Abstract
The cholesterol-dependent cytolysins (CDCs) are a family of bacterial toxins that are important virulence factors for a number of pathogenic Gram-positive bacterial species. CDCs are secreted as soluble, stable monomeric proteins that bind specifically to cholesterol-rich cell membranes, where they assemble into well-defined ring-shaped complexes of around 40 monomers. The complex then undergoes a concerted structural change, driving a large pore through the membrane, potentially lysing the target cell. Understanding the details of this process as the protein transitions from a discrete monomer to a complex, membrane-spanning protein machine is an ongoing challenge. While many of the details have been revealed, there are still questions that remain unanswered. In this review, we present an overview of some of the key features of the structure and function of the CDCs, including the structure of the secreted monomers, the process of interaction with target membranes, and the transition from bound monomers to complete pores. Future directions in CDC research and the potential of CDCs as research tools will also be discussed.
Keywords: Cholesterol-binding protein, Cholesterol-dependent cytolysin, Membrane-protein interactions, Pore-forming toxin
Introduction
Bacteria secrete a wide variety of peptides and proteins into their environment to carry out a broad range of functions, ranging from the scavenging of precious metabolic precursors to defensive actions against potential competitors and predators. The makeup of this milieu of bacterial molecules, termed the secretome, varies from species to species and with external stimuli, assisting with bacterial adaptation to particular environments (Tommassen and Arenas 2017). Bacterial proteins evolved in response to a specific pressure (for example, as defensive weapons against predation by eukaryotic single-cell organisms) can be seconded into other uses during opportunistic infection of animals. An important class of these bacterial proteins, which act as virulence factors and toxins in human disease, are the pore-forming toxins (PFTs) that have evolved to convert from inactive soluble monomers to complex protein machines in the presence of a target cell membrane.
PFTs are categorised into two groups based on the structure of the pore; the α-PFTs, where the pore consists of helical hairpins, and the β-PFTs, where extended β-strands line the pore (Gouaux 1997). Many of the α-PFTs, while highly diverse structurally, retain a common motif of a pair of buried hydrophobic helices which become exposed on interaction with a membrane surface in what is called the ‘umbrella’ model (Parker et al. 1990). In this model, the protective outer helices spread on to the membrane, allowing the concealed helices to insert into the lipid bilayer. Originally identified from the structure of colicin (Parker et al. 1989), this motif was subsequently found in key virulence factors including diphtheria toxin (Choe et al. 1992), delta endotoxin (Li et al. 1991) and cytolysin A (Mueller et al. 2009). As with the α-PFTs, the β-PFTs are structurally diverse but have the common feature of a pore formed from β-hairpins. The first β-PFT for which a structure was determined was aerolysin (Parker et al. 1994), which was the forerunner for studies of several subsequent β-PFTs of medical and technological importance including Staphylococcus α-toxin (Song et al. 1996), anthrax toxin (Petosa et al. 1997) and gasdermin (Ruan et al. 2018). While all these proteins form pores from assembled β-hairpins, the different numbers of hairpins and the structural divergence of the proteins imply that each β-pore evolved independently (Reboul et al. 2016) and has required separate studies of each system to understand the mechanistic differences in pore formation.
A widespread family of β-PFTs are the cholesterol-dependent cytolysins (CDCs) which are found in multiple genera of Gram-positive bacteria including Bacillus, Streptococcus, Clostridium and Listeria. The CDCs act as virulence factors and toxins in human diseases including food poisoning, gangrene and pneumonia. Uniquely for β-PFTs, pore formation by the CDCs is strictly dependent on the presence of high levels of cholesterol in target cellular membranes. They are secreted as soluble, stable monomers which bind to cholesterol-rich membranes where they oligomerise into higher-order complexes, including large (30+ monomers) rings. These pre-pore rings undergo a major conformational change which unfurls shielded helical segments to form membrane-spanning β-hairpins, puncturing the target to produce very large (250 Å or more) pores (Fig. 1). The study of CDC structure and pore-forming mechanism has established the basis to understanding how a wide variety of pore-forming proteins function, which include the membrane attack complex/perforin (MACPF) family of proteins (Hadders et al. 2007; Rosado et al. 2007; Slade et al. 2008; Leung et al. 2017), as well as a toxin family found in the venom of the stonefish family of venomous fish (Ellisdon et al. 2015). It is likely that these proteins are ancient relatives of the CDC family of toxins that comprise a superfamily of the CDC/MACPF proteins.
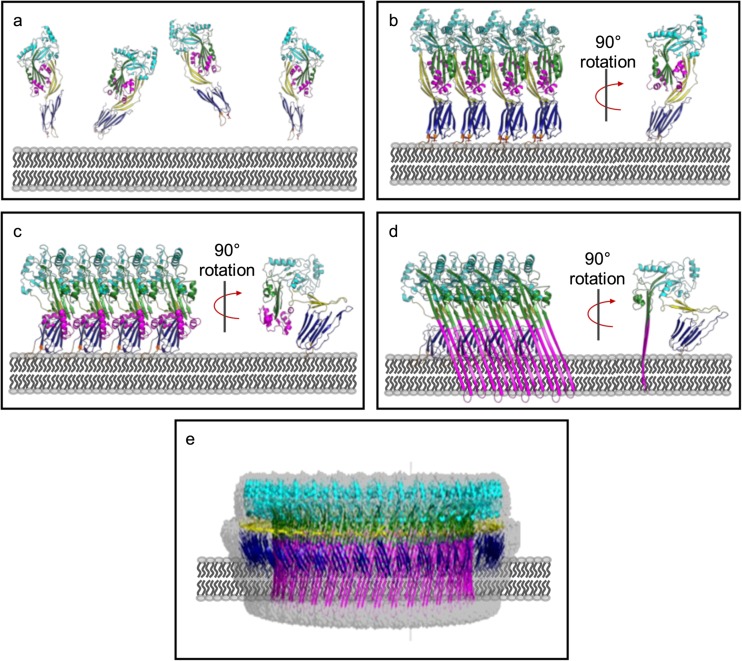
Fig. 1.
