Abstract
Tarantula’s leg muscle thick filament is the ideal model for the study of the structure and function of skeletal muscle thick filaments. Its analysis has given rise to a series of structural and functional studies, leading, among other things, to the discovery of the myosin interacting-heads motif (IHM). Further electron microscopy (EM) studies have shown the presence of IHM in frozen-hydrated and negatively stained thick filaments of striated, cardiac, and smooth muscle of bilaterians, most showing the IHM parallel to the filament axis. EM studies on negatively stained heavy meromyosin of different species have shown the presence of IHM on sponges, animals that lack muscle, extending the presence of IHM to metazoans. The IHM evolved about 800 MY ago in the ancestor of Metazoa, and independently with functional differences in the lineage leading to the slime mold Dictyostelium discoideum (Mycetozoa). This motif conveys important functional advantages, such as Ca2+ regulation and ATP energy-saving mechanisms. Recent interest has focused on human IHM structure in order to understand the structural basis underlying various conditions and situations of scientific and medical interest: the hypertrophic and dilated cardiomyopathies, overfeeding control, aging and hormone deprival muscle weakness, drug design for schistosomiasis control, and conditioning exercise physiology for the training of power athletes.
Keywords: Myosin interacting-heads motif, Muscle disease, Myosin filaments, Muscle evolution, Hypertrophic cardiomyopathy, Tarantula
Introduction
Muscle acquisition was a fundamental step during animal evolution. Muscle is a specialized contractile tissue made up of two major proteins, actin and myosin II. According to the sliding-filament model of muscle contraction (Huxley and Niedergerke 1954; Huxley and Hanson 1954), the interaction of these two proteins within each sarcomere causes the sarcomere to shorten, generating force and movement.
The myosin II molecule, one of the several members of the myosin superfamily (Coluccio 2008), is composed of two heavy chains (MHCIIs), two essential light chains (ELCs), and two regulatory light chains (RLCs) (Sellers 1999; Coluccio 2008). The two MHCIIs assemble on a long tail and two globular heads, each head with a ELC and RLC bound.
The analysis of tarantula’s leg muscle thick filaments has made them the ideal organism for the study of the structure and function of skeletal muscle thick filaments, leading to the myosin interacting-heads motif (IHM) discovery. Cryo-electron microscopy (cryo-EM) allowed the visualization of the two heads of the myosin heavy meromyosin (HMM) in 2D crystals from chicken smooth muscle, which were found to form an asymmetric arrangement, suggesting how the relaxed (switched OFF) state was achieved (Wendt et al. 1999, 2001; Liu et al. 2003). Cryo-EM allowed the visualization of both heads together with its sub-fragment 2 (S2)—missing in the 2D crystals—in native thick filament of relaxed striated muscle of tarantula Aphonopelma sp., suggesting that this “interacting-head” structure underlies the relaxed state of thick filaments in both smooth and myosin-regulated striated muscles over a wide range of species (Woodhead et al. 2005). This single molecular entity was called the myosin IHM (Alamo et al. 2008), whose structure and function is reviewed in Alamo et al. (2017a). This structural motif plays a key role in tarantula myosin filament’s cooperative phosphorylation activation (CPA) mechanism (via swaying constitutively phosphorylated free head), potentiation (via phosphorylated blocked head), and post-tetanic potentiation (Brito et al. 2011). Here, we review the studies revealing the presence of the IHM in thick filaments of many species in different muscle types and conservation of the IHM intramolecular interactions (Alamo et al. 2016). We also analyze the evolutionary relationship among the taxa possessing the IHM and propose a time-frame for its evolution. Finally, we present some recent findings on the consequences of the IHM structure and function in human physiology and pathologies.
Lesson I: the IHM is present in all muscle types and many species in both thick filaments and HMM
The striking similarity between the arrangement of the two heads of a myosin molecule in the thick filament of vertebrate smooth muscle as compared with the one of an invertebrate striated muscle lead to the suggestion that the IHM structure may underlie the relaxed state of thick filaments, in both smooth and myosin-regulated striated muscles, over a wide range of species (Woodhead et al. 2005), and with important functional differences in regards to the types of myosin-linked regulation, such as RLC phosphorylation and direct ELC Ca2+ binding (Jung et al. 2008a).
The IHM is present in thick filaments across bilaterian taxa and muscle types
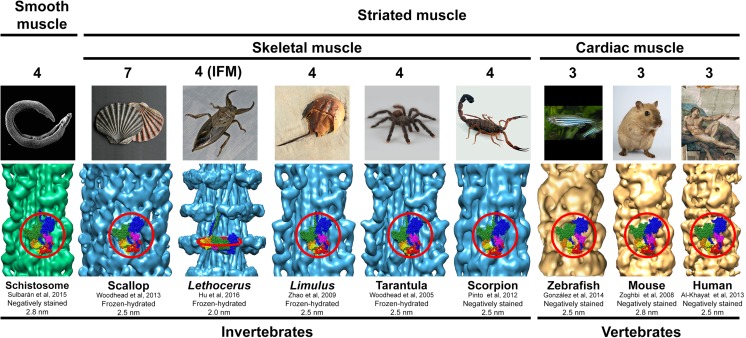
The finding of the IHM in frozen-hydrated tarantula striated muscle thick filaments promoted electron microscopy (EM) studies to search for its presence on thick filaments of other muscle types and bilaterian taxa (Fig. 1). Further cryo-EM studies confirmed its presence in striated muscle thick filaments from the horseshoe crab Limulus polyphemus (Arthropoda, Merostomata) (Zhao et al. 2009) and Placopecten magellanicus (Mollusca) (Woodhead et al. 2013) (Fig. 1). It has been confirmed to be present in negatively stained striated muscle thick filaments from scorpion Tityus discrepans (Arthropoda, Arachnida) (Pinto et al. 2012) and the scallop P. magellanicus (Al-Khayat et al. 2009), in cardiac muscle from zebrafish Danio rerio (Actinopterygii) (González-Solá et al. 2014), mouse Mus musculus (Zoghbi et al. 2008), rabbit Oryctolagus cuniculus (Al-Khayat et al. 2008), and human Homo sapiens (Al-Khayat et al. 2013), and smooth muscle from the blood-fluke Schistosoma mansoni (Platyhelminthes) (Sulbarán et al. 2015a, b) (Fig. 1). These additional studies confirmed that, depending on the lineage, the IHMs are organized in three, four, or seven coaxial helices: three helices for skeletal and cardiac muscle in Vertebrata, four helices for striated muscle in Arthropoda and smooth muscle in Platyhelminthes, and seven helices for striated muscle in Mollusca (Fig. 1). In these thick filaments, the IHMs are located approximately parallel to the filament axis, standing over (but separated) from the thick filament backbone surface. They establish interconnecting intermolecular interactions along these helices and between adjacent crowns, either between all IHMs (as in Arthropoda and Platyhelminthes), only between some crowns (as in the vertebrate’s cardiac muscle), or within each crown (as in Mollusca). By using a 3D reconstruction of frozen-hydrated Lethocerus indicus (Arthropoda, Insecta), the indirect flight muscle (IFM) thick filament (Fig. 1) (Hu et al. 2016) has been recently reported with a global resolution of 0.62 nm and with varying resolutions on the thick filament region between ~1.2 and 2.1 nm in the myosin heads region, 0.55 nm for the region where the myosin tails are packed, and 1.0 nm for the filament central paramyosin (PM) core. The heads region densities show an IHM structure perpendicularly oriented to the filament axis, in contrast to all the other muscle types studied so far, where they are parallel to the filament axis (Fig. 1). Therefore, the angle of the IHMs above the thick filament backbone seem to be variable, either approximately perpendicular (Lethocerus) or parallel (all other muscles studied so far). In addition, the Lethocerus IFM IHM appears to be freestanding with no intermolecular interactions between neighboring IHMs along the helices or within each crown (Fig. 1) and without interactions with its myosin subfragment 2 (S2). It has been recently reported that a lack of negative charge in S2 may explain why Lethocerus thick filaments display an IHM perpendicular to the filament axis, rather than a parallel one as seen in tarantula (Fee et al. 2017). Further studies are required to clarify whether this particular perpendicular angle is specific to species with S2 lacking negative charges as in Lethocerus or if it is a general characteristic only of insect IFM.
Fig. 1.
