Abstract
Genetically triggered thoracic aortic aneurysms (TAAs) account for 30% of all TAAs and can result in early morbidity and mortality in affected individuals. Epigenetic factors are now recognised to influence the phenotype of many genetically triggered conditions and have become an area of interest because of the potential for therapeutic manipulation. Major epigenetic modulators include DNA methylation, histone modification and non-coding RNA. This review examines epigenetic modulators that have been significantly associated with genetically triggered TAAs and their potential utility for translation to clinical practice.
Keywords: Thoracic aortic aneurysm, Aortic dilatation, Epigenetics, Non-coding RNA, MicroRNA
Introduction
Thoracic aortic aneurysm (TAA) is a genetically and phenotypically diverse condition characterised by the progressive permanent dilatation of the thoracic aorta, predisposing to aortic rupture and dissection (Goldfinger et al. 2014). TAA is predominantly clinically silent and develops on a background of adverse remodelling of the aortic wall, which may be inherited or acquired.
Genetically triggered TAAs account for up to 30% of all TAAs (Clouse et al. 1998; Albornoz et al. 2006) and are often consequent upon pathological variants in genes encoding key proteins in either vascular smooth muscle cells (VSMCs), the extracellular matrix (ECM) or transforming growth factor-beta (TGF-β) signalling. Non-genetic TAA is usually observed in older patients with comorbidities including hypertension and atherosclerosis (Goldfinger et al. 2014). Genetically triggered TAA is observed in the clinical syndromes of Marfan (MFS), Loeys-Dietz (LDS) and vascular Ehlers-Danlos (vEDS), and also in association with a bicuspid aortic valve (BAV). In other individuals, TAA may occur in the absence of other clinically discernible features (non-syndromal Thoracic Aortic Aneurysm and Dissection (nsTAAD)). Current medical therapy is variably effective at slowing the rate of dilatation, with increasing dimensions conferring an increase in the risk of dissection, which carries a mortality rate of up to 50% (Melvinsdottir et al. 2016). Most patients will ultimately require surgical repair of the aneurysm, with mortality risk ranging from 1 to 5% for elective repair (Kallenbach et al. 2013) and up to 12% for emergency intervention (Goldfinger et al. 2014).
Phenotype variability is well-documented in all forms of genetically triggered TAA and is particularly highlighted in cases where an identical pathogenic gene variant results in different clinical manifestations (De Backer et al. 2007; Loeys 2016). This phenotype variability significantly impacts the ability to predict clinical outcomes, resulting in uncertainty for both clinicians and patients. Therefore, there is a need to develop more sophisticated risk profiling that aims to provide a precision-medicine model for patient management.
Epigenetics: regulators of phenotype
The emerging characterisation of the “epigenome”, comprising endogenous mediators that regulate gene expression independent of the DNA sequence, has revolutionised our understanding of health and disease beyond traditional Mendelian genetics. “Epigenetics” refers to mechanisms of gene expression regulation by DNA methylation, histone modification and non-coding RNA, such as microRNA (miRNA) and long non-coding RNA (lncRNA) (Tollefsbol 2018). The activity of these mediators is dynamically influenced by environmental stimuli, leading to stable changes in gene expression that are maintained during cell division and can be passed through generations. Accordingly, aberrant epigenetic changes are associated with many disease states (Esteller 2011; Tollefsbol 2018). Characterisation of these changes has resulted in improved understanding of pathological disease mechanisms, as well as the development of disease-specific biological profiles. Importantly, epigenetic mediators have been demonstrated to be modifiable therapeutic targets (Conway et al. 2016), thus providing enormous clinical potential, with the capacity to improve diagnosis, monitoring and treatment of a wide range of diseases.
With epigenetics increasingly recognised in other genetic conditions as modulators of penetrance and expressivity (Feinberg 2007), there is a strong theoretical basis for these mechanisms also occurring in genetically triggered TAA. Detection of aberrant epigenetic changes in TAA will enable a deeper understanding of pathogenesis and may provide explanation for the observed phenotype variation among individuals. In turn, such knowledge may also assist the clinical decision-making process with regard to the optimal time to undertake surgical repair and also provide a stronger mechanistic basis for new drug design towards slowing progression and mitigate the need for surgery. This review provides an update on epigenetic changes found to be associated with genetically triggered TAA, with a summary of progress in translation to clinical practice.
Pathogenesis of thoracic aortic aneurysm
Normal aortic wall architecture
The aortic wall consists of three distinct layers: the outermost adventitia, which contains connective tissue and ECM for tensile support, in addition to local blood and neural supply; the media, which consists of contractile lamellar units comprised of concentric layers of contractile VSMCs interspersed between elastic fibres and surrounded by ECM; and the intima, containing the endothelial surface and subendothelial connective tissue. Collectively, these components enable the aortic wall to tolerate the large changes in pressure that are associated with ventricular contraction, with graded contraction of the VSMCs responsible for reducing the highly pulsatile ventricular flow to a steady, continuous propagation through the vasculature (Belz and Belz 1995). The mechanical properties of the aorta are also maintained through diverse signalling processes within and between the three aortic wall layers, including cell-cell and cell-ECM interactions, mechanotransduction complexes that sense and communicate aortic stretch and responses to external signals, such as neural and hormonal stimuli (Humphrey et al. 2015).
Gene variants
Thoracic aortic aneurysm formation has been linked to variants in a number of genes that are involved in homeostasis of the aortic wall. These include genes that encode proteins responsible for (a) ECM regulation (FBN1, COL3A1, LOX, MFAP5, BGN), (b) the VSMC contractile apparatus (MYH11, ACTA2, MYLK, FLNA, PRKG1) or (c) TGF-β signalling (TGFB2, TGFB3, TGFBR1, TGFBR2, SMAD3) (Verhagen et al. 2018).
Histopathology
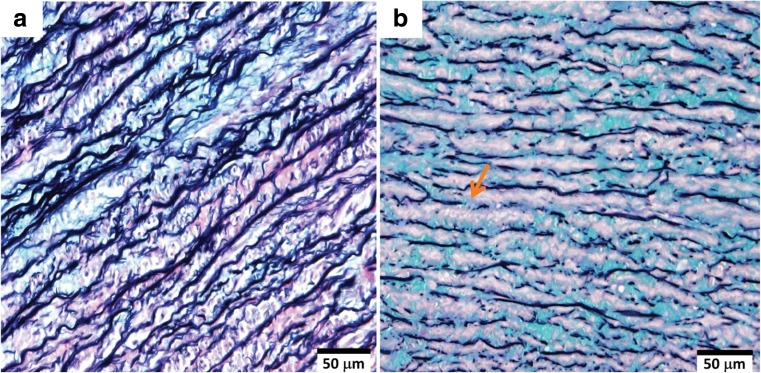
Regardless of the gene affected, there is a unifying histopathological change in the aortic wall in genetically triggered TAAs, defined as medial degeneration (Fig. 1) (Halushka et al. 2016). The most prominent feature is the fragmentation, disorganisation and loss of elastic fibres. This has been associated with a pathological imbalance of the molecules that regulate the ECM, primarily the induction of matrix metalloproteinases (MMPs) or reduction of their endogenous regulators, tissue inhibitors of metalloproteinases (TIMPs) (Rabkin 2014). The fragmentation of elastic fibres, along with increased collagen deposition decreases the compliant nature of the aorta, reducing its recoiling ability and increasing wall stiffness (Emmott et al. 2016). In addition, there is loss of VSMCs due to apoptosis or necrosis and focal accumulation of mucoid ECM material, resulting in the loss and/or disruption of aortic contractile units. Together, the loss of structural integrity and mechanical strength in the aortic wall reduces its ability to manage normal haemodynamic loads, consequently predisposing to aortic dissection (Humphrey et al. 2015).
Fig. 1.
Histology of the aortic wall medial layer from (a) normal aorta and (b) a patient with BAV and TAA, stained with Movat pentachrome. The normal aortic media (a) has long, intact parallel bands of contractile lamellar units comprising elastic lamellae (black) interspersed with VSMCs (purple), with minimal accumulation of ECM material (blue). In contrast, the BAV-TAA (b) media shows severe fragmentation of and loss of elastin fibres (arrow), loss of VSMC nuclei and increased accumulation of mucoid ECM, characteristic of medial degeneration. BAV, bicuspid aortic valve; ECM, extracellular matrix; TAA, thoracic aortic aneurysm; VSMC, vascular smooth muscle cell. Images were provided by M. Emami
Conditions associated with genetically triggered TAA
Marfan syndrome
Marfan syndrome (MFS) is characterised by multisystem features affecting the cardiovascular, ocular and skeletal systems (Loeys et al. 2010), with an estimated prevalence of 6.5 per 100,000 (Groth et al. 2015). TAA is the most common life-threatening complication in MFS, affecting up to 90% of patients (Hiratzka et al. 2010), with aortic dissection being a major cause of mortality (Faivre et al. 2007).
MFS is caused by pathological variants in FBN1, which encodes fibrillin-1, a major ECM glycoprotein in connective tissue distributed throughout the body (Dietz et al. 1991). The underlying mechanism of TAA pathogenesis in MFS remains unclear. Possible mechanisms include the variant fibrillin-1 protein causing an intrinsic impairment of structural integrity or altered force mechanotransduction and adverse remodelling of the aortic wall (Yu and Jeremy 2018). In addition, variants in FBN1 have been associated with altered TGF-β bioavailability and increased sensitivity to angiotensin II signalling in TAA formation (Yu and Jeremy 2018). While variant FBN1 is responsible for MFS, there is important phenotypic heterogeneity among individuals who harbour the same FBN1 variant, including within members of the same family (De Backer et al. 2007; Li et al. 2016b), indicating a role for other genetic or epigenetic mechanisms in determining phenotype.
Loeys-Dietz syndrome
Loeys-Dietz syndrome (LDS) has widespread manifestations in the craniofacial, neurocognitive, skeletal and cardiovascular systems and is characterised by extensive vascular abnormalities including arterial tortuosity and TAA or dissection (Loeys et al. 2010). The prevalence is unknown but appears to be less than MFS (Loeys and Dietz 2008). Aggressive thoracic aortic disease is the most clinically significant event, affecting over 95% of patients (Loeys et al. 2006). LDS is caused by pathological variants in TGF-β signalling pathway genes: the TGF-β ligands (TGFΒ2 and TGFΒ3), its receptors (TGFΒR1 and TGFΒR2) or its key intracellular signalling mediators Smad2 and Smad3 (SMAD2 and SMAD3) (Schepers et al. 2018).