CDCs are secreted as soluble monomers (a), which bind to cholesterol on the surface of target cell membranes and oligomerise to form an early prepore structure (b). This involves slight insertion of the undecapeptide (UDP) loops in D4 into the lipid bilayer. CDC molecules in the early pre-pore undergo a 40 Å vertical collapse in D2, following disruption of the D3 interface (c). Full membrane insertion requires each monomer to contribute two TMH regions, which unfurl and insert in the membrane as β-hairpins (d). These β-hairpins form the β-barrel pore, which is typically about 300 Å in diameter and consists of between 30 and 50 individual monomers (e). All figures generated in Pymol (Schrodinger 2015). a, b Generated in Pymol using PDB: 5AOD. c is a model generated from PDB: 5AOD and PDB: 5LY6. d, c Generated using PDB 5LY6, the fitted model for the cryo-EM structure of the PLY pore complex
Monomer structure
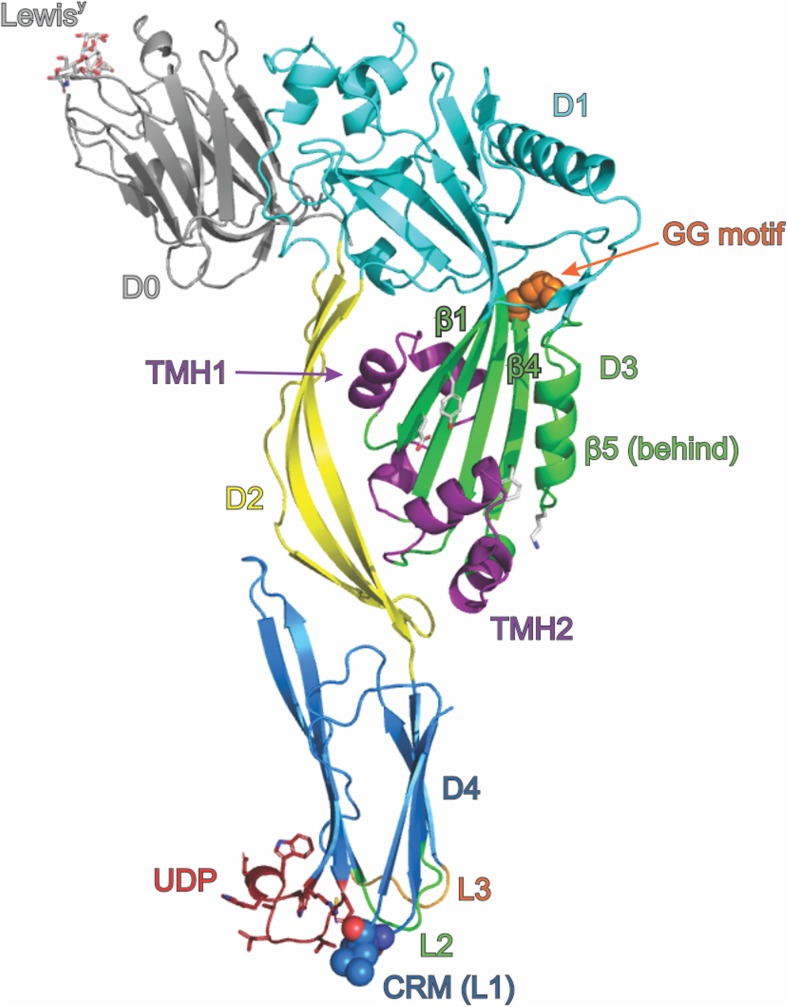
The first structure of a CDC, that for the clostridial toxin perfringolysin O (PFO), had a major impact in understanding how the β-PFTs interact with and penetrate cellular membranes (Rossjohn et al. 1997). The structure revealed an elongated (115 Å × 30 Å × 55 Å), four-domain protein (Fig. 2). Three of the four domains are discontinuous, with only the C terminal domain 4 (D4), a compact β-sandwich, being a discrete section of the protein primary structure. Domain 1 (D1) is primarily a seven-stranded β-sheet structure, with peripheral α-helices. Domain 2 (D2), a set of long β-strands, links the rest of the protein with D4. Domain 3 (D3) is an α/β/α-layered bundle sitting between D1 and D4. Correlating PFO’s structure with its pore-forming ability required significant further experimental work, as initial inspection of the structure gave only tantalising hints into how the protein punched holes in membranes. Extensive mutagenesis and functional analysis revealed that D4 contains the target identification activity, recognising cholesterol-rich membranes through the combination of a membrane-sensing 11 residue loop (the ‘undecapeptide’ or UDP) and a two-residue cholesterol recognition motif (CRM) (Farrand et al. 2010). It was also discovered that D3 is the pore-forming warhead and carries the trans-membrane β-hairpins (TMH1 and TMH2) folded away as helical segments in the soluble protein (Shepard et al. 1998; Shatursky et al. 1999). D1 and D2 provide the scaffold to tie target recognition and the toxin warhead together. In the wider context of the CDC-related MACPF proteins, it is becoming accepted practice to refer to the D1 + D3 unit as a single ‘MACPF/CDC domain’ (Rosado et al. 2007); however, the four domain nomenclature remains prevalent in studies of the CDCs themselves.
Fig. 2.
Diagrammatic representation of the structure of a CDC based on the structure of PFO (PDB: 1PFO) and D0 from LLY complexed with LewisY (PDB: 4GWI). The protein is shown as a cartoon, coloured by domain, with D0 grey, D1 cyan, D2 yellow, D3 green and D4 blue. D0, present in LLY, is shown with LewisY occupying the lectin-binding site. Key features of the structure are indicated, including the undecapeptide (UDP, shown as sticks), the cholesterol recognition motif (CRM, shown as spheres), the di-glycine motif that provides a pivot point during oligomerisation and pore formation (shown as orange spheres) and D3 residues mentioned in the text Y181, E183, F318 and K336 (shown as white sticks, unlabelled). The two transmembrane hairpins TMH1 and TMH2 are shown in their helical bundle state, coloured purple. Other labelled features are β-strands β1, β4 and β5 (discussed in the text) and the L2 and L3 loops of D4
Several other CDC structures have now been determined, including the structures of intermedilysin (Polekhina et al. 2005; Lawrence et al. 2016), anthrolysin O (Bourdeau et al. 2009), suilysin (Xu et al. 2010), streptolysin O (Feil et al. 2014), listeriolysin O (Koster et al. 2014), pneumolysin (Lawrence et al. 2015; Marshall et al. 2015; Park et al. 2016), and vaginolysin (Lawrence et al. 2016) (Table 1). The structural features of the CDCs are highly conserved, which is reflected in sequence identities of around 40% across the family. There are key sequence motifs characteristic of the CDCs, including the D4 UDP and strictly conserved cholesterol recognition motifs (Fig. 2) (Dowd et al. 2012; Hotze et al. 2013). Other highly conserved regions of the CDC sequence are present in D1–3: a strictly conserved diglycine, the trans-membrane hairpins (TMH1 and TMH2) and some short regions of unknown function (Hotze et al. 2013) (Table 2).
Table 1.
Structures of cholesterol-dependent cytolysins available in the PDB
| PDB code | Resolution (Å) | Year | Notes | Reference | |
|---|---|---|---|---|---|
| Perfringolysin O (PFO) Clostridium perfringens |
1PFO | 2.2 | 1997 | Rossjohn et al. (1997) | |
| 1M3J | 3.0 | 2002 | Crystal form II | Rossjohn et al. (2007) | |
| 1MEI | 2.9 | 2002 | Crystal form III | Rossjohn et al. (2007) | |
| 5DHL | 2.67 | 2015 | Mutant N197 W | Unpublished | |
| 5DIM | 2.8 | 2015 | Mutant N197 W, crystal form II | Unpublished | |
| Intermedilysin (ILY) Streptococcus intermedius |
1S3R | 2.6 | 2005 | Polekhina et al. (2005) | |
| 4BIK | 3.5 | 2013 | Monomer locked mutant, CD59-bound | Johnson et al. (2013) | |
| 5IMW | 2.89 | 2016 | Monomer locked mutant | Lawrence et al. (2016) | |
| 5IMT | 2.7 | 2016 | Monomer locked mutant, CD59-bound | Lawrence et al. (2016) | |
| Anthrolysin (ALO) Bacillus anthracis |
3CQF | 3.1 | 2009 | Bourdeau et al. (2009) | |
| Suilysin (SLY) Streptococcus suis |
3HVN | 2.85 | 2010 | Xu et al. (2010) | |
| Streptolysin O (SLO) Streptococcus pyogenes |
4HSC | 2.1 | 2014 | Feil et al. (2014) | |
| Listeriolysin O (LLO) Listeria monocytogenes |
4CDB | 2.15 | 2014 | Koster et al. (2014) | |
| Pneumolysin (PLY) Streptococcus pneumoniae |
4QQA | 2.8 | 2014 | Park et al. (2016) | |
| 4ZGH | 2.9 | 2015 | Lawrence et al. (2015) | ||
| 5CR6 | 1.98 | 2015 | Marshall et al. (2015) | ||
| 5AOD | 2.4 | 2015 | van Pee et al. (2016) | ||
| 4QQQ | 2.5 | 2015 | Mannose complexed | Park et al. (2016) | |
| 5AOE | 2.5 | 2016 | Mutant D168A | van Pee et al. (2017) | |
| 5AOF | 2.45 | 2016 | Deletion mutant Δ146/147 | van Pee et al. (2017) | |
| Vaginolysin (VLY) Gardnerella vaginalis |
5IMY | 2.4 | 2016 | Monomer locked mutant, CD59-bound | Lawrence et al. (2016) |
Table 2.
