Abstract
Structural DNA nanotechnology, in which Watson-Crick base pairing drives the formation of self-assembling nanostructures, has rapidly expanded in complexity and functionality since its inception in 1981. DNA nanostructures can now be made in arbitrary three-dimensional shapes and used to scaffold many other functional molecules such as proteins, metallic nanoparticles, polymers, fluorescent dyes and small molecules. In parallel, the field of dynamic DNA nanotechnology has built DNA circuits, motors and switches. More recently, these two areas have begun to merge—to produce switchable DNA nanostructures, which change state in response to their environment. In this review, we summarise switchable DNA nanostructures into two major classes based on response type: molecular actuation triggered by local chemical changes such as pH or concentration and external actuation driven by light, electric or magnetic fields. While molecular actuation has been well explored, external actuation of DNA nanostructures is a relatively new area that allows for the remote control of nanoscale devices. We discuss recent applications for DNA nanostructures where switching is used to perform specific functions—such as opening a capsule to deliver a molecular payload to a target cell. We then discuss challenges and future directions towards achieving synthetic nanomachines with complexity on the level of the protein machinery in living cells.
Keywords: DNA origami, DNA nanotechnology, Nanomachines, Switchable nanostructures, Actuation
Introduction
In biological systems, complex tasks are achieved by sophisticated molecular machines, such as linear and rotary protein motors, ion pumps and the DNA replication machinery (Alberts 1998). In the emerging field of synthetic DNA nanotechnology (Seeman 1982), a range of nanoscale machines and devices have been developed, which are both inspired by the protein machinery of the cell and assembled from biomolecules. These DNA nanomachines have a range of potential applications including: targeted drug delivery (Douglas et al. 2012; Perrault and Shih 2014; Li et al. 2018b), molecular computation (Zhang and Seelig 2011; Qian and Winfree 2011; Cherry and Qian 2018), in vitro diagnostics (Rinker et al. 2008; Godonoga et al. 2016), tools for biophysical measurement of proteins (Rajendran et al. 2012; Derr et al. 2012; Iwaki et al. 2016; Omabegho et al. 2018), templates for nano-electronic (Maune et al. 2010; Knudsen et al. 2015) or plasmonic devices (Kuzyk et al. 2012; Gopinath et al. 2016; Lee et al. 2018), and even as molecular assembly lines (Gu et al. 2010; Thubagere et al. 2017).
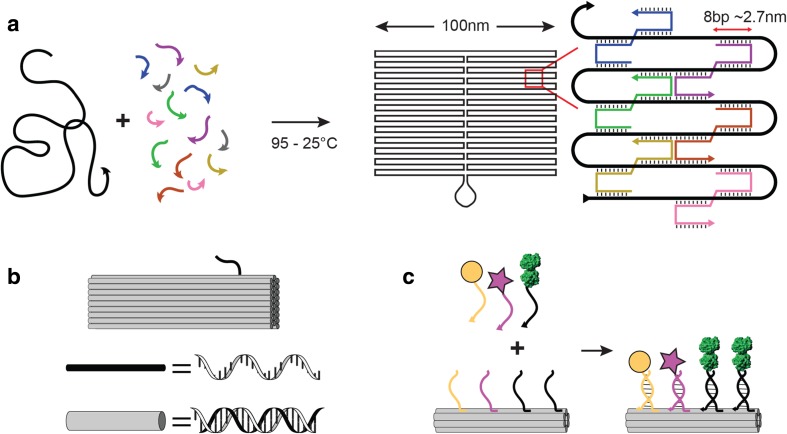
In structural DNA nanotechnology, base pairing drives the formation of self-assembling nanostructures with arbitrary shapes (Seeman 1982). DNA is an ideal material for nanoscale construction because DNA hybridisation is programmable, the rules of base-pairing are predictable, and DNA is easily synthesised and chemically modified. ‘DNA origami’ is a robust method for making custom DNA nanostructures in which a long single-stranded DNA ‘scaffold’ strand is folded into a desired shape using shorter ‘staple’ strands (Rothemund 2006) (Fig. 1a). DNA-origami nanostructures can be folded in three dimensions with complex curvature (Douglas et al. 2009; Dietz et al. 2009), and up to Gigadalton-scale (Wagenbauer et al. 2017). Functionalization of DNA-origami nanostructures is achieved by covalent linkage of the guest molecule to a single-stranded DNA (ssDNA) ‘anti-handle’, which hybridises to a complementary ssDNA ‘handle’ on the surface of the DNA-origami nanostructure (Fig. 1c). Each staple strand has a unique nucleotide sequence, so available handle sites on the DNA-origami nanostructure are uniquely addressable. This allows for control of the number and geometry of functional molecules, with precision of ~ 6 nm (Rothemund 2006). DNA-origami nanostructures have been used to scaffold many other functional molecules such as nanoparticles (Lee et al. 2018), aptamers (Rinker et al. 2008; Godonoga et al. 2016), proteins (Derr et al. 2012; Bell and Keyser 2016), fluorescent dyes (Yurke et al. 2000) and small molecules (Zhao et al. 2012).
Fig. 1.
DNA-origami nanostructures. a In DNA Origami, a long single-stranded DNA scaffold (black) is folded up into a double-stranded DNA shape, which is cross-linked by ~ 300 short ‘staple’ oligonucleotide strands (coloured). b DNA-origami nanostructures are often depicted by representing each DNA duplex with a rigid cylinder of width 2.6 nm (grey) and single-stranded DNA regions with a flexible line (black). c DNA-origami nanostructures can be functionalised by adding single-stranded DNA ‘handles’ to the end of staple strands on the surface of the structure (coloured). Each staple sequence is unique, so handle locations are uniquely addressable. Guest molecules, such as metallic nanoparticles (yellow), fluorophores (pink) or proteins (green), are covalently linked to complementary ‘anti-handle’ sequences. On incubation with the DNA-origami nanostructure, guest molecules are scaffolded by the origami with precision of up to ~ 6 nm
Switchable DNA nanostructures change from one state to another in response to changes in their environment. This switch can be used to activate nanostructures to perform specific functions, such as opening a capsule to deliver a molecular payload to a target cell (Douglas et al. 2012), bringing proteins together to measure their interaction (Ke et al. 2016) or changing the chirality of a plasmonic device (Kuzyk et al. 2014). Switching can be triggered by a local change in the molecular environment. For example, addition of DNA oligonucleotides to trigger a strand displacement reaction (Yurke et al. 2000; Andersen et al. 2009; Song et al. 2017; Thubagere et al. 2017), change in pH (Surana et al. 2011; Burns et al. 2018) or ionic concentration (Mao et al. 1999; Sannohe et al. 2010; Gerling et al. 2015) or addition of proteins (Douglas et al. 2012; Godonoga et al. 2016; Li et al. 2018b) or other small molecules (Zadegan et al. 2017). Recently, progress has been made towards achieving switchable DNA nanostructures that respond to external changes, such as light (Derr et al. 2012; Kuzyk et al. 2016; Willner et al. 2017; Liu et al. 2018a), and electric (Kroener et al. 2017; Kopperger et al. 2018) or magnetic fields (Lauback et al. 2018). External actuation has the potential to provide remote control of nanostructures, independent of their chemical environment, inside cells or tissue. In this review, we give an overview of both molecular and external actuation methods used to switch DNA-origami nanostructures and their potential applications. We also discuss future directions in this field and fundamental limits and challenges towards in vivo implementation.