Vascular Ehlers-Danlos syndrome
The vascular type of Ehlers-Danlos syndrome (vEDS) is characterised by a generalised vasculature and integumentary tissue fragility, with cardinal features of thin, translucent skin, easy bruising, characteristic facial appearance and ruptures of the arterial system, intestines or uterus (Beighton et al. 1998). vEDS is a rare subtype of Ehlers-Danlos syndrome, with estimated prevalence between 1:50,000 and 1:200,000 (Germain 2007). It is caused by pathological variants in COL3A1 that encodes the type III collagen alpha chain, which is highly expressed in the arterial system and hollow organs (Beighton et al. 1998). Aortic dissection or rupture is the leading cause of death, with a median survival of 50 years (Germain 2007).
Bicuspid aortic valve
Bicuspid aortic valve (BAV) is the most common congenital cardiac abnormality, characterised by abnormal fusion of the normally tri-leaflet aortic valve cusps which produces a valve with two cusps that are usually asymmetrical (Osler 1886). It affects 1–2% of the population with a male predominance (Siu and Silversides 2010). TAA is the most common non-valvular comorbidity in BAV, occurring in up to 45% of affected individuals (Tzemos et al. 2008; Siu and Silversides 2010).
The aetiology of BAV is unknown, but it is likely polygenetic, with causative genes identified in less than 4% of cases (Garg et al. 2005; McKellar et al. 2007). Causative gene discovery is further complicated by significant variability in penetrance and associated non-valvular manifestations (Andelfinger et al. 2016). Susceptibility towards TAA in BAV also remains poorly understood. The two predominant theories that aim to explain the increased prevalence are the “haemodynamic theory” and the “genetic theory”. The “haemodynamic theory” broadly suggests that TAA arises from a background of increased aortic wall stress consequent on turbulent blood flow from the malformed BAV (Sievers et al. 2016; Roman et al. 2017). The “genetic theory” suggests that underlying gene variants are responsible for the increased predisposition to both BAV and BAV-TAA (Prakash et al. 2014; Andelfinger et al. 2016). The relative contributions of genetics and haemodynamics remain unclear, but they are likely interrelated in a complex pathological process. Overall, the risk of aortic dissection is very low (< 1% 10-year dissection risk) compared to that of MFS (~ 7.9% per 10 years) and nsTAAD (~ 3.6% per 10 years) (Tzemos et al. 2008; Michelena et al. 2011; Sherrah et al. 2016); however, it is still approximately eight times higher than the general population (Michelena et al. 2011).
Non-syndromal thoracic aortic aneurysm and dissection
Non-syndromal thoracic aortic aneurysm and dissection (nsTAAD) is characterised by an inherited predisposition to TAA and dissection, without any other physical features (Milewicz et al. 2013). As TAA is often asymptomatic, presentation usually occurs at a later age than the syndromic forms, with surgical repair most commonly performed at age 51–60, compared to MFS at age 31–40 (Robertson et al. 2016). Calculated 10-year mortality due to dissection is approximately 7.8% for patients under clinical surveillance; however, with the inclusion of undiagnosed patients, true mortality is likely much higher and possibly up to 50% (Melvinsdottir et al. 2016). Due to the asymptomatic nature and absence of external physical signs, the population incidence is unknown; however, up to 16% of patients undergoing aortic surgery have characteristic pathology consistent with nsTAAD (Robertson et al. 2016). Additionally, up to 50% of nsTAAD probands have a first-degree relative with TAA (Elefteriades and Farkas 2010; Robertson et al. 2016), suggesting that nsTAAD may be relatively common.
The aetiology of nsTAAD is largely unknown, with causative gene variants identified in only 20% of cases. nsTAAD has an autosomal dominant inheritance pattern with increased male penetrance and variable age of onset and progression (Albornoz et al. 2006; Sherrah et al. 2016). The known affected genes are generally involved in the VSMC contractile apparatus (ACTA2, MYH11, MYLK, PRKG1) or TGF-β signalling pathway (SMAD3, TGFΒR1, TGFΒR2) (Milewicz et al. 2013). As with MFS, there is considerable phenotypic variation in severity of TAA among family members sharing the same causative gene variant (Robertson et al. 2016), indicating a role for other genetic or epigenetic modifiers.
Epigenetics in TAA
MicroRNA
The miRNAs are a class of small (~ 22 nucleotide) highly conserved non-coding RNA molecules that modulate gene expression by inhibiting mRNA translation to maintain homeostasis in a variety of physiological processes (Bartel 2004). They dynamically regulate over 60% of human protein-coding genes, in a manner that is highly tissue- and context-dependent (Ha and Kim 2014). Dysregulated miRNA expression is widely implicated in a variety of disease processes (Gommans and Berezikov 2012).
The synthesis and maturation process of miRNA has been extensively covered elsewhere (Ha and Kim 2014). Briefly, miRNA genes are transcribed from intra- or inter-genic regions of the genome and undergo a number of cleavage and maturation steps in the nucleus before export to the cytoplasm, and incorporation with the RNA-induced silencing complex (RISC) (Bartel 2004). The miRNA sequence guides the RISC complex to target mRNA transcripts by imperfect sequence complementarity between the miRNA and the 3’UTR of the target mRNA (Lewis et al. 2005). The target mRNA is then cleaved or translationally repressed, resulting in reduced protein product (Bartel 2004).
Importantly, the imperfect sequence recognition enables each miRNA to regulate the expression of multiple mRNA genes. This gives miRNAs the capability to extensively modify gene expression profiles and dynamically regulate homeostasis. However, despite their functional redundancy, it has become apparent that changes in the expression of a single miRNA and the consequence on protein expression are an important factor in the pathogenesis of many disease states (Gommans and Berezikov 2012).
Several different mechanisms and mediators regulate the expression of miRNAs. These include miRNA transcription being influenced by host gene expression in cases where the promoter sequence is shared, or alternatively miRNA expression may be altered by epigenetic mechanisms, transcription factors and non-coding RNA (Gulyaeva and Kushlinskiy 2016). Genomic variants that result in changes to either mediators of miRNA transcription or other proteins involved in miRNA biogenesis may also affect miRNA function (Gulyaeva and Kushlinskiy 2016). Exogenous factors, such as environmental carcinogens, diet, smoking and alcohol consumption, and endogenous factors, including stress, hormones and changes to the tissue microenvironment, such as hypoxia, have also been shown to induce changes in miRNA expression (Gulyaeva and Kushlinskiy 2016). Single-nucleotide polymorphisms in the miRNA promoter can also modulate miRNA expression (Hrdlickova et al. 2014 and have been documented to be influential in several disease contexts (Yue et al. 2018).
Dysregulated miRNA expression is implicated in vascular pathologies such as atherosclerosis, hypertension, TAA and abdominal aortic aneurysm (AAA) (De Lucia et al. 2017; Li and Maegdefessel 2017), with emerging evidence that it also contributes to genetically triggered TAA. There are relatively few studies focusing on genetically triggered TAA, which is reflective of their low incidence combined with the inherently difficult nature of aortic tissue sample acquisition. The predominant miRNAs that have been associated with genetically triggered TAA to date are the miR-29 family, miR-143/145 and miR-17.
miR-29 family
The miR-29 family, comprising miR-29a, miR-29b and miR-29c, has been the most extensively studied miRNA in genetically triggered TAA (Table 1). The family is transcribed from two independent bi-cistronic clusters as miR-29a/miR-29b1 and miR-29b2/miR-29c, with miR-29b1 and miR-29b2 identical in sequence (Boon et al. 2011). They are considered important regulators of ECM homeostasis, with gene targets in elastin (ELN) (van Rooij et al. 2008), several collagens and MMPs (Liu et al. 2010).
Table 1.
