Abstract
This work describes the performance of a sequencing batch reactor (SBR) and the involvement of a novel reconstituted bacterial consortium in olive mill wastewater (OMW) treatment. The organic loading rate applied to the SBR was serially increased in terms of initial COD from 10 to 75 g L−1 to allow gradual acclimatization of activated sludge to high concentrations of toxic compounds in OMW. After the acclimatization period, up to 60% of the total COD content were effectively biodegraded from OMW at 75 g L−1 COD within 30 day hydraulic retention time. The diversity and community composition of cultivable bacteria participating in the aerobic process of treating OMW were further assessed. A total of 91 bacterial strains were isolated from the reactor and analyzed by amplification of the 16S-23S rRNA internal transcribed spacer (ITS) region and by 16S rRNA gene sequencing. The most abundant phylum was Firmicutes (57.1%) followed by Proteobacteria (35.2%) and Actinobacteria (7.7%). The use of the Biolog® Phenotype Microarray system to evaluate the ability of isolated strains to utilize OMW phenolic compounds is reported in this work for the first time. Interestingly, results showed that all species tested were able to utilize phenolics as sole carbon and energy sources. The removals of COD and phenolics from undiluted OMW by the reconstituted bacterial consortium were almost similar to those obtained by the acclimatized activated sludge, which suggest that cultivable bacteria play the major role in OMW biodegradation. Phytotoxicity assays using tomato seeds showed a significant improvement of seed germination values for treated OMW. Our overall results suggest that the novel developed bacterial consortium could be considered as a good prospect for phenolics-rich wastewaters bioremediation applications.
Keywords: Acclimatized aerobic consortium, Biolog ® phenotyping, Olive mill wastewater, Phenolic compounds, Phytotoxicity, Sequencing batch reactor
Introduction
Olive mill wastewater (OMW), generated from olive oil production process, is a dark brown effluent composed of soft tissues of the olive fruit, residual oil and processing waters (Lanciotti et al. 2005). Annually, around 30 million m3 of OMW are generated in Mediterranean countries which account for almost 95% of the worldwide olive oil production, highlighting Spain, Italy, Greece, Tunisia and Portugal (McNamara et al. 2008). This wastewater is characterized by high chemical and biological oxygen demand values and a high organic pollutant load owing to the presence of biodegradable and recalcitrant compounds, i.e., carbohydrates, polysaccharides, fatty acids, polyalcohols, pectins, tannins, anthocyanins, phenolic compounds and catechol–melaninic polymers (Obied et al. 2007). Hence, when disposed untreated into the environment, OMW creates considerable environmental problems such as pollution of surface and ground waters, alterations in soil quality and microbial populations, plant growth inhibition, toxicity on species from several trophic levels, as well as, air pollution through phenol and sulfur dioxide emissions (Aggelis et al. 2003).
As required by legislation in Tunisia, OMW has to be discharged into evaporation ponds to mitigate its impact on the environment (Jarboui et al. 2010). Currently, several physicochemical, biological and even combined treatment processes have been proposed for the treatment of OMW aiming at removing the complex organic load from this effluent to make it suitable for discharge to the environment (Jaouani et al. 2005a, b). Physicochemical methods, including natural and forced evaporation, flocculation and coagulation, ultrafiltration, reverse osmosis and advanced oxidation processes (AOP) (Chatzisymeon et al. 2013; El-Abbassi et al. 2014; Kavvadias et al. 2010; Michael et al. 2014), require costly investment and maintenance. In the field of biological treatments, many investigations are available on the application of the anaerobic digestion as a promising technology for both OMW decontamination and methane production. The main limitation of OMW anaerobic digestion is the growth inhibition of methanogenic bacteria by phenolic compounds and certain organic acids (Beccari et al. 1996). Among biological methods, aerobic processes with selected microorganisms and composting are indeed the most environmentally friendly and the least expensive (Chiavola et al. 2014; Paredes et al. 2005). Aerobic methods use a more robust biomass to degrade the polluting effluent charge (Neifar et al. 2012). In the past, researchers have used single microbial species for organic matter biodegradation which may limit their field applications as a wide range of contaminants is present in OMW (Aissam et al. 2007; Ammar et al. 2005). Several studies have been performed with bacterial species or fungi, using either free or immobilized cell cultures under laboratory controlled conditions (Neifar et al. 2012; Tziotzios et al. 2007; Zerva et al. 2016). There are, however, few reports which deal with the application of microbial consortia in discontinuous bioreactors for OMW biological treatment despite their ability of efficiently degrading a variety of recalcitrant compounds. For instance, in previous studies carried out by our group, the adapted aerobic consortium was found to be more efficient in COD removal from OMW when compared to both free and immobilized cells of the white rot fungus Coriolopsis polyzona (Jaouani et al. 2005b; Neifar et al. 2012). Nevertheless, the knowledge of the microbial aspect of OMW- acclimatized aerobic consortium is still incomplete.
The main purposes of the present investigation were (a) to evaluate the performance of activated sludge acclimatized to high OMW concentrations in a Sequencing Batch Reactor (SBR) for the treatment of OMW; (b) to determine the bacterial diversity from OMW after acclimatization period using a culture-dependent approach; (c) to examine their ability to utilize OMW phenolic compounds as sole carbon sources; and (d) to assess the capacity of a novel reconstituted bacterial consortium for phenol and COD removal from OMW.
Materials and methods
Seed sludge and olive mill wastewater (OMW)
Sequencing batch reactor (SBR) was inoculated by an activated sludge collected from a municipal wastewater treatment plant located in Tunis (Tunisia).
Fresh OMW was collected from a three-phase decanter olive mill in the region of Tunis (Tunisia) and was immediately stored at − 20 °C to avoid spontaneous fermentation. Before use, OMW was de-frozen, vigorously stirred and decanted. Due to low concentrations of total phosphorus and total nitrogen in OMW, K2HPO4 and (NH4)2SO4 were added to maintain the ratio COD:N:P around 100:5:1. The mixture was then used to feed the SBR.
The reactor was started-up and fed with raw OMW diluted with tap water in several ratios to give liquid media with various initial COD concentrations (10 g L−1, 25 g L−1, 50 g L−1 and 75 g L−1).
SBR start-up and operation
The laboratory-scale reactor used in this study consisted of a 1 L glass tank with 800 mL working volume. The oxygen was supplied by an aquarium air pump and the complete mixture was achieved with a magnetic stirrer. The bioreactor was operated at room temperature (≈ 25 °C).
The SBR plant was first seeded with a sample of activated sludge and fed with diluted OMW at 10 g L−1 COD. During the start-up period, the reactor was operated with a hydraulic retention time of 30 days. An improvement of the removal performance was noticed with time during start-up, as a result of the progressive enrichment/selection of microbial community capable of biodegrading OMW constituents to use them as carbon sources. The biomass, collected from the SBR reactor, was then stepwise acclimatized towards high COD content present in the influent wastewater (OMW) using a batch mode operation with increasing COD concentrations (25 g L−1, 50 g L−1 and 75 g L−1). For medium renewal, the aeration pump and the magnetic mixer were switched off to allow the suspended activated sludge to settle in the reactor (60 min). One hundred mL of settled biomass were transferred into 700 mL of diluted OMW at 25 g L−1 COD. When the reactor achieved pseudo steady-state in terms of effluent COD, settled sludge was consecutively adapted to higher COD concentrations by the same procedure. Analyses of COD on OMW samples from the SBR plant were carried out periodically.
To monitor the stability of the consortium, the acclimatized sludge (100 mL) was used to inoculate 4 successive batch reactors at 75 g L−1 COD.
Sampling, isolation and identification of bacterial strains
After acclimatization period, OMW samples were withdrawn at the end of SBR operation cycle and were serially diluted (1:10) in sterile physiological saline (0.9% w/v, NaCl). A volume of 100 µL from each dilution was evenly spread onto the Tryptic soy agar (TSA) plates containing 10% (v/v) of centrifuged OMW. After 24–48 h of incubation at 30 °C, colonies with distinct morphological features, i.e., color, shape, size, rough or smooth surface were picked and purified by repeatable streaking on another agar plate of the same culture medium. Liquid cultures of the isolates were maintained as frozen stocks at − 80 °C in 20% glycerol.
