibrosis, Nodules in primary tumor lobe.
After choosing the semantic features, the correlations (Pearson correlation coefficient) of each semantic feature with deep features were calculated and we chose the 5 most correlated deep features. We substituted the correlated deep features in place of the semantic feature to see if it was possible to obtain the same classification result obtained using all 13 features (83.7838%).
Our objective was to substitute each semantic feature by one or more deep features and achieve the same classification result. For each semantic feature, we removed one feature from the chosen semantic feature subset (13 features) and substituted that semantic feature by the most correlated deep feature column and, then substituted by the two most correlated deep features and similarly continued substituting until the five most correlated deep features were included and computed the accuracy using a random forests classifier with each feature set using 10-fold cross-validation.
We analyzed deep features from the Vgg-s pre-trained CNN and our designed CNN architecture separately.
By substituting deep features obtained from the Vgg-s pretrained CNN for the following semantic features; long axis diameter, lobulation, concavity, spiculation, texture, cavitation, vascular convergence, peripheral fibrosis; we obtained the same original classification performance (83.7838%). By replacing our trained CNN features for the following semantic features; long axis diameter, concavity, cavitation, nodules in primary tumor lobe; we got back the same original classification performance. Three semantic features (long axis diameter, concavity and cavitation) were explained by both Vgg-s CNN and our designed CNN. One semantic feature (Nodules in primary lobe) was explained only by our designed CNN. That makes sense as Vgg-s would not have any of real lung data where lobes could be discerned. Vgg-s, which trained on much more data, was better at explaining texture, spiculation, lobulation, vascular convergence and peripheral fibrosis. The first 3 and last one seem likely to be a result of lots of data. In total, nine semantic features were represented with their corresponding deep feature columns.
That means, those semantic features could be represented by one or more deep feature columns and the same classification performance could be achieved by replacing them with deep feature(s). In Table 4 we show classification performance after removing each semantic feature one at a time from our chosen subset of 13 features. There is always at least a slight reduction in accuracy. In Table 4 we only show some features out of the 13 chosen features that can be replaced by deep feature columns.
TABLE 4.
Classification performance after removing each of these features one at a time from our chosen subset of 13 features
| Semantic Features | Accuracy (AUC) |
|---|---|
| Long axis Diameter | 82.7027 (0.82) |
| Lobulation | 82.7027 (0.83) |
| Concavity | 83.2432 (0.83) |
| Spiculation | 83.2432 (0.83) |
| Texture | 82.7027 (0.834) |
| Cavitation | 82.7027 (0.828) |
| Vascular Convergence | 83.2432 (0.84) |
| Peripheral fibrosis | 82.7027 (0.83) |
| Nodules in primary lobe | 81.6216 (0.83) |
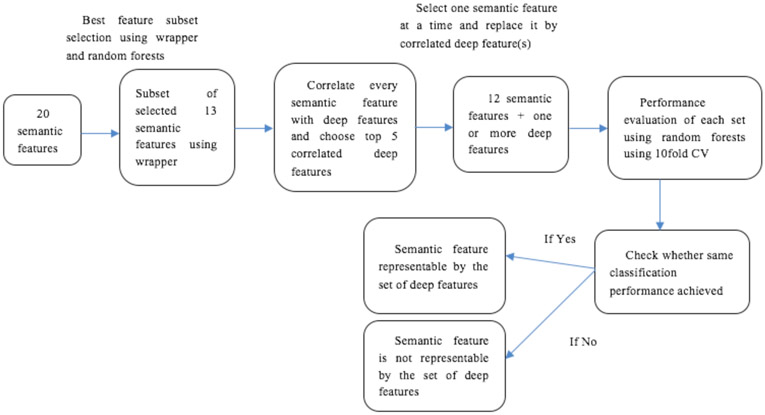
In Table 5 we show the analysis of the semantic features and their corresponding deep features. By using deep feature columns in place of their corresponding semantic feature, we obtained similar performance. For example, long axis diameter, could be represented by feature column numbers 3353 and 2135 using features from the Vgg-s network to get the same classification performance of 83.78%. The correlation value of feature column 3353 and 2135 with long axis diameter was 0.4334 and 0.42 respectively. Long axis diameter could be replaced by feature column 230 by using our designed CNN to obtain similar performance of 83.78%. Correlation of long axis diameter with feature column 230 was 0.3035. In the same way, Concavity can be represented by five deep features columns (column 3534, 2975, 1372, 2111 and 3246) from the Vgg-s network. Concavity was also represented by two deep feature columns (column 547 and 440) from our designed CNN. In most cases, only 1 or 2 features were needed to explain a semantic feature. Fig. 3 demonstrates the approach taken for this analysis.
TABLE 5.