miR-29 family expression in aortic pathology
| Condition | Expression change | Species | Cell/tissue | Effects | Reference |
|---|---|---|---|---|---|
| miR-29a | |||||
| BAV-TAA | ↑ CCV vs control CCV ↑ CCV vs CVX (within-patient) |
Human | AscAo wall | Regional miRNA expression changes induced by haemodynamic stress | Albinsson et al. (2017) |
| BAV-TAA | ↑ | Human | AscAo wall | / | Ikonomidis et al. (2013) |
| TAA | ↑ | Mouse (Fibulin-4R/R) | AscAo wall | / | Boon et al. (2011) |
| Non-genetic TAA | ↓ | Human | AscAo wall In vitro aortic VSMCs |
↑ MMP-2 | Jones et al. (2011) |
| TAD | ↓ | Human | AscAo wall | ↑ collagen and fibrosis | Liao et al. (2011) |
| miR-29b | |||||
| BAV-TAA | ↑ | Human | AscAo wall | / | Boon et al. (2011) |
| Non-genetic TAA | ↑ | Human | AscAo wall | / | Boon et al. (2011) |
| MFS | ↑ | Mouse (Fbn1C1039G/+) | AscAo wall | ↑ MMP-2, ↓ elastin; ↑ elastin fragmentation, ↑ apoptosis Induced by TGF-β1 |
Merk et al. (2012) Okamura et al. (2017) |
| TAA | ↑ | Mouse (Fibulin-4R/R) |
AscAo wall | / | Boon et al. (2011) |
| Normal aorta | ↑ with ageing | Mouse | Aorta (region undefined) | Gradual repression of ECM genes sensitises aorta to aneurysm with increasing age | Boon et al. (2011) |
| Atherosclerosis | ↑ by ox-LDL | Human Mouse (HFD) |
In vitro aortic VSMCs | ↑ MMP-2, ↑ MMP-9 | Chen et al. (2011) Chen et al. (2012) |
| AAA | ↑ | Mouse (Ang-II) | Abdo Ao wall | ↓ elastin, ↓ collagen; ↑ ECM fragmentation | Boon et al. (2011) |
| AAA | ↓ | Human | Abdo Ao wall | ↑ elastin, ↑ collagens (COL1A1, COL3A1, COL5A1) | Maegdefessel et al. (2012) |
| AAA | ↓ | 2 x mouse (PPE-C57BL/6) and (AngII-ApoE−/−) | Abdo Ao wall | ↑ elastin, ↑ collagens; ↑ fibrosis; ↑ AAA diameter | Maegdefessel et al. (2012) |
| Aortic VSMCs | miR-29b inhibition | Human | In vitro aortic VSMC | miR-29b inhibition: ↑ elastin, ↑ collagens (COL1A1, COL3A1) | Maegdefessel et al. (2012) |
| Aortic adventitial fibroblasts | miR-29b inhibition | Human | In vitro aortic VSMC | miR-29b inhibition: ↑ collagens (COL1A1, COL3A1) miR-29b ↓ by TGF-β1 |
Maegdefessel et al. (2012) |
| miR-29c | |||||
| BAV-TAA | ↑ CCV vs control CCV | Human | Asc Ao wall | Regional miRNA expression changes induced by haemodynamic stress | Albinsson et al. (2017) |
| Non-genetic TAA, BAV-TAA | ↑ | Human | AscAo wall | ↓ collagens, ↓ elastin; ↑ MMP-9 | Licholai et al. (2016) |
| TAA | ↑ | Mouse (Fibulin-4R/R) | AscAo wall | / | Boon et al. (2011) |
| TAD | ↓ | Human | AscAo wall | ↑ collagen and fibrosis | Liao et al. (2011) |
AAA, abdominal aortic aneurysm; Abdo, abdominal; AngII, angiotensin II; Ao, aortic; ApoE, apolipoprotein E; AscAo, ascending aortic; BAV, bicuspid aortic valve; CCV, aortic concavity; CVX, aortic convexity; COL1A1, collagen 1 alpha-1 chain; COL3A1, collagen 3 alpha-1 chain; COL5A1, collagen 5 alpha-1 chain; ECM, extracellular matrix; Fbn1, fibrillin-1; HFD, high fat diet; LCA, left coronary artery; MFS, Marfan syndrome; MI, myocardial infarction; MMP, matrix metalloproteinase; ox-LDL, oxidized low-density lipoprotein; PPE, porcine pancreatic elastase; TAA, thoracic aortic aneurysm; TAD, thoracic aortic dissection; TGF-β, transforming growth factor-beta; VSMC, vascular smooth muscle cell
miR-29a
An increase in miR-29a expression was observed in BAV-TAA tissue compared to controls (Ikonomidis et al. 2013; Albinsson et al. 2017), with demonstrated regional differences in miRNA expression. It was also shown to be increased in the aortic concavity (inner curvature) but decreased in the convexity (outer curvature) (Albinsson et al. 2017), which is typically under greater stress (Barker et al. 2012). These data suggest that miR-29a plays a role in adapting the aortic wall to the haemodynamic stress that arises from the BAV (Albinsson et al. 2017). In non-genetic TAA, miR-29a was decreased in aortic tissue obtained during prophylactic replacement surgery both before (Jones et al. 2011) and after dissection (Liao et al. 2011). Further studies using cultured human aortic VSMCs have demonstrated that miR-29a is a direct regulator of MMP-2 expression (Jones et al. 2011), indicating that its primary role may be to regulate ECM homeostasis and that this may differ depending on the underlying genetic aetiology and pathological process.
miR-29b
An increase in miR-29b was observed in aortic tissue of patients with BAV-TAA (Boon et al. 2011) and non-genetic TAA (Jones et al. 2011); however, it was decreased in aortic tissue from abdominal aortic aneurysms (AAA) (Maegdefessel et al. 2012). A murine MFS (Fbn1C1039G/+) model (Merk et al. 2012) and two other mouse models of aortic dilatation (Ang-II/ApoE−/− and Fibulin-4R/R) (Boon et al. 2011) have also demonstrated increased miR-29b expression in aneurysmal aortic tissue. These murine studies showed histological evidence of reduced elastin expression and increased ECM fragmentation and apoptosis. Lending further support to a pathogenic role of miR-29b in aneurysm progression, blockade of miR-29b in vivo was shown to reduce ECM degradation and prevented aneurysmal development in both the MFS mice (Merk et al. 2012) and Ang-II mice (Maegdefessel et al. 2012). Subsequent studies in the MFS mice have shown that while miR-29b blockade prevented TAA formation when administered from birth, it did not reduce aneurysm size once formed (Okamura et al. 2017), indicating a potential limitation in translation to human therapy.
Cell culture studies have demonstrated that miR-29b represses elastin and collagen gene expression and promotes MMP expression, which would result in a reduction of ECM deposition and increased degradation (Chen et al. 2011; Chen et al. 2012). In human aortic VSMCs and adventitial fibroblasts, miR-29b was shown to target COL1A1 and COL3A1, with ELN additionally regulated by miR-29b in VSMCs but not fibroblasts (Maegdefessel et al. 2012). Furthermore, miR-29b is selectively modulated by TGF-β1 depending on the cell type. In human aortic fibroblasts (Maegdefessel et al. 2012) and cardiac fibroblasts (van Rooij et al. 2008), TGF-β1 reduces miR-29b and increases collagen and elastin gene expression, altogether promoting a fibrotic response that is well-established for TGF-β. However, in aortic VSMCs, TGF-β1 has no effect on miR-29b levels (Maegdefessel et al. 2012; Merk et al. 2012).
The discordance in miR-29b expression among different tissue samples suggests that it may be differentially regulated depending on aneurysm context. It is increased in TAA associated with MFS, non-genetic TAA and BAV, while it is decreased in AAA. This may be due to the inherent differences in the tissue at the different locations or differences in the underlying pathogenesis. There is a general observation of elevated TGF-β in genetically triggered TAA as well as in AAA; however, AAA is associated with more significant tissue fibrosis. This may mean that fibroblasts have a reduced role in TAAs and a greater role in AAA, and this may dictate the cell-specific effects of TGF-β and overall miR-29b expression in the aorta.
miR-29c
miR-29c was shown to be increased in BAV-TAA aortic tissue (Licholai et al. 2016; Albinsson et al. 2017), in addition to the aortic tissue taken from a murine TAA model (Fibulin-4R/R) (Boon et al. 2011). Conversely, miR-29c was decreased in acute thoracic aortic dissection tissue samples, similarly to miR-29a (Liao et al. 2011).
Overall, the miR-29 family appears to adopt a cell- and tissue-specific role in ECM regulation. There is a trend towards increased expression in TAAs of different aetiology, which overall favours degradation of the ECM. This is likely mediated through multiple transcriptional programmes that require further elucidation as to their relevance to genetically triggered TAA pathogenesis. Combined, these studies illustrate the important role of the miR-29 family in ECM regulation and its potential for therapeutic modulation.
miR-143/145
miR-143 and miR-145 are two of the best characterised cardiovascular-related miRNAs, which play an essential role in VSMC differentiation and phenotype (Boettger et al. 2009; Cordes et al. 2009). They are transcribed together from a large primary pri-miR-143/145 bi-cistronic gene cluster that is then enzymatically cleaved into miR-143 and miR-145 (Rangrez et al. 2011). They are under transcriptional control by many SMC transcription factors, including TGF-β, in a feed-forward mechanism that maintains VSMCs in a contractile phenotype (Cordes et al. 2009; Boucher et al. 2011). They are also influenced by local environment changes, such as vascular injury, which often represses their transcription in favour of ECM repair (Liu et al. 2013b).
In the normal aortic wall, VSMCs are a phenotypically diverse population, ranging along a spectrum from contractile/differentiated to synthetic/dedifferentiated (Rensen et al. 2007). The majority of aortic VSMCs are contractile, characterised by the expression of a contractile gene profile, and are predominantly composed of actin and myosin filaments. Synthetic VSMCs are usually a minor population in the aortic wall and primarily function in maintenance and repair by promoting expression of ECM genes, such as collagen and elastin (Rensen et al. 2007). miR-143/145 collaboratively maintain contractile VSMC populations in the aorta, with miR-145 promoting contractile gene expression and miR-143 suppressing synthetic gene expression (Cordes et al. 2009).
Loss of miR-143/145 is associated with impaired aortic contractility and is associated with TAA regardless of the underlying aetiology (Elia et al. 2009; Albinsson et al. 2017), in addition to other conditions of vascular damage and disease (Liu et al. 2013b; Quiat and Olson 2013). Decreased miR-143/145 expression was observed in the aortic wall of patients with BAV-TAA that had mild aortic dilatation (< 4.5 cm), in correlation with immunohistochemical evidence of reduced contractile gene expression (Alajbegovic et al. 2017). Additional work has demonstrated that there is decreased miR-143 but increased miR-145 expression together with increased miR-29a in BAV-TAA aortic tissue, and this was in conjunction with a plasma profile showing increased miR-133a and miR-145 and altered MMP/TIMP expression (Ikonomidis et al. 2013). The mechanism of the divergence between miR-143/145 expression is not currently understood, but it may be the result of differing post-translational modifications or differences in miRNA stability.
Other work in BAV-TAA has identified regional differences in miRNA expression, with decreased miR-143 in aortic tissue obtained from the convex compared to concave regions, indicating that haemodynamic changes from the malformed valve may influence local miRNA expression (Albinsson et al. 2017). BAVs with a right-left cusp fusion pattern are significantly associated with increased aortic wall shear stress in the aortic convexity (Barker et al. 2012); therefore, stress-induced degenerative changes may be associated with loss of miR-143/145 expression, which further propagates loss of function that accompanies aortic dilatation.
Murine models of TAA (ApoE−/− and transverse aortic constriction) are concordant with the human tissue studies, showing that miR-143/145 are decreased in TAA (Elia et al. 2009). miR-143/145 knockout rodent models exhibit histological evidence of dedifferentiated VSMCs, reduced medial density, inflammation, fibrosis and a loss of myogenic tone (Xin et al. 2009; Holmberg et al. 2018).
Human cell culture studies have demonstrated the therapeutic potential of miR-145 to protect against vascular damage. In aortic VSMCs, pre-treatment with a miR-145 mimic has been shown to prevent inflammation-induced vascular injury through modulation of CD40 (Li et al. 2016a) and tumour necrosis factor-alpha (TNF-α) (Guo et al. 2016). Activation of CD40 receptors on VSMCs and endothelial cells triggers inflammatory cascades that lead to vascular diseases, such as atherosclerosis (Song et al. 2012). Increased miR-145 expression was shown to suppress CD40 activation and interrupt VSMC proliferation and dedifferentiation in response to platelet-derived growth factor-BB (PDGF-BB) (Li et al. 2016a) and TNFα (Guo et al. 2016), which are both upregulated in early vascular damage. In addition, simultaneous blockade of miR-145 with aspirin treatment abolished the protective effect of aspirin and reversed the associated gene expression profiles (Guo et al. 2016). miR-145 may therefore mediate part of the anti-inflammatory effects of aspirin treatment by blocking TNF-α-mediated activation of CD40.