The diversity of the cultivable bacterial consortium was analyzed by amplification of the internal transcribed spacers between the 16S and the 23S rRNA genes (ITS-PCR) and by 16S rRNA sequencing. Following total genomic DNA extraction, the 16S-23S ITS region and the 16S rRNA gene were amplified using, respectively, the universal primers ITSF/ITSR and 16F27N/16R1525. The amplified 16S rRNA fragments were sequenced and compared with those available at the national center for Biotechnology Information (NCBI) database using the BLAST algorithm. The phylogenetic tree was then inferred using the neighbour joining method (Saitou and Nei 1987) and tree topology was evaluated by performing bootstrap analysis of 1000 data sets using MEGA version 6.0 (Tamura et al. 2007).
Assessment of phenol biodegradation capacity of the isolates using the Phenotype MicroArray technology of Biolog®
The metabolism of OMW phenolics was assessed by means of 96-well Microplates. This test was performed on the following representative strains of the species: Bacillus amyloliquefaciens strain OM48; Klebsiella oxytoca strain OM84; Pseudomonas aeruginosa strain OM88; Cellulosimicrobium cellulans strain OM79; Lysinibacillus macroids strain OM73; Bacillus cereus strain OM43; Rhodococcus zopfii strain OM33; Bacillus thuringencis strain OM60; Rhodococcus pyridinivorans strain OM22; Bacillus nealsonii strain OM56; Ochrobactrum tritici strainOM24; Ochrobactrum tritici strain OM26; Ochrobactrum haematophilum strain OM14; Bacillus thioparans strain OM9; Roseomonas mucosa strain OM18; Kocuria rosea strain OM61; Paenibacillus xylanilyticus strain OM8 and Brevibacillus laterosporus strain OM50. Each well was added with 100 µL of minimal growth medium having the following composition (w/v): (NH4)2SO4, 0.625%; K2HPO4, 0.25%; KH2PO4, 0.125%; MgSO4, 0.05%; CaCl2, 0.0025%; FeSO4, 0.00025%, yeast extract 0.025%. The initial pH value of the medium was adjusted to 7.0 ± 0.1. A volume of 50 µL from each carbon sources, i.e., glucose (as control carbon source) (200 mg L−1 final concentration), OMW phenolics (extracted as mentioned in the following Sect. 6) (200 mg L−1 final concentration) and glucose plus OMW phenolics (200 mg L−1 final concentration each), was then applied in triplicate to the wells of the 96 wells microplate. Cell suspensions were prepared by transferring bacterial colonies from the plate surface with a sterile cotton swab to 10 mL of sterile physiological saline (0.85% w/v, NaCl). They were then adjusted to achieve a 90% of transmittance using a Biolog® turbidimeter. A volume of 100 µL of the suspension was transferred into microplate wells. Appropriate controls were set up for each isolate by loading the cell suspension into the wells (100 µL) without any carbon substrate and loading sterilized water instead (50 µL). The tetrazolium dye which is reduced to formazan during bacterial respiration (producing a purple color) was used as an indicator of cell growth.
Inoculated microplates were then incubated at 26 °C in an Omnilog Reader/Incubater (Biolog) for 10 days. Microplates were read every 15 min with a computer-controlled Microplate reader. At the end of the incubation period, the reduction of tetrazolium dye was expressed as OmniLog units. To compare the utilization of different metabolic compounds, the raw data, including the integrated surface area under the curve, the maximum value, and the slope, were exported and exploited as excel file.
Aerobic treatment of OMW by mixed indigenous cultures
The strains previously tested for their phenol biodegradation ability were mixed and assessed for their capacity to treat OMW.
Experiments were conducted in duplicate in 250-mL Erlenmeyer flasks, containing 50 mL of OMW-based medium at 75 g L−1 COD and supplemented with (NH4)2SO4, K2HPO4, and MgSO4. 7H2O to provide a stoichiometric ratio of dissolved-COD:N:P:Mg of 100:5:1:1. The initial pH value of the medium was adjusted to 7.0 ± 0.1.
After sterilization (at 120 °C for 20 min), flasks were inoculated with equal volumes of the different mid-exponential growth phase pre-cultures on Tryptic Soy Broth (TSB) medium to obtain a final inoculum size of 5% (v/v).
After 30 days of incubation at 120 rpm and 28 °C, spent OMW-based medium was separated from bacterial cells by centrifugation (8000 rpm, 15 min, 4 °C) and then analyzed with regard to its COD and phenolics as described in the following section.
Analytical methods
Chemical oxygen demand (COD), total suspended solids (TSS) and total Nitrogen Kjeldhal (TNK) were determined according to Standard Methods for the Examination of Water and Wastewater (APHA 1998). The phosphorus content was measured colorimetrically using the AFNOR method (1983). OMW phenolics were extracted by ethyl acetate method as described by Jaouani et al. (2005b) and the total phenolic content was assessed by the Folin–Ciocalteu method against a gallic acid calibration curve (Swain and Hillis 1959).
Phytotoxicity bioassay
Phytotoxicity was evaluated by measuring the seed germination index (GI) of tomato (Solanum lycopersicum) as described by Ntougias et al. (2012) with slight modifications. Briefly, 10 tomato seeds were placed in a petri dishes lined with a Whatman No. 1 filter paper and were watered by the untreated or treated OMW at various dilutions in tap water 1/2, 1/4, 1/10, 1/25 and 1/50 (v/v) (or by tap water for the control). Petri dishes containing seeds were incubated for 5 days in the dark at 25 °C, and then their germination index was calculated according to the following equation:
| 1 |
Statistical analysis of data
All experiments in this work were performed in duplicate, and mean values were presented. Experimental data were statistically analyzed using the one-way analysis of variance (ANOVA) followed by a Bonferroni post hoc test using IBM SPSS Statistics 21 software package.
Results and discussion
Olive mill wastewater characterization
The OMW analysis was carried out and the average values of the main parameters are shown in Table 1. OMW is characterized by a high level of organic matter expressed in term of COD content (75.14 g L−1) and high concentrations of total suspend solids (12.07 g L−1). The relatively low biodegradability of this effluent is due to its high amount of total phenolic compounds (3.5 g L−1) and its acidic pH (5.3). However, total nitrogen and phosphorus concentrations were extremely low, at 0.62 g L−1 and 0.84 g L−1, respectively.
Table 1.
Physicochemical characteristics of the olive mill wastewater (OMW) used in this study (mean values of two separate analysis ± standard deviation)
| Parameters | Data |
|---|---|
| pH (25 °C) | 5.3 |
| COD (g L−1) | 75.14 ± 1.21 |
| Total suspended solids (TSS) (g L−1) | 12.07 ± 0.05 |
| Total phenols (g L−1) | 3.50 ± 0.08 |
| Total nitrogen kjeldhal (TNK) (g L−1) | 0.62 ± 0.04 |
| Phosphorus (g L−1) | 0.84 ± 0.10 |
Assessment of the adapted microbial consortium stability and process performance
In many cases, biological processes have been proved to be effective and cost-efficient in OMW treatment. However, the presence of inhibitory compounds in OMW, i.e., lipids, tannins and phenolic compounds requires an acclimatization period for the microorganisms to increase their tolerance to toxicants and to improve their capacity for OMW treatment (Jaouani et al. 2005b; Özgün et al. 2016). High COD contents can be tolerated only if the process operates at a long hydraulic retention time (HRT) and/or with high recycle ratios. If an adaptation period for the microorganisms is performed, the COD reduction achieved by aerobic treatment may be up to 85% with HRT 20–25 days (Paraskeva and Diamadopoulos 2006).
