Abstract
In the last 30 years, ceramides have been found to mediate a myriad of biological processes. Ceramides have been recognized as bioactive molecules and their metabolizing enzymes are attractive targets in cancer therapy and other diseases. The molecular mechanism of action of cellular ceramides are still not fully established, with insights into roles through modification of lipid rafts, creation of ceramide platforms, ceramide channels, or through regulation of direct protein effectors such as protein phosphatases and kinases. Recently, the ‘Many Ceramides’ hypothesis focuses on distinct pools of subcellular ceramides and ceramide species as potential defined bioactive entities. Traditional methods that measure changes in ceramide levels in the whole cell, such as mass spectrometry, fluorescent ceramide analogues, and ceramide antibodies, fail to differentiate specific bioactive species at the subcellular level. However, a few ceramide binding proteins have been reported, and a smaller subgroup within these, have been shown to translocate to ceramide-enriched membranes, revealing these localized pools of bioactive ceramides. In this review we want to discuss and consolidate these works and explore the possibility of defining these binding proteins as new tools are emerging to visualize bioactive ceramides in cells. Our goal is to encourage the scientific community to explore these ceramide partners, to improve techniques to refine the list of these binding partners, making possible the identification of specific domains that recognize and bind ceramides to be used to visualize the ‘Many Ceramides’ in the cell.
Where are bioactive ceramides located? The pathways of ceramide and their subcellular topology.
Sphingolipids comprise a large family of lipids, including sphingosine and related sphingoid bases, ceramides, sphingomyelins, glycosphingolipids, and additional modifications to their structure such as phosphorylation, degree of unsaturation and/or length of the acyl chain, variations in the sphingoid backbone and others (reviewed in [1–3]), with important roles in cellular structure and signaling [1, 2, 4, 5]. Amongst all of these structures, ceramides have become a key point in the sphingolipid network, not only because they serve as the branching point to different classes of sphingolipids, but also because ceramide functions as a bioactive intermediate in signal transduction. To understand ceramide regulation, it is important to understand its subcellular topology and its central position in the global sphingolipid metabolic network. Although sphingolipids are interconnected to many other metabolic pathways, this network has one single point of entrance and one single exit point.
The biosynthesis of sphingolipids has been extensively covered in many informative reviews [1, 3]. Briefly, and focusing on ceramide subcellular topology, the biosynthesis of sphingolipids starts in the endoplasmic reticulum. The enzyme serine palmitoyl transferase (SPT) is the only entrance point of the sphingolipid network. It catalyzes the condensation of the amino acid serine (glycine and alanine also found in atypical/non-canonical sphingolipids) and palmitoyl-CoA to generate 3-ketodihydrosphingosine, which is reduced to dihydrosphingosine. Six distinct ceramide synthases (CerS) acylate dihydrosphingosine to generate diverse dihydroceramides which can be distinguished by the length of their acyl chains [6]. Still in the ER, a desaturase introduces a double bond in position C4 of the sphingoid backbone to generate ceramide [5]. This pool of ceramides in the ER is then shipped into the Golgi apparatus using both vesicular and protein-mediated transfer; the latter by the ceramide transport protein (CERT). In the Golgi, ceramides act as a regulated branching point, and they can be further metabolized to sphingomyelin, ceramide-1-phosphate, complex glycosphingolipids or become hydrolyzed to sphingosine [1]. Complex sphingolipids in the Golgi are then shipped to other membranes, and cellular trafficking will control their cellular distribution (see figure 1).
Figure 1. Ceramide pools and their metabolic connection.
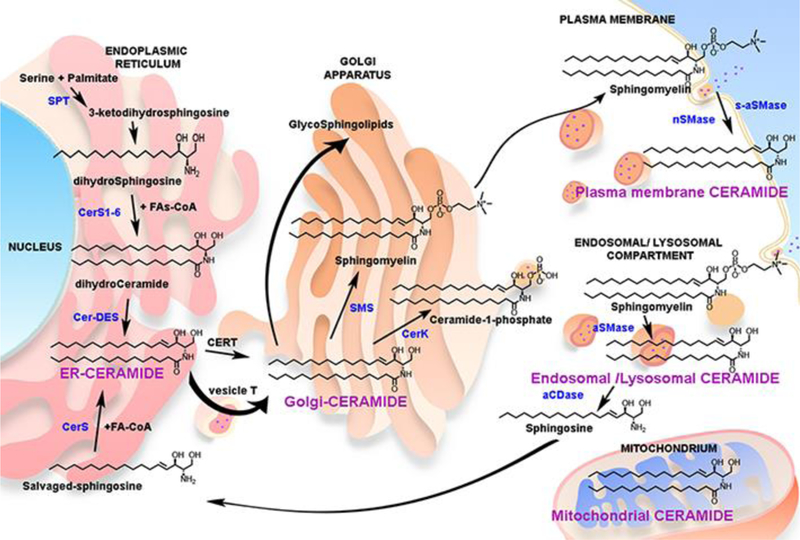
Biosynthetic pathway and metabolism of ceramide (black) – see main text for pathway description. The figure shows metabolizing enzymes (blue) and identified bioactive pools of ceramide (purple). SPT- Serine Palmotoyl Transferase; CerS- (dihydro-)Ceramide Synthase; Cer-DES- Dihydroceramide Desaturase; SMS- Sphingomyelin Synthase; nSMase- Neutral Sphingomyelinase; aSMase- Acid Sphingomyelinase; s-secreted form; aCDase- Acid Ceramidase.
Besides the de novo pathway, ceramide can also be generated from hydrolysis of complex sphingolipids. In this second pathway, ceramide is generated by the breakdown of sphingomyelin or hexosyl-ceramides, catalyzed by sphingomyelinases and hexosyl-ceramidases, respectively. Sphingomyelinases have been found in the ER/Golgi complex, plasma membrane, mitochondria, endosomal/lysosomal compartments, and secreted to the media. Similar to other lipid signaling molecules, this pathway is triggered by degradation of more abundant membrane lipid components. In contrast to de novo pathway that requires the function of several enzymes, this second pathway can rapidly increase ceramides by several fold in specific membrane compartments. The rapid response of this pathway is due to the abundance of sphingomyelin and the requirement for activation of one single enzyme. Thus, this pathway of ceramide generation, has been widely described to occur in response of cellular stress, for example in response of the tumor necrosis factor (TNF) alpha treatment [7, 8].
A third pathway to generate ceramide was named the salvage pathway [9]. Lysosomal (or late endosomal) sphingolipids are degraded to sphingosine by the action of acid sphingomyelinase, (or glucocerebrosidase/galactocerebrosidase), followed by the action of acid ceramidase. This sphingosine can be then recycled to the ER to regenerate ceramide by the previously mentioned CerS [10]. It is not clear if this recycled ceramide is topologically/biologically different from the ceramide generated in the de novo pathway, also located in the ER, but the salvage pathway has been shown to mediate several biologies such as inflammatory processes, modulate cytokine levels (Interleukin-6) and activate specific p38delta-MAPK pathways [11, 12].
Ceramide can also be generated by the reverse action of ceramidases. This has been shown in yeast with the alkaline ceramidases and in mammalian cells with neutral ceramidase [13]. Novgorodov et al provided evidence for the operation of the reverse activity of neutral ceramidase in [14].
Specifically, the generation of ceramide in mitochondria has been shown to trigger apoptosis; however, the source of this ceramide is still uncertain. Several ceramide synthases (CerS) have been shown to localize in purified mitochondria from mouse brain, and they have been related to elevation of ceramide during brain damage [15, 16]. Mitochondrial associated neutral sphingomyelinase (ma-nSMase) [17] and neutral ceramidase (nCDase) [14], by reverse ceramidase activity, have been also proposed to generate ceramide in the mitochondria [16].
How do bioactive ceramides work?
Ceramides are structural constituents of biological membranes, and they have been also shown to exert bioactive functions. Sphingolipids were recognized as bioactive players after the finding that sphingosine inhibits protein kinase C (PKC) [18]. This finding, together with the idea that many breakdown products of major classes of lipids are involved in signal transductions, encouraged further investigations [19]. This led to the understanding that ceramide , the breakdown product of sphingomyelin, was found to mediate cell death by apoptosis in response of TNF alpha [20]. his discovery represented the driving force for the study of ceramide as a bioactive molecule, and ceramide was rapidly shown to be involved in a variety of biologies, including cell differentiation, redox metabolism, cytokine release, inflammation, growth arrest, and cell proliferation, just to mention the most cited ones [1, 21]. Moreover, during the last two decades, many of the enzymes in the sphingolipid pathway have been shown to play critical roles in several pathological conditions, including cancer biology and resistance to cancer treatments [22].
Ceramide is not limited to a single membrane. Instead, ceramide and ceramide-metabolizing enzymes have been found in many compartments, and these spatially different pools of ceramide appear to be involved in different signaling pathways. For instance, using a battery of recombinant sphingomyelinases targeted to different organelles, the Obeid’s laboratory showed for the first time that mitochondrial ceramide, but not ceramide in other compartments, induced cellular apoptosis [23].