Semantic features and corresponding deep feature(s)
| Semantic features |
Deep feature(s) from Vggs which explain semantic feature, with correlation value |
Deep feature(s) from our designed CNN, which explain semantic feature with correlation value |
|||||
|---|---|---|---|---|---|---|---|
| Long axis Diameter |
3353 0.4334 |
2135 0.42 |
230 0.3035 |
||||
| Lobulation | 3534 0.5742 |
1372 0.5614 |
2975 0.5611 |
2111 0.5520 |
NA | ||
| Concavity | 3534 0.5 |
2975 0.4839 |
1372 0.4837 |
2111 0.475 |
3246 0.4612 |
547 0.1776 |
440 0.1514 |
| Spiculation | 2811 0.411 |
NA | |||||
| Texture | 1201 −0.3119 |
3350 0.2936 |
NA | ||||
| Cavitation | 3353 0.388 |
526 0.3551 |
395 0.2748 |
||||
| Vascular Convergence |
1464 0.7052 |
2115 0.701 |
NA | ||||
| Peripheral fibrosis |
3305 0.2076 |
3064 0.2043 |
NA | ||||
| Nodules in primary lobe |
NA | 425 0.1871 |
57 0.1836 |
||||
Fig. 3.
Overview of the approach taken in this study
VI. Discussions and conclusions
In this work, we showed that one or more deep feature column(s) could explain a semantic feature. Semantic features are created by radiologists and represent different nodule characteristics such as, size (long axis diameter, short axis diameter), nodules wall or edge specification (spiculation, border definition), shape (contour, lobulation, concavity) etc. semantic features give valuable information about the nodule which can be used effectively for cancer prognosis and diagnosis. That deep features can replace them also indicates the semantic features cover or explain the related deep features.
Deep features were extracted from a convolutional neural network. In this study, using transfer learning, we analyzed deep features from two different pre-trained CNNs: Vgg-s CNN which was trained on ImageNet and our designed small CNN architecture which was trained on lung nodule images. The Vgg-s architecture was a deeper architecture with 5 convolutions and 3 fully connected layers, but trained on ImageNet (color camera images of objects in scenes). Its features matched several low-level semantic features (texture/size/shape) which could be used effectively for lung nodule classification [9]. on the other hand, our designed CNN [7] was a smaller architecture trained on lung nodule images, which also gave us effective classification performance.
Deep features are denoted by their column number in a row vector representing the hidden layer from which they were extracted. In this study, we tried to represent one or more deep feature column(s) by association with semantic features. There were 20 semantic features defined by radiologists. Backward wrapper based feature selection was performed on the semantic features to generate a subset of features which gave the maximum accuracy. A subset of 13 features with an accuracy of 83.78% was selected. A correlation coefficient was calculated between each of the selected semantic features and deep feature vectors. For each semantic feature, the top five most correlated deep features were selected. From the chosen subset of features, we removed one semantic feature and substituted it by the most correlated deep feature and, calculated the classification performance. If we obtained the same classification performance then stop, otherwise substitute it by the two most correlated deep features and continue substituting until the five most correlated deep features have been used.
Our objective was to substitute each semantic feature by one more deep features and achieve the same classification result using the chosen subset. Nine semantic features can be represented by deep feature(s) using the proposed approach. From this, we can say that those deep feature sets behave same as their corresponding semantic feature. Hence, we argue that they have identifiable semantic meaning.
We extracted only the nodule region from the CT slice for this experiment. So, the information about attachment of the nodule to a vessel or fissure or pleural wall or the lobe location is not available, and we couldn’t represent those features using deep features. We performed data augmentation by rotation and flipping to train our CNN, and it helped in obtaining comparable accuracy to using semantic features and provided features to explain one location based feature (nodules in primary tumor lobe). We also noticed that, due to the extraction of the nodule region only, the features that are related to the margin, size, attenuation and shape can be represented by deep feature(s).
In our future work, we will work on representing semantic features using radiomics features (i.e. traditional features).
Acknowledgments
This research partially supported by the National Institute of Health under grants (NIH U01 CA143062), (NIH U24 CA180927) and (NIH U01 CA200464), National Science Foundation under award number 1513126 and by the State of Florida Dept. of Health under grant (4KB17).