Molecular actuation of DNA-origami nanostructures
DNA strand displacement
If a DNA strand is hybridised to a shorter strand, an overhanging ‘toehold’ will remain as ssDNA. When a fully complementary DNA strand is added, this will bind to the toehold and displace the shorter strand—in a process known as toehold-mediated DNA strand displacement (Fig. 2b, i). The first switchable DNA device triggered by strand displacement was a DNA tweezer, assembled from three DNA strands, which opens or closes on manual addition of DNA ‘fuel’ or ‘anti-fuel’ strands (Yurke et al. 2000). Similarly, the first switchable DNA-origami nanostructure, a three-dimensional DNA-origami box, was opened by strand displacement (Andersen et al. 2009). DNA strand displacement has been used to implement a variety of molecular systems, including autonomous DNA motors that navigate complex tracks (Wickham et al. 2012; Thubagere et al. 2017; Li et al. 2018a), DNA-origami switches (Marini et al. 2011; Torelli et al. 2014; Zhou et al. 2015) and to create complex logic circuits such as a DNA-based winner-take-all neural network for molecular pattern recognition (Cherry and Qian 2018). DNA strand displacement reactions are a powerful method for sophisticated actuation and can accommodate many different molecular triggers in parallel. However, they require DNA fuel strands as input and generate waste DNA fragments.
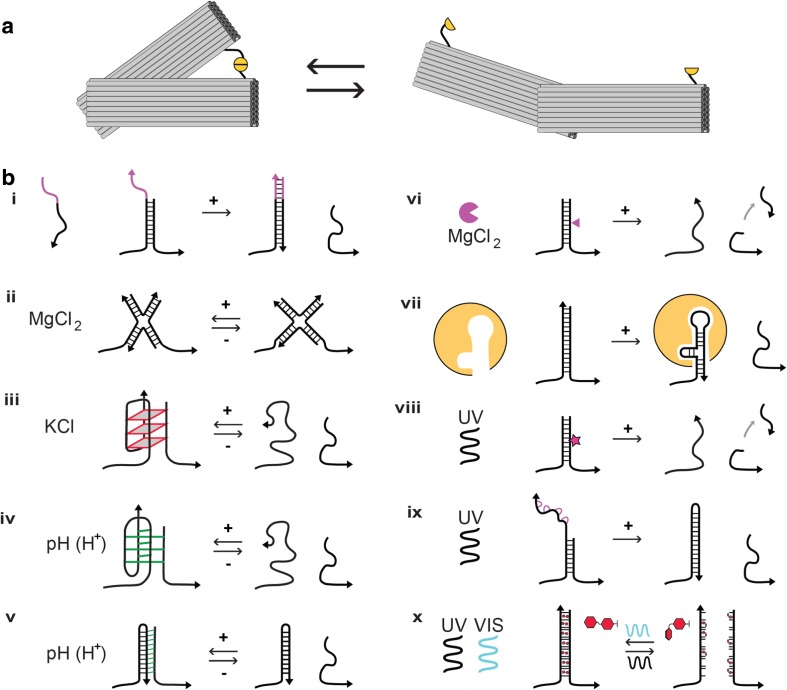
Fig. 2.
Switchable DNA-origami nanostructures actuated by local molecular signals and externally by light. a DNA-origami nanostructures can be designed with flexible regions and a number of possible states. For example, a DNA-origami tweezer, or calliper, can be either closed (left) or open (right), depending on the state of a molecular ‘lock’ that links the two arms (yellow). b A range of methods can be used to open the lock, actuating the DNA-origami nanostructure. i toehold-mediated DNA strand displacement reaction on addition of ssDNA ‘trigger’, ii stacking interaction change on addition of MgCl2, iii G-quadruplex formation on addition of KCl, iv i-motif formation on decrease in pH, v DNA triplex formation on decrease in pH, vi enzymatic cleavage of DNA backbone, vii ligand-stabilisation of DNA aptamer structure, viii photocleavage of modified DNA backbone with UV light, ix release of photocaged based by UV light, x reversible photoswitching of azobenzene modified DNA bases by UV and Visible light
Ionic concentration
One of the earliest nanomechanical DNA devices was driven by changing ionic concentration (Mao et al. 1999). High salt concentrations and low temperature can cause dsDNA with the sequence (CG)n to be switched from the usual right-handed helix (B-DNA) to a left-handed conformation (Z-DNA), and this change in twist was used to drive rotary motion. The fundamental four-way (Holliday) junction that forms the basis of DNA nanotechnology is also sensitive to divalent cation concentration, adopting an open square planar geometry in the absence of cations and a stacked X-shaped structure, with an angle of ~ 60°, above ~ 0.1 mM MgCl2 (Lilley 2000). However, DNA-origami nanostructures are not generally stable below ~ 2 mM MgCl2, so this is not a generalisable switching strategy (Hahn et al. 2014). Blunt end stacking of DNA helices can be used for hierarchal assembly of DNA-origami nanostructures into larger ‘crystals’, based on shape-complementarity (Gerling et al. 2015). This stacking interaction is sensitive to the concentration of counter ions in solution because of the repulsion between the negatively charged surfaces of the DNA-origami. This effect has been used to create both discrete DNA-origami nanostructures and extended DNA-origami lattices that change shape in response to change in magnesium concentration. Similarly, rearrangement of DNA stacking interactions can be exploited to propagate information transfer across a DNA-origami nanostructure and has been directly observed by atomic force microscopy (AFM) (Song et al. 2017). Alternatively, G-quadruplexes are built from the stacking of successive G-tetrads, which are cyclic Hoogsteen bonded square planar alignments of four guanines that are stabilised by bound monovalent Na+ and K+ cations (Fig. 3b) (Patel et al. 2007). Two guanine-rich strands can be incorporated into the DNA nanostructure, which form an interstrand G-quadruplex structure in the presence of K+ and disassemble on removal of K+, which can be used to trigger switching (Fig. 2b, iii) (Sannohe et al. 2010; Kuzuya et al. 2011).
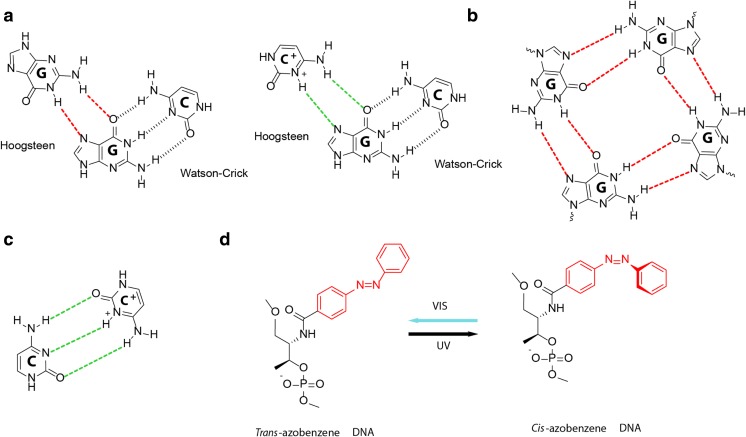
Fig. 3.