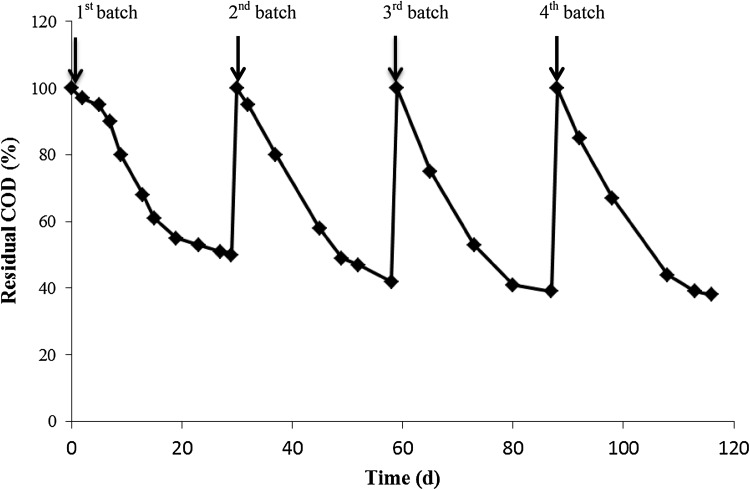
Several investigations have been done on the biological treatment of OMW. Nevertheless, most of these studies deal with treating OMW after making considerable dilutions or proceeding pretreatment steps. In the present study, an attempt to gradually acclimatize the microbial consortium to higher concentrations of OMW by successive stepwise transfers from medium having a COD concentration of 10 g L−1 to one containing 75 g L−1 was carried out, as described in “Material and Methods”. To confirm the stability of the aerobic microbial consortium, the acclimatized consortium was used to successively inoculate new batches containing a COD concentration of 75 g L−1. Similar COD removal efficiencies have been achieved (about 60%) after 30 day HRT, indicating the presence of a stable microbial consortium (Fig. 1). In the same line, Benitez et al. (1997) investigated the aerobic treatment of OMW in a completely mixed batch activated sludge reactor following microorganism acclimatization. The yielded maximum COD removal efficiency was found to be in the range of 58–68% for corresponding initial CODs of 98–65 g L−1.
Fig. 1.
COD removal during repetitive batch experiments carried out at 75 g L−1 COD. Each batch run was for 30 days
We have to mention here that SBR may be considered as a potentially promising solution for OMW treatment, since high removal rates as high as 1.5 g COD L−1 D−1 have been achieved without any prior treatment or dilution.
Bacterial community structure
According to colony and cell morphologies, 91 different cultivable bacterial strains were isolated from OMW samples after acclimatization period.
The diversity of this collection was analyzed by amplification of the ITSs between the 16S and the 23S rRNA genes (ITS-PCR) followed by 16S rRNA sequencing. ITS-PCR showed a high level of diversity among the isolated strains with the detection of 22 distinct haplotypes. Each ITS-type was composed by 1 up to 5 reproducible bands showing an apparent molecular weight ranging from 200 to about 900 bp. Haplotypes C and B were the most represented in the collection and were demonstrated, respectively, in 9 and 8 isolates (Table 2). Molecular identification of the isolates was performed by sequencing the 16S rRNA gene of representative isolates of each haplotype group and comparing the generated sequences to those available in the GenBank database using BLAST algorithm.
Table 2.
ITS Haplotype diversity of groups of OMW isolates
| ITS haplotype | Number of ITS bands | Size of ITS bands (bp) | Strains |
|---|---|---|---|
| A | 1 | 300 | OM64–OM67–OM69–OM71–OM74–OM73 |
| B | 1 | 350 | OM44–OM43–OM45–OM62–OM70–OM76–OM77–OM75 |
| C | 1 | 400 | OM17–OM30–OM16–OM26–OM25–OM27–OM23–OM24–OM32 |
| D | 1 | 500 | OM88 |
| E | 1 | 600 | OM31–OM14–OM28–OM89–OM15–OM29 |
| F | 1 | 900 | OM61 |
| G | 2 | 300–400 | OM60–OM59–OM55 |
| H | 2 | 350–500 | OM42–OM41–OM11–OM72 |
| I | 2 | 300–500 | OM12–OM58– OM56–OM13–OM57 |
| J | 2 | 500–600 | OM47–OM49–OM51–OM52–OM53–OM50 |
| K | 2 | 400–600 | OM33 |
| L | 2 | 600–700 | OM22–OM1–OM21–OM20 |
| M | 3 | 200–300–500 | OM40–OM10 |
| N | 3 | 300–400–500 | OM46–OM48–OM54–OM78–OM81 |
| O | 3 | 350–450–500 | OM82–OM84–OM87–OM90–OM91–OM86 |
| P | 3 | 300–500–700 | OM7–OM35–OM34 |
| Q | 3 | 500–700–900 | OM79 |
| R | 4 | 250–350–500–600 | OM37–OM18–OM6 |
| S | 4 | 300–500–600–650 | OM65–OM68–OM66–OM80–OM83–OM85 |
| T | 5 | 500–550–600–700–800 | OM3–OM36–OM5–OM8–OM19–OM38 |
| U | 5 | 400–550–700–800–850 | OM2–OM39–OM9–OM4 |
| V | 5 | 300–550–600–700–900 | OM63 |
From the phylogenetic analysis shown in Fig. 2 and Table 3, it was possible to discriminate three major phyla of bacteria, i.e., Firmicutes with the highest number of species (57.1%), Proteobacteria (35.2%) and Actinobacteria (7.7%). These phyla have been reported to be commonly abundant in wastewater biological treatment systems (Yadav et al. 2014). Firmicutes were as well identified as the predominant bacteria involved in the decomposition of olive mill waste to generate substrates directly utilized by methanogenic Archaea during anaerobic digestion (Rincòn et al. 2008). Nevertheless, Proteobacteria was the most dominant phylum detected in the natural microbiota of OMW from different olive tree varieties (Tsiamis et al. 2012). The dominance of Firmicutes in this work is probably due to the acclimatization of activated sludge to OMW that may promote the occurrence of particular phyla of species more resistant to recalcitrant compounds.
Fig. 2.
Phylogenetic tree based on the 16S rRNA gene of representative isolates of each haplotype group (A–V) and reference sequences from GenBank (http://www.ncbi.nlm.nih. gov/genbank). Phylogenetic relationships among taxa were evaluated by performing bootstrap analysis of 1000 data sets using MEGA version 6.0. Numbers in parentheses represent the sequences accession numbers. Bar, 0.02 changes per nucleotide position
Table 3.