Ceramides actions have been evaluated using two main hypotheses: (i) ceramides as modulating physical properties of biological membranes and (ii) ceramide as a classical bioactive molecule [24]. The first hypothesis considers ceramide as a structural component that triggers signaling events by modulating membrane structure and dynamics. This scenario has been widely studied by investigating how ceramide behaves in model membranes. Addition or generation of ceramides in liposomes causes negative curvature, membrane budding, endocytosis, and formation of rigid domains [25]. Based on these physical properties, it has been hypothesized that ceramides modulate lipid rafts structure and/or form some sort of ceramide-enriched platforms in cells. This, in turn, results in changes in the conformation of membrane receptors, causing alterations in signaling, or changes in the curvature of the membrane, ultimately inducing exosome secretion [26].A special case of signaling through mechanical changes in the membrane is the one suggested by Colombini’s group, where ceramide forms large and stable channels in the mitochondrial outer membrane, facilitating cytochrome C release during apoptosis [16, 27]. These aspects of ceramide function are developed in sister reviews in this volume (one by Erhard Bieberich and another by Masanao Kinoshita).
The second organizing hypothesis considers ceramide as a bioactive lipid. Specific stimuli would result in the formation of ceramide, and the newly formed ceramide would directly interact with specific domains in proteins, modulating their function a priori, independent of any effect on membrane biophysical properties. Supporting this model, early in vitro assays showed that addition of the natural stereoisomers of ceramide in enzymatic reactions modulated the Km and Vmax of protein phosphatases [28], kinases [29, 30] and proteases [31].
However, we have to be prudent on the mechanism of action of ceramide, mostly when we are approaching one by one the different claimed ceramide signaling pathways. Reports disagree on the mechanisms of action, often only literature supporting a single model is cited, and only experiments supporting the desired model are performed. Many of the mechanisms presented here have been also explained or hypothesized to work through lipid rafts. For example, Ras, TNF alpha, transforming growth factor beta (TGF-β), CD95/FAS and IL-1β signaling have been described to activate sphingomyelinase activity and, in turn, modify lipid raft composition resulting in clustering and activation of cell signaling pathways [32, 33].
Visualizing ceramides and bioactive ceramides in situ.
Unlike proteins which are composed of sequential combinations of 20 amino acids, ‘lipids’ can include thousands of chemical structures with different physical and chemical properties, making their study and understanding very challenging. More than thirty thousand lipids have been described, but millions of structures can be predicted using informatic approaches [34]. The development of new techniques such as mass spectrometry, has allowed us to quantify the lipid profile of a cell, tissue, or biologic fluids at a lipidomic level, as well as changes to this profile under disease or treatment [35, 36]. However, knowing the lipid composition is not enough. Distribution and composition of lipids is tightly regulated as evidenced by the asymmetry of cellular membranes [37] and the distinct lipid composition of various subcellular organelles. The composition of biological membranes is dynamic, and it changes by incorporation and degradation of lipids, membrane trafficking, and lipid flip-flopping from one bilayer to the other. Efficient methods of extraction are required in order to detect the less abundant lipids and gain consistency in quantification [38, 39]; however, these organic lipid extraction protocols lose all the information on subcellular localization or membrane topological information. Therefore, in order to completely understand cellular signaling, we have to be able to follow the signaling events in a spatial-temporal manner. In that sense, confocal fluorescence microscopy has emerged as a very useful tool to study where the action is happening at the subcellular level. For instance, fluorescent proteins can be expressed as fusion proteins to follow protein dynamics during live cell imaging. However, lipids cannot be synthetized as already bound to fluorophores by the cell. Therefore, synthetic fluorescents lipids have to be added exogenously [40], which introduces additional difficulties to their delivery, targeting of subcellular compartments, and proper function. Fluorescent analogues of ceramide are metabolized to other sphingolipids, which makes them interesting for flow studies, but might fail when localization is being studied [41]. More recently, ‘click chemistry’ has been applied to sphingolipids [42]. This approach brings improvements to the traditional use of fluorescent sphingolipids, by using just small functional groups (azido) instead of the whole structure of a fluorophore. These compounds are still not optimal for delivery, targeting to the right subcellular compartment, and further metabolism, but they should have less interference in their functions. The reader is referred to Chapter by Izquierdo et al. in this volume.
Immunostaining is another common tool to visualize chemically fixed proteins, as it creates a snapshot picture during signaling processes. Again, immunostaining of lipids is not that straight forward. One of the caveats in cellular lipid visualization, from a histochemistry point of view, is the lack of methods to chemically fix lipids. Several antibodies have been developed to detect and visualize ceramide in cells [43–46]; however, their validation is still an issue, and this is often based on lipid fat blots [45]. While this can define specificity in vitro, additional approaches are required for validation for cell studies. We suggest approaches to specifically manipulate the target lipid, and better still in specific compartments, in order to establish the specificity of these antibodies. Moreover, protocols for antibodies recognizing lipids are often used at low temperatures, causing potential artefacts in lipid distribution.
Lipid-binding proteins can be expressed as recombinant proteins and used as lipid probes. This approach has been used to stain sphingomyelin using recombinant lysenin and equinatoxin [47, 48], and cholera toxin is used to stain the ganglioside GM1 [49]. These are widely used, but again, one has to be cautious about the protocols used, with the same issues encountered when staining lipids with antibodies. The reader is referred to the Chapter by Toshihide Kobayashi in this volume.
Some small molecules bind specific lipids and can also be used to stain such lipids. For example, Filipin is commonly used to stain cholesterol. However, there are no small molecules known to specifically bind sphingolipids.
The last method to visualize lipids is expression of lipid binging domains as fusion proteins to fluorescent tags. For example, the Pleckstrin homology domain (PH domain) of many proteins binds phosphatidylinositol lipids (PIPs), and it is used as a probe to visualize them in cells. The cysteine-rich domain (CRD) of classical PKC binds diacylglycerol (DAG). These domains fused with fluorescent proteins have been used as molecular sensors to visualize and quantify PIPs [50, 51] and DAG [51]. The major benefits of this approach include the ability to stain specific pools of target lipids, and to do so in the right compartment targeted by the parent protein. Unfortunately, when it comes to ceramide, there are no binding proteins or expressed protein domains that are used as a probe. Nevertheless, several proteins have been shown to interact with ceramide, and some of them have even been shown to translocate to ceramide enriched membranes (see figure 2 as a summary of methods used to visualize ceramide signaling). Using these ceramide binding proteins as probes is therefore a possibility, although barely explored.
Figure 2. Current methods to visualize ceramide signaling.
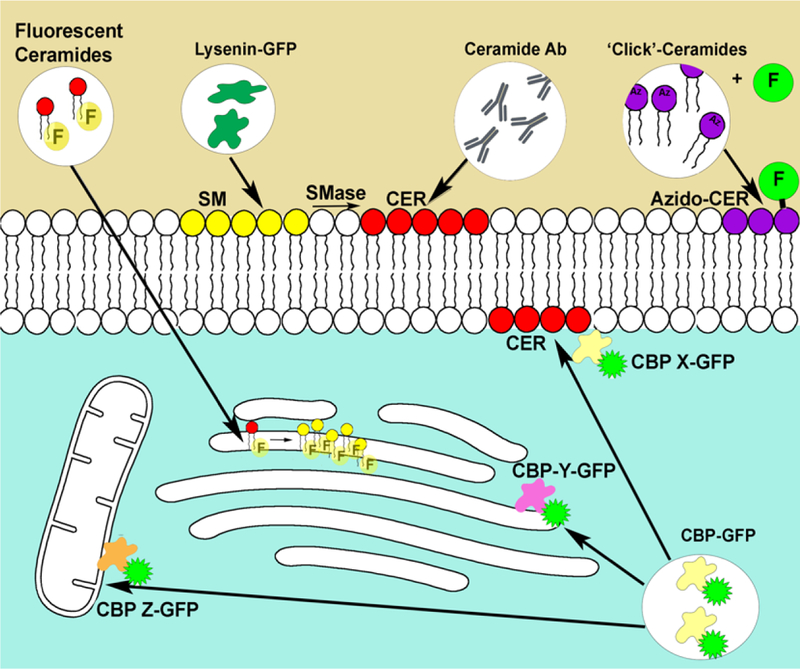
Fluorescent ceramides such as NBD-ceramides have been shown to accumulate in the Golgi apparatus and rapidly transform to NBD-sphingomyelin. Lysenin and other toxins have been shown to selectively bind sphingomyelin, recombinant expression fused to fluorescent proteins are used to visualize sphingomyelin (as ceramide precursor). Several ceramide antibodies have been shown to recognize ceramide. Generation and localization of ceramide in the plasma membrane have been demonstrated. Azido-ceramides and other functionalized ceramides can be combined with click chemistry to label ceramides with fluorophores in situ. Finally, several reported ceramide binding proteins have been shown to translocate to the plasma membrane to bind ceramide. This has been recognized by immunofluorescence or by fusion to the green fluorescent protein. Note that different CBP (X,Y,Z…) would recognize different pools of bioactive ceramide.