DNA structures that are sensitive to pH, ionic concentration and photoactivation. a Non-canonical Hoogsteen hydrogen-bonding patterns for G·G and G·C+ (red and green), shown beside the canonical Watson-Crick hydrogen bonding pattern of G·C (black). These form the basis of G-quadruplex and DNA triplex formation respectively. b The G-tetrad, formed by cyclic Hoogsteen-bonded (red) square planar alignments of four guanines that are stabilised by bound monovalent Na + and K+ cations (not shown). G-quadruplexes are formed by stacking of G-tetrads. c Semiprotonated C·C+ base pairs (green), which are intercalated to form the pH-sensitive i-motif. d trans-cis photoisomerization of azobenzene modification under UV and visible light (Vis) can be used to control DNA strand hybridization (trans) and dissociation (cis)
Protein and aptamer actuation
Another molecular method for DNA-origami actuation is through interaction with proteins. DNA-modifying proteins, as such as restriction enzymes, can be used for site-specific cleavage of the DNA backbone on one side of the double helix. Once cleaved, small DNA fragments (< ~ 5 nucleotides) will dissociate, revealing a ssDNA toehold which is then available for strand displacement. This method has been used to power a number of DNA motors and machines (Fig. 2b, vi) (Bath and Turberfield 2007; Wickham et al. 2012). Binding of DNA aptamers to protein ligands has also been used for actuation (Rinker et al. 2008; Kuzuya et al. 2011; Douglas et al. 2012; Godonoga et al. 2016; Li et al. 2018b). Aptamers are DNA oligonucleotides that bind specific molecular targets (Hamaguchi et al. 2001) and can be used in systems where a complementary DNA strand forms a duplex with the aptamer. Binding of the target molecule destabilises the duplex state, dissociating the aptamer strand from the complementary DNA strand, inducing switching of the DNA-origami structure (Fig. 2b, vii). For example, aptamer switching has been used to release drug payloads on binding to specific cell surface receptors (Douglas et al. 2012; Li et al. 2018b). The use of targeting ligands to drive switching allows the DNA-origami nanostructures to act autonomously, without external intervention, but does not generally allow for mechanisms that are reversible, unless coupled with DNA strand displacement (Kuzuya et al. 2011).
Change in pH
pH sensors are of particular interest as components of switchable DNA-origami nanostructures for in vivo applications, because acidic conditions are found in the endosomal pathway after cell entry, and in cancer tissue (Gerweck and Seetharaman 1996). There are two common pH switches used in DNA nanotechnology (Fig. 2b, iv,v). The first is the i-motif: a quadruplex structure formed from a cytosine-rich ssDNA at an acidic pH (Gehring et al. 1993). This structure consists of intercalated semiprotonated C·C+ base pairs and requires protonation of cytosine at the N3 position (with a pKa below 7) and thus is triggered by change in pH (Fig. 3c). The second is a DNA triplex, formed by pH-sensitive sequence-specific parallel Hoogsteen bonding between a dsDNA strand and a ssDNA strand (Fig. 3a) (Frank-Kamenetskii and Mirkin 1995). These triplexes contain CGC pairings that are only stabilised at acidic pH because they require protonation of the ssDNA cytosine at N3 position pH (pKa ≈ 6.5) and TAT pairings that only destabilise at alkaline pH due to the deprotonation of thymine (pKa ≈ 10). The pH sensitivity of the DNA triplex can be controlled by tuning its CGC/TAT content (Idili et al. 2014). Both i-motifs and triplex DNA switches are convenient because their behaviour is predictable and can be tuned to a narrow range of pH changes.
I-motifs have been used for pH switching of several DNA-origami nanostructures. Kuzuya et al. developed DNA-origami pliers: a 170-nm lever structure decorated with cytosine-rich ssDNAs that form i-motifs at acidic pH (Kuzuya et al. 2014). At slightly alkaline or neutral pH, the pliers take either a cross or antiparallel position, where the ssDNAs are exposed. In slightly acidic conditions (pH 5.6), the i-motifs form and lock the arms in place, resulting a parallel structure. Similarly, Majikes et al. used i-motifs to control the distance between two DNA-origami substructures, with reversible switching (Majikes et al. 2017). More recently, Burns et al. demonstrated protein delivery to cells using a pH switchable DNA-origami box with a lid controlled by i-motifs (Burns et al. 2018). Upon entering the cell, the box encounters the acidic conditions of the endosome, opening the lid and releasing green fluorescent proteins (GFP) from inside the box.
DNA triplex locks have been used for pH actuation of a number of DNA-origami nanostructures, including a cross-like plasmonic DNA nanostructure decorated with two gold nanorods (Kuzyk et al. 2017). At low pH (< 6.5), a duplex on one arm of the cross DNA binds to an ssDNA on the other arm, to form a DNA triplex, locking the origami structure. As the pH is increased, the triplex is unlocked and the origami returns to the relaxed state. A left- or right-handed nanoparticle confirmation can be achieved by manipulating the location of the triplex lock, and pH sensitivity is tuned by changing the TAT content of the lock strand.
A pH-actuated version of the original DNA tweezer has been successfully applied as an in vivo pH sensor. Surana et al. show that an i-motif-based DNA nanomachine coupled with fluorescence resonance energy transfer (FRET) measurements could be used to effectively map spatiotemporal pH changes associated with endosomal maturation in different types of Caenorhabditis elegans. This was the first successful employment and actuation of a DNA nanomachine in an organism (Surana et al. 2011). In subsequent work, a pH responsive I-switch was developed for specific organelles in the cell, such as the trans Golgi network (TGN), cis Golgi (CG) and endoplasmic reticulum (ER), which all have different pH. This allowed for simultaneously live-cell tracking of pH in two different organelles (Modi et al. 2013). An interesting extension of this would be to couple pH measurement to pH-responsive drug delivery to specific organelles.