Despite the wealth of research into miR-143 and miR-145 in vascular disease, there is still a relative paucity of studies in genetically triggered TAA. However, the current studies highlight these miRNA as likely key players in the pathological VSMC dedifferentiation that is commonly observed in genetically triggered TAA.
miR-17
The miR-17/92 cluster, comprising miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a and miR-92a-1, has been extensively characterised in many aspects of human development and disease, with dysregulation in cardiovascular disease well established (Mogilyansky and Rigoutsos 2013).
Increased miR-17 has been observed in BAV-TAA aortic tissue compared to control (Wu et al. 2016). Within individual BAV-TAA patients, miR-17 was increased in less dilated (< 4 cm diameter) aortic tissue segments compared to severely dilated (> 4 cm diameter) segments (Wu et al. 2016), indicating that miR-17 may contribute to early TAA development, but as the aorta dilates this effect diminishes (Wu et al. 2016). Furthermore, increased miR-17 levels correlated with decreased levels of TIMP-1, -2 and -3 and increased MMP-2 levels, indicating that the role of miR-17 is likely to be in the regulation of MMP-mediated ECM degradation.
miR-17/92 expression has not been investigated in MFS or LDS; however, murine palatogenesis studies indicate that the cluster might contribute to palatal manifestations in addition to TAA in these conditions (Wang et al. 2013; Cao et al. 2016; Ries et al. 2017). The cause of palatal deformity is unknown, but it is associated with aberrant TGF-β signalling (Iwata et al. 2011). miR-17/92 has been identified to be essential for normal mouse palatogenesis, with inhibition of the cluster in vivo resulting in palatal deformity (Ries et al. 2017). Decreased miR-17 is accompanied by increased expression of TGF-β receptors and decreased TGF-β ligand in maxillary tissue (Ries et al. 2017).
miR-17 contributes to ECM maintenance by targeting TIMP1 and TIMP2 in human VSMCs, which are repressors of the matrix degrading enzyme, MMP-2 (Wu et al. 2016). Additional miR-17 gene targets identified in non-vascular cell lines include TIMP3 (Yang et al. 2013), TGFBR1 and TGFBR2 (Ries et al. 2017), and the anti-proliferative bone morphogenic protein receptor-2 (BMPR2) (Luo et al. 2014). Other work has described a TGF-β-Smad3-miR-17/92-BMPR2 axis in the promotion of TGF-β-mediated VSMC proliferation (Luo et al. 2014). Increased miR-17/92 and Smad3 have been identified in carotid artery restenosis tissue, which is characterised by excessive TGF-β signalling and VSMC proliferation. Subsequent studies have shown that TGF-β is able to induce miR-17/92 transcription via Smad3 binding at Smad-binding elements in the miRNA promoter, and BMPR2 has been identified as a miR-17/92 target (Luo et al. 2014).
Overall, miR-17/92 may contribute to TAA development by MMP/TIMP-mediated degradation of the aortic ECM, in addition to TGF-β-mediated pathological remodelling and VSMC proliferation.
Circulating miRNA
The identification of changes in miRNA levels in the circulation represents an alternative method of disease investigation, due to their potential utility as relatively non-invasive biomarkers that are economical to measure (Mitchell et al. 2008). miRNAs are stably expressed in plasma and serum and are resistant to degradation by endogenous RNAses (Chen et al. 2008). Their expression within the circulation has been shown to correlate with different physiological states (Gilad et al. 2008) and has led to the development of disease-specific miRNA expression profiles (Mitchell et al. 2008). Development of a similar circulating miRNA profile for TAA may lead to improvements in diagnosis and assist in both prognosis and optimising decisions regarding timing of surgical intervention.
miRNA extracted from whole blood of a small MFS cohort identified 11 miRNAs with significantly different expression compared to a control cohort (Abu-Halima et al. 2018). Significant correlations were seen between increased left ventricular end-diastolic dimension and increased levels of miR-151-5p, miR-24, miR-30e, miR-324-5p, miR-500b, miR-502-3p and miR-627. In addition, miR-331-3p was significantly increased in MFS without mitral valve prolapse (MVP) compared to MFS with MVP (Abu-Halima et al. 2018).
A small number of studies have examined circulating miRNAs in BAV-TAA cohorts. A global miRNA expression profile identified significantly decreased expression of miR-122 and miR-486 and significantly increased expression of miR-130a in plasma compared to a control group (Martinez-Micaelo et al. 2017). Subsequent pathway analysis identified TGF-β signalling as the most enriched pathway, with 32 possible miRNA-gene interactions. Within the BAV cohort, miR-718 expression had a significant inverse correlation with aortic diameter, with predicted gene targets in vascular remodelling and the focal adhesion pathway.
A targeted study evaluating a panel of six miRNAs in a BAV-TAA cohort identified miR-133a and miR-145 to be significantly decreased in the plasma compared to control (Ikonomidis et al. 2013), although these data were not corroborated subsequently (Martinez-Micaelo et al. 2017). The variance may reflect differences in methodology or the known biological heterogeneity that makes BAV-TAA so difficult to study. This is reflected in a more recent study in which the expression of seven miRNAs was analysed according to distinct BAV phenotypic subgroups; those with aortic root dilatation and concomitant insufficiency versus BAV with severe aortic stenosis (Girdauskas et al. 2018a). Within the root dilatation subgroup, circulating miR-17 and 106a were significantly increased in the less dilated (< 50 mm) compared to severely dilated (> 50 mm) aortas; however, when compared overall to the aortic stenosis cohort, significant differences were observed in all seven miRNAs (miR-17, miR-16a, miR19a, miR-20a, miR-21, miR-106a and miR-145) (Girdauskas et al. 2018b). While the lack of a control cohort in this study limits further interpretation, it is clear that there are likely different miRNA profiles associated with different BAV phenotypes, and this should be factored in to future experimental design in BAV studies.
Overall, the development of circulating miRNA profiles in genetically triggered TAAs is still in preliminary stages. More observational studies using larger and well-defined cohorts are required to identify candidate miRNAs that can act as biomarkers of the disease state. In addition, longitudinal studies of the same patient group may provide miRNAs that can be used as markers of TAA progression.
miRNAs in other cardiovascular studies
In addition to studies detailing the role of miRNAs in TAA, there has been extensive characterisation of miRNAs in other cardiovascular pathologies that have been reviewed elsewhere (De Lucia et al. 2017; Li and Maegdefessel 2017). While there are important differences in molecular pathogenesis limiting direct comparison and inference, these studies are informative in terms of understanding miRNA-mRNA regulatory relationships that affect the vasculature. In addition, other studies have characterised the normal function of miRNAs within the vasculature, for example in endothelial cells and VSMCs (Ballantyne et al. 2016), molecules involved in ECM regulation (Hu et al. 2016) and in major signalling pathways, such as TGF-β (Climent et al. 2015; Forte et al. 2016). The relevance of these miRNAs with respect to genetically triggered TAA is yet to be investigated.
Long non-coding RNA
Long non-coding RNAs (lncRNAs) have emerged in the last decade as a large and functionally diverse class of non-protein-coding RNA at least 200 nt in length, which regulate gene expression at the transcriptional and post-transcriptional levels (Engreitz et al. 2016). Research on lncRNA is still emerging, with broad estimations as to the number of lncRNAs ranging from 20,000 to 100,000 (Kopp and Mendell 2018), but very few having been biologically characterised. One of the best established is Xist (X inactive specific transcript), which orchestrates X chromosome inactivation through highly ordered interactions with chromatin scaffolding proteins and targeted binding to X chromosomal DNA, where it mediates transcriptional silencing along the entirety of the chromosome (Wutz et al. 2002). Characterisation of lncRNAs in vascular biology and disease has rapidly progressed, with a number of extensive reviews available (Li and Maegdefessel 2017; Leeper and Maegdefessel 2018; Simion et al. 2018).
MALAT1
MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) is highly expressed in endothelial cells (Simion et al. 2018) and was recently implicated in genetically triggered TAA, with increased expression linked to pathological VSMC dedifferentiation in the aneurysmal aortic wall (Cardenas et al. 2018). In aortic tissue from MFS, nsTAAD and non-genetic TAA, increased expression of MALAT1, the epigenetic repressor histone deacetylase 9 (HDAC9), and the chromatin-remodelling enzyme, BRG1, was observed when compared to normal aortic tissue. These expression changes were also observed in two mutant VSMC cell lines that represent genetically triggered TAA (TGFBR2G357W and ACTA2R179H) and were correlated with decreased contractile gene expression (Cardenas et al. 2018).
MALAT1 is essential to transport BRG1 and HDAC9 into the nucleus, where the promoters of contractile genes are methylated by HDAC9, resulting in pathological VSMC dedifferentiation (Cardenas et al. 2018). These data have been confirmed in a MFS mouse model (Fbn1C1039G/+) where deletion of either MALAT1 or HDAC9 impaired aneurysm formation (Cardenas et al. 2018). These deletions correlated with increased contractile gene expression and decreases in elastin fragmentation, MMP activity, TGF-β signalling and VSMC proliferation.
The cause of the observed upregulation of MALAT1 in genetically triggered TAA tissue is unknown. Its expression can be modulated by factors including hypoxia, p53 transcription factor mutations, miRNA and polymorphisms within the MALAT1 gene (Simion et al. 2018). It may also be influenced by the pathological context. For example, MALAT1 was decreased in human aortic and coronary VSMCs cultured in “ECM stiffening” conditions reflective of arterial disease, and MALAT1 knockdown limited stiffness-induced VSMC proliferation and migration (Yu et al. 2018). Overall, these studies suggest that MALAT1 may play an important role in the regulation of vascular homeostasis.
HOTAIR
Decreased HOTAIR expression has been observed in calcified BAV aortic leaflets, and this was inversely correlated with the expression of two key calcification genes, ALPL, encoding alkaline phosphatase, and BMP2, encoding bone morphogenic protein 2 (Carrion et al. 2014). HOTAIR epigenetically regulates gene expression by promoting histone H3K27 methylation (Gupta et al. 2010).