Ceramide probes: Searching for Ceramide Binding Proteins (CBP).
The first ceramide binding proteins were identified through independent enzymology studies, as ceramide changed the enzymatic properties of the enzymes being investigated. These classical enzymes were protein phosphatases 1[52] and 2A [28, 53], PKC zeta [54], Raf-1 [55], KSR [56, 57], and Cathepsin D [31]. Following the discovery that ceramide could directly activate proteins in vitro, the idea of many other proteins being regulated by ceramide emerged. Soon different strategies to find ceramide binding proteins were explored. Affinity purification strategies with ceramide immobilized onto column-matrix and beads, were developed [58, 59]. The fact that these studies used different cell lines, ceramide species, and protocols, may be the cause of the very low overlapping of putative CBPs (Table 1). Notably, the majority of classical CBPs were not identified in these studies. In order to explain this, it was argued that affinity ceramide beads could be presented differently than in membranes or liposomes [60]. To overcome this limitation, ceramide was prepared within lipid vesicles using a technique that consisted in collecting these vesicles using magnetic beads. By employing this technique, the Bieberich group confirmed that PKC zeta was being pulled-down with ceramide [61]. Another approach useful to confirm already described CBPs consisted in the use of radioactive crosslinkable ceramide analogues followed by pull-down and western blot analysis. Thus, a radioactive and crosslinkable ceramide analogue, [125I] 3-trifluoromethyl-3-(m-iodophenyl)diazirine-ceramide, known as [125I]-TID-ceramide , was used to confirm interaction of ceramide with Raf-1 and Cathepsin D [31, 62]. A further step involved the use of Mass Spectrometry on proteins bound to a similar analogue, [35S]-TDS-ceramide, to identify new proteins as potential CBP. The majority of the proteins identified with this approach were cytoskeletal and stress proteins [63], but again there was no overlapping with previous reports. More recently, the use of bifunctionalized sphingosine (photoactivatable and clickable, called ‘pac’-sphingosine or pacSph ), allowed for the identification of putative sphingolipid partners in addition to the subcellular localization where the interaction occurs. However, since the pacSph was also metabolized to other sphingolipid members or even crossing to other cellular metabolites, pac sphingolipids did not successfully help distinguish which ones were specific for ceramide. Knockdown of S1P -lyase (the only exit point of the sphingolipid pathway) kept the pac function within the sphingolipid family [64]. Interestingly, proteins phosphatases PP1 and PP2A, regulatory subunits of these phosphatases, enzymes of the sphingolipid metabolism, and common hits from ceramide affinity purification methods were identified (Annexin A1, Prohibitin, SSRP1 and MFE-2) [65]. To circumvent metabolism of pac lipids, the same group used pac –ceramide (pacCer), but instead of feeding cells, cytosolic fractions from different cell lines were incubated with liposomes containing pacCer, and bound proteins were purified and identified using mass spectrometry. Again, not much overlap with previous findings was found, but regulatory subunits for protein phosphatase 2A, annexins, some cytoskeletal proteins and the ceramide transfer protein (CERT) could be found [60].
Table 1. CBPs found in 3 ceramide affinity studies and their intersection.
Red numbers indicate the total number of CBP for each study. Common CBPs (gene name) are indicated for each study in intersection with the other ones. The three studies are: A. Kota V 2012; B is unpublished observations (Hama et al.); C. Bidlingmaier S 2016.
| A. C6-CER | B. C16-CER | C. C6-CER | |
|---|---|---|---|
| A. C6-CER | 97 | ||
| B. C16-CER | ANXA1 NAA15 CYFIP1 COPB1 COPG1 |
147 | |
| C. C6-CER | HSD17B4 | 0 | 37 |
There is still much potential in identifying CBPs. There are just a few studies which used ceramide-affinity based assays combined with proteomics. These studies are difficult to compare due to the enormous differences in experimental design (cell lines, ceramide species, conjugation, type of matrix, timeframe and using lysates versus in vivo) and the criteria on how to discard false positive hits and not considering further metabolism of the ceramide affinity probe. The generation of larger list of CBP is still at an early stage. This is currently limited in the cleaning these lists from false positives (choosing better controls), defining groups based on ceramide structure requirements for binding (distinguish between ceramide species or analogue studies), bioinformatics processing for binding sites identification, and mutagenesis analysis for confirmation of the interactions.
How CBPs have been used to visualize ceramide?
The idea of ceramide directly activating certain proteins suggests that these interacting proteins should colocalize with ceramide and that visualization of these proteins could be used as a way of probing bioactive ceramide. Notably, ceramide colocalization with binding partners involves only few independent reports, lacking consistency on their mechanism of binding. These reports are described in the following sections, where a summary of the different works that have reported ceramide interactions and protein translocation in response of ceramide is presented.
• Ceramide Activated Protein Phosphatases
The focus of the study on ceramide activating protein phosphatases started in the early 1990s, when our group found that addition of synthetic ceramide in cellular cytosolic extractions caused phosphatase activity to increase several folds. These phosphatases were designated as CAPP, Ceramide Activated Protein Phosphatases [21, 28].
Further studies showed that PP1 and PP2A are the main groups of proteins studied that are under the regulation of ceramide. Purified subunits of the PP2A [53] and PP1[52] were activated by short and long ceramides in vitro. Activation of phosphatases was stereospecific, and dimeric and heterotrimeric forms of PP2A were more robustly activated than monomeric forms, suggesting that this interaction doesn’t just involve the catalytic subunit [52]. C-Jun was the first suggested endogenous substrate for the ceramide-CAPP axis, and this signaling pathway could be activated by TNF alpha or treatment with bacterial sphingomyelinase [66]. CAPP substrates were rapidly connected to other targets and biologies, including insulin secretion [67] and biologies linked to the sphingomyelin cycle signaling pathways (growth suppression, differentiation, stress responses, cell damage, and apoptosis), mainly those that could be mimicked by TNF alpha or triggered by addition of exogenous ceramides [68]. These substrates included PKC alpha [69], the retinoblastoma gene product (Rb) [70], caspases, Bcl-2 [71], c-Myc, Akt/PKB [72], p53 [73] and SR proteins [74], p38 MAPK [75], beta catenin [76] and ERM proteins [77–79]. Interestingly, a strong body of evidence is emerging from studies in Saccharomyces cerevisiae demonstrating the action of PP2A type phosphatases as downstream targets of ceramide [80, 81].
Despite the growing number of putative substrates for CAPP proteins, there are still several mechanistic issues outstanding, especially defining specific subunits required for the interaction of ceramides with phosphatases and defining specific domains. Moreover, and based on the diversity of substrates, these dephosphorylations could happen in many distinct sub cellular organelles. If the activation was driven by ceramide, ceramide should be generated in a specific compartment to activate the phosphatase. Dephosphorylation of Rb might happen in the nucleus, whereas Bcl proteins are located in the mitochondria. SR proteins are located in ER and nucleus and beta-catenin and ERM proteins, in the plasma membrane. Marchesini et al [76] were the first to visualize how a CAPP was translocated to the cell cortex in response of ceramide production (see figure 3). Neutral sphingomyelinase was found to be translocated to the plasma membrane in MCF-7 cells in response to confluence, resulting in the dephosphorylation of beta-catenin. An siRNAs screening unveiled PP1 gamma as the phosphatase responsible for catalyzing this dephosphorylation. Confluence, as well as addition of exogenous ceramide, resulted in translocation of PP1 gamma to the plasma membrane.
Figure 3.