Table summarizing phylogenetic affiliations of the isolates
| Phylum | Class | Order | Suborder | Family | Genus | Species | Isolates |
|---|---|---|---|---|---|---|---|
| Proteobacteria | Alpha-proteobacteria | Rhodospirillales | Acetobacteraceae | Roseomonas | Roseomonas mucosa | OM37, OM18, OM6, OM11, OM42, OM41, OM72 | |
| Rhizobiales | Brucellaceae | Ochrobactrum | Ochrobactrum haematophilum | OM35, OM34, OM14, OM31, OM28, OM89, OM15, OM29, OM7 | |||
| Ochrobactrum tritici | OM25, OM24, OM23, OM27, OM26, OM16, OM30, OM17, OM32 | ||||||
| Gamma proteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | Pseudomonas aeruginosa | OM88 | ||
| Enterobacteriales | Enterobacteriacea | Klebsiella | Klebsiella oxytoca | OM82, OM84, OM87, OM90, OM91, OM86 | |||
| Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus | Bacillus cereus | OM44, OM43, OM45, OM62, OM70, OM76, OM77, OM75 | |
| Bacillus amyloliquefaciens | OM46, OM65, OM68, OM66, OM80, OM83, OM85, OM48, OM54, OM78, OM81 | ||||||
| Bacillus nealsonii | OM12, OM58, OM56, OM13, OM57, OM10, OM40 | ||||||
| Bacillus thioparans | OM2, OM9, OM39, OM4 | ||||||
| Bacillus thuringencis | OM60, OM59, OM55 | ||||||
| Bacillus subtilis | OM63 | ||||||
| Lysinibacillus | Lysinibacillus macroids | OM64, OM67, OM69, OM71, OM74, OM73 | |||||
| Paenibacillacea | Brevibacillus | Brevibacillus laterosporus | OM47, OM49, OM51, OM52, OM53, OM50 | ||||
| Paenibacillus xylanilyticus | OM3, OM36, OM5, OM8, OM19, OM38 | ||||||
| Actinobacteria | Actinobacteridae | Actinomycetales | Micrococcineae | Micrococcaceae | Kocuria | Kocuria rosea | OM61 |
| Promicromonosporaceae | Cellulosimicrobium | Cellulosimicrobium cellulans | OM79 | ||||
| Corynebacterineae | Nocardiaceae | Rhodococcus | Rhodococcus pyridinivorans | OM1, OM21, OM20, OM22 | |||
| Rhodococcus zopfii | OM33 |
In the present study, Firmicutes were represented exclusively by members of the order Bacillales and more specifically were dominated by the families Bacillaceae characterized by the genera Bacillus and Lysinibacillus and Paenibacillacae characterized by the genera Brevibacillus and Paenibacillus. The Bacillus genus encompassed 34 isolates represented by B. amyloliquefaciens (11 isolates), B. cereus (8 isolates), B. nealsonii (7 isolates), B. thioparans (4 isolates), B. thuringiensis (3 isolates) and B. subtilis (1 isolate). Bacillus species i.e., B. cereus, B. thuringiensis and B. amyloliquefaciens were also the main autochthonous bacterial biota in soil amended by OMW (Naclerio et al. 2010). This was not surprising, since several Bacillus sp. have been reported as capable of using lipids and tannins as sole carbon and energy sources (Ertuğrul et al. 2007; Mondal et al. 2001). Naclerio et al. (2010) reported as well that endospores of Bacillus species isolated from OMW-amended soil exhibit a high biodegradation potential towards OMW phenolic compounds. In fact, according to Henriques and Moran (2000), the nucleoid in the spore core is surrounded by several protective layers that enable the spores to resist external physical and chemical insults and in part determine their exceptional longevity in the environment.
The genus Lysinibacillus was characterized by only a single specie- Lysinibacillus macroids (6 isolates). Lysinibacillus sp. were found to be catabolically versatile with the aptitude to use a wide range of unusual substrates including ethanediol, organophosphorus pesticide malathion, sulfonated azo dyes, fomesafen and dibenzothiophene (Babiak et al. 2011; Saratale et al. 2013; Singh et al. 2012).
Paenibacillacae was represented by Brevibacillus laterosporus (6 isolates) and Paenibacillus xylanilyticus (6 isolates). Though Brevibacillus laterosporus strains have been recognized as eco-friendly, few studies revealed their use in bioremediation. These species have been recently investigated especially in azo dye degradation and textile effluent treatment (Kurade et al. 2016). Isolates belonging to the genus Paenibacillus have been reported to be distinguishable from other aerobic spore-forming species by their ability to grow optimally in 100% (v/v) OMW (Aguilera et al. 2001). These species could have strong biotechnological potential, since they have been recognized as capable to degrade and to metabolize a wide spectrum of aliphatic and aromatic organic pollutants as well as several azo dyes (Johnson et al. 2016; Nawahwi et al. 2013).
Among the phylum Proteobacteria, bacterial species belonging to the class Alphaproteobacteria were the most prominent accounting for 78.1% of Proteobacteria species, followed by Gammaproteobacteria (21.9%). The Alphaproteobacteria were represented by genera Roseomonas and Ochrobactrum, while the Gammaproteobacteria were represented by genera Pseudomonas and Klebsiella. Members of Alphaproteobacteria have been reported to degrade several aromatic substrates. More specifically, Ochrobactrum species possess a broad range of metabolic activities for several petroleum hydrocarbons, insecticides and brominated flame retardants (e.g., tetrabromobisphenol A) (Abraham and Silambarasan 2016; Bezzaa et al. 2015; Zu et al. 2014), while Roseomonas species were proved to process hydrocarbon-degrading abilities (Jain et al. 2010).
The Gammaproteobacteria are a very diverse group characterized by their ability to degrade hydrocarbons, lignin and lignin-related phenolic compounds (Fang et al. 2012; Kostka et al. 2011). In our study, the Gamma subclass of proteobacteria was represented only by Klebsiella oxytoca (6 isolates) and Pseudomonas aeruginosa (1 isolate). These species have been frequently reported as versatile toxic organic compound degraders, and may constitute good candidates for bioremediation processes. In fact, Ammar et al. (2005) showed that indigenous Klebsiella oxytoca strains exhibit important biodegradation capacities towards several monomeric aromatic compounds of OMW (i.e., gentisic, protocatechuic, p-hydroxybenzoic, benzoic, vanillic and ferulic acids), while Hasan and Jabeen (2015) showed that Pseudomonas aeruginosa is capable to degrade up to 400 mg L−1 phenol through an ortho-cleavage pathway.
Actinobacteria were represented entirely by members of the order Actinomycetales and more specifically were dominated by the genus Rhodococcus (5 isolates) followed by Kocuria rosea (1 isolate) and Cellulosimicrobium cellulans (1 isolate). According to Ventura et al. (2007), the presence of Actinobacteria was associated to an efficient degradation of complex organic materials. Rhodococcus species were described as able to degrade and/or convert a wide range of recalcitrant compounds, including aliphatic, monoaromatic-, and polyaromatic hydrocarbons (Kotake et al. 2016; Yang et al. 2014), n-alkanes (Cappelletti et al. 2015), phenols (He et al. 2014), chlorophenols (Hou et al. 2016), sulphonated azo dyes (Heiss et al. 2006), as well as xenobiotic compounds (Khairy et al. 2015) making them suitable in biocatalytic and bioremediation applications. The tolerance of Rhodococcus sp. is associated with their genomic plasticity coding for multiple efflux pumps, combined with a highly versatile metabolism that enables species of this genus to survive under extreme environmental conditions (de Carvalho et al. 2014).
In conclusion, these strains, which have been suggested to be major degraders of several recalcitrant compounds, may constitute potential candidates for bioremediation and can be useful for biotechnological applications.
OMW phenolics biodegradation by the isolated strains
Considering the complex composition of OMW, it was difficult to ascertain precisely whether the isolated bacteria strains could grow only on the aromatic part of the effluent or on other easily biodegradable compounds present in this wastewater. To confirm the aromatic nucleus degradation ability of these isolates, the Biolog Phenotype Microarray technology was used as an alternative method for rapid assessment of OMW phenolics biodegradation. To the best of our knowledge, this is the first study done using this tool to evaluate the ability of microorganisms to utilize phenolic compounds. This technology measures the respiration of cells as a function of time in thousands of microwells simultaneously, and thus allows easier comparison of kinetic plots. For each kinetic curve, key biological information such as the area under the curve, the maximum value, and the slope are calculated on the basis of the raw data points.
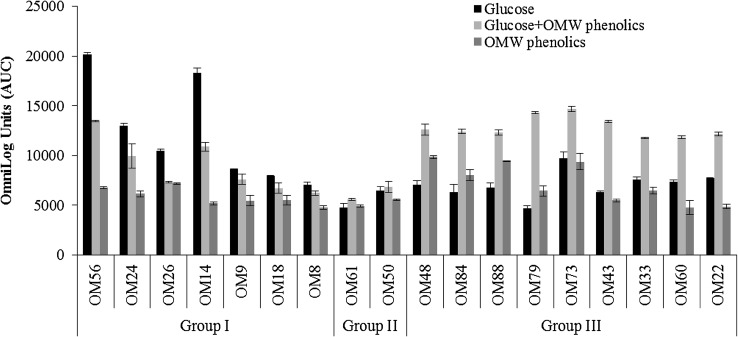
In this study, bacterial respiration kinetics were conducted in the presence of OMW phenolics and glucose, alone or in combination to characterize substrate utilization by the isolated strains. The integrated surface area under the time course kinetic curves (AUC) that reflects the metabolic activity of the bacterial strains was extracted from the software and exploited as excel file (Fig. 3). The respiration kinetics varied considerably according to the different substrates and bacterial species tested. Interestingly, the respiration curves for all strains indicated positive reactions on OMW phenolics, but their respective areas were substantially different. Such result is expected, since the acclimatization of these strains to high OMW concentrations would induce phenolics-degrading enzymes and alleviate inhibition effects to some extent. In the same line, Aissam et al. (2007) and Kumar et al. (2013) reported that phenolic compounds were biodegraded much more readily with phenol-acclimatized microorganisms rather than non-acclimatized ones. The highest respiration rates on OMW phenolics were recorded for Pseudomonas aeruginosa strain OM88, Lysinibacillus macroids strain OM73 and Bacillus amyloliquefaciens strains OM48, while the least respiration rates were observed for Ochrobactrum haematophilum strain OM14, Paenibacillus xylanilyticus strain OM8, Rhodococcus pyridinivorans strain OM22, Kocuria rosea strain OM61, Bacillus thuringencis strain OM60, Brevibacillus laterosporus strain OM50, Roseomonas mucosa strain OM18, Bacillus cereus strain OM43 and Bacillus thioparans strain OM9.