Residues and regions implicated in CDC activity
| Role in pore formation | CDC | Residue/s involved | Reference |
|---|---|---|---|
| Membrane attachment | |||
| Binding to membrane cholesterol (cholesterol recognition motif) | PFO | T490, L491 | Farrand et al. (2010) |
| PLY | T459, L460 | Farrand et al. (2010) | |
| ILY | T517, L518 | Farrand et al. (2010) | |
| SLO | T564, L565 | Farrand et al. (2010) | |
| Membrane anchoring | PFO | L2 loop 398HSGAYVA404 L3 loop 434DKTAH438 Undecapeptide 458ECTGLAWEWWR468 |
Farrand et al. (2010) |
| Membrane specificity | PFO | L3 loop 434DKTAH438 |
Farrand et al. (2015) |
| Receptor binding | |||
| CD59-binding motif | ILY | Y434, Y436, R452, R453, R480 | Lawrence et al. (2016) |
| VLY | Y421, Y423, R438, S439, R467 | Lawrence et al. (2016) | |
| Prepore assembly | |||
| Allosteric coupling of membrane binding with prepore assembly | PFO | R468 | Dowd et al. (2012) |
| PLY | T405 | van Pee et al. (2016) | |
| Salt bridges that stabilise the monomer-monomer interface | PLY | K18, E84 D93, R208 K188, E260 |
Marshall et al. (2015) |
| D1-D1 interactions that stabilise oligomerisation | LLO | K175, S176 | Koster et al. (2014) |
| Hinge around which D3 β5 rotates to expose D3 β4 | PFO | G324, G25 | Ramachandran et al. (2004) |
| Mediates structural transitions in D3 | PFO | W165 | Hotze and Tweten (2012) |
| Flexible linker between D2 and D4, enables movement of D1–3 with respect to D4 | PFO | G392 | Rossjohn et al. (1997) |
| PLY | G361 | van Pee et al. (2016) | |
| Propagates monomer-monomer interactions through π-stacking | PFO | Y181, F318 | Ramachandran et al. (2004) |
| pH sensitivity of pore-formation | LLO | Y206, E247, D208, D320, K316 | Koster et al. (2014) |
| Pore complex and membrane insertion | |||
| Provides the energy required for prepore to pore transition | PFO | E183, K336 | Wade et al. (2015) |
| SLO | E245, K407 | Wade et al. (2015) | |
| β-strands that that insert into the membrane to form the β-barrel | PFO | 189KSQISSALNVNAKVLENSLGVDFNAVANNE218 | Shepard et al. (1998) |
| PFO | 288KDVQAAFKALIKNTDIKNSQQYKD311 | Shatursky et al. (1999) | |
| D4 interactions that stabilise the pore complex | PLY | W433, L461 | van Pee et al. (2017) |
| LLO | W489 | Kozorog et al. (2018) | |
Cholesterol recognition and binding by cholesterol-dependent cytolysins
The distinguishing feature of all CDC family members is recognition of cholesterol via the C-terminal D4. Despite previous expectation that the UDP was responsible for cholesterol recognition due to its high conservation among CDC members, it was found that recognition occurs via two amino acid residues in loop L1 of D4: a Thr-Leu pair (T490 and L491 in PFO) (Farrand et al. 2010). Referred to as the cholesterol recognition motif (CRM), the residue pair is conserved throughout all reported CDCs, and studies reveal mutations to either residue significantly affect the ability of the CDC to bind both cell and model membranes (Farrand et al. 2010; Park et al. 2016). Recognition of cholesterol by the CRM results in membrane insertion of loops L2, L3 and the UDP, providing anchorage of the CDC monomer to the membrane during subsequent pore-formation events. In addition to stabilising the monomer-lipid interaction, the role of the UDP is proposed to be involvement in the pre-pore to pore transition and coupling the initiation of pre-pore assembly to the occurrence of membrane binding (Soltani et al. 2007b; Dowd et al. 2012). The importance of the UDP was highlighted by mutagenesis of UDP residue R468 completely eliminating the pore-forming activity of PFO, with membrane binding by the CDC also diminished (Dowd et al. 2012).
Currently, there are no available structures of a CDC-cholesterol complex, with much of the information regarding cholesterol interaction established from either site-directed mutagenesis or studies investigating binding with various structurally distinct sterols (Ramachandran et al. 2004; Bavdek et al. 2007; Farrand et al. 2015; Savinov and Heuck 2017). Studies based on the latter method established two structural features of cholesterol that are critical for CDC binding: the β-hydroxyl group on ring A, and flexibility of the acyl chain around C20-C22, hypothesised to be necessary to allow the cholesterol molecule to fit into the D4 binding site (Bavdek et al. 2007; Nelson et al. 2008; Savinov and Heuck 2017).
More recently, 19F-NMR spectroscopy was employed to probe the interaction of LLO with cholesterol (Kozorog et al. 2018). Several tryptophan residues, primarily located in D4, displayed significant 19F-NMR chemical shifts upon CDC binding to cholesterol-rich membranes. These chemical shift changes were not present on mixing LLO with cholesterol in solution, indicating that the lipid environment is critical in forming the appropriate interactions. Analysis of the NMR results suggested different roles for the tryptophan residues in D4, with residue W512, located in L1 near the CRM, involved in membrane binding, while W189 (in D1) and W489 were instead important for oligomerisation.
Effect of the lipid environment on cholesterol binding
It is well established that a membrane must have a relatively high level of cholesterol (> 30 mol%) to allow binding and lysis by CDCs (Ohno-Iwashita et al. 1992; Bavdek et al. 2007; Johnson et al. 2012). More recently, investigation into the effect of membrane composition on binding has revealed that the minimum amount of cholesterol required is dependent on the lipid composition of the membrane (Nelson et al. 2008; Flanagan et al. 2009). For example, phospholipids which pack against cholesterol in a tight manner, possessing large hydrophilic head groups, decrease binding in comparison to phospholipids with smaller head groups and looser packing (Nelson et al. 2008). It has also been suggested that the lipid environment may allow different CDCs to exhibit individual degrees of binding specificity for cholesterol (Farrand et al. 2015). This likely occurs via the structure of L3, which provides the CDC with a cellular-specific targeting mechanism via discriminating between lipid environments surrounding the cholesterol to which the CDC can bind (Farrand et al. 2015).
CD59-responsive cholesterol-dependent cytolysins
At present, three CDCs are known to possess the ability to bind the GPI-anchored membrane protein CD59 as a receptor: intermedilysin (ILY), lectinolysin (LLY) and vaginolysin (VLY) (Giddings et al. 2004; Gelber et al. 2008; Wickham et al. 2011). Present on the surface of human cells, CD59 inhibits the membrane attack complex of complement through binding the complement components C8α and C9. Remarkably, ILY binding to CD59 is dependent on nearly the same residues necessary for CD59 to interact with C8α and C9, yet the binding sites for CD59 on these three proteins exhibit no similarity (Wickham et al. 2011). Two additional CDCs, from Streptococcus tigurinus and Streptococcus pseudopneumoniae, are predicted to bind CD59 based on the presence of a conserved D4 motif identified in the known CD59-responsive CDCs listed above (Wickham et al. 2011). Interestingly, despite interaction with CD59, all three of the CD59-responsive CDCs retain a dependence on cholesterol for pore formation. VLY and LLY are able to bind to cholesterol-rich membranes in the absence of CD59, broadening their activity to include non-human cells, although lysis of non-human cells by VLY is approximately 100-fold less efficient than that of human cells (Zilnyte et al. 2015). In contrast, ILY lacks the ability to bind cholesterol-rich membranes directly without the co-expression of CD59 on the cell (Giddings et al. 2004). Thus, the toxin is strictly specific for human cells. Nonetheless, pore formation by ILY is still cholesterol-dependent; the process of pre-pore to pore transition requires the disengagement of the CD59 receptor and so to maintain its interaction with the membrane during this critical transition, the interaction with cholesterol via the CRM is required (LaChapelle et al. 2009).