3D reconstructions from electron micrographs of negatively stained or frozen-hydrated thick filaments for smooth, skeletal, and cardiac muscles of different invertebrate and vertebrate animals. According to the presence of interacting-heads motif (IHM) on the 3D reconstructions for each species, the IHM seem to be present in the bilaterians metazoans. These thick filaments exhibits three, four, or seven helices of IHMs. All these 3D reconstructions show densities corresponding to IHMs (red circles) approximately parallel to the filament axis except the IHM for indirect flight muscle (IFM) of the insect Lethocerus, which is approximately perpendicular. Bare zone location at the top. Zebrafish 3D reconstruction reproduced with permission from González-Solá et al. (2014) and the Biophysical Journal. Images of metazoans from Wikipedia (S. mansoni: David Williams, Illinois State University, Scallop: Manfred Heyde, Lethocerus: Richard Orr, H. sapiens: Michelangelo Buonarroti, The Creation of Adam c. 1512)
The IHM is present in isolated HMM of metazoans
The EM studies on thick filaments (Fig. 1) triggered immediately the interest on understanding the evolutionary origin of the IHM, mainly through the search for the presence of the IHM in taxa lacking muscle and thick filaments, either by EM or small-angle X-ray scattering (SAXS). However, detecting the presence of IHM by 3D reconstructions of frozen-hydrated or even negatively stained isolated thick filaments is very time consuming, so an alternative approach such as the direct detection of IHM in negatively stained isolated HMM molecules was found to be more practical in that regard (Fig. 2). The IHM, which is basically an HMM with a longer S2, can be established in isolated myosin molecules or in thick filaments. In contrast, the IHM in thick filaments can establish anchoring intermolecular interactions with myosin tails in the backbone and through interconnecting intermolecular interactions between neighboring IHMs, constituting helices of IHM on the surface of the backbone.
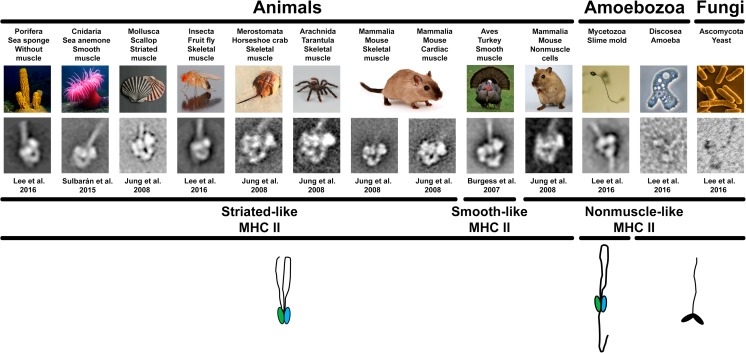
Fig. 2.
Averaged images of heavy meromyosins (HMMs) from different species. The IHM is present in the studied metazoans irrespective of the muscle type (smooth, skeletal, and cardiac muscle), nonmuscle cells, or its absence, as in sponges. The IHM presence was not observed in the yeast Schizosaccharomyces pombe (Ascomycota, Fungi) and the Acanthamoeba castellanii (Discosea, Amoebozoa) (images shown are not averaged), while a differently folded tail with extra tail–IHM interactions was observed in Dictyostelium discoideum (Mycetozoa, Amoebozoa). The presence of the IHM in sponges suggests that the IHM is present in the metazoans. The drawings at the bottom show the folded inhibited form of the myosin heads (blocked head green, free head blue) or not (black heads) and the different ways the tail pack on the two folded myosin heads. The span across species of the three MHCII types (striated-like, smooth-like, and nonmuscle-like) is shown. Electron micrographs of turkey HMM reproduced with permission from Burgess et al. (2007) and the Journal of Molecular Biology. Images of organisms from Wikipedia (sponge: Nick Hobgood, Anemone: Attrattorestrano at Italian Wikipedia, Drosophila: André Karwath, S. Pombe: David O. Morgan, A. castellanii: Lorenzo-Morales et al. 2015)
Negatively stained EM studies of HMM
Using this more practical approach, the IHM was found in the smooth muscle of turkey Meleagris gallopavo (Aves) (Burgess et al. 2007) and in the scallop Pecten maximus (Mollusca) (Jung et al. 2008a, b) (Fig. 2). The IHM was also detected in the HMM from mammals’ (mouse) skeletal, cardiac, and nonmuscle cells, as well as in tarantula (Aphonopelma sp.), horseshoe crab, and scallops (Fig. 2), suggesting that myosin head-head and head–S2 interactions are a general mechanism for switching off myosin II activity in cells (Jung et al. 2008b) and showing that the IHM can be established without additional intermolecular interactions with neighbor myosin tails. Further studies showed that the IHM is present in the sea anemone Condylactis gigantea (Cnidaria) smooth muscle HMM (Sulbarán et al. 2015a, b) (Fig. 2) extending the IHM presence to the non-bilaterian eumetazoans. More recently, the IHM was found in the HMM of animals lacking muscle, such as the sponge Cinachyra alloclada (Porifera) (Fig. 2), extending the IHM presence to the ensemble of metazoans. In contrast, the IHM is absent in the yeast Schizosaccharomyces pombe (Fungi) and the amoebozoan Acanthamoeba castellanii (Discosea), while a completely different myosin tail folding and a variation of the IHM structure was detected in the amoebozoan slime mold Dictyostelium discoideum (Mycetozoa) (Lee et al. 2016, Lee et al. in preparation) (Fig. 2).
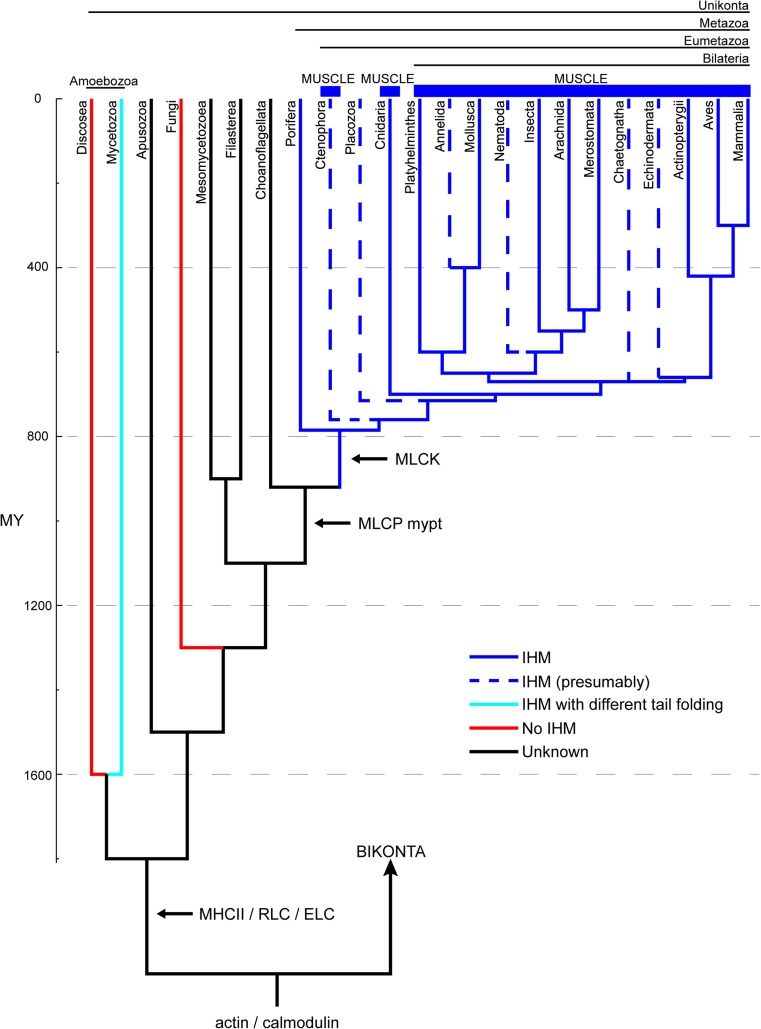
In summary, as shown in Figs. 1 and 3, the presence of the IHM in thick filaments has been confirmed in vertebrates, i.e., bony fishes, mammals, and birds; arthropods, i.e., insects, spiders, scorpions, and horseshoe crab; other invertebrates, i.e., blood-flukes and molluscs; and, as shown in Fig. 2, in the HMM of the sea anemone C. gigantea and the sponge C. alloclada (solid blue lines in Fig. 3). The IHM has not been studied in Ctenophora, Placozoa, and some bilaterian phyla, all of which representing groups that deserve to be studied by EM or SAXS (dashed blue lines in Fig. 3). However, given the phylogenetic distribution of lineages with IHM among the Metazoa, it is hypothesized here that these groups would eventually be shown to exhibit the IHM. The ensemble of the available evidence suggests that the IHM appeared with the evolution of the Metazoa, with and without muscle. The specific evidence that Porifera have the IHM suggests that either the IHM structure evolved once at the origin of Metazoa, or twice—independently—in Porifera and in Eumetazoa (Fig. 3). Further EM or SAXS studies on Ctenophora and Placozoa should help clarify this point. Additional evidence in support of the idea of the emergence of the IHM concomitantly with the evolution of metazoans comes from the fact that the IHM seems to be absent in other Unikonta, such as the fungus S. pombe and the amoebozoan A. castellanii (red lines in Fig. 3). However, analyses of the IHM presence and structure in some of Metazoa’s closest relatives, most particularly in Choanoflagellata, Filasterea, and Mesomycetozoea, will be necessary to confirm the relation between IHM and Metazoa.
