Abstract
Various human activities lead to the pollution of ground, drinking, and wastewater with toxic metals. It is well known that metal ions preferentially bind to DNA phosphate backbones or DNA nucleobases, or both. Foreman et al. (Environ Toxicol Chem 30(8):1810–1818, 2011) reported the use of a DNA-dye based assay suitable for use as a toxicity test for potable environmental water. They compared the results of this test with the responses of live-organism bioassays. The DNA-based demonstrated that the loss of SYBR Green I fluorescence dye bound to calf thymus DNA was proportional to the toxicity of the water sample. However, this report raised questions about the mechanism that formed the basis of this quasi-quantitatively test. In this review, we identify the unique and preferred DNA-binding sites of individual metals. We show how highly sensitive and selective DNA-based sensors can be designed that contain multiple binding sites for 21 heavy metal cations that bind to DNA and change its structure, consistent with the release of the DNA-bound dye.
Keywords: Heavy metals, Binding, Water, Toxicity, DNA
Introduction
Global water pollution by metals originates from natural as well as human sources such as treated and untreated domestic sewerage, mining and land development, industrial and solid waste disposal and agricultural runoff (Destouni and Jarsjö 2018). In living organisms including humans, metals and other chemical toxicants bioaccumulate as a result of consuming contaminated drinking water and food, particularly fish and molluscs, with toxicant concentrations increasing as they pass along the food chain (Gruber 1989).
Heavy metals constitute a group of about 40 elements with a density greater than five (Passow et al. 1961). “Toxicity” describes the degree of ill-health effects in populations of organisms (Tchounwou et al. 2012) and the resulting responses across a broad spectrum of organisms resulting from a complex interplay of biochemical, physical, individual and population/community processes in response to an introduced toxicant. Metal ions must be bioavailable (i.e. able to move across biological membranes) before they can initiate an adverse effect (Batley et al. 2004), and low pH typically increases metal bioavailability (Hyne et al. 2005). It has been hypothesised that metal ion-induced toxicity inactivates vital processes, inhibits metabolic pathways (Corner and Sparrow 1956) and directly or indirectly displaces essential metals from the active sites of macromolecules and/or disrupts depolymerisation or repair of nucleotide bases with subsequent errors in protein synthesis (Tchounwou et al. 2012).
In this review, we describe the structure and biology of DNA/RNA, followed by its interaction with heavy metals causing toxicity. We then describe the effects of 25 of common heavy metals and how DNA can be used to screen for these metals in the environment.
Structure of DNA and RNA
The following brief discussion of DNA structure is highly relevant to its interaction between heavy metal ions, so we present it for the benefit of readers who are not familiar with this area. DNA is a complex polynucleotide comprising three main constituents:
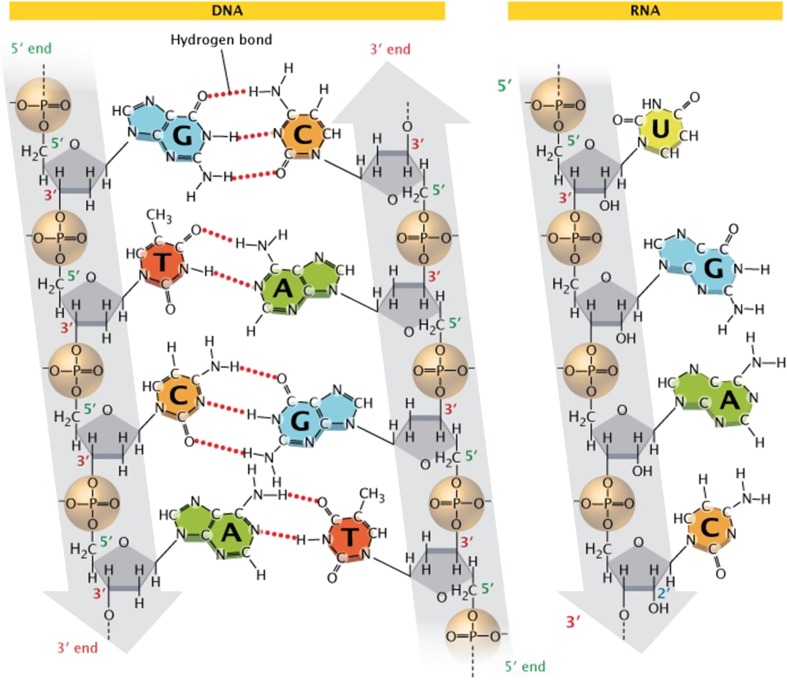
A cyclic sugar (deoxyribose in DNA or ribose in RNA whereby the H- group on the 2′ carbon in DNA is replaced by an OH– group in RNA)
A purine or pyrimidine base attached to the 1′ carbon of the sugar group via an N-glycosydic bond. The purine bases are adenine (A) and guanine (G) and the pyrimidine bases are thymine (T) and cytosine (C)
A phosphate moiety, responsible for the strong negative charge of nucleotides, attached to the 5′ carbon of the sugar by a phosphoester bond (Watson and Crick 1953). RNA contains these same bases with the exception that uracil (U) replaces T (see Fig. 1). The linear sequence of nucleotides is referred to as the primary structure of DNA or RNA.