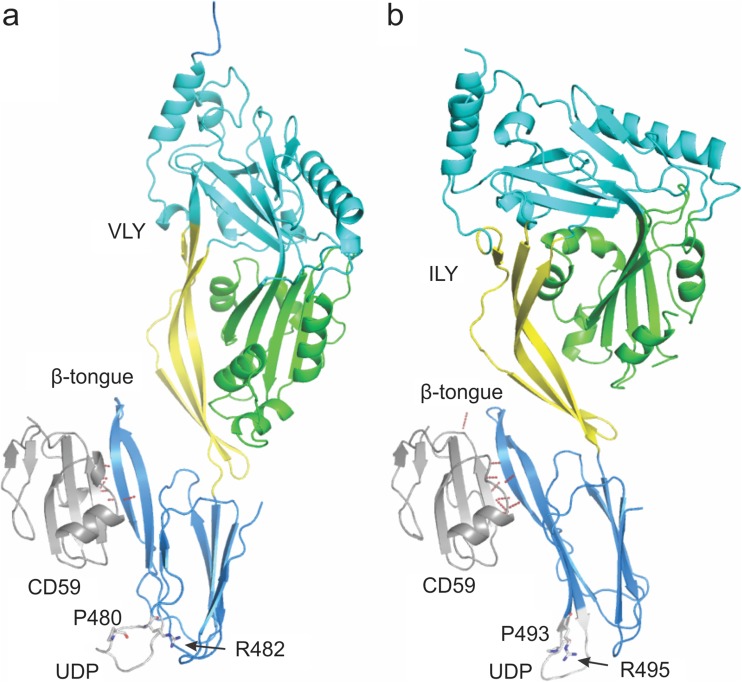
Both ILY and VLY have been crystallised in complex with a mutant of CD59, CD59D22A, and their structures reveal that binding to CD59 occurs in a comparable manner (Lawrence et al. 2016). Both interact primarily with CD59 via the β-tongue of D4 (Fig. 3), with backbone hydrogen bonds between the second strand of the β-hairpin in D4 and the last β strand in the CD59 structure, forming an extended β sheet between the two molecules. A further similarity is the amount of protein surface buried as a result of the interaction, being 680 and 682 Å2 for the VLY and ILY complexes, respectively. Finer details, however, including the exact position of binding on the β tongue, location of CD59 in respect to the D4 β tongue and the orientation of particular CDC residues, and thus the nature of interactions involving these residues, differ between the two different CD59-binding CDCs.
Fig. 3.
Diagrammatic representation of the structures of the a VLY-CD59 and b ILY-CD59 complexes. The proteins are shown as cartoons coloured by domain with D1 cyan, D2 yellow, D3 green and D4 blue for the CDC and CD59 in grey. Hydrogen bonds identified between CD59 and the associated CDC are shown as dashed red lines. The β-tongue, UDP, conserved UDP arginine and unusual UDP proline are labelled. The structures were oriented after being superimposed on the CD59 molecules to emphasise the position of the D4 β-tongue interaction with CD59
The structures of ILY alone or complexed with CD59 displayed the UDP in a conformation strikingly different to that observed in non-CD59-dependent CDCs (Polekhina et al. 2005; Johnson et al. 2013; Lawrence et al. 2016). While the ILY structure displays the UDP residue R495 pointing out into solution, other CDCs, including PFO, show the equivalent arginine residue pointing towards the backbone of the L1 loop. Modelling suggested interaction between this residue and L1 ensures the correct conformation for cholesterol binding. CD59-responsive CDCs have a strictly conserved proline in the UDP motif instead of the tryptophan found in other CDCs. It is likely that this proline residue indirectly prevents cholesterol binding, thereby enhancing CD59 binding, by preventing the arginine residue interacting with L1. Interestingly, two distinct conformations were observed to be adopted by the UDP in the two unique molecules found in the crystal lattice of the VLY-CD59 complex; one correlates closely with that seen in the ILY-CD59 structure, the other conformation is not dissimilar to that observed in the non-CD59-responsive PFO. This observation may explain how VLY can initially bind to either CD59 or cholesterol as a receptor (Zilnyte et al. 2015).
Membrane carbohydrates as receptors for cholesterol-dependent cytolysins
A unique member of the CDC family is LLY, in that it shares characteristic CDC domains (D1–D4) but also possesses an additional N-terminal fucose-binding lectin domain (Farrand et al. 2008; Feil et al. 2012) termed domain 0 (D0, Fig. 2). Characterisation of LLY revealed a unique recognition for the difucosylated glycans Lewis b (Leb) and Lewis y (Ley), found respectively on blood cells and some epithelial cells, in particular those of a cancerous phenotype (Farrand et al. 2008). The molecular basis of this recognition was determined via X-ray crystal structures of the lectin domain in complex with L-fucose, Leb and Ley (Feil et al. 2012). It has also been proposed that the D4 domains of a further two CDCs may bind sugars found on red blood cells: PLY to the blood antigen sialyl-LewisX and SLO to the glycan lacto-N-neotetraosyl ceramide (Shewell et al. 2014).
Prepore complex assembly
Formation of a functional membrane-inserted pore requires that the TMH regions 1 and 2 disengage from the rest of the CDC molecule, unfurl to form β-strands that insert into membranes to form the β-barrel pore. In the monomer, the TMH regions are part of D3 and are stabilised by their interactions with D2 (Fig. 2). Furthermore, the membrane-bound monomer places the TMH regions approximately 40 Å above the membrane (Fig. 1). This means that to insert into membranes, the TMH regions need to move a significant distance towards the membrane plane. CDCs therefore pass through several intermediate stages accompanied by structural rearrangements when transitioning from a soluble monomer to membrane-inserted pore complex.
Binding to the membrane via D4 initiates pore formation by mediating distal structural changes that enable CDC oligomerisation (Ramachandran et al. 2004; Soltani et al. 2007a; Dowd et al. 2012). An analysis of different conformational states of PFO by X-ray crystallography suggests that membrane binding leads to a movement of D4 around a central β-sheet in D2. This in turn causes D2 to bend in such a way that most of the contacts between D2 and D3 are lost, thus allowing D3 to move away from the rest of the molecule (Rossjohn et al. 2007). Initial interactions between monomers are transient and weak, but progress in a synergistic manner. The crystal structures of ILY, LLO, and PLY (see Table 1) reveal a linear arrangement of protein molecules in the crystal lattice and provide some insight into the potential arrangement of CDCs in the early stages of oligomerisation. A similar linear arrangement was observed by electron microscopy for a PFO mutant where W165 was replaced by non-aromatic residues (Hotze and Tweten 2012) and likely represents an early transient stage in pore complex assembly. Inspection of the proposed monomer-monomer interface revealed shape and charge complementarity, suggesting the interface contributes to the stability of the assembled oligomers by excluding water (Marshall et al. 2015). It is also likely that electrostatic interactions between residues at the monomer-monomer interface play a key role in oligomer stability (Lawrence et al. 2015). The same study, comparing ILY, LLO and PLY, showed a progressive increase in both the number of contacts as well as the buried surface area at the monomer-monomer interface, confirming further the synergistic nature of monomer-monomer interactions. Given that the residues at the monomer-monomer interface are conserved among the different CDCs, this initial mode of contact is likely to be conserved among CDCs (Marshall et al. 2015).