Fig. 3.
Consensus phylogenetic relationships of Unikonta, i.e., Metazoa, Fungi, Amoebozoa, and some related unicellular eukaryotes, for which the IHM structure has been studied directly via electron microscopy (EM) and small-angle X-ray scattering (SAXS) (like the squid Loligo pealeii). Phylogenetic relations and node ages are based on recent studies (Parfrey et al. 2011; Cavalier-Smith et al. 2014; Erwin 2015; Telford et al. 2015). Solid blue lines: myosin II establishing the IHM structure (dashed blue lines represent unstudied metazoans presumably with IHM); cyan line: myosin II with different tail folding exhibiting extra tail–IHM interactions (as found in D. discoideum); red lines: no IHM detected; black lines: unstudied taxa. The emergence of MHCII, RLC, ELC, MLCK and MCLP mypt genes (Steinmetz et al. 2012) are shown with horizontal arrows (cf. Mycetozoa (Griffith et al. 1987 see text).
When did the IHM emerge?
It has previously been noted that the IHM appears to be shared by taxa separated by >600 MY (Jung et al. 2008a) or >500 MY (Gillilan et al. 2013) (cf. Peterson et al. 2004). However, according to the phylogenetic tree shown in Fig. 3, the IHM evolved about 800 MY ago in the ancestor of Metazoa (Fig. 3, blue lines). The evolution of this particular IHM quaternary conformation seems, therefore, to be related somehow to the emergence of coordinated movement in these multicellular organisms, and the presence of its more complex form is related with the apparition of muscle in Eumetazoa and particularly in Bilateria (i.e., sarcomeres with thick and thin filaments), which required synchronized Ca2+ regulation as provided by the related emergence of MLCK and MLCP enzymes, as well as the Ca2+ binding troponin complex (see below). It should be noted that cnidarians and ctenophores have striated muscle with considerable structural similarity to the one found in bilaterians, but lacking titin and the troponin complex, crucial components of bilaterian striated muscles (Seipel and Schmid 2005; Schuchert et al. 1993). This has been suggested as evidence for an independent and convergent evolution of striated muscle in these lineages and in bilaterians (Steinmetz et al. 2012). According to this view, the evolution of striated muscles in Cnidaria/Ctenophora and Bilateria would have occurred through the addition of different sets of proteins to a common contractile apparatus present in the ancestor of metazoans. The existence of a fully functional IHM structure in animals without true muscle, such as sponges, provides indirect evidence in support for this idea. As indicated above, analyses of Metazoa’s closest relatives, Choanoflagellata, Filasterea, and Mesomycetozoea, will, however, be necessary to confirm this and to set a precise time-frame for the evolution of the IHM. Finally, the dissimilar myosin tail folding observed in D. discoideum, implying an IHM with different tail–IHM interactions and particularly the lack of the IHM structure in other amoebozoans such as A. castellanii, strongly suggest that it represents an older and independent evolution of the IHM structure exhibiting functional differences (Bosgraaf and van Haastert 2006; Wang et al. 2011) as compared with the IHM that evolved later in Metazoa. Supporting this proposal, Fig. 2 shows that D. discoideum, in which MLCK and MLCP have been reported to be present (Griffith et al. 1987) (cf. Steinmetz et al. 2012), exhibits a completely different myosin tail folding and a variation of the IHM structure (Fig. 3, cyan line). Further investigations on the presence and structure of IHM and the presence of MLCK and MLCP in Mycetozoa and other slime molds are very important to clarify this point, as well as achieving a quasi-atomic model of the D. discoideum IHM.
Lesson II: the evolution of myosin IHM: implications of its intramolecular interactions and evolutionary advantages
In this section, we review the evidences of the IHM presence shown in Figs. 1 and 2, aiming to understand (i) its evolutionary advantages, (ii) the emerging of the components needed for its assembling, and (iii) the conservation of its intramolecular interactions.
Paired myosin heads constituting an IHM provides evolutionary advantages over unpaired heads
The myosin II molecule is the only myosin molecule capable of generating filaments (Dulyaninova and Bresnick 2013; Sellers 2000). Besides myosin II, the main components of these myosin thick filaments are paramyosin, myosin binding protein C (MyBP-C), filagenins, titin, etc., depending on the species (Craig and Woodhead 2006). Therefore, the study of the sequences of the myosin II molecule components, MHCII, ELC, and RLC, is very important to understand the relationship between the conservation of certain myosin sequences and the IHM functioning, as well as the association between mutations on these sequences and certain muscle malfuctioning and pathologies.
Each head of the myosin II molecule carries out three basic functions: (1) binding to actin, (2) hydrolyzing ATP, and (3) producing force when attached to actin. The myosin II is, therefore, essential for force production and movement. On the other hand, the arrangement of these two heads into an IHM asymmetric structure produces important additional properties that are not observed in single myosin heads, such as: (4) ATPase self-inhibition on both heads, (5) free head swaying, (6) blocked head force potentiation, and (7) post-tetanic potentiation (Brito et al. 2011). These four additional functions endorse the organisms that can establish functional IHM molecules, i.e., which possess MHCII, ELC, and RLC with key additional evolutionary advantages, such as the helical organization of the IHMs in thick filaments, allowing the movement of actin filaments towards the center of the myosin thick filament, with the resulting generation of force and force potentiation. This configuration is, therefore, related to muscle evolution as proposed below (Sulbarán et al. 2014, 2015, b; Alamo et al. 2016; Lee et al. 2016). Besides, this configuration appears to form transient filaments in different locations needed for the functioning of simpler interdigitating arrays of actin and myosin with a more irregular organization, such as in vertebrate smooth muscle or nonmuscle cells (Seipel and Schmid 2005). Furthermore, mutations in the myosin II gene could produce slightly altered MHCIIs with different intramolecular interactions (Alamo et al. 2017b), which, in turn, could be responsible for IHM structural variations, such as striated-like, smooth-like, and nonmuscle-like MHCII sequences, or IHM structure variations like in D. discoideum. In addition, mutations in the ELC and RLC genes could be responsible for the structural modifications endorsing the IHM with direct ELC–Ca2+ binding or RLC phosphorylation regulation (Trybus 1994).
It has been suggested that the IHM may have emerged as a small, self-inhibiting unit before the evolution of thick filaments (Sulbarán et al. 2013). The emergence of the IHM self-relaxation mechanism should have endorsed evolutionary advantages to the myosin molecule by itself, like in nonmuscle cells, allowing simple force production and movement. Later, when assemblies of myosin molecules became organized as thick filaments and sarcomeres, further evolutionary advantages were achieved, including cooperativity between the different myosin heads. An additional advantage of the emergence of IHM, based on the conservation of IHM intramolecular interactions across metazoans and the presence of IHM in bilaterians, would be the saving of ATP in skeletal, cardiac, and smooth muscle achieved through the super-relaxed (SRX) state (Alamo et al. 2016), reviewed in (Alamo et al. 2017a).