External actuation of DNA-origami nanostructures
Photoactuation
Three techniques are used for photoactuation in DNA nanotechnology: photoswitching, photocleavage and photocaging. The most common method is the photoswitching of azobenzene attached to the DNA backbone (Fig. 3d). Asanuma et al. show that the formation of an azobenzene-modified DNA duplex can be photoregulated using ultraviolet (UV) and visible light (Asanuma et al. 2007). Azobenzene takes a cis-form when exposed to UV light (300-400 nm), which inhibits DNA hybridisation. In contrast, the azobenzene takes a trans-form when exposed to visible light (> 400 nm), which allows formation of a stable DNA duplex. Photocleavage is achieved by incorporating a photocleavable (PC) linker into the DNA backbone. The linker is cleaved when exposed to UV light, thereby breaking the DNA strand. Photocleavage of oligonucleotides has been demonstrated with several chemical modifications, including o-nitrobenzyl ethers (Ordoukhanian and Taylor 1995), 2-nitrobenzyl (Bai et al. 2003; Li et al. 2003) and 7-diethylaminocoumarin (Weyel et al. 2017). Photocaging refers to the act of ‘caging’ active molecules with a photolabile group, thereby preventing activity of the molecule. Upon exposure to light of the correct wavelength, the molecule is freed of the ‘cage’ and is active again. Photocaged DNA strands have been used for the photoregulation of DNA function (Liu and Deiters 2014), DNA aptamer activation (Heckel and Mayer 2005), DNAzyme activation (Hwang et al. 2014) and for DNA computation using logic gates (Prokup et al. 2012).
Both photoswitching and photocleavage have been used for DNA-origami nanostructure actuation. Yang et al. developed photoresponsive DNA-origami hexagons by connecting them to azobenzene-modified oligonucleotides (Azo-ODNs) (Yang et al. 2012). By manipulating the number and positions of the Azo-ODNs on the origami units, different origami hexagons could be linked together via photoactuation of the Azo-ODNs to form larger two-dimensional structures of different shapes. Azo-ODNs have also been used to form photoswitchable DNA-origami nanocapsules (Takenaka et al. 2014), which open when exposed to UV light and close when exposed to visible light. More recently, reversible photoactuation of cross-shaped DNA-origami nanostructures was achieved by photoisomerisation of azobenzene modifications in a ‘lock’ duplex (Kuzyk et al. 2016; Willner et al. 2017). When exposed to UV light, the azobenzenes transform to cis-form, resulting in dehybridisation of the lock strands and opening of the structure. However, the reversibility of azobenzene-modified DNA is temperature sensitive (Samai et al. 2017), and reversible switching requires a high number of modifications—in these examples, 7 modifications in a 10 or 20 base-pair duplex (Kuzyk et al. 2016; Willner et al. 2017).
Photocleavable linkers have been used for payload release from DNA-origami nanostructures (Kohman et al. 2016). In this example, proteins were loaded onto a rectangular DNA-origami structure via an o-nitrobenzyl-based PC linker and an alkyne oligo. Upon exposure to UV light, the PC linker was cleaved resulting in the release of the protein. Similarly, the linear protein motors dynein and kinesin, which move in opposite directions along microtubules, have been conjugated to a DNA-origami ‘chassis’ by PC linkers (Derr et al. 2012). UV light was used to cleave the PC linkers, giving selective detachment of one type of motor protein, which controlled the subsequent direction of the DNA-origami chassis along the microtubule. While photocaging has not been used for DNA-origami nanostructure actuation, it has recently been used for actuation of a simple DNA tweezer (Liu et al. 2018a). While irreversible, this actuation was measured to be ~ 60 times faster than strand displacement, and simulations predicted that forces of up to 46 pN were generated.
Overall, photoactuation is a simple, fast and effective method for structural switching. Currently, photocleavage and photocaging do not allow for reversible actuation, while photoswitching only has limited reversibility. Another limitation of photoactuation is that it relies on UV radiation, which damages DNA, most commonly through formation of cyclobutane pyrimidine dimers (CPDs) between neighbouring pyrimidine residues (Vink and Roza 2001). UV damage of DNA is wavelength dependent and is particularly dominant at UVB (280–315 nm) and UVC (200–280 nm) wavelengths. Chen et al. (2017) showed that UVB and UVC cause a ‘flattening’ effect in DNA-origami nanostructures, as the UV radiation relieves internal stress in the origami. At high UV doses, the DNA-origami nanostructures are damaged. UVA (315–400 nm), on the other hand, showed no conformational damage to the DNA-origami nanostructures. Another limitation of UV radiation is that it has low penetration depth (< 1 mm) and therefore is less useful for the photoactuation of DNA-origami nanostructures in vivo (Anderson and Parrish 1981).
Electrical actuation
The phosphate backbone of DNA results in DNA-origami nanostructures that have a net negative charge. Thus, electric fields can be used to actuate them. Kroener et al. demonstrated electrical switching of a 100-nm long DNA-origami nanolever on a gold electrode (Kroener et al. 2017). Fluorescence intensity was used to measure the angle between the lever and the electrode, as the surface acts to quench fluorescence (Chance et al. 2007). An alternating low frequency voltage of ± 200 mV was applied. At negative potentials, the negatively charged levers were repelled into a standing position, while at positive potentials, they were attracted to the surface, with a response time of less than 100 μs.
In more recent work, Kopperger et al. demonstrated the electrical actuation of a flexible DNA-origami arm on a DNA-origami platform (Kopperger et al. 2018) (Fig. 4b). The platform was decorated with multiple ssDNA ‘latches’, complementary to an ssDNA ‘catch’ on the arm. The catch on the arm hybridises to a complementary latch on the base platform at a position that is determined by the direction of the external electric field. As the applied electric field is rotated, the arm moves from one position to another in less than a millisecond. The arm can also be used to transport gold nanorods from one side of the platform to the other. In the future, this could be coupled to cargo-transfer mechanisms (Kassem et al. 2016; Thubagere et al. 2017) to result in computer-directed arrangement of molecules using DNA origami. A challenge in applying electrical switching in vivo is that the electric field can cause heating and damage and must be kept below 30–40 kV/m (Menachery and Pethig 2005).
Fig. 4.
Externally actuated DNA-origami nanostructures by electric and magnetic fields. a A DNA-origami ‘lever’ (green), 1–10 μm in length and with 56-helix cross-section (inset), is attached to a surface by a freely rotating linker. The other end is decorated with a 1-μm diameter magnetic nanoparticle. Rotation of the applied external magnetic field () drives rotation of the nanolever. (Lauback et al. 2018) b DNA-origami nanostructures are negatively charged and respond to applied electric fields. A 25-nm 6-helix rod (blue) is linked to a 55 nm × 55 nm DNA-origami platform (grey), by a flexible single-stranded DNA linker (black). A ‘latch’ strand on the underside of the rod (orange) can hybridise to a number of complementary ‘catch’ strands on the platform (orange). The position of the rod is controlled by the direction of the applied electric field () (Kopperger et al. 2018)
Magnetic actuation
While DNA-origami nanostructures are not intrinsically magnetic, they can be functionalised with magnetic nanoparticles. Generally, to actuate a DNA-origami nanostructure require a force and torque of ~ 1 pN and 10–50 pN nm, respectively (Marras et al. 2015). However, to achieve forces of this magnitude with a magnetic field requires superparamagnetic beads of 1 μm or larger (Xu et al. 2016). In a recent advance, Lauback et al. bridged this difference in scale by using a highly stiff microscale lever as the link between a microscale magnetic nanoparticle and a nanoscale DNA-origami device (Lauback et al. 2018) (Fig. 4a). The system consists of three components: a lever, a rotor and a hinge, all constructed from 56-helix DNA-origami nanobricks. The lever was designed to be both mechanically stiff and long enough (1 μm) for coupling to a micron-scale magnetic nanoparticle. The other end of the lever was fixed to the surface, and an externally applied magnetic field was used to drive rotation of the bead and in turn, of the lever. The lever was connected to the platform by either a DNA-origami rotor, to achieve continuous rotational motion, or a DNA-origami hinge, to achieve finite angular motion. Rotational motion was controlled to a frequency of 2 Hz with up to 80 pN nm of torque, while the angular motion was specified with accuracy of ± 8°.