Fig. 3.
Comparison of Area Under the Respiration Curve (AUC) data from the Biolog bacterial phenotypic microarray. Group I: (OM56) Bacillus nealsonii strain OM56; (OM24) Ochrobactrum tritici strain OM24; (OM26) Ochrobactrum tritici strain OM26; (OM14) Ochrobactrum haematophilum strain OM14; (OM9) Bacillus thioparans strain OM9; (OM18) Roseomonas mucosa strain OM18; (OM8) Paenibacillus xylanilyticus strain OM8; Group II: (OM61) Kocuria rosea strain OM61; (OM50) Brevibacillus laterosporus strain OM50; Group III: (OM48) Bacillus amyloliquefaciens strain OM48; (OM84) Klebsiella oxytoca strain OM84; (OM88) Pseudomonas aeruginosa strain OM88; (OM79) Cellulosimicrobium cellulans strain OM79; (OM73) Lysinibacillus macroids strain OM73; (OM43) Bacillus cereus strain OM43; (OM33) Rhodococcus zopfii strain OM33; (OM60) Bacillus thuringencis strain OM60; (OM22) Rhodococcus pyridinivorans strain OM22
Based on respiration behavior on OMW phenolics as glucose co-substrates, it was possible to discriminate 3 distinct groups. Group I includes strains having lower respiration rates (statistically significant at p ≤ 0.05) in the presence of OMW phenolics and glucose compared to glucose alone (i.e., Bacillus nealsonii strain OM56; Ochrobactrum tritici strain OM24; Ochrobactrum tritici strain OM26; Ochrobactrum haematophilum strain OM14; Bacillus thioparans strain OM9; Roseomonas mucosa strain OM18; Paenibacillus xylanilyticus strain OM8). For these strains, glucose appeared to be more preferentially used as it supported higher respiration rates than OMW phenolics alone. These results indicate that OMW phenolic acids had a negative effect on growth of the latter strains. The inhibitory effect of phenolic compounds may be explained by adsorption to cell membranes, interaction with cell enzymes, carbohydrates and proteins by hydrogen bonding and metal ion deprivation (Scalbert 2012). However, Group II encompasses strains that demonstrated tolerance to the presence of OMW phenolics, since their respiration rates were quite similar whatever the substrate used. As for the rest of strains (Group III), the respiration rates were significantly stimulated (p < 0.05) by the mixture of glucose and OMW phenolics (i.e., Bacillus amyloliquefaciens strain OM48; Klebsiella oxytoca strain OM84; Pseudomonas aeruginosa strain OM88; Cellulosimicrobium cellulans strain OM79; Lysinibacillus macroids strain OM73; Bacillus cereus strain OM43; Rhodococcus zopfii strain OM33; Bacillus thuringencis strain OM60; Rhodococcus pyridinivorans strain OM22). Microorganisms in nature show the preference for simple carbon sources such as glucose, and unless it is completely exhausted, the complex carbon sources like aromatic compounds are not degraded (Collier et al. 1996). This phenomenon of carbon catabolic repression has been shown to occur in several microorganisms, while in the present study, strains of Group III seem to be able to co-metabolize both OMW phenolic compounds and glucose. A similar trend was observed for Pseudomonas putida when grown on an aromatic compound and glucose (Collier et al. 1996).
When the respiration rate was examined on each substrate separately, different behaviors were seen among strains of Group III. The species Bacillus thuringencis strain OM60 and Rhodococcus pyridinivorans strain OM22 showed higher respiration rates on glucose compared to OMW phenolics alone, which is explained by the fact that glucose is an easily biodegradable compound, whereas phenolics are not. However, the strains Bacillus cereus strain OM43, Rhodococcus zopfii strain OM33 and Lysinibacillus macroids strain OM73 utilize with almost equal efficiency both glucose and phenolics. The remaining strains (i.e., Bacillus amyloliquefaciens strain OM48; Klebsiella oxytoca strain OM84; Pseudomonas aeruginosa strain OM88; Cellulosimicrobium cellulans strain OM79) were able to utilize phenolics more efficiently than glucose, since higher respiration rates were recorded in the presence of OMW phenolics as sole carbon sources. According to Basu and Phale (2006), the low utilization of glucose may be attributed to the regulation of glucose metabolizing enzymes and/or to the transport process. The unusual carbon source preference by the latter strains provides opportunities for bioremediation of aromatic compounds even in the presence of simple carbon substrates in the environment.
These findings suggest that most of bacterial strains isolated from OMW can be promising for effectively treating wastewaters containing phenolic compounds and their derivatives.
Aerobic treatment of OMW using indigenous bacterial mixed cultures
The native bacteria always display a good adaptability to the natural environment. Accordingly, it is a cost-effective and highly efficient method to reuse the above isolated indigenous microorganisms for organic matter and phenolics removal from OMW. Several biodegradation studies have focused on the use of defined single microorganisms, while the present research attempts to treat OMW using mixed bacterial consortium.
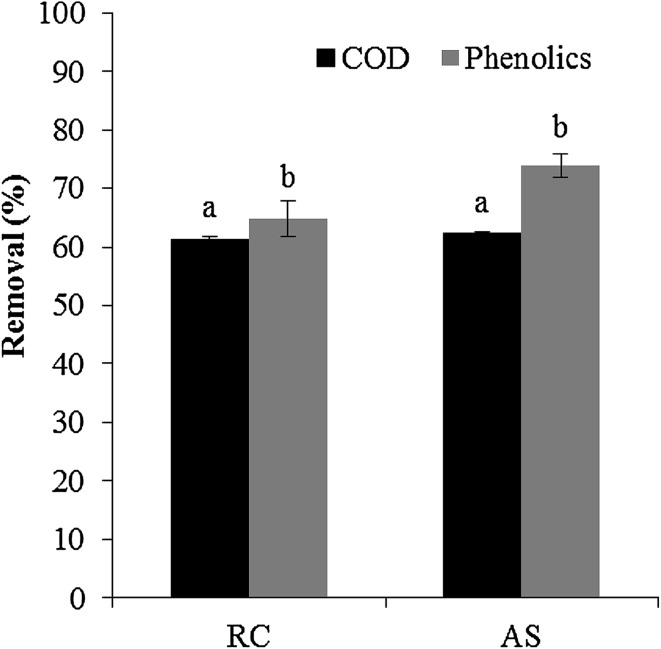
The degradation ability of the indigenous bacterial mixed cultures has been examined using OMW at 75 g L−1 COD. The reconstituted bacterial consortium showed COD and phenolics removal efficiencies of up to 61% and 64%, respectively, which are comparable to those obtained with acclimatized activated sludge (Fig. 4). Although both cultivable and non-cultivable microorganisms take part in degradation, our results suggest that cultivable bacteria play the major role in OMW biodegradation. As a general rule, it appeared that the combination of a wider range of bacterial species with a broader enzymatic profile has a greater ability to degrade mixed pollutants in OMW. Several studies have utilized bacterial consortia for bioremediation of OMW and some promising results have been obtained with consortia originated from activated sludge (Benitez et al. 1997), commercial communities (Ranalli 1992), wastewaters and soil samples (Zouari and Ellouz 1996). For instance, Zouari and Ellouz (1996) reported COD removal rate of 50% and degradation of almost all of the simple aromatics in undiluted OMW with reconstituted bacterial mixtures, while Benitez et al. (1997) showed up to 58–84% removal efficiency for corresponding initial CODs of 98–20 g L−1 as well as an intense reduction in the total phenolic content (up to 90%) and a complete removal of some simple phenolics.