Intramolecular interactions and assembling of the IHM
The assemblage of an IHM requires a certain number of intramolecular interactions (Woodhead et al. 2005; Alamo et al. 2008, 2016). Specifically, five of such intramolecular interactions, the priming/forming “f” and “g”, and the stabilizing “a”, “d”, and “e”, must be fully established for the IHM to be formed (Alamo et al. 2016). In thick filaments, the IHM also establishes intermolecular interactions with neighbor myosin heads and tails, which are important for the generation of the IHM helices on thick filament backbone surface. However, the IHM can be established in the HMM without these additional intermolecular interactions with neighbor myosin heads and tails (Fig. 2). According to the comparison of MHCII sequence alignments between selected lineages within Unikonta and the tarantula Aphonopelma sp., Alamo et al. (2016) have suggested that the required amino acid sequences at the stabilizing (“d”) and priming forming (“f”) IHM intramolecular interactions are present in metazoans but not in fungi or amoebozoans. Indeed, using this approach, Alamo et al. (2016) predicted that the IHM should be present across metazoans such as D. melanogaster, the sea anemone C. gigantea, and the sponge Amphimedon sp., but not in the yeast S. pombe or the amoebozoans D. discoideum and A. castellanii. In addition, the conservation of intramolecular interactions across animal species and the presence of IHM in bilaterians suggested that an SRX state should be maintained playing a role in saving ATP in muscle (Alamo et al. 2016). In agreement with these predictions, the IHM has been detected in isolated embryonic and adult HMM molecules of D. melanogaster IFM, the sea anemone C. gigantea, and the sponge C. alloclada (Lee et al. in preparation) (Fig. 2). Furthermore, as predicted by Alamo et al. (2016), the IHM is absent in the amoebozoan A. castellanii and the yeast S. pombe (Lee et al. 2016) (Fig. 2), confirming that these lineages lack the proper charged amino acids in the sequences involved in the intramolecular interactions necessary for the IHM quaternary conformation assemblage. However, a different myosin tail folding and a variation of the IHM structure were detected in the amoebozoan slime mold D. discoideum (Lee et al. 2016) (Fig. 2). As shown in Fig. 2, the myosin tail folding in this species is very different to the simple myosin tail folding observed in metazoans, possibly exhibiting different intramolecular interactions and additional tail–IHM interactions not considered in the analysis done by Alamo et al. (2016). The presence of the IHM in metazoans and its absence in the yeast S. pombe and the amoebozoan A. castellanii, all containing the IHM protein core set (see below), support the proposal that the actual assembly of the quaternary IHM structure requires the establishment of five appropriate intramolecular interactions (Alamo et al. 2016), which appears to be absent in D. discoideum as a consequence of additional tail–IHM interactions and very different intramolecular interactions.
An assembled IHM requires ELC and RLC to incorporate Ca2+ regulation in a functional IHM
In addition to the intramolecular interactions mentioned above, the assembling of a functional IHM requires the presence of MHCII, RLC, and ELC proteins (i.e., the IHM protein core set). Endorsing an assembled IHM with regulation by Ca2+ requires either an ELC capable of binding Ca2+, as in the anterior byssus retractor muscle of some Mollusca or the presence of MLCK and calmodulin to allow the myosin RLC phosphorylation, and the presence of MLCP to allow its dephosphorylation, so the assembled IHM can interact with actin to produce force and regulate it. Therefore, for assembling and making a Ca2+ regulated IHM, the IHM protein core set should include actin and MHCII with the required appropriate IHM intramolecular interactions, and, depending on the Ca2+ regulation type, either an ELC capable of Ca2+ binding or a phosphorylatable RLC, calmodulin, MLCK, and MLCP. However, it seems that these necessary genes did not appear at the same time but progressively: actin and calmodulin were present in the ancestor of Bikonta and Unikonta, while MHCII, RLC, and ELC appeared only in the stem Unikonta (an MHCII homolog has been found in Naegleria gruberi (Bikonta, Excavata) (Fritz-Laylin et al. 2010), but whether this represents a deeper ancestry or a case of horizontal gene transfer remains to be evaluated). MLCP myosin binding subunit gene (mypt) appeared in the common ancestor of Choanoflagellata and Metazoa, MLCP protein phosphatase 2 (pp1) gene in Unikonta and Bikonta and the MLCK gene in Metazoa (Steinmetz et al. 2012). Therefore, based only on the temporal emergence of the IHM protein core set genes, we conclude that, in spite that MHCII, RLC, ELC, actin, and calmodulin are present in the ensemble of Unikonta, only metazoans are capable of building an IHM with Ca2+ regulation via RLC phosphorylation (Fig. 3). Supporting this conclusion, the results in Fig. 2 show that the IHM is present in metazoans but absent in the yeast S. pombe and the amoebozoan A. castellanii, which lack the MLCP mypt and MLCK genes.
The MHCII sequence appears to convey the assembly and type of myosin/thick filament
The three types of MHCII sequences, striated-like, smooth-like, and nonmuscle-like (Fig. 2), have been discussed in relation to the presence of the IHM and thick filament type (Alamo et al. 2016). The nonmuscle type seems to be present in all Unikonta, while the smooth-like and striated-like types seem to be associated with the emergence of muscle (Hooper and Thuma 2005). The nonmuscle-like MHCII assembles as minifilaments in the amoebozoans D. discoideum and A. castellanii, but not in the sponge C. alloclada. The striated-like MHCII make bipolar thick filaments in striated and smooth muscle thick filaments (e.g., anemone, blood-fluke) (Sulbarán et al. 2015a, b) but not in the sponge (that lack muscle and thick filaments) or vertebrate smooth muscle that make specialized side-polar thick filaments (Alamo et al. 2016). Therefore, the type of MHCII does not only convey the specific amino acid sequences required for establishing the IHM intramolecular interactions, but also appears to convey the tail packing associated with the filament presence and filament type (Alamo et al. 2016).
Lesson III: IHM implications in human diseases
Recently, an increasing interest has emerged on the human IHM structure and its associated SRX energetics consequences for understanding the structural basis of human muscle diseases, like some cardiomyopathies and muscle loss associated with aging and hormone deprival, muscle force conditioning enhancement by exercise training, the design of drugs targeting the IHM and SRX to alleviate obesity and type 2 diabetes, and parasitic diseases control.
Human cardiomyopathies
Hypertrophic cardiomyopathy (HCM) and dilated cardiomyopathies (DCM) are diseases of the human myocardium caused by variants in genes encoding sarcomere proteins (Geisterfer-Lowrance et al. 1990; Seidman and Seidman 1991; Konno et al. 2010). Three of these human genes (MYH7, MYL2, and MYL3) encode respectively the three human cardiac IHM components, β-MHCII, ELC, and RLC. The proposition that HCM is linked to variants that would have an effect on the formation of IHM has been proposed in several studies that have mapped HCM variants on the IHM structure. Using a model built with the human S2 crystal model PDB 2FXM (Blankenfeldt et al. 2006) and the chicken smooth muscle myosin PDB 1I84 model (Liu et al. 2003) fitted to the tarantula thick filament 3D map (Woodhead et al. 2005), it was firstly pointed out the involvement of HCM charge-inverting variants E924K, E927K, E930K, and E935K were located in the negatively charged Ring 1 of S2 (Blankenfeldt et al. 2006). Using the tarantula IHM PDB 3DTP quasi-atomic model, the charge-changing (R403Q/L/W, E224K, E927K, E930K, E935K) HCM variants were found to be involved in the intermolecular interaction “a” between the free head and S2, suggesting that HCM charge-changing variants could be involved specifically in altering IHM intramolecular interactions (Alamo et al. 2008). It has been proposed that this interaction is likely to be important in maintaining the relaxed conformation of myosin in the thick filament, and variants in the S2 negatively charged Ring 1 may lead to more disordered heads (Alamo et al. 2008), and perhaps some dysregulation of contraction (Colegrave and Peckham 2014). As the negatively charged Ring 2 is likely to be a site of interaction for MyBP-C (Gruen and Gautel 1999), it has been proposed that variants on it could potentially affect the interaction of MyBP-C with S2, modulating contraction by MyBP-C (Colegrave and Peckham 2014). Ten HCM and 27 DCM variants were mapped to the tarantula IHM PDB 3DTP, suggesting a random variants distribution rather than a disease-specific clustering (Waldmüller et al. 2011). The 3DTP model was fitted against the mouse cardiac muscle thick filament 3D map EMD-1465 (Moore et al. 2012), showing that the R403Q variant phenotype (cf. Alamo et al. 2008) could result from altered head–head interactions, suggesting that potential effects of the variants on relaxed head–head and head–tail interactions are probably important for understanding the mutant phenotype (Moore et al. 2012). In consequence, Moore et al. (2012) suggested that: (1) HCM variants map to regions that could alter the IHM, (2) variant-induced disruption of thick filament regulation could alter heart energy consumption, and (3) variants could induce a disruption of the distribution of heads between DRX and SRX states, potentially leading to hypertrophy via an altered energetic state of the heart (Ashrafian et al. 2003).