Fig. 1.
The chemical structure of DNA (left) and RNA (right). In double-stranded DNA, complementary base pairs bind via hydrogen bonds. Sugar phosphate backbones (grey) are aligned 3′ to 5′ in an anti-parallel fashion as shown. These sugars are deoxyribose and ribose in DNA and RNA, respectively. Furthermore, thymine is replaced by uracil in RNA, but both are capable of forming base pairs to adenine. Although RNA may form transient secondary structures that are double stranded, it is typically single-stranded. Copyright (2018) Nature Education
Double-stranded DNA (dsDNA) is formed when A pairs with T (via two hydrogen bonds) and C pairs with G (via three hydrogen bonds). The stereochemistry of the bases linked by hydrogen bonds results in a dsDNA helix with the bases buried in the hydrophobic centre and the sugar-phosphate backbone on the outside. The molar concentration of A = T and G = C, from which also it is can be deduced that A + G = T + C (pyrimidines = purines). This same principle is true in RNA except T is replaced by U so that A = U. This interaction between BPs is referred to as the secondary structure of the DNA or RNA.
Segments of dsDNA are capable of interacting with other segments to form helix structures and minor and major grooves. These structures are referred to as the tertiary structure of DNA. There are three biologically active forms of dsDNA helices, namely α-, β- and Z- helices. The most common, β-DNA, forms a right-handed helix, at neutral pH and physiological salt concentrations, with sugar base pairs (BPs) oriented at right angles to the helix. α-DNA is also a right-handed helix, but in contrast to β-DNA, α-DNA is wider and shorter, its BPs are not arranged at right angles to the helix, its structure is more compact with more BPs per turn, and there are fewer structural similarities between its major and minor grooves. α-DNA typically is formed during conditions of DNA dehydration, such as that used to promote crystallisation of dsDNA. The Z-helix (so named because of its zigzag-shaped backbone) is distinct from both α- or β-DNA in structure on account of its left-handed helix. Double-strand RNA (dsRNA) has 11 base pairs (BPs) per turn (compared to 10 in β-DNA) with its bases tilted 30° with respect to the helical axis (Lescrinier et al. 2003).
The twisting of dsDNA when a helix is formed creates minor (smaller) and major (larger) grooves. The size and exposed regions of DNA within these grooves dictate the type and strength of bonds with free molecules that are present in the surrounding solution. Indeed, many proteins and drugs target a specific type of minor or major groove (Savreux-Lenglet et al. 2015). Furthermore, changes in the minor groove width are likely critical in allowing changes in DNA structure that occur when DNA binds to monovalent cations (Savelyev and MacKerell 2015).
Heavy metal ion toxicity
Metal cations bound to DNA (at electron-dense rich sites) are classified as exclusively phosphate-binding (alkali and alkaline earth metals such as K(I), Ca(II), Mg(II) Ba(II)), exclusively base-binding (Ag(I) and Hg(II)), or those that bind to both components of DNA with affinities in the order Co(II)~Ni(II) < Mn(II)~Zn(II) < Cd(II) < Pb(II) < Cu(II) (Duguid et al. 1993; Izatt et al. 1971; Shin and Eichhorn 1968; Sissoeff et al. 1976). The exclusive binding of metal cations to DNA bases is based on the formation of energetically favourable linear coordination complexes. In contrast, metal cations that bind simultaneously to the base and phosphate moieties of DNA are governed by the nucleophilicity of the binding site and the coordination geometry of the metal cations (Moldrheim et al. 1998). Fluctuations in nucleophilicity of the most reactive DNA binding site, G-N7, is thought to be a consequence of variation in π-stacking interactions between base residues along the dsDNA (Šponer et al. 2008).
Whilst the DNA conformation determines the number of binding sites (Sissoeff et al. 1976), binding of metal cations deforms the DNA structure via mechanisms that include degradation, chain lengthening (Sissoeff et al. 1976) and helix-to-coil transitions. Metals with strong base affinities disrupt hydrogen bonding between BPs to destabilise β-DNA structures and cause alternative conformations (Izatt et al. 1971). Metals with strong phosphate affinities stabilise β-DNA via charge neutralisation of the sugar-phosphate backbone (Duguid et al. 1993). Alkaline earth metals bind BPs with lower affinity than transition metals, possibly due to altered electronic distributions within the DNA (Duguid et al. 1993). After a critical fraction of the DNA phosphate charge has been neutralised by the adsorption of cations, competition occurs between cations that induce DNA condensation (3+ or 4+) and reversing (1+ or 2+), as predicted by Manning’s (1978) theory of atmospheric cation binding to DNA (Pack 1982). The intrinsically electrostatic forces between DNA chains are adsorbed by metal cations above a critical concentration leading to a condensed phase of DNA (Koltover et al. 2000).