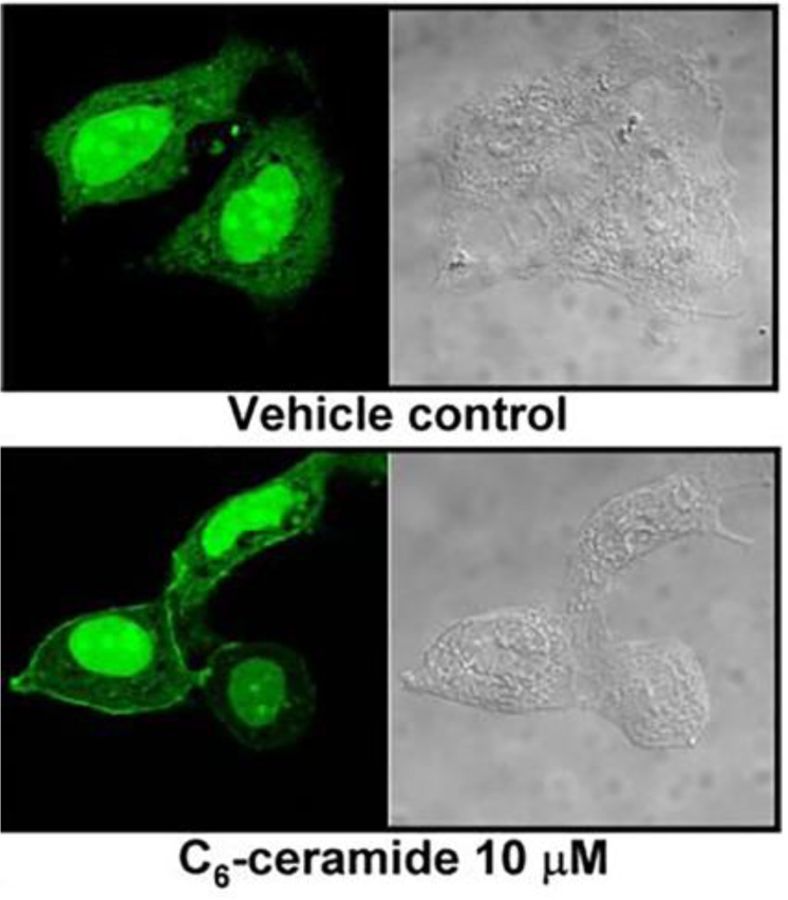
Visualization of ceramide-PP1 gamma interaction. [From Marchesisni et al 2007]
Although several hundred kinases regulate protein phosphorylation, only a small number of phosphatases catalyze Ser/Thr dephosphorylation. PPP family of protein phosphatases are small catalytic enzymes about 37KDa, encoded by only 13 genes. However, hundreds of interacting proteins direct specificity of substrate and localization for the catalytic subunits. In that sense, ceramide could play a role in regulating the interaction between the catalytic subunit of the phosphatase and a regulatory subunit. In support of this hypothesis, inhibitor-2 for PP2A has been suggested to bind ceramide. This association would interfere with the interaction between inhibitor-2 and PP2A, releasing an active PP2A phosphatase. The authors first showed that inhibitor-2 bound biotin-labeled d-erythro-C6-ceramide (interestingly, cathepsin D was also detected to bind ceramide in this study). Ceramide treatments of cells expressing inhibitor-2 affected phosphatase activity in situ, but it did not change the subcellular localization of the inhibitor-2 nor that of the phosphatase [82].
The studies showing protein phosphatases being regulated in vitro by ceramide, their affinity to ceramide probes, and translocation studies have led many authors to consolidate the ceramide-dependent activation of CAPPs in different signaling pathways. However, neither the interaction at molecular level, nor mechanistic studies on the translocation of CAPPs have being addressed and defined yet.
• Atypical Protein Kinase C zeta
Classical Protein Kinase C was the first enzyme reported to be regulated by sphingolipids, with the finding that sphingosine inhibits the enzyme directly [18]. Ceramide was also shown to inhibit PKC in cells, but that inhibition was indirect by activating a PP1 phosphatase which dephosphorylated and deactivated PKC [69, 83]. By then, the discovery that TNF alpha and other cytokines activated the sphingomyelin pathway [19, 84] prompted the investigation of the relationship between ceramide and TNF alpha biologies. One of these pathways that were explored involved the role of PKC zeta in activating the nuclear factor kappa B (NF-KappaB) in mitosis. Moscat’s group found that PKC zeta could be activated in vitro by ceramide, and in vivo by adding bacterial sphingomyalinase to the cell media [54]. In vitro binding studies showed interaction between ceramide and PKCzeta, and also ceramide-enhanced interaction between PKC zeta and a regulatory partner PAR-4 [30]. Following these studies, ceramide was reported to interfere with the interaction between PKCzeta and protein 14–3-3, resulting in Akt phosphorylation by the released PKC zeta [85].These results implicated ceramide in the PKC zeta, PAR-4,14–3-3, Akt and PKA pathway of apoptosis [86].
As several reports began to consolidate this interaction, the next step was to define where on PKC zeta ceramide was binding. he first idea came by analogy to DAG and classical and novel PKCs. PKC proteins contain a pseudosubstrate that inhibits kinase activity. DAG and calcium pre-activate classical PKC, which, following autophosphorylation, creates a stearic impediment for the pseudosubstrate, revealing the catalytic site. Because atypical PKCs contain modified C1 domains that are unable to bind DAG (C1 domain), it was hypothesized that the modified C1 domain in atypical PK s could bind ceramide [87]. In an effort to find the ceramide binding region on PKC zeta, Bieberich’s lab analyzed proteolytic fragments on ceramide overlay assays and vesicles containing ectopically expressed protein. These assays found that the last C-carboxy 20KDa sequence bound to C16-ceramide, and not the hypothesized C1 domain, located in the N-terminal region of PKCs [88]. Ceramide was reported to mediate the binding between the atypical PKC zeta and PAR-4 during apoptosis [86]. The studies on ceramide and aPKC interaction in vivo were further expanded when a new ceramide antibody was developed by the same group and used in subcellular localization studies [43]. The ceramide antibody colocalized with aPKC at the apical membrane, and when applied to the media, the antibody resulted in aPKC re-distribution from the cell cortex to the cytosol and nucleus [88]. In this study, the use of ceramide antibody was proposed to deplete inner leaflet ceramide by bringing this ceramide pool to the outer leaflet ceramide. Migrating cells presented the ceramide antibody and showed aPKC colocalization on plasma membrane protrusions, all along with Cdc42, tubulin and beta-catenin [88]. Through cell studies involving a mechanism of secretion of acid sphingomyelinase, PKC zeta was found to perfectly colocalize with ceramide at the base of the primary cilia in MDCK cells [89]. The same authors showed that in embryonic stem cells, aPKC also interacted with ceramide during ciliogenesis. Surprisingly in this study, a strong correlation between anti-ceramide antibody staining and acetylated tubulin during mitotic spindle formation was reported [89].
• PKC delta and epsilon
Since PKC alpha was the first enzyme shown to be regulated by sphingolipids, and specifically inhibited by sphingosine, it was then also evaluated by ceramide. In this early work, it had already been shown that ceramide had no effect on in vitro activity of PKC alpha [18, 19]. On the other hand, Huwiler et al showed that PKC alpha was able to bind to TID-ceramide and responded to ceramide by translocating to the plasma membrane one hour after stimulation [90].
In this same study, the authors found that TID-ceramide also bound to PKC delta, but not to the zeta or epsilon isoforms of PKC. Furthermore, TID-ceramide had different effects on PKC alpha and delta. Ceramide increased activity of the alpha isoform whereas it decreased authophosphorylation of the delta isoform [90]. Although no translocation was observed for delta in this previous study, another group described that the delta and epsilon isoforms responded to exogenous ceramide (C2-ceramide 10μM 2 min-60 min), reverse translocating from the plasma membrane to the cytosol within 5 min after ceramide treatment. Most importantly, generation of endogenous ceramide using sphingomyelinase, TNF alpha and FasL, also resulted in translocation of these isoforms [91]. Studies that aimed to identify the ceramide binding site were again based on the cysteine rich domain of PKCs. In these studies, the authors found that the C1B domain in delta and epsilon PKCs was required for translocation of PKC to the Golgi apparatus in response of ceramide stimulation (C6-ceramide 10 μM 20–60 min), whereas cells stimulated with DAG or phorbol esters resulted in translocation to the plasma membrane [87].
In summary, the contradictory studies on PKCs binding ceramide, might be due to additional requirements for regulation of this interaction, as shown previously for different cell lines. The promising theory of ceramide binding C1 domains in PKCs and other C1-containing proteins, seemed to lose strength by the finding of ceramide binding the most C-terminal region, and not the atypical C1 domain of PKC zeta. Moreover, the need of C1 domains in ceramide-induced translocation of delta and epsilon isoforms in vivo but TID-ceramide only binding delta but not epsilon nor zeta isoforms, suggests in vivo regulation of these PKCs by ceramide, but questions a biological significance of direct interaction through the C1 domain, which could occur in vitro, but might not be the ceramide binding motif in vivo.
• EGFR-Ras-Raf-MEK signaling pathway
○ Kinase suppressor of Ras
The Kinase suppressor of Ras (KSR), together with CAPPs and PKC zeta, was described early on as an effector of ceramide signaling. Ceramide was found to induce epidermal growth factor receptor phosphorylation, and kinase activity towards an EGFR peptide was increased in cell lysates upon addition of exogenous ceramide. These findings prompted a search for a ceramide activated protein kinase [56]. This ceramide-activated protein kinase (CAP kinase) was a membrane-associated, proline-directed, serine/threonine kinase, and sequential chromatography found it to be 97KDa in size. Ceramide increased the 97KDa protein kinase in vitro activity and TNF alpha increased the activity in cellulo [57]. It was then suggested that TNF alpha signaling pathway resulting in ERK activation required ceramide generated from the sphingomyelin hydrolysis. Sphingomyelin hydrolysis was required for Raf-1 phosphorylation, which in turn triggered ERK signaling, moving CAP Kinase substrate to Raf-1, and identifying the Kinase Suppressor of Ras (KSR) as the CAP Kinase [29, 92]. This system might be more complicated since KSR is now known not to have kinase activity but to rather function as a scaffolding protein enhancing Raf/MAPK signaling [93, 94]. Very interesting, recombinant KSR and Raf-1 co-immunoprecipitation dramatically increased in the presence of ceramide (C2-ceramide 50μM). Natural ceramides were also efficient on these in vitro assays, where other lipids (DAG, PS, PC) failed in the reconstitution between Raf and KSR [29]. Therefore, this model could suggest that ceramide, rather than directly activating the “enzyme”, could modulate the interaction with other partners. Similarly, PKCs and CAPPs are heavily regulated by binding partners, and ceramide could have similar functions regulating these enzymes.