Decreased HOTAIR expression has also been identified in the aortic tissue of a small non-genetic TAA cohort, which was negatively correlated with aortic diameter (Guo et al. 2017). Knockdown in human aortic VSMCs resulted in decreased expression of collagens I and III and increased apoptosis, indicating HOTAIR is a potential mediator of ECM regulation in the aorta (Guo et al. 2017).
HOTAIR expression is down-regulated by the WNT/β-catenin pathway in cultured human aortic valvular interstitial cells subjected to cyclic mechanical stretch (Carrion et al. 2014). Increased WNT/β-catenin signalling is implicated in MFS (Chopra et al. 2017) and in other aneurysm formation and vascular pathology in general (Mill and George 2012), implicating HOTAIR as another mediator of pathological WNT/β-catenin signalling, thus potentially driving aortic and valvular pathology in genetically triggered TAA.
HIF1A-AS1
HIF1A-AS1 (hypoxia inducible factor 1 alpha antisense RNA 1) is implicated in TAA in association with BRG1-mediated VSMC apoptosis (Wang et al. 2015). BRG1 is a major regulator of vascular development through chromatin remodelling (Griffin et al. 2008), and its expression was increased in aortic tissue of a non-genetic TAA cohort (Wang et al. 2015). In human aortic VSMCs, BRG1 knockdown resulted in VSMC apoptosis and reduced proliferation, which was mediated through HIF1A-AS1 (Wang et al. 2015). HIF1A-AS1 expression in TAA tissue has not been investigated; however, increased levels were detected in the serum of a non-genetic TAA cohort (Zhao et al. 2014), indicating a potential role for this lncRNA in aneurysm pathogenesis.
DNA methylation
DNA methylation is a physiological process that involves the addition of methyl groups to DNA, which results in repression of gene transcription without changing the DNA sequence (Curradi et al. 2002). It is a somatically heritable trait that is also influenced by environment. Both cytosine and adenine nucleotides can be methylated; however, DNA methylation is found almost exclusively in CpG dinucleotides, which are segments of DNA sequence comprising a cytosine followed by a guanine nucleotide in a 5′ to 3′ direction with a phosphate in between (Curradi et al. 2002). In mammals, approximately 70–80% of all CpG cytosines are methylated (Jabbari and Bernardi 2004). Certain CpG regions within the DNA, known as CpG islands, serve as regulatory units of transcription. These islands are typically 200–1500 base pairs long, with 50% located in the promoter regions of constitutively active genes, and 25% within regions that serve as alternative promoters (Larsen et al. 1992). These CpG islands typically are methylated at very low levels, thus allowing the expression of the resident gene (Curradi et al. 2002).
Tissue-specific, genome-wide DNA methylation profiling performed in aortic tissue from BAV-TAA has identified nine novel genes with significant differential methylation and differential gene expression, when compared to TAA tissue without a known genetic trigger (Shah et al. 2015). The strongest finding was the hypermethylation and decreased expression of PTPN22 in BAV-TAA tissue. PTPN22 (protein tyrosine phosphatase, non-receptor type 22) has an important role in regulating immune responses, with decreases in its encoded protein, lymphoid tyrosine phosphatase (Lyp), shown to increase T cell signalling (Burn et al. 2011). Activated T cells in the aortic wall can lead to VSMC apoptosis and degradation of the ECM (He et al. 2008); thus, hypermethylation of PTPN22 in the aorta may contribute to medial degeneration.
Genes previously associated with TAA, including ACTA2, and genes associated with cardiovascular development, such as GATA4, were shown to be differentially methylated but not differentially expressed in BAV-TAA (Shah et al. 2015). As DNA methylation is important for embryological development, it has been suggested that altered DNA methylation in these genes may contribute to abnormalities in aortic valve development and aortic cell migration during embryogenesis and, therefore, may not necessarily be altering gene expression at the time of tissue analysis.
Epigenetic mechanisms may also contribute to the altered MMP expression that is observed in genetically triggered TAA. There are extensive CpG islands throughout MMP9 (Chernov and Strongin 2011), and hypomethylation of the MMP9 promoter has been correlated with increased MMP9 transcriptional activity in lymphoma cells (Chicoine et al. 2002). MMP3 gene expression was increased in a colon cancer cell line after dual knockout of key DNA methyltransferase genes (DNMT1 and DNMT3b) (Couillard et al. 2006). In human VSMCs, miR-29b has been shown to increase MMP2 and MMP9 expression through the suppression of the DNA methyltransferase genes, DNMT3a and DNMT3b (Takada et al. 2009; Chen et al. 2011).
The TET2 DNA demethylase is highly expressed in VSMCs and is an important regulator of VSMC phenotype (Liu et al. 2013a). Decreased TET2 expression has been identified to occur in proliferative vascular pathologies, such as atherosclerosis, and knockdown in human VSMCs resulted in loss of contractile phenotype and increased expression of synthetic VSMC markers (Liu et al. 2013a). Overexpression of TET2 was shown to be sufficient to induce the contractile phenotype, and local injection into rat arteries prevented the intimal hyperplasia that is associated with pathological VSMC switching (Liu et al. 2013a). The role of TET2 in TAA development is a current area of investigation.
Histone modifications
Histones are positively charged proteins that associate with DNA to form nucleosomes, the basic structural unit of chromatin. Nucleosomes contain a small length of DNA wrapped tightly around eight core histone proteins: two dimers of H2A/H2B and two dimers of H3/H4 (Luger et al. 1997). Each core histone protein contains a modifiable amino acid tail. Various post-transcriptional modifications occur at the N-termini of these tails, including acetylation, methylation, phosphorylation and ubiquitination (Kouzarides 2007). These modifications can alter DNA-histone interactions, ultimately changing the chromatin structure and modulating the accessibility of transcriptional machinery to DNA binding sites to produce changes in gene transcription (Webster et al. 2013).
Two well-characterised histone modifications associated with transcriptional activation are H3 acetylation at Lys 9 (H3K9) and H3 methylation at Lys 4 (H3K4) (Verdone et al. 2005; Greer and Shi 2012). Increased acetylation and methylation at these specific histone residues in the SMAD2 promoter was associated with increased Smad2 protein in VSMCs isolated from aortic tissue samples from individuals with MFS, BAV and non-genetic TAA (Gomez et al. 2011). SMAD2 overexpression in these VSMCs was subsequently shown to be modulated by histone acetyltransferases (p300/PCAF) (Gomez et al. 2013). Thus, the dysregulated TGF-β signalling that is frequently observed in genetically triggered TAA may be contributed to by aberrant histone modulations affecting SMAD2 transcription.
Histone acetylation is also able to modulate VSMC phenotype by altering chromatin accessibility and SMC transcription factor binding (Liu et al. 2014). Contractile VSMC gene expression is induced by the binding of myocardin/serum response factor (SRF) to CArG (CC(A/T)6GG) DNA sequence elements in SMC gene promoter regions (McDonald et al. 2006), and the binding capacity of SRF to CArG-elements has been shown to rely heavily on preceding histone hyperacetylation (Qiu and Li 2002).
Aberrant histone modifications affecting VSMCs in TAA may be a result of the altered biomechanical environment that accompanies aortic dilatation. It has been suggested that the increased wall strain and altered mechanotransduction cascades result in changes to VSMC chromatin state that subsequently alters downstream gene transcription profiles (Chen et al. 2013; Michel et al. 2018). Altered histone acetylation has been observed in AAA, with increased expression of histone acetyltransferases in human AAA tissue compared to normal aortic tissue (Han et al. 2016); however, this has not yet been explored in TAA.
Epigenetics as a therapeutic target
The use of non-coding RNAs as therapeutic molecules is in the early stages of development, with many currently in preclinical stages but few having reached clinical trials. Miravirsen, a miR-122 inhibitor for the treatment of chronic hepatitis C infection, has progressed to phase 2a studies and has demonstrated efficacy with few moderate-severe adverse events (Janssen et al. 2013). A small number of miRNA therapeutics are in phase 1 studies, mostly for the treatment of various cancers (Mellis and Caporali 2018); however, one trial was suspended due to multiple immune-related severe adverse events (Beg et al. 2017; Chakraborty et al. 2017).
An injectable hyaluronan-based hydrogel enabling targeted miRNA delivery to the heart has demonstrated preclinical efficacy in a murine model of myocardial infarction (MI). miR-29b that had been incorporated into the hydrogel and injected into the MI border zone demonstrated beneficial ECM changes and maintenance of cardiac function compared to control, decreasing the area of myocardial fibrosis (Monaghan et al. 2018). Similarly, administration of miR-302 to promote cardiomyocyte proliferation post-MI demonstrated local and sustained proliferation for two weeks after a single injection, indicating this method may have long-acting therapeutic potential (Wang et al. 2017).
In AAA, preclinical efficacy has been established for miR-126 in suppressing inflammation associated with aneurysm development. Using a dual-targeted approach in a murine Ang-II model, miR-126 conjugated with a VCAM-1-specific antibody was incorporated into ultrasound-guided microbubbles, which were subsequently delivered to inflamed endothelial cells of the AAA in weekly intravenous injections (Wang et al. 2018). VCAM-1 adhesion molecules are upregulated in AAA, thus binding allowed the targeted release of miR-126 to the affected endothelium to exert its anti-inflammatory effects. Amelioration of aneurysmal growth was observed after a period of four weeks.
Delivery of miRNA therapeutics to the aortic tissue in genetically triggered TAA may be more challenging than in AAA, given that the pathology in TAA predominantly occurs in the media, as opposed to the intima in AAA, in which intravenous agents benefit from having direct access. This may be resolved with novel techniques, such as a hydrogel-based injection, that will undoubtedly continue to emerge over the coming years.
Therapeutics aimed at methylation and histone-based modifications is complicated by their non-specific effects, which typically result in broad changes in gene expression (Yang et al. 2014). A number of DNA methyltransferase inhibitors (DNMTs) and histone deacetylase inhibitors (HDACi) are either in use or in clinical trials for cancer treatment (Jones et al. 2016) and are being explored for the treatment of various cardiovascular diseases; however, this has not progressed beyond preclinical studies (Schiattarella et al. 2018).