Discussion
An ideal actuation method for a nanomachine would be fast, reversible, able to process many inputs independently, produce both digital and analogue responses, allow both local and external control and not generate waste heat or by-products. While a range of methods have been used to actuate DNA-origami nanostructures that cover this range of properties, as yet no single method is able to provide all. Currently, the specific choice of method depends on the application.
DNA strand displacement is still the best method for multiplexing many trigger molecules in parallel, and is reversible, but requires external DNA strand input and generates waste DNA fragments. In comparison, both aptamer and pH actuation allow for autonomous switching in in vivo settings, as no external interaction is required after delivery into an organism. However, these methods currently lack reversibility and their response cannot be modified externally after delivery. In the future, the ability to link multiple pH or aptamer switches together to produce DNA-origami nanostructures that are activated in response to a more complex set of environmental triggers would increase the utility of these methods. For example, by incorporating multiple i-motifs that are each tuned to a different pH into the same DNA-origami nanostructure so that it is activated only within a certain pH window and can reversibly switch off at either higher or lower pH. Another example would be an aptamer actuated DNA nanocapsule that is only activated after processing a complex set of local conditions, such as upregulation of one set of cell-surface receptors above a certain threshold and downregulation of another set.
For applications that require greater control of DNA-origami nanostructures, the use of external actuation that can be more easily controlled at the macroscale (such as light, electric and magnetic field) shows great potential. Among these, light-based switching of DNA-origami nanostructures is the most well-developed. A limitation of current photoactuation methods is the use of wavelengths that damage cells (UV), and do not penetrate tissue (UV-Vis). The development of photoswitching systems that utilise tissue-penetrating photons, which typically exists in the near-infrared (NIR) wavelength range of 630–950 nm, would increase the utility of this approach (Weissleder 2001; Frangioni 2003). For example, if recently developed NIR photoswitching molecules (Yang et al. 2014) were conjugated to DNA, it may allow photoswitching of DNA-origami nanostructures in vivo. Another promising future direction is DNA-origami nanostructures that can be driven through a multi-state switching cycle by sequential activation at a number of different wavelengths. To achieve this requires development of a range of photoswitching molecules that can be spectrally multiplexed.
While photoactuation is currently useful for applications that require digital switching of DNA-origami nanostructures between discrete states, electrical or magnetic switching can be more appropriate for analogue switching through a range of states. The use of applied electric and magnetic fields also allows for reversibility, does not generate waste and results in a fast response time. Sub-second response times have been demonstrated with current electrical and magnetic switching in vitro. Electrical switching does not require the attachment of DNA-origami nanostructures to a ‘switching molecule’ as the DNA-origami nanostructure itself is negatively charged. Magnetic switching on the other hand requires the attachment of the DNA-origami nanostructure to a superparamagnetic bead.
In our view, recent advances in using electric and magnetic fields to switch DNA-origami nanostructures will promote rapid development of these structures for use in vivo. While this area has much potential, more generally, there are some fundamental limits to the application of switchable DNA-origami nanostructures. In vivo applications are limited by biostability and biocompatibility. The structural integrity of DNA-origami nanostructures is compromised in physiological fluids due to nuclease degradation and low levels of salt (Hahn et al. 2014). Promisingly, the stabilisation of DNA-origami nanostructures has been demonstrated using lipid-wrapping (Perrault and Shih 2014) and coating with an oligolysine-polyethylene (PEG) shell (Ponnuswamy et al. 2017). DNA-origami nanostructures are also sensitive to both mechanical and thermal damage. The force required to melt DNA is in the pN region (Santosh and Maiti 2009) and so is the force needed for the rupture of DNA-origami nanostructures (Engel et al. 2018). This limits the external force that can be placed on these systems by electric and magnetic fields. In comparison, the force required to rupture liposomes is three orders of magnitude larger (nN) (Wang et al. 2012). Similarly, external fields applied should be low enough in power that bulk heating of the solution does not melt the DNA-origami nanostructures, which begins at approximately 40 °C, or damage cells, which for electric fields occurs above 30–40 kV/m (Menachery and Pethig 2005). UV cross-linking agents can be used to increase thermal stability of DNA-origami nanostructures up to 85 °C (Rajendran et al. 2011) but do not protect the surrounding tissue.
Overall, switchable DNA-origami nanostructures have been used to construct functional nanomachines that can respond to either their local chemical environment or be remotely controlled. However, the complexity of these synthetic devices and their applications is still far below that of even the simplest protein machinery found in living cells. To achieve the goal of functional nanomachines will require both the development of new actuation methods and the enhancement of existing methods. The combination of multiple methods across scales also has the potential to rapidly increase the complexity and utility of these devices. For example, for in vivo applications, magnetic switching could be used to trigger drug delivery at a target site on the macroscale, coupled with pH-gated activation at the cellular level. Development of new multiscale frameworks to represent, design and program such systems will facilitate this. Rapid advances in high spatial and time-resolution molecular imaging (Schueder et al. 2017; Liu et al. 2018b) will also drive the development of complex DNA machinery, by allowing direct observation of the dynamics of nanostructures actuation both in vitro and in vivo. The ultimate goal of this would be to make real-time observations of complex DNA-origami nanodevices changing shape at the single-molecule level in live organisms.
Acknowledgments
We thank Dr Alice Williamson for helpful discussion and comments on the manuscript.
Funding
This work was supported by Australian Research Council Discovery Early Career Research Fellowship DE180101635 (SW), University of Sydney Nano Institute Scholarship (JKDS, MTL).
Author contributions
S.W. and J.K.D.S. prepared the figures and wrote and edited the manuscript together, and M.T.L. and A. A. contributed to the manuscript editing and figures.