Fig. 4.
Average of phenolics and COD removal efficiencies of OMW at 75 g L−1 COD by the reconstituted bacterial consortium (RC) and acclimatized activated sludge (AS). Data are presented as mean values from duplicate experiments. a, b Means without a common letter differ (p < 0.05)
In conclusion, our research study succeeded in establishing a stable cultivable bacterial consortium that may be used for bioremediation of OMW or similar phenolics-rich wastewaters. Despite experiments being performed at the Erlenmeyer scale and under sterile conditions, they can provide a good insight into the efficiency of the novel reconstituted bacterial consortium on OMW treatment. Further research on the scale-up of this consortium at bioreactor scale is required to ascertain whether the results achieved by the current study can be turned into viable process.
Phytotoxicity assessment
The impact of biotreatment with the reconstituted bacterial consortium in decreasing the concomitant OMW toxicity was determined through germination assays using tomato seeds. The test was performed on different treated and untreated OMW (at 75 g L−1 COD) dilutions, i.e., 1/1, 1/2, 1/4, 1/10, 1/25 and 1/50. Results of the study showed that phytotoxicity of treated and untreated OMW decreased significantly following dilution (Fig. 5). Indeed, no germination was registered for the dilution 1/2 (v/v), 1/4 (v/v) and when undiluted OMW was used. The seeds watered with treated and untreated OMW germinated only when the dilution exceeded 1/10 (v/v). Similarly, several researchers have previously reported the strong prohibition of seeds and seedling growth in the presence of high concentration of OMW (Daâssi et al. 2014; Komilis et al. 2005). The germination index increased proportionally with the increase of dilution ratio as maximum percentages were registered at 1/50 (v/v) treated and untreated OMW. This decrease in the phytotoxicity of untreated OMW could be attributed to the reduction in the levels of phenols, salinity and other phytotoxic compounds following dilution (Rusan et al. 2015). Treated OMW showed a significant improvement in tomato seeds GIs compared to the untreated OMW at the same dilution ratios. Specifically, the GI achieved up to 110% at the dilution of 1/50 (v/v) which is significantly higher than that found in untreated OMW and in tap water (p < 0.05). This is mainly attributed to the lower amounts of phenolic compounds in treated OMW and to the significant amounts of nutrients that may promote seed germination and primary root elongation of tomato seedlings (Mekki et al. 2017). The phytotoxicity effect of treated OMW is expected to be much lower in the field as later stages of growth are less sensitive to salinity and other stress conditions and owing to the buffer capacity of the soil. Accordingly, lower OMW dilution ratios might be applied to enhance the economic viability and environmental sustainability of irrigated agriculture.
Fig. 5.
Effect of OMW dilution and treatment with the reconstituted bacterial consortium on germination index of S. lycopersicum seeds. Untreated OMW (UOMW), treated OMW (TOMW), number under the slash indicates the dilution ratio. Data are presented as mean values from duplicate experiments. a–e Means without a common letter differ (p < 0.05)
Although the COD content of the treated OMW far exceeds the limits imposed by the Tunisian standards for the reuse of treated wastewater for irrigation (INNORPI, NT 106.03, 1989), the treated OMW seemed to possess good fertilization properties.
Conclusions
In this study, the performance and microbial feature of the biomass in the SBR treating OMW were investigated. After acclimatization of the activated sludge to high OMW concentrations, the average COD removal efficiency reached up to 60%. Microbial analysis of the biomass showed the presence of a core set of bacterial species with phenol degrading properties that could be used for bioremediation purposes. Interestingly, the reconstituted bacterial consortium was found to possess comparable OMW bioremediation potential to that of acclimatized activated sludge. According to phytotoxicity assays, the treated OMW may be applied on agricultural soils, but it requires dilution to reduce further the residual phytotoxic compounds. In all, this study provided preliminary but important data on a novel bacterial consortium that could be used for treatment of phenolics-contaminated wastewaters.
Acknowledgements
This work was supported by the Ministry of Higher Education and Scientific Research of the Tunisian Republic and the MADFORWATER Horizon 2020 Research and Innovation Program [grant agreement No. 688320].
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- Abraham J, Silambarasan S. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol using a novel bacterium Ochrobactrum sp. JAS2: a proposal of its metabolic pathway. Pestic Biochem Phys. 2016;126:13–21. doi: 10.1016/j.pestbp.2015.07.001. [DOI] [PubMed] [Google Scholar]
- AFNOR (1983) Recueil de norme française: eau, méthodes d’essai, 2éme édition, Paris
- Aggelis G, Iconomou D, Christou M, Bokas D, Kotzailias S, Christou G, Tsagou V, Papanikolaou S. Phenolic removal in a model olive oil mill wastewater using Pleurotusostreatus in bioreactor cultures and biological evaluation of the process. Water Res. 2003;37:3897–3904. doi: 10.1016/S0043-1354(03)00313-0. [DOI] [PubMed] [Google Scholar]
- Aguilera M, Monteoliva-Sánchez M, Suárez A, Guerra V, Lizama C, Bennasar A, Ramos-Cormenzana A. Paenibacillus jamilae sp. nov., an exopolysaccharide-producing bacterium able to grow in olive-mill wastewater. Int J Syst Evol Micr. 2001;51:1687–1692. doi: 10.1099/00207713-51-5-1687. [DOI] [PubMed] [Google Scholar]
- Aissam H, Penninckx MJ, Benlemlih M. Reduction of phenolics content and COD in olive oil mill wastewaters by indigenous yeasts and fungi. World J Microb Biot. 2007;23:1203–1208. doi: 10.1007/s11274-007-9348-0. [DOI] [Google Scholar]
- Ammar E, Nasri M, Medhioub K. Isolation of Enterobacteria able to degrade simple aromatic compounds from the wastewater from olive oil extraction. World J Microb Biot. 2005;21:253–259. doi: 10.1007/s11274-004-3625-y. [DOI] [Google Scholar]
- APHA . Standard methods for the examination of water and wastewater. 12. Washington: American Public Health Association; 1998. [Google Scholar]
- Babiak P, Kyslíková E, Stepánek V, Vale Sová R, Palyzová A, Maresová H, Hájícek J, Kyslík P. Whole-cell oxidation of omeprazole sulfide to enantiopure esomeprazole with Lysinibacillus sp. B71. Bioresour Technol. 2011;102:7621–7626. doi: 10.1016/j.biortech.2011.05.052. [DOI] [PubMed] [Google Scholar]
- Basu A, Phale PS. Inducible uptake and metabolism of glucose by the phosphorylative pathway in Pseudomonas putida CSV86. FEMS Microbiol Lett. 2006;259:311–316. doi: 10.1111/j.1574-6968.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Beccari M, Bonemazzi F, Majone M, Riccardi C. Interaction between acidoge-nesis and metanogenesis in the anaerobic treatment of olive oil mill effluent. Water Res. 1996;30:183–189. doi: 10.1016/0043-1354(95)00086-Z. [DOI] [Google Scholar]