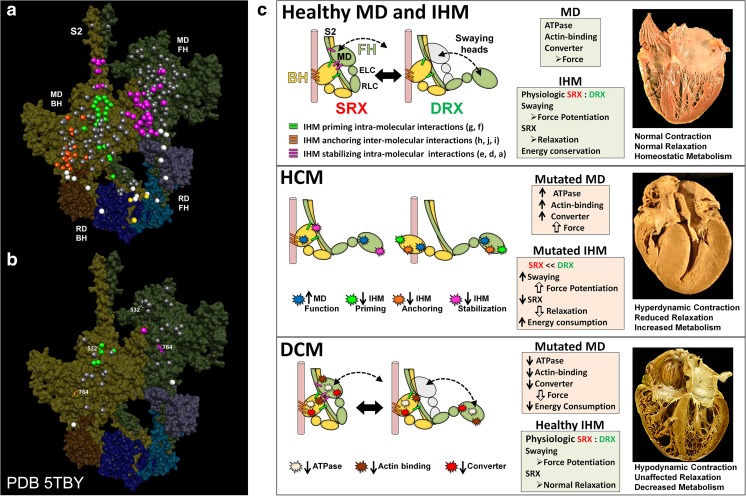
Direct evidences for these earlier suggestions have been provided recently by the homologous human β-cardiac IHM structure (PDB 5TBY) (Fig. 4a, b), based on the tarantula PDB 3JBH IHM model, which has allowed mapping the variants from 6112 HCM and 1315 DCM patients and 33,370 controls (Alamo et al. 2017b). The mapping (Fig. 4a, b) showed that these variants cluster on the IHM intramolecular interactions: HCM variants (72% that changed electrostatic charges) disproportionately altered IHM interaction residues (expected 23%; HCM 54%, p = 2.6 × 10−19; DCM 26%, p = 0.66; controls 20%, p = 0.23). The location of the HCM variants (Fig. 4a) predict impaired IHM formation and stability and attenuation of the SRX state, accounting for altered contractility, reduced diastolic relaxation, and increased energy consumption, which fully characterize HCM pathogenesis (Fig. 4c). This study demonstrated that HCM and DCM variants differentially impact myosin motor domain (MD) functions and specific IHM interactions, suggesting the structural mechanisms for failed contractile power in DCM and the markedly compromised relaxation and energetics in HCM (Fig. 4c) (Ashrafian et al. 2003, 2011). These conclusions suggested that the identification of IHM residues in PDB 5TBY involved in relaxation could provide new opportunities to target the IHM with small molecules like MYK-461 (Green et al. 2016) to improve HCM pathophysiology and other heart diseases with diastolic dysfunction and by extending these analyses to skeletal myopathies so that further insights into critical roles of the highly conserved IHM in the relaxation of striated muscles could be expected. With that in mind, two very important advances in the future would be the solving the human β-cardiac HMM atomic structure by X-ray crystallography and achieving the human β-cardiac HMM and IHM near-atomic structure by cryo-EM (cf. Yang et al. 2017).
Fig. 4.
Assessment of the effects of hypertrophic (HCM) and dilated (DCM) variants according to their localization on the same positions on the blocked (BH) and free (FH) heads but exposed to different environments. Mapping of pathogenic and likely pathogenic HCM (a) and DCM (b) variants on the homologous human myosin β-cardiac IHM PDB 5TBY, based on the tarantula PDB 3JBH quasi-atomic model. Each variant appears as a pair, one located on or associated with the blocked head (olive) and one on the free head (green). Essential light chain ELC (BH, brown; FH, purple), regulatory light chain RLC (BH, dark blue; FH, light blue). a HCM variants that alter residues involved in IHM interactions (73/135 variants, 54%) are represented by colored balls: priming, green (“f” and “g”, see interaction colors code on c top panel); anchoring, orange (“i” and “j”); stabilizing, magenta (“a”, “d”, and “e”); scaffolding, white (ELC–MHC and RLC–MHC); RLC–RLC interface, yellow. Variants that do not alter residues involved in IHM interactions are shown in gray. b DCM variants (7/27; 26%) colored as described above. c The molecular pathogenesis of HCM and DCM assessed in the context of the IHM paradigm. Myosin interactions involved in IHM assembly and myosin motor domain (MD) functions that are altered by variants are depicted. Top panel: Relaxed healthy cardiac muscle contains myosin heads populations in the super-relaxed (SRX) state (left) with lowest ATP consumption and a disordered relaxed (DRX) state (right) with swaying free heads that generate force with higher ATP consumption (see Alamo et al. 2016). The population of cardiac myosins in SRX is more stable than in skeletal muscle (Hooijman et al. 2011), which supports physiologic contraction and relaxation, energy conservation, and normal cardiac morphology. Middle panel: HCM myosin variants (colored stars) both alter residues involved in MD functions (causing increased biophysical power, Tyska et al. 2000) and destabilize IHM interactions (particularly those with altered electrostatic charge). Reduced populations of myosins in the SRX state and increased populations of myosins in DRX as well as enhanced MD properties will result in increased contractility, decreased relaxation, and increased ATP consumption, the three major phenotypes observed in HCM hearts. Compensatory signals may promote ventricular hypertrophy. Bottom panel: MYH7 DCM variants (colored stars) have modest effects on IHM interactions but substantially reduce MD functions, particularly nucleotide binding, resulting in reduced ATP consumption and sarcomere power (Schmitt et al. 2006), with minimal impact on relaxation and overall diminished contractility. Compensatory signals result in ventricular dilatation to maintain circulatory demands in DCM hearts. Reproduced with permission from Alamo et al. (2017b) and eLife
Recently, Nag et al. (2017) built a homology model for a human β-cardiac myosin IHM using the earlier tarantula PDB 3DTP (Alamo et al. 2008) and the human β-cardiac motor domain crystal structure PDB 4P7H, as well as a human cardiac MyBP-C homology model based on the known structures of some of its domains. It was found that four myosin HCM-causing mutations—but not two other mutations—weakened the IHM S1–S2 interaction. It was proposed that clinical hypercontractility caused by HCM and cMyBP-C mutations is due to an increase in the number of myosin heads (S1) that are accessible for force production. This hypothesis was based on: (1) the demonstration of myosin tail (S2)-dependent functional regulation of actin-activated human β-cardiac myosin ATPase, (2) that both S2 and MyBP-C bind to S1 and that phosphorylation of either S1 or MyBP-C weakens these interactions. In addition, it was proposed a model involving multiple interactions, including those with myosin’s own S2 and MyBP-C, that hold myosin in a sequestered state.
Muscle loss associated with aging and hormone deprival
Sarcopenia, the loss of skeletal muscle mass and function with advancing age, is a significant cause of disability and loss of independence in the elderly and, thus, represents a formidable challenge for the aging population (Christensen et al. 2009). The molecular mechanism of sarcopenia-associated muscle dysfunction remains poorly understood, but previous studies have shown that sarcopenia-associated muscle dysfunction is associated with decreased force production, maximal shortening velocity, and Ca2+ sensitivity of force (Gregorich et al. 2016). As previously mentioned, all these processes are strongly associated to the myosin activation mechanism, orchestrated by the RLC phosphorylation and the underlying IHM structure. Particularly, the loss of strength on female skeletal muscle as a consequence of aging and loss of ovarian hormone production and the disruption of the myosin SRX by estradiol deficiency has led to the suggestion that SRX (through the IHM) could be targetable for strategies to offset weakness and metabolic changes that occur with age in human female muscle (Colson et al. 2015).
Conditioning exercise training of power athletes
Understanding the conditions in which a single maximal isometric contraction affects the contractile performance of the human plantar flexor muscle is important for the training of power athletes (Gago et al. 2014b). Post-activation potentiation (PAP), an acute enhancement of muscle contractile properties and performance subsequent to a maximal conditioning contraction, is linked to RLC phosphorylation (Gago et al. 2014a, 2017). As these authors reported, PAP observed after a conditioning task coexists with fatigue and depends on strength level, muscle fiber type, and age. The training conditioning should be performed in a position such that full activation should be easy to achieve and tailored to mach an athlete or group of athlete’s current status and characteristics, maximizing performance in a specific sporting event (Gago 2016). The IHM CPA mechanism should help yield a better understanding of the human PAP and its use on conditioning exercise training of Olympic power athletes.