Future directions
Our understanding of the role of epigenetics in the pathophysiology of complex genetic diseases continues to expand at a rapid pace. With respect to genetically triggered TAA, epigenetics may provide insights into the observed phenotypic variation and, importantly, may assist in the development of personalised clinical management. Epigenetic mediators, such as miRNAs, are effector biological molecules whose expression can be altered through targeted mechanisms, presenting new opportunities for therapeutic strategies. However, large cohort studies capable of yielding robust clinical correlates to identify potential targets are still required. The mechanisms by which epigenetic mediators are regulated remain poorly understood and warrant further investigation. This would subsequently enable better understanding of the possible causative role of epigenetics in pathogenesis and strengthen the basis for exploring epigenetics-based therapeutics. These future research aims would significantly benefit from international multi-site collaborations and the establishment of a large international database and biobank.
Conclusion
Genetically triggered TAA is a complex condition with phenotypic heterogeneity that results in early morbidity and mortality in affected individuals. Epigenetic changes appear to play an important role in the pathogenesis and phenotypic heterogeneity of TAA and may represent feasible targets for future therapy.
Acknowledgements
The authors would like to thank Morvarid Emami for providing the histology images.
Conflict of interest
Stefanie Portelli declares that she has no conflict of interest. Elizabeth Robertson declares that she has no conflict of interest. Cassandra Malecki declares that she has no conflict of interest. Kiersten Liddy declares that she has no conflict of interest. Brett Hambly declares that he has no conflict of interest. Richmond Jeremy declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Abu-Halima M, Kahraman M, Henn D, Radle-Hurst T, Keller A, Abdul-Khaliq H, Meese E (2018) Deregulated microRNA and mRNA expression profiles in the peripheral blood of patients with Marfan syndrome. J Transl Med 16 10.1186/s12967-018-1429-3 [DOI] [PMC free article] [PubMed]
- Alajbegovic A, et al. Regulation of microRNA expression in vascular smooth muscle by MRTF-A and actin polymerization. Biochim Biophys Acta, Mol Cell Res. 2017;1864:1088–1098. doi: 10.1016/j.bbamcr.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Albinsson S, et al. Patients with bicuspid and tricuspid aortic valve exhibit distinct regional microRNA signatures in mildly dilated ascending aorta. Heart Vessel. 2017;32:750–767. doi: 10.1007/s00380-016-0942-7. [DOI] [PubMed] [Google Scholar]
- Albornoz G, Coady MA, Roberts M, Davies RR, Tranquilli M, Rizzo JA, Elefteriades JA. Familial thoracic aortic aneurysms and dissections—incidence, modes of inheritance, and phenotypic patterns. Ann Thorac Surg. 2006;82:1400–1405. doi: 10.1016/j.athoracsur.2006.04.098. [DOI] [PubMed] [Google Scholar]
- Andelfinger G, Loeys B, Dietz H. A decade of discovery in the genetic understanding of thoracic aortic disease. Can J Cardiol. 2016;32:13–25. doi: 10.1016/j.cjca.2015.10.017. [DOI] [PubMed] [Google Scholar]
- Ballantyne MD, McDonald RA, Baker AH. lncRNA/microRNA interactions in the vasculature. Clin Pharmacol Ther. 2016;99:494–501. doi: 10.1002/cpt.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker AJ, et al. Bicuspid aortic valve is associated with altered wall shear stress in the ascending aorta. Circ Cardiovasc Imaging. 2012;5:457–466. doi: 10.1161/CIRCIMAGING.112.973370. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Beg MS, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beighton P, De Paepe A, Steinmann B, Tsipouras P, Wenstrup RJ. Ehlers-Danlos syndromes: revised nosology, Villefranche, 1997. Am J Med Genet. 1998;77:31–37. doi: 10.1002/(SICI)1096-8628(19980428)77:1<31::AID-AJMG8>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Belz GG, Belz GG. Elastic properties and Windkessel function of the human aorta. Cardiovasc Drugs Ther. 1995;9:73–83. doi: 10.1007/BF00877747. [DOI] [PubMed] [Google Scholar]
- Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/jci38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon RA, et al. MicroRNA-29 in aortic dilation: implications for aneurysm formation. Circ Res. 2011;109:1115–1119. doi: 10.1161/circresaha.111.255737. [DOI] [PubMed] [Google Scholar]
- Boucher JM, Peterson SM, Urs S, Zhang C, Liaw L. The miR-143/145 cluster is a novel transcriptional target of Jagged-1/Notch signaling in vascular smooth muscle cells. J Biol Chem. 2011;286:28312–28321. doi: 10.1074/jbc.M111.221945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn GL, Svensson L, Sanchez-Blanco C, Saini M, Cope AP. Why is PTPN22 a good candidate susceptibility gene for autoimmune disease? FEBS Lett. 2011;585:3689–3698. doi: 10.1016/j.febslet.2011.04.032. [DOI] [PubMed] [Google Scholar]
- Cao H, et al. A new plasmid-based microRNA inhibitor system that inhibits microRNA families in transgenic mice and cells: a potential new therapeutic reagent. Gene Ther. 2016;23:527–542. doi: 10.1038/gt.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas CLL, et al. An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat Commun. 2018;9:1009–1014. doi: 10.1038/s41467-018-03394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion K, Dyo J, Patel V, Sasik R, Mohamed SA, Hardiman G, Nigam V. The long non-coding HOTAIR is modulated by cyclic stretch and WNT/β-CATENIN in human aortic valve cells and is a novel repressor of calcification genes. PLoS One. 2014;9:e96577. doi: 10.1371/journal.pone.0096577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee S-S. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol Ther Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K-C, Liao Y-C, Hsieh IC, Wang Y-S, Hu C-Y, Juo S-HH. OxLDL causes both epigenetic modification and signaling regulation on the microRNA-29b gene: novel mechanisms for cardiovascular diseases. J Mol Cell Cardiol. 2012;52:587–595. doi: 10.1016/j.yjmcc.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Chen KC, Wang YS, Hu CY, Chang WC, Liao YC, Dai CY, Juo SH. OxLDL up-regulates microRNA-29b, leading to epigenetic modifications of MMP-2/MMP-9 genes: a novel mechanism for cardiovascular diseases. FASEB J. 2011;25:1718–1728. doi: 10.1096/fj.10-174904. [DOI] [PubMed] [Google Scholar]
- Chen LJ, Wei SY, Chiu JJ. Mechanical regulation of epigenetics in vascular biology and pathobiology. J Cell Mol Med. 2013;17:437–448. doi: 10.1111/jcmm.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Chernov A, Strongin A. Epigenetic regulation of matrix metalloproteinases and their collagen substrates in cancer. Biomol Concepts. 2011;2:135–147. doi: 10.1515/bmc.2011.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicoine E, Esteve PO, Robledo O, Van Themsche C, Potworowski EF, St-Pierre Y. Evidence for the role of promoter methylation in the regulation of MMP-9 gene expression. Biochem Biophys Res Commun. 2002;297:765–772. doi: 10.1016/S0006-291X(02)02283-0. [DOI] [PubMed] [Google Scholar]
- Chopra S, Al-Sammarraie N, Lai Y, Azhar M. Increased canonical WNT/β-catenin signalling and myxomatous valve disease. Cardiovasc Res. 2017;113:6–9. doi: 10.1093/cvr/cvw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Climent M, Quintavalle M, Miragoli M, Chen J, Condorelli G, Elia L. TGFβ triggers miR-143/145 transfer from smooth muscle cells to endothelial cells, thereby modulating vessel stabilization. Circ Res. 2015;116:1753–1764. doi: 10.1161/circresaha.116.305178. [DOI] [PubMed] [Google Scholar]
- Clouse WD, Hallett JW, Jr, Schaff HV, Gayari MM, Ilstrup DM, Melton LJ., 3rd Improved prognosis of thoracic aortic aneurysms: a population-based study. JAMA. 1998;280:1926–1929. doi: 10.1001/jama.280.22.1926. [DOI] [PubMed] [Google Scholar]
- Conway SJ, Woster PM, Greenlee WJ, Georg G, Wang S. Epigenetics: novel therapeutics targeting epigenetics. J Med Chem. 2016;59:1247–1248. doi: 10.1021/acs.jmedchem.6b00098. [DOI] [PubMed] [Google Scholar]
- Cordes KR, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705+. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillard J, Demers M, Lavoie G, St-Pierre Y. The role of DNA hypomethylation in the control of stromelysin gene expression. Biochem Biophys Res Commun. 2006;342:1233–1239. doi: 10.1016/j.bbrc.2006.02.068. [DOI] [PubMed] [Google Scholar]
- Curradi M, Izzo A, Badaracco G, Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. 2002;22:3157–3173. doi: 10.1128/MCB.22.9.3157-3173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Backer J, Loeys B, Leroy B, Coucke P, Dietz H, De Paepe A. Utility of molecular analyses in the exploration of extreme intrafamilial variability in the Marfan syndrome. Clin Genet. 2007;72:188–198. doi: 10.1111/j.1399-0004.2007.00845.x. [DOI] [PubMed] [Google Scholar]
- De Lucia C, et al. MicroRNA in cardiovascular aging and age-related cardiovascular diseases. Front Med. 2017;4:74. doi: 10.3389/fmed.2017.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz HC, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- Elefteriades JA, Farkas EA. Thoracic aortic aneurysm: clinically pertinent controversies and uncertainties. JACC. 2010;55:841–857. doi: 10.1016/j.jacc.2009.08.084. [DOI] [PubMed] [Google Scholar]
- Elia L, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott A, et al. Biomechanics of the ascending thoracic aorta: a clinical perspective on engineering data. Can J Cardiol. 2016;32:35–47. doi: 10.1016/j.cjca.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17:756–770. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Faivre L, et al. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am J Hum Genet. 2007;81:454–466. doi: 10.1086/520125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Forte A, Galderisi U, Cipollaro M, De Feo M, Della Corte A. Epigenetic regulation of TGF-beta1 signalling in dilative aortopathy of the thoracic ascending aorta. Clin Sci (Lond) 2016;130:1389–1405. doi: 10.1042/cs20160222. [DOI] [PubMed] [Google Scholar]