In 1998, considering how phorbol esters and DAG interacted with the cysteine rich domain (CDR) in classical and novel PKCs, Wim J. Van Blitterswijk hypothesized that the atypical CDR domain of atypical PKC, Raf and KSR could bind ceramide. Previously, these atypical-CDR-containing proteins were reported to bind or being activated by ceramide, it was therefore possible that instead of binding DAG, these analogous CDRs were binding ceramide [95]. Raf-1 and aPKC contain CDR domains. In cells, protein 14–3-3 blocks the CDR in Raf-1 and PAR-4 in aPKCs, in this paper it was also hypothesized that release of these factors would allow the binding of ceramide and activation of Raf-1 and aPKCs. In 2009 the domain A3 of KSR1 was tested to bind ceramide. KSR1-CA3 domain is homologous to the atypical C1 domain of PKC zeta. Using an ELISA assay, it was shown that purified GST-KSR1-CA3 bound ceramide but no other control lipids, including sphingomyelin, DAG, dihydroceramide, PC and GM1. Mutation of two cysteines on this domain attenuated ceramide binding and blocked translocation to the plasma membrane of the full length protein in response to EGF, where it was found to colocalize with ceramide antibody [96].
○ Raf-1
Motivated by recent studies on ceramide production linked to MAPK pathway activation in cells, Huwiler et al. were the first to fish for CBPs using a radioiodinated photoaffinity labeling analog of ceramide ([125I]-TID-ceramide). After immunoprecipitation of several components of the MAPK pathway, only Raf-1 was found to be crosslinked to the ceramide analogue. In vitro evaluation using Immunoprecipitated Raf-1, also showed activation of Raf-1 kinase activity by C6- and C16- ceramides [62].
Subsequent studies from the same group used TID-ceramide to identify other ceramide binding proteins. These were PKC alpha and delta (but not epsilon or zeta) [90], cPLA2 and Cathepsin D [97]. Interestingly, Raf1, PKCs, and cPLA2 interactions were identified in domains with homology to the C1 domain of PKCs. Importantly, the authors described this interaction as cell type dependent. For instance, the ceramide analogue binds cRaf and MEKK1, but when endothelial and mesangial cells are compared, Raf-1 binds ceramide only in mesangial, but not in endothelial cells, whereas MEKK1 binds ceramide only in endothelial cells. The interaction is therefore much more complex, as new players are brought to the picture, establishing a connection to the hypothesis postulated by Blitterswijk, where PAR-4, 14–3-3 or other factors such as Hsp70 and Hsp90 regulate the interaction to ceramide [55].
• Annexin A1
Annexins are Ca2+ sensitive proteins that bind negatively charged phospholipids, and they are involved in different biologies including trafficking, permeability, and fusion of membranes. In 2008, Draeger’s laboratory found that Annexin A1 bound membranes enriched in ceramides, whereas other members of the annexin family did not. The interaction of Annexin A1 with ceramide-enriched membranes was Ca2+ dependent, and ceramide decreased the amount of Ca2+ needed for this interaction. However, ceramide alone was not sufficient for the binding to occur, since the presence of phospholipids was still a requirement. The authors proposed Annexin A1 as a tool to stain (visualize) ceramide enriched membranes in cells, and a ceramide antibody was used to validate ceramide staining. Since this interaction depends on Ca2+, Jurkat cells were treated with Ca2+/ionomycin for 30 minutes, and discrete regions on the plasma membrane presented perfect localization between ceramide antibody and Annexin A1-GFP [98]. The same group found this interaction useful to study plasma membrane repair after perforation by toxins that result in an increase of intracellular calcium [99]. Using Annexin–GFP to stain ceramide platforms, the authors also concluded that, during apoptosis, plasma membrane ceramide is transferred by direct contact to mitochondria in a process they named “kiss-of-death” (see figure 4) [100].
Figure 4.

Visualization of ceramide in mitochondria in response to increased levels of intracellular calcium. [From Babichuck et al. 2011].
• Cathepsin D
Cathepsin is an aspartyl protease that functions in the degradation of lysosomal protein cargo and in the activation of precursors of active proteins in lysosomal compartments. In 1999, Schütze’s lab focused its attention on the putative activation of proteases by ceramide during apoptosis [101]. To investigate the interaction between ceramide and proteases, the authors studied the interaction of lysate proteins from U937 cells with a ceramide (C6-ceramide) affinity sepharose column. Contrary to previous hypotheses, caspases 2 and 3 were not detected, but cathepsin D did bind ceramide. In vitro assays using TID-ceramide and recombinant cathepsin D showed that the interaction was specific for ceramide and to a lesser degree for sphingosine, but no binding was detected for dihydro forms of ceramide, DAG, sphingomyelin, or palmitate. Moreover, ceramide interaction with Cathepsin D resulted in its activation by autocatalytic proteolysis. In agreement with their finding, the lack of lysosomal ceramide in cells without acid sphingomyelinase was associated with decreased activity of cathepsin D [31]. No further studies on the ceramide binding site of Cathepsin D have been provided.
• LAPTM4
In 2015, Tom et al. found that the Lysosome Associated Protein Transmembrane 4B (LAPTM4B) regulated ceramide levels in late endosomes. Using fluorescent and the cross linkable analogue, ceramide-BP-Bodipy, it was concluded that LAPTM4B protein was interacting with ceramide [102]. By studying the binding of protein p24 to C18-sphingomyelin, the authors were able to suggest a putative motif for this interaction ([V/I/T/L]XX[[V/I/T/ L][V/I/T/L]XX[V/I/T/L][F/W/Y])[103]. Screening for proteins containing this motif (other filters were applied), 615 unique proteins were found to contain this motif. Almost 50% of these were plasma membrane proteins, and around 20% were GPCRs. In 2018, Zhou K and col. found that a putative sphingolipid binding motif was present in two of the four transmembrane regions of the LAPTM4B protein, and one of these transmembrane domains was able to bind the fluorescent ceramide-bodipy, acting as a ceramide-regulated element in the endosomal protein. t is not clear how the sphingomyelin motif that the authors identified was used for ceramide binding. This motif should be further explored and validated for in vitro and in vivo studies, in order to define the specificity of ceramide vs sphingomyelin.
Summary and Conclusions
Ceramides have been established as bioactive lipids, and many biologies mediated or triggered by ceramide have been reported. These include cell death, cell differentiation, inflammation, and cell motility [1]. Different methodologies have been employed to support the hypothesis of ceramides as bioactive lipids. The majority of the studies involving ceramide have used exogenous ceramides, often short chains, or have exogenously applied sphingomyelinases that hydrolyze sphingomyelin in the plasma membrane to produce ceramide [79]. Physiological production of ceramide has been achieved by stimulating the cell with cytokines such as TNF, CD95L or IL-1β shown to activate hydrolysis of sphingomyelin, ultimately producing ceramides [7]. More recently, cells have been treated with nanoliposomes containing the desired species of ceramides [104]. Expression and regulation of enzymes in the ceramide metabolism pathway have also been used to manipulate ceramide levels [5, 105]. For instance, acid ceramidase, an enzyme responsible for the hydrolysis of ceramide, has been shown to be involved in prostate cancer cells’ acquired resistance to radiation therapy. Pharmacological inhibition of these enzymes resulted in ceramide accumulation and re-sensitized cancer cells to death upon radiation [106].