Initial monomer-monomer interactions are followed by more stable interactions between adjacent CDCs that places each monomer in the correct geometry with respect to each other and commits the oligomers to form a complete, circular pre-pore structure on the membrane surface. D4 binding to cholesterol containing membranes facilitates changes in the monomer such that the β5 strand in D3 moves away from β4 around the diglycine linker (G324-G325), opening up β4 to interact with β1 from the adjacent molecule (Fig. 2). The interaction between β4 and β1 in turn opens up β4 in the second molecule to bind a third. These movements within D3 helps establish a π-stacking interaction between Y181 in β1 and F318 in β4, which has been hypothesised to stabilise the growing oligomer by locking β4 and β1 in position. At this point, CDCs are committed to the pre-pore state in a non-reversible manner (Ramachandran et al. 2004; Hotze and Tweten 2012).
The pre-pore structure is characterised by a circular ring made up of 34–37 monomers that surround an intact lipid membrane (Ramachandran et al. 2004; Dang et al. 2005; Tilley et al. 2005; van Pee et al. 2016; van Pee et al. 2017) (Table 3). Atomic force microscopy (AFM) experiments have shown that pre-pore complexes resembling arcs and incomplete rings also exist in PLY (Sonnen et al. 2014), LLO (Koster et al. 2014; Mulvihill et al. 2015; Podobnik et al. 2015; Ruan et al. 2016), SLO (Bhakdi et al. 1985) and SLY (Leung et al. 2014) (arcs and incomplete rings are discussed further in a section below). The pre-pore complexes have been shown to extend 110–120 Å above the surface of the membrane by AFM (Czajkowsky et al. 2004; Leung et al. 2014; Podobnik et al. 2015; Ruan et al. 2016; van Pee et al. 2016), consistent with more recent Cryo-ET data for the pre-pore complex (van Pee et al. 2017), and with the crystal structures of the linear oligomers of PLY (Lawrence et al. 2015; Marshall et al. 2015; van Pee et al. 2016). AFM of PFO (Czajkowsky et al. 2004), PLY (van Pee et al. 2016), SLY (Leung et al. 2014), cryo-electron tomography (Cryo-ET) on PLY (van Pee et al. 2017) and single particle cryo-EM of PLY (Tilley et al. 2005) and SLY (Leung et al. 2014) show that prepore complex assembly, whether as complete rings, incomplete rings or arcs, precedes pore formation in agreement with previous biochemical evidence (Hotze and Tweten 2012).
Table 3.
Structural studies on the pre-pore and pore complexes
| PDB/EMD code | Resolution (Å) | Year | Method | Notes | Reference | |
|---|---|---|---|---|---|---|
| Pneumolysin (PLY) Streptococcus pneumoniae |
– | 33 | 1999 | Cryo-EM | Pore complex | Gilbert et al. (1999) |
| EMD-1106 | 28 | 2005 | Cryo-EM | Pre-pore complex | Tilley et al. (2005) | |
| EMD-1107 | 29 | 2005 | Cryo-EM | Pore comples | Tilley et al. (2005) | |
| EMD-2611 EMD-2612 EMD-2613 EMD-2614 EMD-2615 |
Cryo-ET | Pre-pore complex | Sonnen et al. (2014) | |||
| EMD-2616 EMD-2617 EMD-2618 EMD-2619 |
Cryo-ET | Pore complex | Sonnen et al. (2014) | |||
| EMD-4118 (PDB: 5ly6) |
4.5 | 2017 | Cryo-EM | Wt pore complex | van Pee et al. (2017) | |
| – | 22 | 2017 | Cryo-ET | Pre-pore complex | van Pee et al. (2016) | |
| – | 27 | 2017 | Cryo-ET | Pore complex | van Pee et al. (2016) | |
| Suilysin (SLY) Streptococcus suis |
EMD-2983 | 2014 | Cryo-EM | Pore complex | Leung et al. (2014) | |
| EMD-2979 | 15 | 2014 | Cryo-EM | Pre-pore complex | Leung et al. (2014) |
The transition from the prepore to a pore complex
Pre-pore to pore transitions require the disengagement of D3 from the D1,2 interface, the collapse of the prepore complex by 40 Å and the unfurling of the α-helices in TMH1 and TMH2 into β-strands followed by their insertion into membranes. In the case of CDCs such as PFO and SLO, the rearrangement of the β-strands in D3 and the associated tilt of the molecule with respect to the plane of the membrane positions residues E183 and K336, on adjacent monomers, into close proximity. This enables the spontaneous formation of a strong salt bridge between the two residues, drawing the two strands β1 and β4 closer. This movement in turn pulls α-helices that form TMH1 away from the D1,2 interface. Formation of the salt bridge is exothermic and provides the energy required to transition from a pre-pore to pore (Wade et al. 2015). In the case of PLY and related CDCs, K336 is replaced by a glycine residue. Given that the residues required for the π-stacking interaction are also absent in these CDCs, it is likely that the D3-D1,2 interface is disrupted via a different mechanism. The crystal structure of PLY showed that D2 was more flexible suggesting that these CDCs have a smaller energy barrier to overcome than PFO when transitioning from pre-pore to pore (Lawrence et al. 2015).
The significant reduction in height from 110 to 80 Å for the protein above the membrane surface observed during pre-pore to pore transition (Czajkowsky et al. 2004; Ramachandran et al. 2005; Leung et al. 2014; Reboul et al. 2014; van Pee et al. 2016, 2017) brings the TMH helices sufficiently close to the membrane so that when they unfurl and insert, they span the width of the membrane. Initial experiments suggested that this was accomplished by the bending of D2 (Tilley et al. 2005). However, molecular dynamic studies (Reboul et al. 2014) and cryo-EM microscopy experiments on PLY (van Pee et al. 2017) and SLY (Leung et al. 2014) reveal that the decrease in height is due to the rotation of D2 by 90° towards the plane of the membrane, around a flexible glycine linker between D2 and D4. Given that D2 connects D4 with the rest of the molecule, this movement results in the placement of D1 and D3 closer to the membrane surface. It is likely that at this stage, the TMH helices are in a partially unfolded state (Sato et al. 2013). Time lapse AFM experiments also identified a transient late pre-pore phase where D2 had collapsed but the TMH regions had not yet inserted into the membrane bilayer (van Pee et al. 2016).
The final stage of pore formation is the insertion of the TMH regions into the membrane, although the molecular details of the sequence of events is not entirely clear, i.e. is the β-barrel assembled prior to membrane insertion or upon membrane insertion? A mechanism whereby the initial unfurling of a single TMH region triggers the unfurling of neighbouring TMH regions has been suggested (van Pee et al. 2017). Upon membrane insertion, the four helices that constitute the TMH1 and TMH2 regions unfurl to form four β-strands that insert into the membrane at a 20° incline from the central axis of the pore to form the β-barrel (Sato et al. 2013; Leung et al. 2014; van Pee et al. 2017). The membrane-inserted region of the pore has a hydrophobic side that faces membrane lipids while the opposite side is hydrophilic, consisting of a central negatively charged patch made up of Asp and Glu residues, flanked above and below by positively charged residues. Interestingly, a previously uncharacterised α-barrel that lines the inside of the β-barrel pore was also observed in the pore complex structure by cryo-EM (van Pee et al. 2017). This is formed by the rearrangement of the β5 strand, and an adjacent loop in D3, into a helix-turn-helix motif. Another notable feature in the pore complex was the reduced distance between neighbouring D4 domains in comparison to the linear PLY oligomers. Hydrogen bonds, surface and charge complementarity between adjacent monomers, and ionic interactions between adjacent membrane-inserted β-strands, stabilise the pore structure. The final stage of pore formation is the elimination of the lipid bilayer from within the pore complex. It has been hypothesised that the charged residues that line the inside of the membrane pore repel the hydrophobic lipids and likely displace them away from the forming pore into the rest of the membrane (van Pee et al. 2017), whereas others have suggested that the lipid is lost as a micelle or small vesicle (Bonev et al. 2001; Leung et al. 2014).