Conflict of interest
Shelley Wickham declares that she has no conflict of interest. Jasleen Kaur Daljit Singh declares that she has no conflict of interest. Ali Abbas declares that he has no conflict of interest. Minh Tri Luu declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/S0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- Anderson RR, Parrish JA. The optics of human skin. J Invest Dermatol. 1981;77:13–19. doi: 10.1111/1523-1747.ep12479191. [DOI] [PubMed] [Google Scholar]
- Asanuma H, Liang X, Nishioka H, Matsunaga D, Liu M, Komiyama M. Synthesis of azobenzene-tethered DNA for reversible photo-regulation of DNA functions: hybridization and transcription. Nat Protoc. 2007;2:203–212. doi: 10.1038/nprot.2006.465. [DOI] [PubMed] [Google Scholar]
- Bai X, Li Z, Jockusch S, Turro NJ, Ju J. Photocleavage of a 2-nitrobenzyl linker bridging a fluorophore to the 5′ end of DNA. Proc Natl Acad Sci. 2003;100:409–413. doi: 10.1073/pnas.242729099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J, Turberfield AJ. DNA nanomachines. Nat Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- Bell NAW, Keyser UF. Digitally encoded DNA nanostructures for multiplexed, single-molecule protein sensing with nanopores. Nat Nanotechnol. 2016;11:645–651. doi: 10.1038/nnano.2016.50. [DOI] [PubMed] [Google Scholar]
- Burns JR, Lamarre B, Pyne ALB, Noble JE, Ryadnov MG. DNA origami inside-out viruses. ACS Synth Biol. 2018;7:767–773. doi: 10.1021/acssynbio.7b00278. [DOI] [PubMed] [Google Scholar]
- Chance R. R., Prock A., Silbey R. Advances in Chemical Physics. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2007. Molecular Fluorescence and Energy Transfer Near Interfaces; pp. 1–65. [Google Scholar]
- Chen H, Li R, Li S, Andréasson J, Choi JH. Conformational effects of UV light on DNA origami. J Am Chem Soc. 2017;139:1380–1383. doi: 10.1021/jacs.6b10821. [DOI] [PubMed] [Google Scholar]
- Cherry KM, Qian L. Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks. Nature. 2018;559:370–376. doi: 10.1038/s41586-018-0289-6. [DOI] [PubMed] [Google Scholar]
- Derr ND, Goodman BS, Jungmann R, Leschziner AE, Shih WM, Reck-Peterson SL. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science. 2012;338:662–665. doi: 10.1126/science.1226734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- Engel MC, Smith DM, Jobst MA, Sajfutdinow M, Liedl T, Romano F, Rovigatti L, Louis AA, Doye JPK. Force-induced unravelling of DNA origami. ACS Nano. 2018;12:6734–6747. doi: 10.1021/acsnano.8b01844. [DOI] [PubMed] [Google Scholar]
- Frangioni JV. In vivo near-infrared fluorescence imaging. Curr Opin Chem Biol. 2003;7:626–634. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetskii MD, Mirkin SM. Triplex DNA structures. Annu Rev Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- Gehring K, Leroy J-L, Guéron M. A tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature. 1993;363:561–565. doi: 10.1038/363561a0. [DOI] [PubMed] [Google Scholar]
- Gerling T, Wagenbauer KF, Neuner AM, Dietz H. Dynamic DNA devices and assemblies formed by shape-complementary, non–base pairing 3D components. Science. 2015;347:1446–1452. doi: 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]
- Gerweck LE, Seetharaman K. Cellular pH gradient in tumor versus normal tissue: potential exploitation for the treatment of cancer. Cancer Res. 1996;56:1194–1198. [PubMed] [Google Scholar]
- Godonoga M, Lin T-Y, Oshima A, Sumitomo K, Tang MSL, Cheung Y-W, Kinghorn AB, Dirkzwager RM, Zhou C, Kuzuya A, Tanner JA, Heddle JG (2016) A DNA aptamer recognising a malaria protein biomarker can function as part of a DNA origami assembly. Sci Rep 6:21266. 10.1038/srep21266 [DOI] [PMC free article] [PubMed]
- Gopinath A, Miyazono E, Faraon A, Rothemund PWK. Engineering and mapping nanocavity emission via precision placement of DNA origami. Nature. 2016;535:401–405. doi: 10.1038/nature18287. [DOI] [PubMed] [Google Scholar]
- Gu H, Chao J, Xiao S-J, Seeman NC. A proximity-based programmable DNA nanoscale assembly line. Nature. 2010;465:202–205. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J, Wickham SFJ, Shih WM, Perrault SD. Addressing the instability of DNA nanostructures in tissue culture. ACS Nano. 2014;8:8765–8775. doi: 10.1021/nn503513p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi N, Ellington A, Stanton M. Aptamer beacons for the direct detection of proteins. Anal Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- Heckel A, Mayer G. Light regulation of aptamer activity: an anti-thrombin aptamer with caged thymidine nucleobases. J Am Chem Soc. 2005;127:822–823. doi: 10.1021/ja043285e. [DOI] [PubMed] [Google Scholar]
- Hwang K, Wu P, Kim T, Lei L, Tian S, Wang Y, Lu Y. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew Chem Int Ed Engl. 2014;53:13798–13802. doi: 10.1002/anie.201408333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idili A, Vallée-Bélisle A, Ricci F. Programmable pH-triggered DNA nanoswitches. J Am Chem Soc. 2014;136:5836–5839. doi: 10.1021/ja500619w. [DOI] [PubMed] [Google Scholar]
- Iwaki M, Wickham SF, Ikezaki K, Yanagida T, Shih WM. A programmable DNA origami nanospring that reveals force-induced adjacent binding of myosin VI heads. Nat Commun. 2016;7:13715. doi: 10.1038/ncomms13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem S, Lee ATL, Leigh DA, Markevicius A, Solà J. Pick-up, transport and release of a molecular cargo using a small-molecule robotic arm. Nat Chem. 2016;8:138–143. doi: 10.1038/nchem.2410. [DOI] [PubMed] [Google Scholar]
- Ke Y, Meyer T, Shih WM, Bellot G. Regulation at a distance of biomolecular interactions using a DNA origami nanoactuator. Nat Commun. 2016;7:10935. doi: 10.1038/ncomms10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen JB, Liu L, Bank Kodal AL, Madsen M, Li Q, Song J, Woehrstein JB, Wickham SFJ, Strauss MT, Schueder F, Vinther J, Krissanaprasit A, Gudnason D, Smith AAA, Ogaki R, Zelikin AN, Besenbacher F, Birkedal V, Yin P, Shih WM, Jungmann R, Dong M, Gothelf KV. Routing of individual polymers in designed patterns. Nat Nanotechnol. 2015;10:892–898. doi: 10.1038/nnano.2015.190. [DOI] [PubMed] [Google Scholar]
- Kohman RE, Cha SS, Man H-Y, Han X. Light-triggered release of bioactive molecules from DNA nanostructures. Nano Lett. 2016;16:2781–2785. doi: 10.1021/acs.nanolett.6b00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopperger E, List J, Madhira S, Rothfischer F, Lamb DC, Simmel FC. A self-assembled nanoscale robotic arm controlled by electric fields. Science. 2018;359:296–301. doi: 10.1126/science.aao4284. [DOI] [PubMed] [Google Scholar]