Obesity and type 2 diabetes
An important consequence of the IHM asymmetric structure is that the ATPases activities of its two heads are switched off producing the myosin SRX state, reviewed in Alamo et al. (2017a), and detected with a very slow release of ATP in tarantula muscle (Naber et al. 2011). The transition from low ATPase activity in the SRX state to the higher ATPase activity in the DRX state increases the skeletal muscle metabolic rate, with implications for the treatment of some diseases, as described by Cooke (2011) on the role of the IHM OFF state in whole body metabolism, and IHM as a therapeutic target for obesity. A key factor for achieving the SRX state is that the RLCs of the blocked and free heads should bind together, forming the RLC–RLC interface. The tarantula IHM 3DTP and 3JBH structures have been used for searching small molecules that could destabilize this interface, aiming to develop drugs to alleviate some human diseases. Spectroscopic studies of the rabbit skeletal muscle SRX state support the presence of this RLC–RLC putative interface, showing that the divalent cation bound to the RLC and the RLC NTE are involved in the SRX state stability (Nogara et al. 2016b). These authors found a probe that allows measuring the SRX state population, enabling high-throughput screens for finding potential pharmaceuticals targeting this interface. Their results using this probe suggest that other small molecules bound to this interface could also destabilize the SRX state and provide a screening method for searching small molecules that destabilize this interface. As the SRX to DRX transition increases the muscle metabolic rate, finding small molecules that destabilize the RLC–RLC interface could lead to potentially effective treatments for obesity and type 2 diabetes, as the SRX state destabilization produced by these small molecules should increase the metabolic rate of resting skeletal muscle, increasing the calories consumption. Using this screening method, these authors identified that piperine destabilizes the SRX state (Nogara et al. 2016a). Piperine, a black pepper alkaloid, can mitigate obesity and type 2 diabetes in rodent models. In consequence, these authors concluded that up-regulation of resting muscle metabolism could treat obesity and type 2 diabetes and that piperine would provide a useful lead compound for the development of these therapies.
Parasitic diseases control
Schistosomes like S. mansoni and the Chinese liver fluke Clonorchis sinensis are human parasites. Schistosomiasis is the third most devastating tropical disease globally, infecting 240 million humans (Ross et al. 2017). C. sinensis is endemic to Japan, China, Taiwan, and Southeast Asia, causing clonorchiasis, infecting more than 15 million humans (Lai et al. 2016). Currently, a small molecule, praziquantel (PZQ), is the unique drug in use against schistosomiasis (Bergquist et al. 2017) and clonorchiasis (Lai et al. 2016). PZQ produces a spam of the S. mansoni smooth muscle, that, together with host immune response, induces its death (Doenhoff et al. 2008) and has been reported to bind the S. mansoni myosin RLC (Gnanasekar et al. 2009). In view of the emerging resistance of schistosomes to PZQ (Bergquist et al. 2017), it should be important to disclose the molecular basis of the PZQ action on the schistosome RLC, aiming to further improve the PZQ molecule, modifying it to increase its pharmacological potency, or new anti-schistosomal drug discovery. The structure of the thick filaments of the smooth muscle of S. mansoni and the striated muscle of tarantula are very similar, exhibiting the presence of the IHM (PDB 3JAX homologously based on the tarantula 3DTP IHM model) (Fig. 1) (Sulbarán et al. 2015a, b). Additionally, the S. mansoni RLC exhibits a long NTE with MLCK and PKC consensus sequences, similarly to in tarantula (Alamo et al. 2008), exhibiting a 100% sequence conservation between them (cf. 40.3% in C. sinensis). These similarities suggest the possible presence of myosin-linked RLC phosphorylation regulation in S. mansoni. Consequently, further structural research on their IHM should elucidate whether PZQ is involved in the stability of the RLC–RLC interface. Apart from tarantula, S. mansoni, and C. sinensis, there are only seven more species reported to exhibit long RLC NTEs (Alamo et al. 2008), as all the remaining species exhibit short NTEs (Brito et al. 2011). Interestingly, the other species with long RLC NTEs include three transmitting-disease mosquitoes of the genus Aedes (dengue fever, yellow fever, Zika, Chikungunya, and lymphatic filariasis), Culex (West Nile virus, Japanese and St. Louis encephalitis, filariasis, and avian malaria), and Anopheles (human malaria), apart from three insects (D. melanogaster, Lethocerus, cricket Gryllotalpa) and the giant tube worm Riftia pachyptila (Annelida) not involved in human disease. The RLC similarity between these long RLC NTEs species suggests that the IHM and its RLC–RLC interface of these mosquitoes should be further studied in this context.
Conclusions and perspectives
The myosin interacting-heads motif (IHM) appears to be oriented parallel to the filament axis in the thick filaments of most species. The IHM evolved about 800 MY ago in the ancestor of Metazoa and independently and most probably with functional differences in the lineage leading to the slime mold Dictyostelium discoideum (Mycetozoa). Future studies will allow assessing the IHM presence by electron microscopy (EM) and small-angle X-ray scattering (SAXS) in Apusozoa, Choanoflagellata, Filasterea, and Mesomycetozoea. Hopefully, IHM studies will aid in the amelioration of some human diseases like hypertrophic (HCM) and dilated (DCM) cardiomyopathies, muscle loss associated with aging and hormone deprival, the design of drugs targeting the IHM and super-relaxed (SRX) state to alleviate obesity and type 2 diabetes, and parasitic diseases control.
Acknowledgements
We thank Dr. Gustavo Márquez for help with the manuscript. Molecular graphics images were produced using the UCSF Chimera package (Pettersen et al. 2004) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by the National Institutes of Health grant P41 RR-01081). This work was supported in part by Centro de Biología Estructural del Mercosur (http://www.cebem-lat.org) (to R.P.) and the Howard Hughes Medical Institute (to R.P.).
We dedicate this review to the memory of Dr. Hugh E. Huxley.
Conflict of interest
Lorenzo Alamo declares that he has no conflicts of interest. Antonio Pinto declares that he has no conflicts of interest. Guidenn Sulbarán declares that he has no conflicts of interest. Jesús Mavárez declares that he has no conflicts of interest. Raúl Padrón declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on ‘Latin America’ edited by Pietro Ciancaglini and Rosangela Itri.