Arcs and incomplete rings
Although CDCs form complete ring-like pre-pore and pore structures (Czajkowsky et al. 2004; Tilley et al. 2005; van Pee et al. 2016, 2017), CDCs including SLO (Bhakdi et al. 1985), PLY (Sonnen et al. 2014), SLY (Leung et al. 2014) and LLO (Koster et al. 2014; Mulvihill et al. 2015; Podobnik et al. 2015; Ruan et al. 2016) have also been shown to form arcs, slits and incomplete rings. These structures are able to fuse to form larger rings or interlocked arcs and can apparently insert into membranes to form pores of varying size and morphology (Leung et al. 2014; Mulvihill et al. 2015). In some instances, arcs and incomplete rings have been associated with mutant forms of CDCs as in the case of certain PLY and PFO mutants (Hotze and Tweten 2012; van Pee et al. 2016). In other instances, arcs and incomplete ring structures in PFO and PLY did not appear to insert as efficiently into membranes as normal ring-shaped pre-pores, with only a small proportion becoming membrane spanning (Hotze and Tweten 2012; Sonnen et al. 2014).
The physiological significance of arcs and rings is a subject of much debate. The propensity for CDCs to form arcs and incomplete rings may be a consequence of experimental conditions (e.g. protein concentration, lipid environment, incubation time and temperature) or in response to particular molecular cues (Leung et al. 2014; Podobnik et al. 2015; Gilbert and Sonnen 2016; Ruan et al. 2016). It could also be a specialisation of certain CDCs such as LLO, which has been implicated in a variety of different cellular roles such as activating host-signalling pathways, histone modifications and promoting protein degradation (Osborne and Brumell 2017); these atypical roles may not require the formation of large circular pores. Further experimental data needs to be obtained before a conclusion on the physiological role of these CDC structures can be made.
Future research directions and applications
Since 1997, when the first structure of a CDC was determined (Rossjohn et al. 1997), a wealth of additional structural and biological data have been revealed regarding CDC function. However, there are details still missing from our understanding. The process of assembly from membrane-anchored monomers to oligomers and hence to pre-pores is only clear in broad terms; the molecular level detail is yet to be determined. Similarly, the trigger that converts a pre-pore assembly into a final trans-membrane pore is unknown, as is the basis by which D4 recognition of cholesterol controls the activity of CDCs that bind to membranes via CD59, independently of cholesterol. The application of techniques such as cryo-ET and cryo-EM, as well as large-scale molecular dynamics simulations, promises to address many of these questions.
Our understanding of CDC structure-function, while imperfect, is still good enough to allow some technological applications of CDCs. Exploiting the specificity of D4-membrane interaction to the presence of cholesterol allowed the development of cholesterol biosensors (Liu et al. 2014, 2017). A series of site-specific mutations in the loops of these fluorescent PFO D4-based biosensors tuned their cholesterol requirements to allow optimal binding at cholesterol levels ranging from the base 48% of wild-type PFO down to 3% (Liu et al. 2017). These biosensors can be used in live cells to follow relative cholesterol concentrations of both leaflets of cellular membranes in real time. Mutagenesis of another CDC, LLO, allowed the modification of its sensitivity to pH, producing a toxin that could be specifically activated by the introduction of a slightly acidic buffer (Kisovec et al. 2017). Further technological uses for this fascinating class of molecular machines are sure to be developed as our insight into their mechanism of action continues to mature.
Acknowledgments
M.W.P. is a National Health and Medical Research Council of Australia Research Fellow.
Funding information
This work was supported by an Australian Research Council Discovery Grant to M.W.P and by an NIH grant R37-AI037657 to R.K.T. Funding was from the Victorian Government Operational Infrastructure Support Scheme to St Vincent’s Institute.
Conflict of interest
M.P. Christie declares that she has no conflict of interest. B.A. Johnstone declares that she has no conflict of interest. R.K. Tweten declares that he has no conflict of interest. M.W. Parker declares that he has no conflict of interest. C.J. Morton declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Contributor Information
Michael W. Parker, Phone: +61 3 8344 2211, Email: mwp@unimelb.edu.au
Craig J. Morton, Phone: +61 3 8344 8751, Email: craig.morton@unimelb.edu.au
References
- Bavdek A, et al. Sterol and pH interdependence in the binding, oligomerization, and pore formation of listeriolysin O. Biochemistry. 2007;46:4425–4437. doi: 10.1021/bi602497g. [DOI] [PubMed] [Google Scholar]
- Bhakdi S, Tranum-Jensen J, Sziegoleit A. Mechanism of membrane damage by streptolysin-O. Infect Immun. 1985;47:52–60. doi: 10.1128/iai.47.1.52-60.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonev BB, Gilbert RJ, Andrew PW, Byron O, Watts A. Structural analysis of the protein/lipid complexes associated with pore formation by the bacterial toxin pneumolysin. J Biol Chem. 2001;276:5714–5719. doi: 10.1074/jbc.M005126200. [DOI] [PubMed] [Google Scholar]
- Bourdeau RW, et al. Cellular functions and X-ray structure of anthrolysin O, a cholesterol-dependent cytolysin secreted by Bacillus anthracis. J Biol Chem. 2009;284:14645–14656. doi: 10.1074/jbc.M807631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Bennett MJ, Fujii G, Curmi PM, Kantardjieff KA, Collier RJ, Eisenberg D. The crystal structure of diphtheria toxin. Nature. 1992;357:216–222. doi: 10.1038/357216a0. [DOI] [PubMed] [Google Scholar]
- Czajkowsky DM, Hotze EM, Shao Z, Tweten RK. Vertical collapse of a cytolysin prepore moves its transmembrane beta-hairpins to the membrane. EMBO J. 2004;23:3206–3215. doi: 10.1038/sj.emboj.7600350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang TX, Hotze EM, Rouiller I, Tweten RK, Wilson-Kubalek EM. Prepore to pore transition of a cholesterol-dependent cytolysin visualized by electron microscopy. J Struct Biol. 2005;150:100–108. doi: 10.1016/j.jsb.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Dowd KJ, Farrand AJ, Tweten RK. The cholesterol-dependent cytolysin signature motif: a critical element in the allosteric pathway that couples membrane binding to pore assembly. PLoS Pathog. 2012;8:e1002787. doi: 10.1371/journal.ppat.1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisdon AM, et al. Stonefish toxin defines an ancient branch of the perforin-like superfamily. Proc Natl Acad Sci U S A. 2015;112:15360–15365. doi: 10.1073/pnas.1507622112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrand S, et al. Characterization of a streptococcal cholesterol-dependent cytolysin with a Lewis y and b specific lectin domain. Biochemistry. 2008;47:7097–7107. doi: 10.1021/bi8005835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrand AJ, LaChapelle S, Hotze EM, Johnson AE, Tweten RK. Only two amino acids are essential for cytolytic toxin recognition of cholesterol at the membrane surface. Proc Natl Acad Sci U S A. 2010;107:4341–4346. doi: 10.1073/pnas.0911581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrand AJ, Hotze EM, Sato TK, Wade KR, Wimley WC, Johnson AE, Tweten RK. The cholesterol-dependent cytolysin membrane-binding interface discriminates lipid environments of cholesterol to support beta-barrel pore insertion. J Biol Chem. 2015;290:17733–17744. doi: 10.1074/jbc.M115.656769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil SC, et al. Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its Lewis antigen specificity. Structure. 2012;20:248–258. doi: 10.1016/j.str.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil SC, Ascher DB, Kuiper MJ, Tweten RK, Parker MW. Structural studies of Streptococcus pyogenes streptolysin O provide insights into the early steps of membrane penetration. J Mol Biol. 2014;426:785–792. doi: 10.1016/j.jmb.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan John J., Tweten Rodney K., Johnson Arthur E., Heuck Alejandro P. Cholesterol Exposure at the Membrane Surface Is Necessary and Sufficient to Trigger Perfringolysin O Binding. Biochemistry. 2009;48(18):3977–3987. doi: 10.1021/bi9002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelber SE, Aguilar JL, Lewis KL, Ratner AJ. Functional and phylogenetic characterization of Vaginolysin, the human-specific cytolysin from Gardnerella vaginalis. J Bacteriol. 2008;190:3896–3903. doi: 10.1128/JB.01965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings KS, Zhao J, Sims PJ, Tweten RK. Human CD59 is a receptor for the cholesterol-dependent cytolysin intermedilysin. Nat Struct Mol Biol. 2004;11:1173–1178. doi: 10.1038/nsmb862. [DOI] [PubMed] [Google Scholar]
- Gilbert RJ, Sonnen AF. Measuring kinetic drivers of pneumolysin pore structure. Eur Biophys J. 2016;45:365–376. doi: 10.1007/s00249-015-1106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert RJ, et al. Two structural transitions in membrane pore formation by pneumolysin, the pore-forming toxin of Streptococcus pneumoniae. Cell. 1999;97:647–655. doi: 10.1016/S0092-8674(00)80775-8. [DOI] [PubMed] [Google Scholar]
- Gouaux E. Channel-forming toxins: tales of transformation. Curr Opin Struct Biol. 1997;7:566–573. doi: 10.1016/S0959-440X(97)80123-6. [DOI] [PubMed] [Google Scholar]
- Hadders MA, Beringer DX, Gros P. Structure of C8alpha-MACPF reveals mechanism of membrane attack in complement immune defense. Science. 2007;317:1552–1554. doi: 10.1126/science.1147103. [DOI] [PubMed] [Google Scholar]
- Hotze EM, Tweten RK. Membrane assembly of the cholesterol-dependent cytolysin pore complex. Biochim Biophys Acta. 2012;1818:1028–1038. doi: 10.1016/j.bbamem.2011.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotze EM, Le HM, Sieber JR, Bruxvoort C, McInerney MJ, Tweten RK. Identification and characterization of the first cholesterol-dependent cytolysins from Gram-negative bacteria. Infect Immun. 2013;81:216–225. doi: 10.1128/IAI.00927-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BB, Moe PC, Wang D, Rossi K, Trigatti BL, Heuck AP. Modifications in perfringolysin O domain 4 alter the cholesterol concentration threshold required for binding. Biochemistry. 2012;51:3373–3382. doi: 10.1021/bi3003132. [DOI] [PubMed] [Google Scholar]
- Johnson S, Brooks NJ, Smith RA, Lea SM, Bubeck D. Structural basis for recognition of the pore-forming toxin intermedilysin by human complement receptor CD59. Cell Rep. 2013;3:1369–1377. doi: 10.1016/j.celrep.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisovec M, et al. Engineering a pH responsive pore forming protein. Sci Rep. 2017;7:42231. doi: 10.1038/srep42231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster S, et al. Crystal structure of listeriolysin O reveals molecular details of oligomerization and pore formation. Nat Commun. 2014;5:3690. doi: 10.1038/ncomms4690. [DOI] [PubMed] [Google Scholar]
- Kozorog M, et al. (19) F NMR studies provide insights into lipid membrane interactions of listeriolysin O, a pore forming toxin from Listeria monocytogenes. Sci Rep. 2018;8:6894. doi: 10.1038/s41598-018-24692-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaChapelle S, Tweten RK, Hotze EM. Intermedilysin-receptor interactions during assembly of the pore complex: assembly intermediates increase host cell susceptibility to complement-mediated lysis. J Biol Chem. 2009;284:12719–12726. doi: 10.1074/jbc.M900772200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence SL, et al. Crystal structure of Streptococcus pneumoniae pneumolysin provides key insights into early steps of pore formation. Sci Rep. 2015;5:14352. doi: 10.1038/srep14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence SL, et al. Structural basis for receptor recognition by the human CD59-responsive cholesterol-dependent cytolysins. Structure. 2016;24:1488–1498. doi: 10.1016/j.str.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C, et al. Stepwise visualization of membrane pore formation by suilysin, a bacterial cholesterol-dependent cytolysin. Elife. 2014;3:e04247. doi: 10.7554/eLife.04247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C, et al. Real-time visualization of perforin nanopore assembly. Nat Nanotechnol. 2017;12:467–473. doi: 10.1038/nnano.2016.303. [DOI] [PubMed] [Google Scholar]
- Li JD, Carroll J, Ellar DJ. Crystal structure of insecticidal delta-endotoxin from Bacillus thuringiensis at 2.5 A resolution. Nature. 1991;353:815–821. doi: 10.1038/353815a0. [DOI] [PubMed] [Google Scholar]
- Liu SL, et al. Simultaneous in situ quantification of two cellular lipid pools using orthogonal fluorescent sensors. Angew Chem Int Ed Engl. 2014;53:14387–14391. doi: 10.1002/anie.201408153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, et al. Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat Chem Biol. 2017;13:268–274. doi: 10.1038/nchembio.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JE, et al. The crystal structure of Pneumolysin at 2.0 Å resolution reveals the molecular packing of the pre-pore complex. Sci Rep. 2015;5:13293. doi: 10.1038/srep13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller M, Grauschopf U, Maier T, Glockshuber R, Ban N. The structure of a cytolytic alpha-helical toxin pore reveals its assembly mechanism. Nature. 2009;459:726–730. doi: 10.1038/nature08026. [DOI] [PubMed] [Google Scholar]
- Mulvihill E, van Pee K, Mari SA, Muller DJ, Yildiz O. Directly observing the lipid-dependent self-assembly and pore-forming mechanism of the cytolytic toxin listeriolysin O. Nano Lett. 2015;15:6965–6973. doi: 10.1021/acs.nanolett.5b02963. [DOI] [PubMed] [Google Scholar]
- Nelson LD, Johnson AE, London E. How interaction of perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH: insights into the origin of perfringolysin O-lipid raft interaction. J Biol Chem. 2008;283:4632–4642. doi: 10.1074/jbc.M709483200. [DOI] [PubMed] [Google Scholar]
- Ohno-Iwashita Y, Iwamoto M, Ando S, Iwashita S. Effect of lipidic factors on membrane cholesterol topology—mode of binding of theta-toxin to cholesterol in liposomes. Biochim Biophys Acta. 1992;1109:81–90. doi: 10.1016/0005-2736(92)90190-W. [DOI] [PubMed] [Google Scholar]
- Osborne Suzanne E., Brumell John H. Listeriolysin O: from bazooka to Swiss army knife. Philosophical Transactions of the Royal Society B: Biological Sciences. 2017;372(1726):20160222. doi: 10.1098/rstb.2016.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SA, Park YS, Bong SM, Lee KS. Structure-based functional studies for the cellular recognition and cytolytic mechanism of pneumolysin from Streptococcus pneumoniae. J Struct Biol. 2016;193:132–140. doi: 10.1016/j.jsb.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Parker MW, Pattus F, Tucker AD, Tsernoglou D. Structure of the membrane-pore-forming fragment of colicin A. Nature. 1989;337:93–96. doi: 10.1038/337093a0. [DOI] [PubMed] [Google Scholar]