Although ceramides have been related to many biologies, not much is known about the molecular mechanisms of action of ceramide. There are still many unanswered questions such as 1. Is there a conserved ceramide binding motif? It is difficult not to make a reasonable comparison to DAG and PKCs. Ceramide and DAG are structurally similar, and both act as a hubs in the metabolism of other bioactive and structural lipids. They even share parallel chemical modification and, often, functions (SM/PC, C1P/PA, S1P/LPA, etc). This similarity led to the idea that C1 domains from atypical PKC could be binding ceramide instead of DAG. Even some novel PKCs could show ceramide binding in addition to binding DAG. Although some studies have shown binding of ceramide to novel PKCs, it is not clear if this could be sufficient to activate PKC in vivo, or if this mechanism is actually real. Moreover, Bieberich group found that it is not the C1 domain of PKC zeta that binds ceramide, rather, it is the last C-terminal region of the protein. Other ceramide binding proteins contain versions of this C1 domain, but it is still not clear if this is a specific interaction or simply the result of unspecific binding due to the structural similarity to DAG. In that sense, there are no crystal structures showing ceramide bound to any C1 domain. Activation of PPPs is supported by multiple studies, but these have not yet identified binding domains. In the case of PP2A, the catalytic subunit is activated by ceramide, but this activation is much more dramatic when the dimeric and trimeric forms are stimulated with ceramide. Since not all dephosphorylations downstream of PP1/PP2A are stimulated by ceramide, and catalytic subunits partner in regulatory subunits, rather than existing as free monomers, the idea of ceramide requiring both (or more) subunits gains strength. From the different proteomic screenings on ceramide binding proteins, there are no studies that aim to identify common motifs, which could be useful to find a ceramide binding motif. Such proteomics studies have to be improved, since they still have several issues (metabolism of the probes, proper controls, validations, etc) probably as a result of too much background of false positives. 2. Are different species of ceramide acting on different targets? Recently the bioactive roles of ceramides have been reviewed, proposing the ‘Many ceramide’ hypothesis, where ceramide cannot be treated as a unique entity, but each species of ceramide could have specific functions [4]. The vast majority of studies on ceramide signaling still consider ceramide as an entity, whereas just a few studies have started to point to single species as key ceramide species in certain biologies [107, 108]. These studies are still in early stages, and certainly not sufficient to decipher how important the ceramide acyl chain length and saturation degree are in signaling. Moreover, it is still not clear if a specific species is responsible for a specific biology because of its structure, or a result of casually being found in the right compartment. 3. Where are these bioactive pools located in the cell? Ceramide is found in all subcellular membranes, and most of these different pools of ceramide have been proved to trigger signaling. Classically, increases in ceramide are measured in the whole cell lysate, causing all information from different pools to get lost. Furthermore, possible local changes in active ceramide might not be detected by dilution of larger non-active pools or active towards other biologies. To answer the question of where the bioactive pools are, we need to develop techniques to detect ceramide in cells and in sub-cellular compartments. There are some clues suggesting that very little ceramide is present in the plasma membrane, but this dramatically increase during signaling [79]. Fluorescent and exogenous ceramides are rapidly metabolized and accumulate in the Golgi apparatus, making it very difficult to visualize plasma membrane pools. Only CBPs have been selectively localized in specific membranes in response to ceramide generation (plasma membrane, Golgi, mitochondria, and ER), and not staining any pool of cellular ceramide as ceramide antibodies or ceramide probes would do. However, only one or few reports describe these findings, and tools to visualize these pools of ceramide still need to be understood and developed.
In the last years, studies on ceramide and other sphingolipid binding partners using proteomics have started to appear in the literature. We expect more tuned studies with better controlled conditions in order to start defining which proteins show real affinity and specificity for ceramide. We need to know how proteins bind ceramide, and if it is possible to define and express ceramide binding domains. It would be ideal to have ceramide crosslinkable structures that cannot be metabolized to other members of the sphingolipid family, as well as methods that allow to identify the lipid-protein combo, assigning specific proteins to specific lipids. These studies should also allow us to distinguish between ceramide species and discern whether specific species with specific binding partners exist. The identification of new CBPs will also help differentiate between different subcellular bioactive pools of ceramide.
Highlights.
There are no available tools to visualize bioactive ceramide
Few Ceramide Binding Proteins (CBPs) have been reported.
There is a need to improve screening methods to identify larger lists of CBPs
CBPs could lead to find a ceramide binding motif (or different ones)
These motifs could be used to visualize different bioactive pools of ceramide
Few CBPs have already been used to stain bioactive pools of ceramide localized in different compartments
Acknowledgements
This work was supported by NIH grant CA218678.
Abbreviations
- CAPP
Ceramide activated protein phosphatase
- CBP
Ceramide binding protein
- C1P
Ceramide 1 phosphate
- DAG
Diacylglycerol
- ER
Endoplasmic reticulum
- KSR
Kinase suppressor Ras
- LPA
Lysophosphatidic acid
- PA
Phosphatidic acid
- PC
Phosphatidylcholine
- PKC
Protein kinase C
- PPP
Protein phosphatase PPP family
- PS
Phosphatidylserine
- TNF
Tumor necrosis factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hannun YA and Obeid LM, Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol, 2008. 9(2): p. 139–50. [DOI] [PubMed] [Google Scholar]
- 2.Hannun YA and Obeid LM, Sphingolipids and their metabolism in physiology and disease. Nature Reviews Molecular Cell Biology, 2018. 19(3): p. 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merrill AH Jr., Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem Rev, 2011. 111(10): p. 6387–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hannun YA and Obeid LM, Many ceramides. J Biol Chem, 2011. 286(32): p. 27855–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adada M, Luberto C, and Canals D, Inhibitors of the sphingomyelin cycle: Sphingomyelin synthases and sphingomyelinases. Chem Phys Lipids, 2016. 197: p. 45–59. [DOI] [PubMed] [Google Scholar]
- 6.Mullen TD, Hannun YA, and Obeid L, Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem J, 2012. 441(3): p. 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez-Corbacho MJ, et al. , Tumor Necrosis Factor-alpha (TNFalpha)-induced Ceramide Generation via Ceramide Synthases Regulates Loss of Focal Adhesion Kinase (FAK) and Programmed Cell Death. J Biol Chem, 2015. 290(42): p. 25356–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaipia A, et al. , Tumor necrosis factor-alpha and its second messenger, ceramide, stimulate apoptosis in cultured ovarian follicles. Endocrinology, 1996. 137(11): p. 4864–70. [DOI] [PubMed] [Google Scholar]
- 9.Becker KP, et al. , Selective inhibition of juxtanuclear translocation of protein kinase C betaII by a negative feedback mechanism involving ceramide formed from the salvage pathway. J Biol Chem, 2005. 280(4): p. 2606–12. [DOI] [PubMed] [Google Scholar]
- 10.Kitatani K, Idkowiak-Baldys J, and Hannun YA, The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell Signal, 2008. 20(6): p. 1010–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perry DM, et al. , Defining a role for acid sphingomyelinase in the p38/interleukin-6 pathway. J Biol Chem, 2014. 289(32): p. 22401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wada M, et al. , P38 delta MAPK promotes breast cancer progression and lung metastasis by enhancing cell proliferation and cell detachment. Oncogene, 2017. 36(47): p. 6649–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Bawab S, et al. , Biochemical characterization of the reverse activity of rat brain ceramidase. A CoA-independent and fumonisin B1-insensitive ceramide synthase. J Biol Chem, 2001. 276(20): p. 16758–66. [DOI] [PubMed] [Google Scholar]