- Benitez FJ, Beltran-Heredia J, Torregrosa J, Acero JL, Cercas V. Aerobic degradation of olive mill wastewaters. Appl Microbiol Biot. 1997;47:185–188. doi: 10.1007/s002530050910. [DOI] [PubMed] [Google Scholar]
- Bezzaa FA, Beukes M, Nkhalambayausi Chirwaa EM. Application of biosurfactant produced by Ochrobactrum intermedium CN3 for enhancing petroleum sludge bioremediation. Process Biochem. 2015;50:1911–1922. doi: 10.1016/j.procbio.2015.07.002. [DOI] [Google Scholar]
- Cappelletti M, Presentato A, Milazzo G, Turner RJ, Fedi S, Frascari D, Zannoni D. Growth of Rhodococcus sp. strain BCP1 on gaseous n-alkanes: new metabolic insights and transcriptional analysis of two soluble di-iron monooxygenase genes. Front Microbiol. 2015;6:393–407. doi: 10.3389/fmicb.2015.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzisymeon E, Foteinis S, Mantzavinos D, Tsoutsos T. Life cycle assessment of advanced oxidation processes for olive mill wastewater treatment. J Clean Prod. 2013;54:229–234. doi: 10.1016/j.jclepro.2013.05.013. [DOI] [Google Scholar]
- Chiavola A, Farabegoli G, Antonetti F. Biological treatment of olive mill wastewater in a sequencing batch reactor. Biochem Eng J. 2014;85:71–78. doi: 10.1016/j.bej.2014.02.004. [DOI] [Google Scholar]
- Collier DN, Hager PW, Phibbs PVJr. Catabolite repression control in the pseudomonads. Res Microbiol. 1996;147:551–561. doi: 10.1016/0923-2508(96)84011-3. [DOI] [PubMed] [Google Scholar]
- Daâssi D, Belbahri L, Vallat A, Woodward S, Nasri M, Mechichi T. Enhanced reduction of phenol content and toxicity in olive mill wastewaters by a newly isolated strain of Coriolopsis gallica. Environ Sci Pollut R. 2014;21:1746–1758. doi: 10.1007/s11356-013-2019-9. [DOI] [PubMed] [Google Scholar]
- de Carvalho CC, Costa SS, Fernandes P, Couto I, Viveiros M. Membrane transport systems and the biodegradation potential and pathogenicity of genus Rhodococcus. Front Physiol. 2014;5:133–145. doi: 10.3389/fphys.2014.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Abbassi A, Kiai H, Raiti J, Hafidi A. Application of ultrafiltration for olive processing wastewaters treatment. J Clean Prod. 2014;65:432–438. doi: 10.1016/j.jclepro.2013.08.016. [DOI] [Google Scholar]
- Ertuğrul S, Dönmez G, Takaç S. Isolation of lipase producing Bacillus sp. from olive mill wastewater and improving its enzyme activity. J Hazard Mater. 2007;149:720–724. doi: 10.1016/j.jhazmat.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Fang W, Fang Z, Peng Z, Chang F, Hong Y, Zhang X, Peng H, Xiao Y. Evidence for lignin oxidation by the giant panda fecal microbiome. PLoS One. 2012;7:1–10. doi: 10.1371/journal.pone.0050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SA, Jabeen S. Degradation kinetics and pathway of phenol by Pseudomonas and Bacillus species. Biotechnol Biotech Equ. 2015;29:45–53. doi: 10.1080/13102818.2014.991638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Niu C, Lu Z. Individual or synchronous biodegradation of di-n-butyl phthalate and phenol by Rhodococcusruber strain DP-2. J Hazard Mater. 2014;273:104–109. doi: 10.1016/j.jhazmat.2014.03.033. [DOI] [PubMed] [Google Scholar]
- Heiss GS, Gowan B, Dabbs ER. Cloning of DNA from a Rhodococcus strain conferring the ability to decolorize sulfonated azo dyes. FEMS Microbiol Lett. 2006;99:221–226. doi: 10.1111/j.1574-6968.1992.tb05571.x. [DOI] [PubMed] [Google Scholar]
- Henriques AO, Moran CPJr. Structure and assembly of the bacterial endospore coat. Methods. 2000;20:95–110. doi: 10.1006/meth.1999.0909. [DOI] [PubMed] [Google Scholar]
- Hou J, Liu F, Wu N, Ju J, Yu B. Efficient biodegradation of chlorophenols in aqueous phase by magnetically immobilized aniline-degrading Rhodococcus rhodochrous strain. J Nanobiotechnol. 2016;14:1–8. doi: 10.1186/s12951-016-0158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institut National De La Normalisation Et De La Propriete Industrielle (1989) Environment protection—use of reclaimed water for agricultural purposes—physical, chemical and biological specifications (in French), Tunisian standards, INNORPI, NT 106.03
- Jain PK, Gupta VK, Pathak H, Lowry M, Jaroli DP. Characterization of 2T engine oil degrading indigenous bacteria, isolated from high altitude (Mussoorie) India World J Microb Biotechnol. 2010;26:1419–1426. doi: 10.1007/s11274-010-0316-8. [DOI] [Google Scholar]
- Jaouani A, Guillen F, Penninckx MJ, Martinez AT, Martinez MJ. Role of Pycnoporus coccineus laccase in the degradation of aromatic compounds in olive oil mill wastewater. Enzyme Microb Techol. 2005;36:478–486. doi: 10.1016/j.enzmictec.2004.11.011. [DOI] [Google Scholar]
- Jaouani A, Vanthournhout M, Penninckx MJ. Olive oil mill wastewater purification by combination of coagulation- flocculation and biological treatments. Environ Technol. 2005;26:633–641. doi: 10.1080/09593330.2001.9619503. [DOI] [PubMed] [Google Scholar]
- Jarboui R, Sellami F, Azri C, Gharsallah N, Ammar E. Olive mill wastewater evaporation management using PCA method: case study of natural degradation in stabilization ponds (Sfax, Tunisia) J Hazard Mater. 2010;176:992–1005. doi: 10.1016/j.jhazmat.2009.11.140. [DOI] [PubMed] [Google Scholar]
- Johnson J, Shah B, Jain K, Parmar N, Hinsu A, Patel N, Chaitanya GJ, Madamwara D. Draft genome sequence of Paenibacillus sp. strain DMB5, acclimatized and enriched for catabolizing anthropogenic compounds. Am Soc Microbiol. 2016;4:e00211–e00216. doi: 10.1128/genomeA.00211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavvadias V, Doulaa MK, Komnitsas K, Liakopouloua N. Disposal of olive oil mill wastes in evaporation ponds: effects on soil properties. J Hazard Mater. 2010;182:144–155. doi: 10.1016/j.jhazmat.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Khairy H, Wübbeler JH, Steinbüchel A. Biodegradation of the organic disulfide 4, 4-dithiodibutyric acid by Rhodococcus spp. Appl Environ Microbiol. 2015;81:8294–8306. doi: 10.1128/AEM.02059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komilis DP, Karatzas E, Halvadakis CP. The effect of olive mill wastewater on seed germination after various pretreatment techniques. Australas J Environ Manag. 2005;74:339–348. doi: 10.1016/j.jenvman.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Kostka JE, Prakash O, Overholt WA, Green SJ, Freyer G, Canion A, Delgardio J, Norton N, Hazen TC, Huettel M. Hydrocarbon degrading bacteria and the bacterial community response in gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl Environ Microbiol. 2011;77:7962–7974. doi: 10.1128/AEM.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake T, Matsuzawa J, Suzuki-Minakuchi C, Okada K, Nojiri H, Iwata K. Purification and partial characterization of the extradioldioxygenase, 2- carboxy-2, 3-dihydroxybiphenyl 1, 2-dioxygenase, in the fluorene degradation pathway from Rhodococcussp. strain DFA3. Biosci Biotechnol Biochem. 2016;80:1–7. doi: 10.1080/09168451.2015.1123605. [DOI] [PubMed] [Google Scholar]
- Kumar S, Mishra VK, Kumar U, Kumar A, Varghese S. Biodegradation of phenol by bacterial strains and their catalytic ability. Intl J Agric Environ Biotechnol. 2013;6:108–115. [Google Scholar]