- Garg V, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Germain DP. Ehlers-Danlos syndrome type IV. Orphanet J Rare Dis. 2007;2:32–32. doi: 10.1186/1750-1172-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad S, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girdauskas E, et al. Evaluation of microribonucleic acids as potential biomarkers in the bicuspid aortic valve-associated aortopathy. Interact Cardiovasc Thorac Surg. 2018;27:60–66. doi: 10.1093/icvts/ivy033. [DOI] [PubMed] [Google Scholar]
- Girdauskas E, et al. Novel approaches for BAV aortopathy prediction—is there a need for cohort studies and biomarkers? Biomolecules. 2018;8:58. doi: 10.3390/biom8030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfinger JZ, Halperin JL, Marin ML, Stewart AS, Eagle KA, Fuster V. Thoracic aortic aneurysm and dissection. JACC. 2014;64:1725–1739. doi: 10.1016/j.jacc.2014.08.025. [DOI] [PubMed] [Google Scholar]
- Gomez D, Coyet A, Ollivier V, Jeunemaitre X, Jondeau G, Michel J-B, Vranckx R. Epigenetic control of vascular smooth muscle cells in Marfan and non-Marfan thoracic aortic aneurysms. Cardiovasc Res. 2011;89:446–456. doi: 10.1093/cvr/cvq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez D, Kessler K, Michel J-B, Vranckx R. Modifications of chromatin dynamics control the Smad2 pathway activation in aneurysmal smooth muscle cells. Circ Res. 2013;113:881–890. doi: 10.1161/CIRCRESAHA.113.301989. [DOI] [PubMed] [Google Scholar]
- Gommans WM, Berezikov E. Controlling miRNA regulation in disease. Methods Mol Biol. 2012;822:1–18. doi: 10.1007/978-1-61779-427-8_1. [DOI] [PubMed] [Google Scholar]
- Greer E, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135:493–500. doi: 10.1242/dev.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth KA, et al. Prevalence, incidence, and age at diagnosis in Marfan syndrome. Orphanet J Rare Dis. 2015;10:153. doi: 10.1186/s13023-015-0369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Transl Med. 2016;14:143. doi: 10.1186/s12967-016-0893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Chang Q, Pei H, Sun X, Qian X, Tian C, Lin H. Long non-coding RNA–mRNA correlation analysis reveals the potential role of HOTAIR in pathogenesis of sporadic thoracic aortic aneurysm. J Vasc Surg. 2017;66:1305–1305. doi: 10.1016/j.jvs.2017.08.013. [DOI] [PubMed] [Google Scholar]
- Guo X, Yu L, Chen M, Wu T, Peng X, Guo R, Zhang B. miR-145 mediated the role of aspirin in resisting VSMCs proliferation and anti-inflammation through CD40. J Transl Med. 2016;14:211. doi: 10.1186/s12967-016-0961-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, et al. Long noncoding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Halushka MK, et al. Consensus statement on surgical pathology of the aorta from the Society for Cardiovascular Pathology and the Association For European Cardiovascular Pathology: II. Noninflammatory degenerative diseases—nomenclature and diagnostic criteria. Cardiovasc Pathol. 2016;25:247–257. doi: 10.1016/j.carpath.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Han Y, et al. Histone acetylation and histone acetyltransferases show significant alterations in human abdominal aortic aneurysm. Clin Epigenetics. 2016;8:3. doi: 10.1186/s13148-016-0169-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, et al. Characterization of the inflammatory cells in ascending thoracic aortic aneurysms in patients with Marfan syndrome, familial thoracic aortic aneurysms and sporadic aneurysms. J Thorac Cardiovasc Surg. 2008;136:922–929.e921. doi: 10.1016/j.jtcvs.2007.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratzka LF, et al. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with thoracic aortic disease. A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology, American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons, and Society for Vascular Medicine. J Am Coll Cardiol. 2010;55:e27–e129. doi: 10.1016/j.jacc.2010.02.015. [DOI] [PubMed] [Google Scholar]
- Holmberg J, et al. Loss of vascular myogenic tone in miR-143/145 knockout mice is associated with hypertension-induced vascular lesions in small mesenteric arteries. Arterioscler Thromb Vasc Biol. 2018;38:414–424. doi: 10.1161/ATVBAHA.117.310499. [DOI] [PubMed] [Google Scholar]
- Hrdlickova B, de Almeida RC, Borek Z, Withoff S. Genetic variation in the non-coding genome: involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta Mol basis Dis. 2014;1842:1910–1922. doi: 10.1016/j.bbadis.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Hu JZ, et al. The angiogenic effect of microRNA-21 targeting TIMP3 through the regulation of MMP2 and MMP9. PLoS One. 2016;11:e0149537. doi: 10.1371/journal.pone.0149537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey JD, Schwartz MA, Tellides G, Milewicz DM. Role of mechanotransduction in vascular biology: focus on thoracic aortic aneurysms and dissections. Circ Res. 2015;116:1448–1461. doi: 10.1161/CIRCRESAHA.114.304936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomidis JS, et al. Plasma biomarkers for distinguishing etiologic subtypes of thoracic aortic aneurysm disease. J Thorac Cardiovasc Surg. 2013;145:1326–1333. doi: 10.1016/j.jtcvs.2012.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Parada C, Chai Y. The mechanism of TGF-beta signaling during palate development. Oral Dis. 2011;17:733–744. doi: 10.1111/j.1601-0825.2011.01806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari K, Bernardi G. Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene. 2004;333:143–149. doi: 10.1016/j.gene.2004.02.043. [DOI] [PubMed] [Google Scholar]
- Janssen HLA, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Jones JA, et al. Selective microRNA suppression in human thoracic aneurysms: relationship of miR-29a to aortic size and proteolytic induction. Circ Cardiovasc Genet. 2011;4:605–613. doi: 10.1161/CIRCGENETICS.111.960419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Issa J-PJ, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- Kallenbach K, et al. Treatment of ascending aortic aneurysms using different surgical techniques: a single-centre experience with 548 patients. Eur J Cardiothorac Surg. 2013;44:337–345. doi: 10.1093/ejcts/ezs661. [DOI] [PubMed] [Google Scholar]
- Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-M. [DOI] [PubMed] [Google Scholar]
- Leeper NJ, Maegdefessel L. Non-coding RNAs: key regulators of smooth muscle cell fate in vascular disease. Cardiovasc Res. 2018;114:611–621. doi: 10.1093/cvr/cvx249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li Y, Huang J, Jiang Z, Zhong Y, Xia M, Wang H, Jiao Y. MicroRNA-145 regulates platelet-derived growth factor-induced human aortic vascular smooth muscle cell proliferation and migration by targeting CD40. Am J Transl Res. 2016;8:1813–1825. [PMC free article] [PubMed] [Google Scholar]
- Li Y, Maegdefessel L. Non-coding RNA contribution to thoracic and abdominal aortic aneurysm disease development and progression. Front Physiol. 2017;8:429. doi: 10.3389/fphys.2017.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu J, Chen M, Du B, Li Q, Xing Q, Zhang Y. A FBN1 mutation association with different phenotypes of Marfan syndrome in a Chinese family. Clin Chim Acta. 2016;460:102–106. doi: 10.1016/j.cca.2016.06.031. [DOI] [PubMed] [Google Scholar]
- Liao M, et al. A microRNA profile comparison between thoracic aortic dissection and normal thoracic aorta indicates the potential role of microRNAs in contributing to thoracic aortic dissection pathogenesis. J Vasc Surg. 2011;53:1341–1349.e1343. doi: 10.1016/j.jvs.2010.11.113. [DOI] [PubMed] [Google Scholar]
- Licholai S, Blaz M, Kapelak B, Sanak M. Unbiased profile of microRNA expression in ascending aortic aneurysm tissue appoints molecular pathways contributing to the pathology. Ann Thorac Surg. 2016;102:1245–1252. doi: 10.1016/j.athoracsur.2016.03.061. [DOI] [PubMed] [Google Scholar]
- Liu R, et al. TET2 is a master regulator of smooth muscle cell plasticity. Circulation. 2013;128:2047–2057. doi: 10.1161/CIRCULATIONAHA.113.002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Leslie KL, Martin KA. Epigenetic regulation of smooth muscle cell plasticity. Biochim Biophys Acta, Gene Regul Mech. 2014;1849:448–453. doi: 10.1016/j.bbagrm.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, et al. Flank sequences of miR-145/143 and their aberrant expression in vascular disease: mechanism and therapeutic application. J Am Heart Assoc. 2013;2:e000407. doi: 10.1161/JAHA.113.000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension. 2010;55:974–982. doi: 10.1161/hypertensionaha.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeys B. The search for genotype/phenotype correlation in Marfan syndrome: to be or not to be? Eur Heart J. 2016;37:3291–3293. doi: 10.1093/eurheartj/ehw154. [DOI] [PubMed] [Google Scholar]
- Loeys BL, Dietz HC. Loeys Dietz syndrome. Seattle: University of Washington; 2008. [Google Scholar]
- Loeys BL, et al. The revised Ghent nosology for the Marfan syndrome. J Med Genet. 2010;47:476–485. doi: 10.1136/jmg.2009.072785. [DOI] [PubMed] [Google Scholar]
- Loeys BL, et al. Aneurysm syndromes caused by mutations in the TGF-β receptor. N Engl J Med. 2006;355:788–798. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luo T, Cui S, Bian C, Yu X. Crosstalk between TGF-β/Smad3 and BMP/BMPR2 signaling pathways via miR-17–92 cluster in carotid artery restenosis. Mol Cell Biochem. 2014;389:169–176. doi: 10.1007/s11010-013-1938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maegdefessel L, et al. Inhibition of microRNA-29b reduces murine abdominal aortic aneurysm development. J Clin Invest. 2012;122:497–506. doi: 10.1172/JCI61598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Micaelo N, Beltran-Debon R, Baiges I, Faiges M, Alegret JM (2017) Specific circulating microRNA signature of bicuspid aortic valve disease. J Transl Med 15. 10.1186/s12967-017-1176-x [DOI] [PMC free article] [PubMed]
- McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKellar SH, Tester DJ, Yagubyan M, Majumdar R, Ackerman MJ, Sundt TM., 3rd Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J Thorac Cardiovasc Surg. 2007;134:290–296. doi: 10.1016/j.jtcvs.2007.02.041. [DOI] [PubMed] [Google Scholar]