- 14.Novgorodov SA, et al. , Novel pathway of ceramide production in mitochondria: thioesterase and neutral ceramidase produce ceramide from sphingosine and acyl-CoA. J Biol Chem, 2011. 286(28): p. 25352–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, et al. , JNK3 signaling pathway activates ceramide synthase leading to mitochondrial dysfunction. J Biol Chem, 2007. 282(35): p. 25940–9. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Corbacho MJ, et al. , Sphingolipids in mitochondria. Biochim Biophys Acta, 2017. 1862(1): p. 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajagopalan V, et al. , Critical determinants of mitochondria-associated neutral sphingomyelinase (MA-nSMase) for mitochondrial localization. Biochim Biophys Acta, 2015. 1850(4): p. 628–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannun YA, et al. , Sphingosine inhibition of protein kinase C activity and of phorbol dibutyrate binding in vitro and in human platelets. J Biol Chem, 1986. 261(27): p. 12604–9. [PubMed] [Google Scholar]
- 19.Hannun YA and Bell RM, Functions of sphingolipids and sphingolipid breakdown products in cellular regulation. Science, 1989. 243(4890): p. 500–7. [DOI] [PubMed] [Google Scholar]
- 20.Obeid LM, et al. , Programmed cell death induced by ceramide. Science, 1993. 259(5102): p. 1769–71. [DOI] [PubMed] [Google Scholar]
- 21.Jones J and Hannun Y, Ceramide: Cell Regulation from a Lipid Perspective. . Signaling Pathways in Liver Disease, ed. Calvien J.-F.D.a.P.-A.. Vol. 30 2005: Springer, Heidelberg. [Google Scholar]
- 22.Hannun YA, Bioactive sphingolipids in cancer biology and therapy 2015, New York, NY: Springer Science+Business Media; pages cm. [Google Scholar]
- 23.Birbes H, et al. , Selective hydrolysis of a mitochondrial pool of sphingomyelin induces apoptosis. FASEB J, 2001. 15(14): p. 2669–79. [DOI] [PubMed] [Google Scholar]
- 24.van Blitterswijk WJ, et al. , Ceramide: second messenger or modulator of membrane structure and dynamics? Biochem J, 2003. 369(Pt 2): p. 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holopainen JM, Angelova MI, and Kinnunen PK, Vectorial budding of vesicles by asymmetrical enzymatic formation of ceramide in giant liposomes. Biophys J, 2000. 78(2): p. 830–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trajkovic K, et al. , Ceramide triggers budding of exosome vesicles into multivesicular Endosomes. Science, 2008. 319(5867): p. 1244–1247. [DOI] [PubMed] [Google Scholar]
- 27.Colombini M, Ceramide channels and mitochondrial outer membrane permeability. J Bioenerg Biomembr, 2017. 49(1): p. 57–64. [DOI] [PubMed] [Google Scholar]
- 28.Dobrowsky RT and Hannun YA, Ceramide stimulates a cytosolic protein phosphatase. J Biol Chem, 1992. 267(8): p. 5048–51. [PubMed] [Google Scholar]
- 29.Zhang Y, et al. , Kinase suppressor of Ras is ceramide-activated protein kinase. Cell, 1997. 89(1): p. 63–72. [DOI] [PubMed] [Google Scholar]
- 30.Wang YM, et al. , Atypical PKC zeta is activated by ceramide, resulting in coactivation of NF-kappaB/JNK kinase and cell survival. J Neurosci Res, 1999. 55(3): p. 293–302. [DOI] [PubMed] [Google Scholar]
- 31.Heinrich M, et al. , Cathepsin D targeted by acid sphingomyelinase-derived ceramide. EMBO J, 1999. 18(19): p. 5252–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Resh MD, Membrane targeting of lipid modified signal transduction proteins. Subcell Biochem, 2004. 37: p. 217–32. [DOI] [PubMed] [Google Scholar]
- 33.Bollinger CR, Teichgraber V, and Gulbins E, Ceramide-enriched membrane domains. Biochim Biophys Acta, 2005. 1746(3): p. 284–94. [DOI] [PubMed] [Google Scholar]
- 34.Oresic M, Informatics and computational strategies for the study of lipids. Biochimica Et Biophysica Acta-Molecular and Cell Biology of Lipids, 2011. 1811(11): p. 991–999. [DOI] [PubMed] [Google Scholar]
- 35.Bielawski J, et al. , Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods, 2006. 39(2): p. 82–91. [DOI] [PubMed] [Google Scholar]
- 36.Smith PBW, Snyder AP, and Harden CS, Characterization of Bacterial Phospholipids by Electrospray-Ionization Tandem Mass-Spectrometry. Analytical Chemistry, 1995. 67(11): p. 1824–1830. [DOI] [PubMed] [Google Scholar]
- 37.Murate M and Kobayashi T, Revisiting transbilayer distribution of lipids in the plasma membrane. Chem Phys Lipids, 2016. 194: p. 58–71. [DOI] [PubMed] [Google Scholar]
- 38.Folch J, Lees M, and Stanley GHS, A Simple Method for the Isolation and Purification of Total Lipides from Animal Tissues. Journal of Biological Chemistry, 1957. 226(1): p. 497–509. [PubMed] [Google Scholar]
- 39.Sourkes TL, Thudichum’s successors. Neurochemical Research, 2007. 32(10): p. 1808–1812. [DOI] [PubMed] [Google Scholar]
- 40.Maier O, Oberle V, and Hoekstra D, Fluorescent lipid probes: some properties and applications (a review). Chemistry and Physics of Lipids, 2002. 116(1–2): p. 3–18. [DOI] [PubMed] [Google Scholar]
- 41.Lipsky NG and Pagano RE, Sphingolipid metabolism in cultured fibroblasts: microscopic and biochemical studies employing a fluorescent ceramide analogue. Proc Natl Acad Sci U S A, 1983. 80(9): p. 2608–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Izquierdo E and Delgado A, Click chemistry in sphingolipid research. Chem Phys Lipids, 2018. [DOI] [PubMed]
- 43.Krishnamurthy K, Dasgupta S, and Bieberich E, Development and characterization of a novel anti-ceramide antibody. J Lipid Res, 2007. 48(4): p. 968–75. [DOI] [PubMed] [Google Scholar]
- 44.Cowart LA, et al. , Structural determinants of sphingolipid recognition by commercially available anti-ceramide antibodies. J Lipid Res, 2002. 43(12): p. 2042–8. [DOI] [PubMed] [Google Scholar]
- 45.Glycobiotech. Monoclonal antibody agains ceramide Available from: http://glycobiotech.de/mouse-monoclonal-antibody-against-ceramide.html.
- 46.Rotolo J, et al. , Anti-ceramide antibody prevents the radiation gastrointestinal syndrome in mice. J Clin Invest, 2012. 122(5): p. 1786–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hullin-Matsuda F, et al. , Imaging Lipid Membrane Domains with Lipid-Specific Probes. Lipidomics, Vol 2: Methods and Protocols, 2009. 580: p. 203–220. [DOI] [PubMed] [Google Scholar]
- 48.Yamaji A, et al. , Lysenin, a novel sphingomyelin-specific binding protein. J Biol Chem, 1998. 273(9): p. 5300–6. [DOI] [PubMed] [Google Scholar]
- 49.Fishman PH, Role of membrane gangliosides in the binding and action of bacteria toxins. J Membr Biol, 1982. 69(2): p. 85–97. [DOI] [PubMed] [Google Scholar]
- 50.Ueda Y and Hayashi Y, PIP3 Regulates Spinule Formation in Dendritic Spines during Structural Long-Term Potentiation. Journal of Neuroscience, 2013. 33(27): p. 11040–11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato M, Ueda Y, and Umezawa Y, Imaging diacylglycerol dynamics at organelle membranes. Nature Methods, 2006. 3(10): p. 797–799. [DOI] [PubMed] [Google Scholar]
- 52.Chalfant CE, et al. , Long chain ceramides activate protein phosphatase-1 and protein phosphatase-2A. Activation is stereospecific and regulated by phosphatidic acid. J Biol Chem, 1999. 274(29): p. 20313–7. [DOI] [PubMed] [Google Scholar]
- 53.Dobrowsky RT and Hannun YA, Ceramide-activated protein phosphatase: partial purification and relationship to protein phosphatase 2A. Adv Lipid Res, 1993. 25: p. 91–104. [PubMed] [Google Scholar]
- 54.Lozano J, et al. , Protein kinase C zeta isoform is critical for kappa B-dependent promoter activation by sphingomyelinase. J Biol Chem, 1994. 269(30): p. 19200–2. [PubMed] [Google Scholar]
- 55.Huwiler A, et al. , Differential binding of ceramide to MEKK1 in glomerular endothelial and mesangial cells. Biochim Biophys Acta, 2004. 1636(2–3): p. 159–68. [DOI] [PubMed] [Google Scholar]
- 56.Mathias S, Dressler KA, and Kolesnick RN, Characterization of a ceramide-activated protein kinase: stimulation by tumor necrosis factor alpha. Proc Natl Acad Sci U S A, 1991. 88(22): p. 10009–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, et al. , Renaturation and tumor necrosis factor-alpha stimulation of a 97-kDa ceramide-activated protein kinase. J Biol Chem, 1994. 269(4): p. 3047–52. [PubMed] [Google Scholar]
- 58.Kota V, Szulc ZM, and Hama H, Identification of C6-ceramide-interacting proteins in D6P2T Schwannoma cells. Proteomics, 2012. 12(13): p. 2179–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bidlingmaier S, et al. , Proteome-wide Identification of Novel Ceramide-binding Proteins by Yeast Surface cDNA Display and Deep Sequencing. Molecular & Cellular Proteomics, 2016. 15(4): p. 1232–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bockelmann S, et al. , A search for ceramide binding proteins using bifunctional lipid analogs yields CERT-related protein StarD7. J Lipid Res, 2018. 59(3): p. 515–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bieberich E, Lipid vesicle-mediated affinity chromatography using magnetic activated cell sorting (LIMACS): a novel method to analyze protein-lipid interaction. J Vis Exp, 2011(50). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huwiler A, et al. , Ceramide-binding and activation defines protein kinase c-Raf as a ceramide-activated protein kinase. Proc Natl Acad Sci U S A, 1996. 93(14): p. 6959–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elsen L, et al. , Identification of ceramide binding proteins in neuronal cells: a critical point of view. Neurochem Res, 2002. 27(7–8): p. 717–27. [DOI] [PubMed] [Google Scholar]
- 64.Gerl MJ, et al. , Sphingosine-1-Phosphate Lyase Deficient Cells as a Tool to Study Protein Lipid Interactions. PLoS One, 2016. 11(4): p. e0153009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haberkant P, et al. , Bifunctional Sphingosine for Cell-Based Analysis of Protein-Sphingolipid Interactions. ACS Chem Biol, 2016. 11(1): p. 222–30. [DOI] [PubMed] [Google Scholar]