References
- Alamo Lorenzo, Koubassova Natalia, Pinto Antonio, Gillilan Richard, Tsaturyan Andrey, Padrón Raúl. Lessons from a tarantula: new insights into muscle thick filament and myosin interacting-heads motif structure and function. Biophysical Reviews. 2017;9(5):461–480. doi: 10.1007/s12551-017-0295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo L, Qi D, Wriggers W, Pinto A, Zhu J, Bilbao A, Gillilan RE, Hu S, Padrón R. Conserved intramolecular interactions maintain myosin interacting-heads motifs explaining tarantula muscle super-relaxed state structural basis. J Mol Biol. 2016;428:1142–1164. doi: 10.1016/j.jmb.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo L, Ware JS, Pinto A, Gillilan RE, Seidman JG, Seidman CE, Padrón R. Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife. 2017;6:e24634. doi: 10.7554/eLife.24634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo L, Wriggers W, Pinto A, Bártoli F, Salazar L, Zhao FQ, Craig R, Padrón R. Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J Mol Biol. 2008;384:780–797. doi: 10.1016/j.jmb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khayat HA, Kensler RW, Squire JM, Marston SB, Morris EP. Atomic model of the human cardiac muscle myosin filament. Proc Natl Acad Sci U S A. 2013;110:318–323. doi: 10.1073/pnas.1212708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khayat HA, Morris EP, Kensler RW, Squire JM. Myosin filament 3D structure in mammalian cardiac muscle. J Struct Biol. 2008;163:117–126. doi: 10.1016/j.jsb.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khayat HA, Morris EP, Squire JM. The 7-stranded structure of relaxed scallop muscle myosin filaments: support for a common head configuration in myosin-regulated muscles. J Struct Biol. 2009;166:183–194. doi: 10.1016/j.jsb.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Ashrafian H, McKenna WJ, Watkins H. Disease pathways and novel therapeutic targets in hypertrophic cardiomyopathy. Circ Res. 2011;109:86–96. doi: 10.1161/CIRCRESAHA.111.242974. [DOI] [PubMed] [Google Scholar]
- Ashrafian H, Redwood C, Blair E, Watkins H. Hypertrophic cardiomyopathy: a paradigm for myocardial energy depletion. Trends Genet. 2003;19:263–268. doi: 10.1016/S0168-9525(03)00081-7. [DOI] [PubMed] [Google Scholar]
- Bergquist R, Utzinger J, Keiser J. Controlling schistosomiasis with praziquantel: how much longer without a viable alternative? Infect Dis Poverty. 2017;6:74. doi: 10.1186/s40249-017-0286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenfeldt W, Thomä NH, Wray JS, Gautel M, Schlichting I. Crystal structures of human cardiac beta-myosin II S2-delta provide insight into the functional role of the S2 subfragment. Proc Natl Acad Sci U S A. 2006;103:17713–17717. doi: 10.1073/pnas.0606741103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosgraaf L, van Haastert PJ. The regulation of myosin II in Dictyostelium. Eur J Cell Biol. 2006;85:969–979. doi: 10.1016/j.ejcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Brito R, Alamo L, Lundberg U, Guerrero JR, Pinto A, Sulbarán G, Gawinowicz MA, Craig R, Padrón R. A molecular model of phosphorylation-based activation and potentiation of tarantula muscle thick filaments. J Mol Biol. 2011;414:44–61. doi: 10.1016/j.jmb.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, Knight PJ. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J Mol Biol. 2007;372:1165–1178. doi: 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE, Snell EA, Berney C, Fiore-Donno AM, Lewis R. Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa. Mol Phylogenet Evol. 2014;81:71–85. doi: 10.1016/j.ympev.2014.08.012. [DOI] [PubMed] [Google Scholar]
- Christensen K, Doblhammer G, Rau R, Vaupel JW. Ageing populations: the challenges ahead. Lancet. 2009;374:1196–1208. doi: 10.1016/S0140-6736(09)61460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrave M, Peckham M. Structural implications of beta-cardiac myosin heavy chain mutations in human disease. Anat Rec (Hoboken) 2014;297:1670–1680. doi: 10.1002/ar.22973. [DOI] [PubMed] [Google Scholar]
- Colson BA, Petersen KJ, Collins BC, Lowe DA, Thomas DD. The myosin super-relaxed state is disrupted by estradiol deficiency. Biochem Biophys Res Commun. 2015;456:151–155. doi: 10.1016/j.bbrc.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio LM. Myosin: a superfamily of molecular motors. Dordrecht: Springer; 2008. [Google Scholar]
- Cooke R. The role of the myosin ATPase activity in adaptive thermogenesis by skeletal muscle. Biophys Rev. 2011;3:33–45. doi: 10.1007/s12551-011-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Woodhead JL. Structure and function of myosin filaments. Curr Opin Struct Biol. 2006;16:204–212. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Doenhoff MJ, Cioli D, Utzinger J. Praziquantel: mechanisms of action, resistance and new derivatives for schistosomiasis. Curr Opin Infect Dis. 2008;21:659–667. doi: 10.1097/QCO.0b013e328318978f. [DOI] [PubMed] [Google Scholar]
- Dulyaninova NG, Bresnick AR. The heavy chain has its day: regulation of myosin-II assembly. BioArchitecture. 2013;3:77–85. doi: 10.4161/bioa.26133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin DH. Early metazoan life: divergence, environment and ecology. Philos Trans R Soc Lond Ser B Biol Sci. 2015;370:20150036. doi: 10.1098/rstb.2015.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fee Lanette, Lin Weili, Qiu Feng, Edwards Robert J. Myosin II sequences for Lethocerus indicus . Journal of Muscle Research and Cell Motility. 2017;38(2):193–200. doi: 10.1007/s10974-017-9476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, Shu S, Neupane R, Cipriano M, Mancuso J, Tu H, Salamov A, Lindquist E, Shapiro H, Lucas S, Grigoriev IV, Cande WZ, Fulton C, Rokhsar DS, Dawson SC. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Gago P (2016) Post activation potentiation: modulating factors and mechanisms for muscle performance. Ph.D. thesis. The Swedish School of Sport and Health Sciences, Stockholm
- Gago P, Arndt A, Ekblom MM. Post activation potentiation of the plantarflexors: implications of knee angle variations. J Hum Kin. 2017;57:29–38. doi: 10.1515/hukin-2017-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago P, Arndt A, Tarassova O, Ekblom MM. Post activation potentiation can be induced without impairing tendon stiffness. Eur J Appl Physiol. 2014;114:2299–2308. doi: 10.1007/s00421-014-2945-3. [DOI] [PubMed] [Google Scholar]
- Gago P, Marques MC, Marinho DA, Ekblom MM. Passive muscle length changes affect twitch potentiation in power athletes. Med Sci Sports Exerc. 2014;46:1334–1342. doi: 10.1249/MSS.0000000000000245. [DOI] [PubMed] [Google Scholar]
- Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, Seidman JG. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- Gillilan RE, Kumar VS, O’Neall-Hennessey E, Cohen C, Brown JH. X-ray solution scattering of squid heavy meromyosin: strengthening the evidence for an ancient compact off state. PLoS One. 2013;8:e81994. doi: 10.1371/journal.pone.0081994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasekar M, Salunkhe AM, Mallia AK, He YX, Kalyanasundaram R. Praziquantel affects the regulatory myosin light chain of Schistosoma mansoni. Antimicrob Agents Chemother. 2009;53:1054–1060. doi: 10.1128/AAC.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Solá M, Al-Khayat HA, Behra M, Kensler RW. Zebrafish cardiac muscle thick filaments: isolation technique and three-dimensional structure. Biophys J. 2014;106:1671–1680. doi: 10.1016/j.bpj.2014.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EM, Wakimoto H, Anderson RL, Evanchik MJ, Gorham JM, Harrison BC, Henze M, Kawas R, Oslob JD, Rodriguez HM, Song Y, Wan W, Leinwand LA, Spudich JA, McDowell RS, Seidman JG, Seidman CE. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 2016;351:617–621. doi: 10.1126/science.aad3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorich ZR, Peng Y, Cai W, Jin Y, Wei L, Chen AJ, McKiernan SH, Aiken JM, Moss RL, Diffee GM, Ge Y. Top-down targeted proteomics reveals decrease in myosin regulatory light-chain phosphorylation that contributes to sarcopenic muscle dysfunction. J Proteome Res. 2016;15:2706–2716. doi: 10.1021/acs.jproteome.6b00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LM, Downs SM, Spudich JA. Myosin light chain kinase and myosin light chain phosphatase from Dictyostelium: effects of reversible phosphorylation on myosin structure and function. J Cell Biol. 1987;104:1309–1323. doi: 10.1083/jcb.104.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruen M, Gautel M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J Mol Biol. 1999;286:933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- Hooijman P, Stewart MA, Cooke R. A new state of cardiac myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart. Biophys J. 2011;100:1969–1976. doi: 10.1016/j.bpj.2011.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SL, Thuma JB. Invertebrate muscles: muscle specific genes and proteins. Physiol Rev. 2005;85:1001–1060. doi: 10.1152/physrev.00019.2004. [DOI] [PubMed] [Google Scholar]