- Parker MW, Tucker AD, Tsernoglou D, Pattus F. Insights into membrane insertion based on studies of colicins. Trends Biochem Sci. 1990;15:126–129. doi: 10.1016/0968-0004(90)90205-P. [DOI] [PubMed] [Google Scholar]
- Parker MW, Buckley JT, Postma JP, Tucker AD, Leonard K, Pattus F, Tsernoglou D. Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states. Nature. 1994;367:292–295. doi: 10.1038/367292a0. [DOI] [PubMed] [Google Scholar]
- Petosa C, Collier RJ, Klimpel KR, Leppla SH, Liddington RC. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- Podobnik M, et al. Plasticity of listeriolysin O pores and its regulation by pH and unique histidine [corrected] Sci Rep. 2015;5:9623. doi: 10.1038/srep09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polekhina G, Giddings KS, Tweten RK, Parker MW. Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin. Proc Natl Acad Sci U S A. 2005;102:600–605. doi: 10.1073/pnas.0403229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Tweten RK, Johnson AE. Membrane-dependent conformational changes initiate cholesterol-dependent cytolysin oligomerization and intersubunit beta-strand alignment. Nat Struct Mol Biol. 2004;11:697–705. doi: 10.1038/nsmb793. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Tweten RK, Johnson AE. The domains of a cholesterol-dependent cytolysin undergo a major FRET-detected rearrangement during pore formation. Proc Natl Acad Sci U S A. 2005;102:7139–7144. doi: 10.1073/pnas.0500556102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul CF, Whisstock JC, Dunstone MA. A new model for pore formation by cholesterol-dependent cytolysins. PLoS Comput Biol. 2014;10:e1003791. doi: 10.1371/journal.pcbi.1003791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul CF, Whisstock JC, Dunstone MA. Giant MACPF/CDC pore forming toxins: a class of their own. Biochim Biophys Acta. 2016;1858:475–486. doi: 10.1016/j.bbamem.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Rosado CJ, et al. A common fold mediates vertebrate defense and bacterial attack. Science. 2007;317:1548–1551. doi: 10.1126/science.1144706. [DOI] [PubMed] [Google Scholar]
- Rossjohn J, Feil SC, McKinstry WJ, Tweten RK, Parker MW. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell. 1997;89:685–692. doi: 10.1016/S0092-8674(00)80251-2. [DOI] [PubMed] [Google Scholar]
- Rossjohn J, Polekhina G, Feil SC, Morton CJ, Tweten RK, Parker MW. Structures of perfringolysin O suggest a pathway for activation of cholesterol-dependent cytolysins. J Mol Biol. 2007;367:1227–1236. doi: 10.1016/j.jmb.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Rezelj S, Bedina Zavec A, Anderluh G, Scheuring S. Listeriolysin O membrane damaging activity involves arc formation and lineaction—implication for listeria monocytogenes escape from phagocytic vacuole. PLoS Pathog. 2016;12:e1005597. doi: 10.1371/journal.ppat.1005597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Xia S, Liu X, Lieberman J, Wu H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature. 2018;557:62–67. doi: 10.1038/s41586-018-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Tweten RK, Johnson AE. Disulfide-bond scanning reveals assembly state and beta-strand tilt angle of the PFO beta-barrel. Nat Chem Biol. 2013;9:383–389. doi: 10.1038/nchembio.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savinov Sergey, Heuck Alejandro. Interaction of Cholesterol with Perfringolysin O: What Have We Learned from Functional Analysis? Toxins. 2017;9(12):381. doi: 10.3390/toxins9120381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger LLC (2015) The PyMOL Molecular Graphics System, Version 1.8
- Shatursky O, Heuck AP, Shepard LA, Rossjohn J, Parker MW, Johnson AE, Tweten RK. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: a novel paradigm for pore-forming toxins. Cell. 1999;99:293–299. doi: 10.1016/S0092-8674(00)81660-8. [DOI] [PubMed] [Google Scholar]
- Shepard LA, et al. Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: an alpha-helical to beta-sheet transition identified by fluorescence spectroscopy. Biochemistry. 1998;37:14563–14574. doi: 10.1021/bi981452f. [DOI] [PubMed] [Google Scholar]
- Shewell LK, et al. The cholesterol-dependent cytolysins pneumolysin and streptolysin O require binding to red blood cell glycans for hemolytic activity. Proc Natl Acad Sci U S A. 2014;111:E5312–E5320. doi: 10.1073/pnas.1412703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade DJ, Lovelace LL, Chruszcz M, Minor W, Lebioda L, Sodetz JM. Crystal structure of the MACPF domain of human complement protein C8 alpha in complex with the C8 gamma subunit. J Mol Biol. 2008;379:331–342. doi: 10.1016/j.jmb.2008.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltani CE, Hotze EM, Johnson AE, Tweten RK. Specific protein-membrane contacts are required for prepore and pore assembly by a cholesterol-dependent cytolysin. J Biol Chem. 2007;282:15709–15716. doi: 10.1074/jbc.M701173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltani CE, Hotze EM, Johnson AE, Tweten RK. Structural elements of the cholesterol-dependent cytolysins that are responsible for their cholesterol-sensitive membrane interactions. Proc Natl Acad Sci U S A. 2007;104:20226–20231. doi: 10.1073/pnas.0708104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- Sonnen AF, Plitzko JM, Gilbert RJ. Incomplete pneumolysin oligomers form membrane pores. Open Biol. 2014;4:140044. doi: 10.1098/rsob.140044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley SJ, Orlova EV, Gilbert RJ, Andrew PW, Saibil HR. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell. 2005;121:247–256. doi: 10.1016/j.cell.2005.02.033. [DOI] [PubMed] [Google Scholar]
- Tommassen J, Arenas J. Biological functions of the secretome of Neisseria meningitidis. Front Cell Infect Microbiol. 2017;7:256. doi: 10.3389/fcimb.2017.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pee K, Mulvihill E, Muller DJ, Yildiz O. Unraveling the pore-forming steps of pneumolysin from Streptococcus pneumoniae. Nano Lett. 2016;16:7915–7924. doi: 10.1021/acs.nanolett.6b04219. [DOI] [PubMed] [Google Scholar]
- van Pee K, Neuhaus A, D'Imprima E, Mills DJ, Kuhlbrandt W, Yildiz O (2017) CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin. Elife:6. 10.7554/eLife.23644 [DOI] [PMC free article] [PubMed]
- Wade KR, Hotze EM, Kuiper MJ, Morton CJ, Parker MW, Tweten RK. An intermolecular electrostatic interaction controls the prepore-to-pore transition in a cholesterol-dependent cytolysin. Proc Natl Acad Sci U S A. 2015;112:2204–2209. doi: 10.1073/pnas.1423754112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham SE, et al. Mapping the intermedilysin-human CD59 receptor interface reveals a deep correspondence with the binding site on CD59 for complement binding proteins C8alpha and C9. J Biol Chem. 2011;286:20952–20962. doi: 10.1074/jbc.M111.237446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Huang B, Du H, Zhang XC, Xu J, Li X, Rao Z. Crystal structure of cytotoxin protein suilysin from Streptococcus suis. Protein Cell. 2010;1:96–105. doi: 10.1007/s13238-010-0012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilnyte M, Venclovas C, Zvirbliene A, Pleckaityte M. The cytolytic activity of vaginolysin strictly depends on cholesterol and is potentiated by human CD59. Toxins (Basel) 2015;7:110–128. doi: 10.3390/toxins7010110. [DOI] [PMC free article] [PubMed] [Google Scholar]