- Kroener F, Heerwig A, Kaiser W, Mertig M, Rant U. Electrical actuation of a DNA origami Nanolever on an electrode. J Am Chem Soc. 2017;139:16510–16513. doi: 10.1021/jacs.7b10862. [DOI] [PubMed] [Google Scholar]
- Kuzuya A, Sakai Y, Yamazaki T, Xu Y, Komiyama M. Nanomechanical DNA origami “single-molecule beacons” directly imaged by atomic force microscopy. Nat Commun. 2011;2:449. doi: 10.1038/ncomms1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuya A, Watanabe R, Yamanaka Y, Tamaki T, Kaino M, Ohya Y. Nanomechanical DNA origami pH sensors. Sensors. 2014;14:19329–19335. doi: 10.3390/s141019329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyk A, Schreiber R, Fan Z, Pardatscher G, Roller E-M, Högele A, Simmel FC, Govorov AO, Liedl T. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature. 2012;483:311–314. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- Kuzyk A, Schreiber R, Zhang H, Govorov AO, Liedl T, Liu N. Reconfigurable 3D plasmonic metamolecules. Nat Mater. 2014;13:862–866. doi: 10.1038/nmat4031. [DOI] [PubMed] [Google Scholar]
- Kuzyk A, Yang Y, Duan X, Stoll S, Govorov AO, Sugiyama H, Endo M, Liu N. A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function. Nat Commun. 2016;7:10591. doi: 10.1038/ncomms10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyk A, Urban MJ, Idili A, Ricci F, Liu N. Selective control of reconfigurable chiral plasmonic metamolecules. Sci Adv. 2017;3:e1602803. doi: 10.1126/sciadv.1602803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauback S, Mattioli KR, Marras AE, Armstrong M, Rudibaugh TP, Sooryakumar R, Castro CE. Real-time magnetic actuation of DNA nanodevices via modular integration with stiff micro-levers. Nat Commun. 2018;9:1446. doi: 10.1038/s41467-018-03601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Huh J-H, Kim K, Lee S. DNA origami-guided assembly of the roundest 60–100 nm gold nanospheres into plasmonic metamolecules. Adv Funct Mater. 2018;28:1707309. doi: 10.1002/adfm.201707309. [DOI] [Google Scholar]
- Li Z, Bai X, Ruparel H, Kim S, Turro NJ, Ju J. A photocleavable fluorescent nucleotide for DNA sequencing and analysis. Proc Natl Acad Sci. 2003;100:414–419. doi: 10.1073/pnas.242729199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Jieming, Johnson-Buck Alexander, Yang Yuhe Renee, Shih William M., Yan Hao, Walter Nils G. Exploring the speed limit of toehold exchange with a cartwheeling DNA acrobat. Nature Nanotechnology. 2018;13(8):723–729. doi: 10.1038/s41565-018-0130-2. [DOI] [PubMed] [Google Scholar]
- Li Suping, Jiang Qiao, Liu Shaoli, Zhang Yinlong, Tian Yanhua, Song Chen, Wang Jing, Zou Yiguo, Anderson Gregory J, Han Jing-Yan, Chang Yung, Liu Yan, Zhang Chen, Chen Liang, Zhou Guangbiao, Nie Guangjun, Yan Hao, Ding Baoquan, Zhao Yuliang. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nature Biotechnology. 2018;36(3):258–264. doi: 10.1038/nbt.4071. [DOI] [PubMed] [Google Scholar]
- Lilley DMJ. Structures of helical junctions in nucleic acids. Q Rev Biophys. 2000;33:109–159. doi: 10.1017/S0033583500003590. [DOI] [PubMed] [Google Scholar]
- Liu Q, Deiters A. Optochemical control of deoxyoligonucleotide function via a nucleobase-caging approach. Acc Chem Res. 2014;47:45–55. doi: 10.1021/ar400036a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Jiang S, Loza O, Fahmi NE, Šulc P, Stephanopoulos N. Rapid Photoactuation of a DNA nanostructure using an internal photocaged trigger strand. Angew Chem Int Ed. 2018;57:9341–9345. doi: 10.1002/anie.201804264. [DOI] [PubMed] [Google Scholar]
- Liu T-L, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J, Kohrman AQ, Medwig TN, Dambournet D, Forster R, Cunniff B, Ruan Y, Yashiro H, Scholpp S, Meyerowitz EM, Hockemeyer D, Drubin DG, Martin BL, Matus DQ, Koyama M, Megason SG, Kirchhausen T, Betzig E. Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science. 2018;360:eaaq1392. doi: 10.1126/science.aaq1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majikes JM, Ferraz LCC, LaBean TH. pH-driven actuation of DNA origami via parallel I-motif sequences in solution and on surfaces. Bioconjug Chem. 2017;28:1821–1825. doi: 10.1021/acs.bioconjchem.7b00288. [DOI] [PubMed] [Google Scholar]
- Mao C, Sun W, Shen Z, Seeman NC. A nanomechanical device based on the B–Z transition of DNA. Nature. 1999;397:144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- Marini M, Piantanida L, Musetti R, Bek A, Dong M, Besenbacher F, Lazzarino M, Firrao G. A revertible, autonomous, self-assembled DNA-origami nanoactuator. Nano Lett. 2011;11:5449–5454. doi: 10.1021/nl203217m. [DOI] [PubMed] [Google Scholar]
- Marras AE, Zhou L, Su H-J, Castro CE. Programmable motion of DNA origami mechanisms. Proc Natl Acad Sci. 2015;112:713–718. doi: 10.1073/pnas.1408869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maune HT, Han S, Barish RD, Bockrath M, Iii WAG, Rothemund PWK, Winfree E. Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nat Nanotechnol. 2010;5:61–66. doi: 10.1038/nnano.2009.311. [DOI] [PubMed] [Google Scholar]
- Menachery A, Pethig R. Controlling cell destruction using dielectrophoretic forces. IEE Proc - Nanobiotechnol. 2005;152:145–149. doi: 10.1049/ip-nbt:20050010. [DOI] [PubMed] [Google Scholar]
- Modi S, Nizak C, Surana S, Halder S, Krishnan Y. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat Nanotechnol. 2013;8:459–467. doi: 10.1038/nnano.2013.92. [DOI] [PubMed] [Google Scholar]
- Omabegho T, Gurel PS, Cheng CY, Kim LY, Ruijgrok PV, Das R, Alushin GM, Bryant Z. Controllable molecular motors engineered from myosin and RNA. Nat Nanotechnol. 2018;13:34–40. doi: 10.1038/s41565-017-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordoukhanian P, Taylor J-S. Design and synthesis of a versatile photocleavable DNA building block. application to phototriggered hybridization. J Am Chem Soc. 1995;117:9570–9571. doi: 10.1021/ja00142a028. [DOI] [Google Scholar]
- Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault SD, Shih WM. Virus-inspired membrane encapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano. 2014;8:5132–5140. doi: 10.1021/nn5011914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnuswamy N, Bastings MMC, Nathwani B, Ryu JH, Chou LYT, Vinther M, Li WA, Anastassacos FM, Mooney DJ, Shih WM. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat Commun. 2017;8:15654. doi: 10.1038/ncomms15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokup A, Hemphill J, Deiters A. DNA computation: a photochemically controlled AND gate. J Am Chem Soc. 2012;134:3810–3815. doi: 10.1021/ja210050s. [DOI] [PubMed] [Google Scholar]
- Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- Rajendran A, Endo M, Katsuda Y, Hidaka K, Sugiyama H. Photo-cross-linking-assisted thermal stability of DNA origami structures and its application for higher-temperature self-assembly. J Am Chem Soc. 2011;133:14488–14491. doi: 10.1021/ja204546h. [DOI] [PubMed] [Google Scholar]
- Rajendran A, Endo M, Sugiyama H. Single-molecule analysis using DNA origami. Angew Chem Int Ed. 2012;51:874–890. doi: 10.1002/anie.201102113. [DOI] [PubMed] [Google Scholar]
- Rinker S, Ke Y, Liu Y, Chhabra R, Yan H. Self-assembled DNA nanostructures for distance dependent multivalent ligand-protein binding. Nat Nanotechnol. 2008;3:418–422. doi: 10.1038/nnano.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Samai S, Bradley DJ, Choi TLY, Yan Y, Ginger DS. Temperature-dependent photoisomerization quantum yields for azobenzene-modified DNA. J Phys Chem C. 2017;121:6997–7004. doi: 10.1021/acs.jpcc.6b12241. [DOI] [Google Scholar]
- Sannohe Y, Endo M, Katsuda Y, Hidaka K, Sugiyama H. Visualization of dynamic conformational switching of the G-Quadruplex in a DNA nanostructure. J Am Chem Soc. 2010;132:16311–16313. doi: 10.1021/ja1058907. [DOI] [PubMed] [Google Scholar]
- Santosh M, Maiti PK. Force induced DNA melting. J Phys Condens Matter. 2009;21:034113. doi: 10.1088/0953-8984/21/3/034113. [DOI] [PubMed] [Google Scholar]
- Schueder F, Lara-Gutiérrez J, Beliveau BJ, Saka SK, Sasaki HM, Woehrstein JB, Strauss MT, Grabmayr H, Yin P, Jungmann R. Multiplexed 3D super-resolution imaging of whole cells using spinning disk confocal microscopy and DNA-PAINT. Nat Commun. 2017;8:2090. doi: 10.1038/s41467-017-02028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman NC. Nucleic acid junctions and lattices. J Theor Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- Song J, Li Z, Wang P, Meyer T, Mao C, Ke Y. Reconfiguration of DNA molecular arrays driven by information relay. Science. 2017;357:eaan3377. doi: 10.1126/science.aan3377. [DOI] [PubMed] [Google Scholar]
- Surana S, Bhat JM, Koushika SP, Krishnan Y. An autonomous DNA nanomachine maps spatiotemporal pH changes in a multicellular living organism. Nat Commun. 2011;2:340. doi: 10.1038/ncomms1340. [DOI] [PubMed] [Google Scholar]
- Takenaka T, Endo M, Suzuki Y, Yang Y, Emura T, Hidaka K, Kato T, Miyata T, Namba K, Sugiyama H. Photoresponsive DNA nanocapsule having an open/close system for capture and release of nanomaterials. Chem Eur J. 2014;20:14951–14954. doi: 10.1002/chem.201404757. [DOI] [PubMed] [Google Scholar]
- Thubagere AJ, Li W, Johnson RF, Chen Z, Doroudi S, Lee YL, Izatt G, Wittman S, Srinivas N, Woods D, Winfree E, Qian L. A cargo-sorting DNA robot. Science. 2017;357:eaan6558. doi: 10.1126/science.aan6558. [DOI] [PubMed] [Google Scholar]
- Torelli E, Marini M, Palmano S, Piantanida L, Polano C, Scarpellini A, Lazzarino M, Firrao G. A DNA origami nanorobot controlled by nucleic acid hybridization. Small. 2014;10:2918–2926. doi: 10.1002/smll.201400245. [DOI] [PubMed] [Google Scholar]
- Vink AA, Roza L. Biological consequences of cyclobutane pyrimidine dimers. J Photochem Photobiol B. 2001;65:101–104. doi: 10.1016/S1011-1344(01)00245-7. [DOI] [PubMed] [Google Scholar]
- Wagenbauer KF, Sigl C, Dietz H. Gigadalton-scale shape-programmable DNA assemblies. Nature. 2017;552:78–83. doi: 10.1038/nature24651. [DOI] [PubMed] [Google Scholar]
- Wang X, Shindel MM, Wang S-W, Ragan R. Elucidating driving forces for liposome rupture: external perturbations and chemical affinity. Langmuir. 2012;28:7417–7427. doi: 10.1021/la300127m. [DOI] [PubMed] [Google Scholar]
- Weissleder Ralph. A clearer vision for in vivo imaging. Nature Biotechnology. 2001;19(4):316–317. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- Weyel XMM, Fichte MAH, Heckel A. A two-photon-photocleavable linker for triggering light-induced strand breaks in oligonucleotides. ACS Chem Biol. 2017;12:2183–2190. doi: 10.1021/acschembio.7b00367. [DOI] [PubMed] [Google Scholar]
- Wickham SFJ, Bath J, Katsuda Y, Endo M, Hidaka K, Sugiyama H, Turberfield AJ. A DNA-based molecular motor that can navigate a network of tracks. Nat Nanotechnol. 2012;7:169–173. doi: 10.1038/nnano.2011.253. [DOI] [PubMed] [Google Scholar]
- Willner EM, Kamada Y, Suzuki Y, Emura T, Hidaka K, Dietz H, Sugiyama H, Endo M. Single-molecule observation of the photoregulated conformational dynamics of DNA origami nanoscissors. Angew Chem Int Ed. 2017;56:15324–15328. doi: 10.1002/anie.201708722. [DOI] [PubMed] [Google Scholar]
- Xu H, Jones S, Choi B-C, Gordon R. Characterization of individual magnetic nanoparticles in solution by double nanohole optical tweezers. Nano Lett. 2016;16:2639–2643. doi: 10.1021/acs.nanolett.6b00288. [DOI] [PubMed] [Google Scholar]
- Yang Y, Endo M, Hidaka K, Sugiyama H. Photo-controllable DNA origami nanostructures assembling into predesigned multiorientational patterns. J Am Chem Soc. 2012;134:20645–20653. doi: 10.1021/ja307785r. [DOI] [PubMed] [Google Scholar]
- Yang Y, Hughes RP, Aprahamian I. Near-infrared light activated azo-BF2 switches. J Am Chem Soc. 2014;136:13190–13193. doi: 10.1021/ja508125n. [DOI] [PubMed] [Google Scholar]
- Yurke B, Turberfield AJ, Mills AP, Jr, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- Zadegan RM, Lindau EG, Klein WP, Green C, Graugnard E, Yurke B, Kuang W, Hughes WL. Twisting of DNA origami from intercalators. Sci Rep. 2017;7:7382. doi: 10.1038/s41598-017-07796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DY, Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- Zhao Y-X, Shaw A, Zeng X, Benson E, Nyström AM, Högberg B. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano. 2012;6:8684–8691. doi: 10.1021/nn3022662. [DOI] [PubMed] [Google Scholar]
- Zhou L, Marras AE, Su H-J, Castro CE. Direct design of an energy landscape with bistable DNA origami mechanisms. Nano Lett. 2015;15:1815–1821. doi: 10.1021/nl5045633. [DOI] [PubMed] [Google Scholar]