- Mellis D, Caporali A. MicroRNA-based therapeutics in cardiovascular disease: screening and delivery to the target. Biochem Soc Trans. 2018;46:11–21. doi: 10.1042/BST20170037. [DOI] [PubMed] [Google Scholar]
- Melvinsdottir IH, Lund SH, Agnarsson BA, Sigvaldason K, Gudbjartsson T, Geirsson A. The incidence and mortality of acute thoracic aortic dissection: results from a whole nation study. Eur J Cardiothorac Surg. 2016;50:1111–1117. doi: 10.1093/ejcts/ezw235. [DOI] [PubMed] [Google Scholar]
- Merk DR, et al. miR-29b participates in early aneurysm development in Marfan syndrome. Circ Res. 2012;110:312–324. doi: 10.1161/circresaha.111.253740. [DOI] [PubMed] [Google Scholar]
- Michel J-B, Jondeau G, Milewicz DM. From genetics to response to injury: vascular smooth muscle cells in aneurysms and dissections of the ascending aorta. Cardiovasc Res. 2018;114:578–589. doi: 10.1093/cvr/cvy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelena HI, et al. Incidence of aortic complications in patients with bicuspid aortic valves. JAMA. 2011;306:1104–1112. doi: 10.1001/jama.2011.1286. [DOI] [PubMed] [Google Scholar]
- Milewicz DM, Regalado ES, Shendure J, Nickerson DA, Guo D-c. Successes and challenges of using whole exome sequencing to identify novel genes underlying an inherited predisposition for thoracic aortic aneurysms and acute aortic dissections. Trends Cardiovascul Med. 2013;24:53–60. doi: 10.1016/j.tcm.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill C, George SJ. Wnt signalling in smooth muscle cells and its role in cardiovascular disorders. Cardiovasc Res. 2012;95:233–240. doi: 10.1093/cvr/cvs141. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan MG, et al. Exogenous miR-29B delivery through a hyaluronan-based injectable system yields functional maintenance of the infarcted myocardium. Tissue Eng Part A. 2018;24:57–67. doi: 10.1089/ten.TEA.2016.0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, et al. Long-term miR-29b suppression reduces aneurysm formation in a Marfan mouse model. Phys Rep. 2017;5:e13257. doi: 10.14814/phy2.13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osler W. The bicuspid condition of the aortic valves. Trans Assoc Am Phys. 1886;2:185–192. [Google Scholar]
- Prakash SK, et al. A roadmap to investigate the genetic basis of bicuspid aortic valve and its complications: insights from the International BAVCon (Bicuspid Aortic Valve Consortium) J Am Coll Cardiol. 2014;64:832–839. doi: 10.1016/j.jacc.2014.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, Li L. Histone acetylation and recruitment of serum responsive factor and CREB-binding protein onto SM22 promoter during SM22 gene expression. Circ Res. 2002;90:858–865. doi: 10.1161/01.RES.0000016504.08608.B9. [DOI] [PubMed] [Google Scholar]
- Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123:11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabkin SW. Differential expression of MMP-2, MMP-9 and TIMP proteins in thoracic aortic aneurysm—comparison with and without bicuspid aortic valve: a meta-analysis. VASA. 2014;43:433–442. doi: 10.1024/0301-1526/a000390. [DOI] [PubMed] [Google Scholar]
- Rangrez AY, Massy ZA, Metzinger-Le Meuth V, Metzinger L. miR-143 and miR-145: molecular keys to switch the phenotype of vascular smooth muscle cells. Circ Cardiovasc Genet. 2011;4:197–205. doi: 10.1161/circgenetics.110.958702. [DOI] [PubMed] [Google Scholar]
- Rensen SS, Doevendans PA, van Eys GJ. Regulation and characteristics of vascular smooth muscle cell phenotypic diversity. Neth Hear J. 2007;15:100–108. doi: 10.1007/BF03085963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries RJ, Yu W, Holton N, Cao H, Amendt BA. Inhibition of the miR-17-92 cluster separates stages of palatogenesis. J Dent Res. 2017;96:1257–1264. doi: 10.1177/0022034517716915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson EN, van der Linde D, Sherrah AG, Vallely MP, Wilson M, Bannon PG, Jeremy RW. Familial non-syndromal thoracic aortic aneurysms and dissections—incidence and family screening outcomes. Int J Cardiol. 2016;220:43–51. doi: 10.1016/j.ijcard.2016.06.086. [DOI] [PubMed] [Google Scholar]
- Roman MJ, et al. Aortic dilatation associated with bicuspid aortic valve: relation to sex, hemodynamics, and valve morphology (the National Heart Lung and Blood Institute-sponsored national registry of genetically triggered thoracic aortic aneurysms and cardiovascular conditions) Am J Cardiol. 2017;120:1171–1175. doi: 10.1016/j.amjcard.2017.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers D, et al. A mutation update on the LDS-associated genes TGFB2/3 and SMAD2/3. Hum Mutat. 2018;39:621–634. doi: 10.1002/humu.23407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiattarella Gabriele G., Madonna Rosalinda, Van Linthout Sophie, Thum Thomas, Schulz Rainer, Ferdinandy Peter, Perrino Cinzia. Epigenetic modulation of vascular diseases: Assessing the evidence and exploring the opportunities. Vascular Pharmacology. 2018;107:43–52. doi: 10.1016/j.vph.2018.02.009. [DOI] [PubMed] [Google Scholar]
- Shah AA, et al. Epigenetic profiling identifies novel genes for ascending aortic aneurysm formation with bicuspid aortic valves. Heart Surg Forum. 2015;18:E134–E139. doi: 10.1532/hsf.1247. [DOI] [PubMed] [Google Scholar]
- Sherrah AG, et al. Nonsyndromic thoracic aortic aneurysm and dissection. JACC. 2016;67:618–626. doi: 10.1016/j.jacc.2015.11.039. [DOI] [PubMed] [Google Scholar]
- Sievers HH, Stierle U, Hachmann RM, Charitos EI. New insights in the association between bicuspid aortic valve phenotype, aortic configuration and valve haemodynamics. Eur J Cardiothorac Surg. 2016;49:439–446. doi: 10.1093/ejcts/ezv087. [DOI] [PubMed] [Google Scholar]
- Simion V, Haemmig S, Feinberg MW (2018) LncRNAs in vascular biology and disease. Vasc Pharmacol 10.1016/j.vph.2018.01.003 [DOI] [PMC free article] [PubMed]
- Siu SC, Silversides CK. Bicuspid aortic valve disease. JACC. 2010;55:2789–2800. doi: 10.1016/j.jacc.2009.12.068. [DOI] [PubMed] [Google Scholar]
- Song Z, Jin R, Yu S, Nanda A, Granger DN, Li G. Crucial role of CD40 signaling in vascular wall cells in neointimal formation and vascular remodeling after vascular interventions. Arterioscler Thromb Vasc Biol. 2012;32:50–64. doi: 10.1161/ATVBAHA.111.238329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Berezikov E, Choi YL, Yamashita Y, Mano H. Potential role of miR-29b in modulation of Dnmt3a and Dnmt3b expression in primordial germ cells of female mouse embryos. RNA. 2009;15:1507–1514. doi: 10.1261/rna.1418309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefsbol Trygve O. Epigenetics in Human Disease. 2018. Epigenetics of Human Disease; pp. 3–10. [Google Scholar]
- Tzemos N, et al. Outcomes in adults with bicuspid aortic valves. JAMA. 2008;300:1317–1325. doi: 10.1001/jama.300.11.1317. [DOI] [PubMed] [Google Scholar]
- van Rooij E, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone L, Caserta M, Mauro ED. Role of histone acetylation in the control of gene expression. Biochem Cell Biol. 2005;83:344–353. doi: 10.1139/o05-041. [DOI] [PubMed] [Google Scholar]
- Verhagen JMA, et al. Expert consensus recommendations on the cardiogenetic care for patients with thoracic aortic disease and their first-degree relatives. Int J Cardiol. 2018;258:243–248. doi: 10.1016/j.ijcard.2018.01.145. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. MicroRNA-17-92, a direct Ap-2alpha transcriptional target, modulates T-box factor activity in orofacial clefting. PLoS Genet. 2013;9:e1003785. doi: 10.1371/journal.pgen.1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, et al. Local and sustained miRNA delivery from an injectable hydrogel promotes cardiomyocyte proliferation and functional regeneration after ischemic injury. Nat Biomed Eng. 2017;1:983–992. doi: 10.1038/s41551-017-0157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, et al. BRG1 expression is increased in thoracic aortic aneurysms and regulates proliferation and apoptosis of vascular smooth muscle cells through the long non-coding RNA HIF1A-AS1 in vitro. Eur J Cardiothorac Surg. 2015;47:439–446. doi: 10.1093/ejcts/ezu215. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. Dual-targeted theranostic delivery of miRs arrests abdominal aortic aneurysm development. Mol Ther. 2018;26:1056–1065. doi: 10.1016/j.ymthe.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster ALH, Yan MS-C, Marsden PA. Epigenetics and cardiovascular disease. Can J Cardiol. 2013;29:46–57. doi: 10.1016/j.cjca.2012.10.023. [DOI] [PubMed] [Google Scholar]
- Wu J, et al. Progressive aortic dilation is regulated by miR-17-associated miRNAs. JACC. 2016;67:2965–2977. doi: 10.1016/j.jacc.2016.04.027. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Xin M, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Du WW, Li H, Liu F, Khorshidi A, Rutnam ZJ, Yang BB. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 2013;41:9688–9704. doi: 10.1093/nar/gkt680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Han H, De Carvalho Daniel D, Lay Fides D, Jones Peter A, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Jeremy RW. Angiotensin, transforming growth factor β and aortic dilatation in Marfan syndrome: of mice and humans. Int J Cardiol Heart Vasc. 2018;18:71–80. doi: 10.1016/j.ijcha.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CK, Xu TN, Assoian RK, Rader DJ. Mining the stiffness-sensitive transcriptome in human vascular smooth muscle cells identifies long noncoding RNA stiffness regulators. Arterioscler Thromb Vasc Biol. 2018;38:164–173. doi: 10.1161/ATVBAHA.117.310237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue M, et al. MSDD: a manually curated database of experimentally supported associations among miRNAs, SNPs and human diseases. Nucleic Acids Res. 2018;46:D181–D185. doi: 10.1093/nar/gkx1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Feng G, Wang Y, Yue Y, Zhao W. Regulation of apoptosis by long non-coding RNA HIF1A-AS1 in VSMCs: implications for TAA pathogenesis. Int J Clin Exp Pathol. 2014;7:7643–7652. [PMC free article] [PubMed] [Google Scholar]