- Hu Z, Taylor DW, Reedy MK, Edwards RJ, Taylor KA. Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 Å resolution. Sci Adv. 2016;2:e1600058. doi: 10.1126/sciadv.1600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley H, Hanson J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature. 1954;173:973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]
- Huxley AF, Niedergerke R. Structural changes in muscle during contraction; interference microscopy of living muscle fibres. Nature. 1954;173:971–973. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- Jung HS, Burgess SA, Billington N, Colegrave M, Patel H, Chalovich JM, Chantler PD, Knight PJ. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc Natl Acad Sci U S A. 2008;105:6022–6026. doi: 10.1073/pnas.0707846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Komatsu S, Ikebe M, Craig R. Head–head and head–tail interaction: a general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T, Chang S, Seidman JG, Seidman CE. Genetics of hypertrophic cardiomyopathy. Curr Opin Cardiol. 2010;25:205–209. doi: 10.1097/HCO.0b013e3283375698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai DH, Hong XK, Su BX, Liang C, Hide G, Zhang X, Yu X, Lun ZR. Current status of Clonorchis sinensis and clonorchiasis in China. Trans R Soc Trop Med Hyg. 2016;110:21–27. doi: 10.1093/trstmh/trv100. [DOI] [PubMed] [Google Scholar]
- Lee K, Yang S, Liu X, Korn ED, Sarsoza F, Bernstein SI, Pollard L, Lord MJ, Trybus KM, Craig R. Myosin II head interaction in primitive species. Biophys J. 2016;110(Supplement 1, issue 3):615a. [Google Scholar]
- Liu J, Wendt T, Taylor D, Taylor K. Refined model of the 10S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J Mol Biol. 2003;329:963–972. doi: 10.1016/s0022-2836(03)00516-3. [DOI] [PubMed] [Google Scholar]
- Lorenzo-Morales J, Khan NA, Walochnik J. An update on Acanthamoeba keratitis: diagnosis, pathogenesis and treatment. Parasite. 2015;22:10. doi: 10.1051/parasite/2015010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JR, Leinwand L, Warshaw DM. Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ Res. 2012;111:375–385. doi: 10.1161/CIRCRESAHA.110.223842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naber N, Cooke R, Pate E. Slow myosin ATP turnover in the super-relaxed state in tarantula muscle. J Mol Biol. 2011;411:943–950. doi: 10.1016/j.jmb.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag S, Trivedi DV, Sarkar SS, Adhikari AS, Sunitha MS, Sutton S, Ruppel KM, Spudich JA. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat Struct Mol Biol. 2017;24:525–533. doi: 10.1038/nsmb.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogara L, Naber N, Pate E, Canton M, Reggiani C, Cooke R. Piperine’s mitigation of obesity and diabetes can be explained by its up-regulation of the metabolic rate of resting muscle. Proc Natl Acad Sci U S A. 2016;113:13009–13014. doi: 10.1073/pnas.1607536113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogara L, Naber N, Pate E, Canton M, Reggiani C, Cooke R. Spectroscopic studies of the super relaxed state of skeletal muscle. PLoS One. 2016;11:e0160100. doi: 10.1371/journal.pone.0160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KJ, Lyons JB, Nowak KS, Takacs CM, Wargo MJ, McPeek MA. Estimating metazoan divergence times with a molecular clock. Proc Natl Acad Sci U S A. 2004;101:6536–6541. doi: 10.1073/pnas.0401670101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pinto A, Sánchez F, Alamo L, Padrón R. The myosin interacting-heads motif is present in the relaxed thick filament of the striated muscle of scorpion. J Struct Biol. 2012;180:469–478. doi: 10.1016/j.jsb.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Ross AG, Chau TN, Inobaya MT, Olveda RM, Li Y, Harn DA. A new global strategy for the elimination of schistosomiasis. Int J Infect Dis. 2017;54:130–137. doi: 10.1016/j.ijid.2016.09.023. [DOI] [PubMed] [Google Scholar]
- Schmitt JP, Debold EP, Ahmad F, Armstrong A, Frederico A, Conner DA, Mende U, Lohse MJ, Warshaw D, Seidman CE, Seidman JG. Cardiac myosin missense mutations cause dilated cardiomyopathy in mouse models and depress molecular motor function. Proc Natl Acad Sci U S A. 2006;103:14525–14530. doi: 10.1073/pnas.0606383103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert P, Reber-Müller S, Schmid V. Life stage specific expression of a myosin heavy chain in the hydrozoan Podocoryne carnea. Differentiation. 1993;54:11–18. doi: 10.1111/j.1432-0436.1993.tb00654.x. [DOI] [PubMed] [Google Scholar]
- Seidman CE, Seidman JG. Mutations in cardiac myosin heavy chain genes cause familial hypertrophic cardiomyopathy. Mol Biol Med. 1991;8:159–166. [PubMed] [Google Scholar]
- Seipel K, Schmid V. Evolution of striated muscle: jellyfish and the origin of triploblasty. Dev Biol. 2005;282:14–26. doi: 10.1016/j.ydbio.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Sellers JR. Myosins. 2. Oxford: Oxford University Press; 1999. [Google Scholar]
- Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- Steinmetz PR, Kraus JE, Larroux C, Hammel JU, Amon-Hassenzahl A, Houliston E, Wörheide G, Nickel M, Degnan BM, Technau U. Independent evolution of striated muscles in cnidarians and bilaterians. Nature. 2012;487:231–234. doi: 10.1038/nature11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulbarán G, Alamo L, Pinto A, Márquez G, Méndez F, Padrón R, Craig R. Schistosome muscles contain striated muscle-like myosin filaments in a smooth muscle-like architecture. Biophys J. 2014;106(2):159a. [Google Scholar]
- Sulbarán G, Alamo L, Pinto A, Márquez G, Méndez F, Padrón R, Craig R. An invertebrate smooth muscle with striated muscle myosin filaments. Proc Natl Acad Sci U S A. 2015;112:E5660–E5668. doi: 10.1073/pnas.1513439112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulbarán G, Biasutto A, Alamo L, Riggs C, Pinto A, Méndez F, Craig R, Padrón R. Different head environments in tarantula thick filaments support a cooperative activation process. Biophys J. 2013;105:2114–2122. doi: 10.1016/j.bpj.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulbarán G, Mun JY, Lee KH, Alamo L, Pinto A, Sato O, Ikebe M, Liu X, Korn ED, Padrón R, Craig R. The inhibited, interacting-heads motif characterizes myosin II from the earliest animals with muscle. Biophys J. 2015;108:301a. [Google Scholar]
- Telford MJ, Budd GE, Philippe H. Phylogenomic insights into animal evolution. Curr Biol. 2015;25:R876–R887. doi: 10.1016/j.cub.2015.07.060. [DOI] [PubMed] [Google Scholar]
- Trybus KM. Role of myosin light chains. J Muscle Res Cell Motil. 1994;15:587–594. doi: 10.1007/BF00121066. [DOI] [PubMed] [Google Scholar]
- Tyska MJ, Hayes E, Giewat M, Seidman CE, Seidman JG, Warshaw DM. Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ Res. 2000;86:737–744. doi: 10.1161/01.res.86.7.737. [DOI] [PubMed] [Google Scholar]
- Waldmüller S, Erdmann J, Binner P, Gelbrich G, Pankuweit S, Geier C, Timmermann B, Haremza J, Perrot A, Scheer S, Wachter R, Schulze-Waltrup N, Dermintzoglou A, Schönberger J, Zeh W, Jurmann B, Brodherr T, Börgel J, Farr M, Milting H, Blankenfeldt W, Reinhardt R, Özcelik C, Osterziel KJ, Loeffler M, Maisch B, Regitz-Zagrosek V, Schunkert H, Scheffold T. Novel correlations between the genotype and the phenotype of hypertrophic and dilated cardiomyopathy: results from the German Competence Network Heart Failure. Eur J Heart Fail. 2011;13:1185–1192. doi: 10.1093/eurjhf/hfr074. [DOI] [PubMed] [Google Scholar]
- Wang Y, Steimle PA, Ren Y, Ross CA, Robinson DN, Egelhoff TT, Sesaki H, Iijima M. Dictyostelium huntingtin controls chemotaxis and cytokinesis through the regulation of myosin II phosphorylation. Mol Biol Cell. 2011;22:2270–2281. doi: 10.1091/mbc.E10-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA. Visualization of head–head interactions in the inhibited state of smooth muscle myosin. J Cell Biol. 1999;147:1385–1390. doi: 10.1083/jcb.147.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci U S A. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead JL, Zhao FQ, Craig R. Structural basis of the relaxed state of a Ca2+-regulated myosin filament and its evolutionary implications. Proc Natl Acad Sci U S A. 2013;110:8561–8566. doi: 10.1073/pnas.1218462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padrón R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- Yang S, Lee K, Sato O, Ikebe M, Craig R. 3D Reconstruction of the folded, inhibited form of vertebrate smooth muscle myosin II by single particle analysis. Biophys J. 2017;112:266a. [Google Scholar]
- Zhao FQ, Craig R, Woodhead JL. Head–head interaction characterizes the relaxed state of Limulus muscle myosin filaments. J Mol Biol. 2009;385:423–431. doi: 10.1016/j.jmb.2008.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci U S A. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]