References
- [1].Siegel RL, Miller KD and Jemal A: Cancer statistics, 2015. CA: a cancer journal for clinicians, 65(1), pp.5–29. (2015) [DOI] [PubMed] [Google Scholar]
- [2].Gill RR, Matsusoka S and Hatabu H: Cavities in the lung in oncology patients: imaging overview and differential diagnoses. Applied Radiology, 39(6), p.10 (2010) [Google Scholar]
- [3].Li Y, Swensen SJ, Karabekmez LG, Marks RS, Stoddard SM, Jiang R, Worra JB, Zhang F, Midthun DE, de Andrade M and Song Y: Effect of emphysema on lung cancer risk in smokers: a computed tomography-based assessment. Cancer prevention research, pp.canprevres–0151. (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].MacMahon H, Naidich DP, Goo JM, Lee KS, Leung AN, Mayo JR, Mehta AC, Ohno Y, Powell CA, Prokop M and Rubin GD: Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society 2017. Radiology, 284(1), pp.228–243. (2017) [DOI] [PubMed] [Google Scholar]
- [5].LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W and Jackel LD: Backpropagation applied to handwritten zip code recognition. Neural computation, 1(4), pp.541–551. (1989) [Google Scholar]
- [6].Krizhevsky A, Sutskever I and Hinton GE: Imagenet classification with deep convolutional neural networks. In Advances in neural information processing systems, pp. 1097–1105. (2012) [Google Scholar]
- [7].Paul R, Hawkins SH, Schabath MB, Gillies RJ, Hall LO and Goldgof DB, 2018. Predicting malignant nodules by fusing deep features with classical radiomics features. Journal of Medical Imaging, 5(1), p.011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Thrun S: Is learning the n-th thing any easier than learning the first?. In Advances in neural information processing systems, (pp. 640–646). (1996) [Google Scholar]
- [9].Chatfield K, Simonyan K, Vedaldi A and Zisserman A: Return of the devil in the details: Delving deep into convolutional nets. arXiv preprint arXiv:1405.3531. (2014) [Google Scholar]
- [10].National Lung Screening Trial Research Team.: Reduced lung-cancer mortality with low-dose computed tomographic screening. New England Journal of Medicine, 365(5), pp.395–409. (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Schabath MB, Massion PP, Thompson ZJ, Eschrich SA, Balagurunathan Y, Goldof D, Aberle DR and Gillies RJ: Differences in patient outcomes of prevalence, interval, and screen-detected lung cancers in the CT arm of the National Lung Screening Trial. PloS one, 11(8), p.e0159880 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].User Guide.: Definiens AG; Germany: Definiens developer XD 2.0.4. (2009) [Google Scholar]
- [13].Yip SS, Liu Y, Parmar C, Li Q, Liu S, Qu F, Ye Z, Gillies RJ and Aerts HJ: Associations between radiologist-defined semantic and automatically computed radiomic features in non-small cell lung cancer. Scientific reports, 7(1), p.3519 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu Y, Kim J, Qu F, Liu S, Wang H, Balagurunathan Y, Ye Z and Gillies RJ: CT features associated with epidermal growth factor receptor mutation status in patients with lung adenocarcinoma. Radiology, 280(1), pp.271–280. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li Q, Balagurunathan Y, Liu Y, Qi J, Schabath MB, Ye Z and Gillies RJ: Comparison Between Radiological Semantic Features and Lung-RADS in Predicting Malignancy of Screen-Detected Lung Nodules in the National Lung Screening Trial. Clinical lung cancer, 19(2), pp.148–156. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zeiler MD and Fergus R: Visualizing and understanding convolutional networks In European conference on computer vision (pp. 818–833). Springer, Cham; (2014) [Google Scholar]
- [17].Chollet F, 2017. Keras (2015). [Google Scholar]
- [18].Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M and Ghemawat S: Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv preprint arXiv:1603.04467. (2016) [Google Scholar]
- [19].Kohavi R and Sommerfield D: Feature Subset Selection Using the Wrapper Method: Overfitting and Dynamic Search Space Topology. In KDD (pp. 192–197). (1995) [Google Scholar]
- [20].Ho TK: Random decision forests. In Document analysis and recognition, 1995., proceedings of the third international conference on, 1, pp. 278–282. IEEE; (1995) [Google Scholar]
- [21].Li H, Lin Z, Shen X, Brandt J and Hua G: A convolutional neural network cascade for face detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 5325–5334). (2015) [Google Scholar]
- [22].Tieleman T and Hinton G: Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude. COURSERA: Neural networks for machine learning, 4(2), pp.26–31. (2012) [Google Scholar]
- [23].Ng AY: Feature selection, L 1 vs. L 2 regularization, and rotational invariance In Proceedings of the twenty-first international conference on Machine learning (p. 78). ACM; (2004) [Google Scholar]
- [24].Srivastava N, Hinton G, Krizhevsky A, Sutskever I and Salakhutdinov R: Dropout: A simple way to prevent neural networks from overfitting. The Journal of Machine Learning Research, 15(1), pp. 1929–1958. (2014) [Google Scholar]
- [25].Liu Y, Wang H, Li Q, McGettigan MJ, Balagurunathan Y, Garcia AL, Thompson ZJ, Heine JJ, Ye Z, Gillies RJ and Schabath MB: Radiologic features of small pulmonary nodules and lung cancer risk in the National Lung Screening Trial: A nested case-control study. Radiology, 286(1), pp.298–306. (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu Y, Balagurunathan Y, Atwater T, Antic S, Li Q, Walker RC, Smith G, Massion PP, Schabath MB and Gillies RJ: Radiological image traits predictive of cancer status in pulmonary nodules. Clinical Cancer Research, pp.clincanres–3102. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]