- Kurade MB, Waghmode TR, Khandare RV, Jeon B-H, Govindwar SP. Biodegradation and detoxification of textile dye Disperse Red 54 by Brevibacillus laterosporus and determination of its metabolic fate. J Biosci Bioeng. 2016;121:442–449. doi: 10.1016/j.jbiosc.2015.08.014. [DOI] [PubMed] [Google Scholar]
- Lanciotti R, Gianotti A, Baldi D, Angrisani R, Suzzi G, Mastrocola D, Guerzoni ME. Use of Yarrowia lipolytica strains for the treatment of olive mill wastewater. Bioresour Technol. 2005;96:317–322. doi: 10.1016/j.biortech.2004.04.009. [DOI] [PubMed] [Google Scholar]
- McNamara CJ, Anastasiou CC, O’Flaherty V, Mitchell R. Bioremediation of olive mill wastewater. Int Biodeter Biodegr. 2008;61:127–134. doi: 10.1016/j.ibiod.2007.11.003. [DOI] [Google Scholar]
- Mekki A, Arous F, Aloui F, Sayadi S. Treatment and Valorization of Agro-wastes as Biofertilizers. Waste Biomass Valor. 2017;8:611–619. doi: 10.1007/s12649-016-9620-3. [DOI] [Google Scholar]
- Michael I, Panagi A, Ioannou LA, Frontistis Z, Fatta-Kassinos D. Utilizing solar energy for the purification of olive mill wastewater using a pilot-scale photocatalytic reactor after coagulation-flocculation. Water Res. 2014;60:28–40. doi: 10.1016/j.watres.2014.04.032. [DOI] [PubMed] [Google Scholar]
- Mondal KC, Banerjee D, Jana M, Pati BR. Colorimetric assay method for determination of the tannin acyl hydrolase activity. Anal Biochem. 2001;295:168–171. doi: 10.1006/abio.2001.5185. [DOI] [PubMed] [Google Scholar]
- Naclerio G, Antonio F, Petrella E, Nerone V, Cocco F, Celico F. Potential role of Bacillus endospores in soil amended by olive mill wastewater. Water Sci Technol. 2010;61:2873–2879. doi: 10.2166/wst.2010.082. [DOI] [PubMed] [Google Scholar]
- Nawahwi MZ, Ibrahim Z, Yahya A. Degradation of the Azo Dye Reactive Red 195 by Paenibacillus spp. R2. J Bioremed Biodeg. 2013;4:174–180. doi: 10.4172/2155-6199.1000174. [DOI] [Google Scholar]
- Neifar M, Jaouani A, Martínez MJ, Penninckx MJ. Comparative study of olive oil mill wastewater treatment using free and immobilized Coriolopsis polyzona and Pycnoporus coccineus. J Microbiol. 2012;50:746–753. doi: 10.1007/s12275-012-2079-4. [DOI] [PubMed] [Google Scholar]
- Ntougias S, Baldrian P, Ehaliotis C, Nerud F, Antoniou T, Merhautová V, Zervakis GI. Biodegradation and detoxification of olive mill wastewater by selected strains of the mushroom genera Ganoderma. and Pleurotus. Chemosphere. 2012;88:620–626. doi: 10.1016/j.chemosphere.2012.03.042. [DOI] [PubMed] [Google Scholar]
- Obied HK, Bedgood DR, Prenzler PD, Robards K. Bioscreening of Australian olive mill waste extracts: biophenol content, antioxidant, antimicrobial and molluscicidalactivities. Food Chem Toxicol. 2007;45:1238–1248. doi: 10.1016/j.fct.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Özgün ÖK, Özkök İP, Kutay C, Orhon D. Characteristics and biodegradability of olive mill wastewaters. Environ Technol. 2016;37:1240–1248. doi: 10.1080/09593330.2015.1110204. [DOI] [PubMed] [Google Scholar]
- Paraskeva P, Diamadopoulos E. Technologies for olive mill wastewater (OMW) treatment: a review. J Chem Technol Biotechnol. 2006;81:1475–1485. doi: 10.1002/jctb.1553. [DOI] [Google Scholar]
- Paredes C, Cegarra J, Bernal MP, Roig A. Influence of olive mill wastewater in composting and impact of the compost on a Swiss chard crop and soil properties. Environ Int. 2005;31:305–312. doi: 10.1016/j.envint.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Ranalli A. Microbiological treatment of oil mill waste waters. Grasas Aceites. 1992;43:16–19. doi: 10.3989/gya.1992.v43.i1.1191. [DOI] [Google Scholar]
- Rincòn B, Borja R, Gonzàlez JM, Portillo MC, Sàiz-Jiménez C. Influence of organic loading rate and hydraulic retention time on the performance, stability and microbial communities of one-stage anaerobic digestion of two-phase olive mill solid residue. Biochem Eng J. 2008;40:253–261. doi: 10.1016/j.bej.2007.12.019. [DOI] [Google Scholar]
- Rusan MJ, Albalasmeh AA, Zuraiqi S, Bashabsheh M. Evaluation of phytotoxicity effect of olive mill wastewater treated by different technologies on seed germination of barley (Hordeum vulgare L.) Environ Sci Pollut R. 2015;22:9127–9135. doi: 10.1007/s11356-014-4004-3. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saratale RG, Gandhi SS, Purankar MV, Kurade MB, Govindwar SP, Oh SE, Saratale GD. Decolorization and detoxification of sulfonated azo dye C.I. Remazol Red and textile effluent by isolated Lysinibacillus sp. RGS. J Biosci Bioeng. 2013;115:658–667. doi: 10.1016/j.jbiosc.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Scalbert A. Antimicrobial properties of tannins. Phytochemistry. 2012;30:3875–3883. doi: 10.1016/0031-9422(91)83426-L. [DOI] [Google Scholar]
- Singh B, Kaur J, Singh K. Transformation of malathion by Lysinibacillus sp. isolated from soil. Biotechnol Lett. 2012;34:863–867. doi: 10.1007/s10529-011-0837-8. [DOI] [PubMed] [Google Scholar]
- Swain T, Hillis WE. The quantitative analysis of phenolic constituents of Prunus domestica. J Sci Food Agric. 1959;10:63–68. doi: 10.1002/jsfa.2740100110. [DOI] [Google Scholar]
- Tamura K, Dudley J, Nei M. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tsiamis G, Tzagkaraki G, Chamalaki A, Xypteras N, Andersen G, Vayenas D, Bourtzis K. Olive-Mill wastewater bacterial communities display a cultivar specific profile. Curr Microbiol. 2012;64:197–203. doi: 10.1007/s00284-011-0049-4. [DOI] [PubMed] [Google Scholar]
- Tziotzios G, Michailakis S, Vayenas DV. Aerobic biological treatment of olive mill wastewater by olive pulp bacteria. Int Biodeter Biodegr. 2007;60:209–214. doi: 10.1016/j.ibiod.2007.03.003. [DOI] [Google Scholar]
- Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, Van Sinderen D. Genomics of actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol R. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav TC, Khardenavis AA, Kapley A. Shifts in microbial community in response to dissolved oxygen levels in activated sludge. Bioresour Technol. 2014;165:257–264. doi: 10.1016/j.biortech.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Yang HY, Jia RB, Chen B, Li L. Degradation of recalcitrant aliphatic and aromatic hydrocarbons by a dioxin-degrader Rhodococcus sp. strain p52. Environ Sci Pollut Res. 2014;21:11086–11093. doi: 10.1007/s11356-014-3027-0. [DOI] [PubMed] [Google Scholar]
- Zerva A, Zervakis GI, Christakopoulos P, Topakas E. Degradation of olive mill wastewater by the induced extracellular ligninolytic enzymes of two wood-rot fungi. J Environ Manag. 2016;203:791–798. doi: 10.1016/j.jenvman.2016.02.042. [DOI] [PubMed] [Google Scholar]
- Zouari N, Ellouz R. Microbial consortia for the aerobic degradation of aromatic compounds in olive oil mill effluent. J Ind Microbiol. 1996;16:155–162. doi: 10.1007/BF01569998. [DOI] [Google Scholar]
- Zu L, Xiong J, Li G, Fang Y, An T. Concurrent degradation of tetrabromobisphenol A by Ochrobactrum sp. T under aerobic condition and estrogenic transition during these processes. Ecotoxicol Environ Safe. 2014;104:220–225. doi: 10.1016/j.ecoenv.2014.03.015. [DOI] [PubMed] [Google Scholar]