- 66.Reyes JG, et al. , c-Jun is a downstream target for ceramide-activated protein phosphatase in A431 cells. J Biol Chem, 1996. 271(35): p. 21375–80. [DOI] [PubMed] [Google Scholar]
- 67.Kowluru A and Metz SA, Ceramide-activated protein phosphatase-2A activity in insulin-secreting cells. FEBS Lett, 1997. 418(1–2): p. 179–82. [DOI] [PubMed] [Google Scholar]
- 68.Chalfant CE and Hannun YA, The role of serine/threonine protein phosphatases in ceramide signaling. Ceramide Signaling, ed. Futerman AH. Vol. 49–61 2002, Georgetown, TX: Landes Bioscience. [Google Scholar]
- 69.Lee JY, Hannun YA, and Obeid LM, Ceramide inactivates cellular protein kinase Calpha. J Biol Chem, 1996. 271(22): p. 13169–74. [DOI] [PubMed] [Google Scholar]
- 70.Wolff RA, et al. , Role of ceramide-activated protein phosphatase in ceramide-mediated signal transduction. J Biol Chem, 1994. 269(30): p. 19605–9. [PubMed] [Google Scholar]
- 71.Ruvolo PP, et al. , Ceramide induces Bcl2 dephosphorylation via a mechanism involving mitochondrial PP2A. J Biol Chem, 1999. 274(29): p. 20296–300. [DOI] [PubMed] [Google Scholar]
- 72.Basu S, et al. , BAD enables ceramide to signal apoptosis via Ras and Raf-1. J Biol Chem, 1998. 273(46): p. 30419–26. [DOI] [PubMed] [Google Scholar]
- 73.Chalfant CE, et al. , Analysis of ceramide-activated protein phosphatases. Methods Enzymol, 2000. 312: p. 420–8. [DOI] [PubMed] [Google Scholar]
- 74.Chalfant CE, et al. , FAS activation induces dephosphorylation of SR proteins; dependence on the de novo generation of ceramide and activation of protein phosphatase 1. J Biol Chem, 2001. 276(48): p. 44848–55. [DOI] [PubMed] [Google Scholar]
- 75.Kitatani K, et al. , Protein kinase C-induced activation of a ceramide/protein phosphatase 1 pathway leading to dephosphorylation of p38 MAPK. J Biol Chem, 2006. 281(48): p.36793–802. [DOI] [PubMed] [Google Scholar]
- 76.Marchesini N, Jones JA, and Hannun YA, Confluence induced threonine41/serine45 phospho-beta-catenin dephosphorylation via ceramide-mediated activation of PP1cgamma. Biochim Biophys Acta, 2007. 1771(12): p. 1418–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeidan YH, Jenkins RW, and Hannun YA, Remodeling of cellular cytoskeleton by the acid sphingomyelinase/ceramide pathway. J Cell Biol, 2008. 181(2): p. 335–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Canals D, Roddy P, and Hannun YA, Protein phosphatase 1alpha mediates ceramide-induced ERM protein dephosphorylation: a novel mechanism independent of phosphatidylinositol 4, 5-biphosphate (PIP2) and myosin/ERM phosphatase. J Biol Chem, 2012. 287(13): p. 10145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Canals D, et al. , Differential effects of ceramide and sphingosine 1-phosphate on ERM phosphorylation: probing sphingolipid signaling at the outer plasma membrane. J Biol Chem, 2010. 285(42): p. 32476–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matmati N, et al. , Identification of C18:1-phytoceramide as the candidate lipid mediator for hydroxyurea resistance in yeast. J Biol Chem, 2013. 288(24): p. 17272–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Teixeira V, et al. , Ceramide signaling targets the PP2A-like protein phosphatase Sit4p to impair vacuolar function, vesicular trafficking and autophagy in Isc1p deficient cells. Biochim Biophys Acta, 2016. 1861(1): p. 21–33. [DOI] [PubMed] [Google Scholar]
- 82.Mukhopadhyay A, et al. , Direct interaction between the inhibitor 2 and ceramide via sphingolipid-protein binding is involved in the regulation of protein phosphatase 2A activity and signaling. FASEB J, 2009. 23(3): p. 751–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kitatani K, Idkowiak-Baldys J, and Hannun YA, Mechanism of inhibition of sequestration of protein kinase C alpha/betaII by ceramide. Roles of ceramide-activated protein phosphatases and phosphorylation/dephosphorylation of protein kinase C alpha/betaII on threonine 638/641. J Biol Chem, 2007. 282(28): p. 20647–56. [DOI] [PubMed] [Google Scholar]
- 84.Kim MY, et al. , Identification of sphingomyelin turnover as an effector mechanism for the action of tumor necrosis factor alpha and gamma-interferon. Specific role in cell differentiation. J Biol Chem, 1991. 266(1): p. 484–9. [PubMed] [Google Scholar]
- 85.Fox TE, et al. , Ceramide recruits and activates protein kinase C zeta (PKC zeta) within structured membrane microdomains. J Biol Chem, 2007. 282(17): p. 12450–7. [DOI] [PubMed] [Google Scholar]
- 86.Wang G, et al. , Direct binding to ceramide activates protein kinase Czeta before the formation of a pro-apoptotic complex with PAR-4 in differentiating stem cells. J Biol Chem, 2005. 280(28): p. 26415–24. [DOI] [PubMed] [Google Scholar]
- 87.Kashiwagi K, et al. , Importance of C1B domain for lipid messenger-induced targeting of protein kinase C. J Biol Chem, 2002. 277(20): p. 18037–45. [DOI] [PubMed] [Google Scholar]
- 88.Wang G, et al. , The carboxyl-terminal domain of atypical protein kinase Czeta binds to ceramide and regulates junction formation in epithelial cells. J Biol Chem, 2009. 284(21): p. 14469–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He Q, et al. , Primary cilia in stem cells and neural progenitors are regulated by neutral sphingomyelinase 2 and ceramide. Mol Biol Cell, 2014. 25(11): p. 1715–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huwiler A, Fabbro D, and Pfeilschifter J, Selective ceramide binding to protein kinase C-alpha and -delta isoenzymes in renal mesangial cells. Biochemistry, 1998. 37(41): p. 14556–62. [DOI] [PubMed] [Google Scholar]
- 91.Sawai H, et al. , Ceramide-induced translocation of protein kinase C-delta and -epsilon to the cytosol. Implications in apoptosis. J Biol Chem, 1997. 272(4): p. 2452–8. [DOI] [PubMed] [Google Scholar]
- 92.Yao B, et al. , Phosphorylation of Raf by ceramide-activated protein kinase. Nature, 1995. 378(6554): p. 307–10. [DOI] [PubMed] [Google Scholar]
- 93.Morrison DK, KSR: a MAPK scaffold of the Ras pathway? J Cell Sci, 2001. 114(Pt 9): p. 1609–12. [DOI] [PubMed] [Google Scholar]
- 94.Claperon A and Therrien M, KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene, 2007. 26(22): p. 3143–58. [DOI] [PubMed] [Google Scholar]
- 95.van Blitterswijk WJ, Hypothesis: ceramide conditionally activates atypical protein kinases C, Raf-1 and KSR through binding to their cysteine-rich domains. Biochem J, 1998. 331 (Pt 2): p. 679–80. [PMC free article] [PubMed] [Google Scholar]
- 96.Yin X, et al. , A ceramide-binding C1 domain mediates kinase suppressor of ras membrane translocation. Cell Physiol Biochem, 2009. 24(3–4): p. 219–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huwiler A, et al. , Ceramide binds to the CaLB domain of cytosolic phospholipase A2 and facilitates its membrane docking and arachidonic acid release. FASEB J, 2001. 15(1): p. 7–9. [DOI] [PubMed] [Google Scholar]
- 98.Babiychuk EB, Monastyrskaya K, and Draeger A, Fluorescent annexin A1 reveals dynamics of ceramide platforms in living cells. Traffic, 2008. 9(10): p. 1757–75. [DOI] [PubMed] [Google Scholar]
- 99.Babiychuk EB, et al. , Intracellular Ca(2+) operates a switch between repair and lysis of streptolysin O-perforated cells. Cell Death Differ, 2009. 16(8): p. 1126–34. [DOI] [PubMed] [Google Scholar]
- 100.Walev I, et al. , Streptolysin O-permeabilized granulocytes shed L-selectin concomitantly with ceramide generation via neutral sphingomyelinase. J Leukoc Biol, 2000. 68(6): p.865–72. [PubMed] [Google Scholar]
- 101.Gamen S, et al. , CPP32 inhibition prevents Fas-induced ceramide generation and apoptosis in human cells. FEBS Lett, 1996. 390(2): p. 232–7. [DOI] [PubMed] [Google Scholar]
- 102.Blom T, et al. , LAPTM4B facilitates late endosomal ceramide export to control cell death pathways. Nat Chem Biol, 2015. 11(10): p. 799–806. [DOI] [PubMed] [Google Scholar]
- 103.Contreras FX, et al. , Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature, 2012. 481(7382): p. 525–9. [DOI] [PubMed] [Google Scholar]
- 104.Kitatani K, et al. , Ceramide limits phosphatidylinositol-3-kinase C2beta-controlled cell motility in ovarian cancer: potential of ceramide as a metastasis-suppressor lipid. Oncogene, 2016. 35(21): p. 2801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Canals D, et al. , Drug targeting of sphingolipid metabolism: sphingomyelinases and ceramidases. Br J Pharmacol, 2011. 163(4): p. 694–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Holman DH, et al. , Lysosomotropic acid ceramidase inhibitor induces apoptosis in prostate cancer cells. Cancer Chemother Pharmacol, 2008. 61(2): p. 231–42. [DOI] [PubMed] [Google Scholar]
- 107.Wang Z, et al. , Overexpression of ceramide synthase 1 increases C18-ceramide and leads to lethal autophagy in human glioma. Oncotarget, 2017. 8(61): p. 104022–104036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grammatikos G, et al. , Serum sphingolipidomic analyses reveal an upregulation of C16-ceramide and sphingosine-1-phosphate in hepatocellular carcinoma. Oncotarget, 2016. 7(14): p. 18095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]