Mercury
DNA-Hg(II) binding to DNA bases occurs via preferred Hg(II)-(thymidine)2 complexes that cross-link the dsDNA by binding to alternating d(AT)n polymers (Duguid et al. 1993; Izatt et al. 1971; Katz 1963). Hg is capable of disrupting hydrogen bonds by forming N3-Hg(II)-N3 bonds with structural changes in the DNA that depend on the density of Hg(II) within the DNA (Moldrheim et al. 1998; Sissoeff et al. 1976; Yamane and Davidson 1962). Hg(II) also cross-links base pairs (BPs) in DNA (irrespective of its base composition) via deprotonation of A and C amino groups and T-N3 and G-N1 groups (Moldrheim et al. 1998; Nandi et al. 1965).
Hg(II) binds BPs with the following preference: T > G > > A, C (Gruenwedel and Cruikshank 1990). This allows Hg(II) to significantly stabilise naturally occurring T-T miss-pairing in dsDNA (Tanaka et al. 2007). The ability of the T-Hg(II)-T pair to stabilise hairpin dsDNA flanked by self-complementary sequences results in a more stable and preferred Hg(II)-complex than A-Hg(II)–T cross-linked pairs (Kuklenyik and Marzilli 1996; Miyake et al. 2006).
Hg-BP binding has been reported to occur at N1, N3, N7, N9, C8, and exocyclic NH2 (Onyido et al. 2004). Hg(II) weakens BP hydrogen bonds and replaces them by chelation into σ electron pairs of nitrogen atoms in a linear = N-Hg(II)-N = configuration (Matsuda and Takeuchi 1967; Yamane and Davidson 1962). Although Hg(II) selectively binds to A-T BPs (with a strong 5′-T-pyrimidine bias), Hg-DNA complex formation is driven by energetically favourable linear coordination complexes rather than nucleophicity (Moldrheim et al. 1998). Whilst the secondary structure of DNA remains intact (Onyido et al. 2004), Hg(II) changes β-DNA through C-DNA to Z-DNA (Gruenwedel and Cruikshank 1990).
From the foregoing, it is clear that for DNA-based detection of Hg in water, the DNA secondary structure should be preserved following the removal of Hg(II) from the DNA molecule. Such restoration of conformation has been reported for calf thymus DNA changing from β-DNA, to C-DNA, and then to Z-DNA conformations (Gruenwedel and Cruikshank 1990). Significantly, methyl mercury [CH3Hg(II)] irreversibly converts dsDNA into ssDNA. Therefore, DNA-based sensors that quantify total Hg content of a sample must be able to distinguish between the varying states of Hg on the sensor-signalling mechanism.
Chromium
Environmental Cr exists mostly as Cr(IV) and Cr(III) (Cohen et al. 1993; Zhitkovich et al. 1996). Cr(III) preferentially binds to the exterior of DNA via phosphate moieties to form an octahedral geometry that is impermeable to cell membranes and is non-toxic (Izatt et al. 1971). In contrast, Cr(IV) forms the tetrahedral di-anion, chromate capable of entering cells (Tsapakos and Wetterhahn 1983), where it is reduced into unstable and reactive Cr(V) or Cr(IV) that eventually form Cr(III) (Haight Jr et al. 1971; Tsapakos and Wetterhahn 1983). The oxidative stress involved in such potent intracellular reduction may account for the toxicity and carcinogenesis of Cr(VI) (Kortenkamp et al. 1996a, b; Zhitkovich et al. 1996) through DNA interactions such as strand ligation (Sugden 1999).
Cr(IV) preferentially binds to G-containing BPs and does so via phosphate moieties or the N-7 group of purine BPs in the major groove of B-DNA. Cr(IV) also induces cross-linking between DNA and proteins and strongly prefers ssDNA over dsDNA (Tsapakos and Wetterhahn 1983). This behaviour implies Cr(IV) DNA sensors must avoid self-complementary sequences which may fold to form dsDNA regions. Importantly, the interaction between Cr(VI) and DNA oligos is thermodynamically and kinetically unfavourable between pH 7.0 and pH 7.4, because both species are negatively charged (Tsapakos and Wetterhahn 1983). Thus, without a natural microsomal/NADH or synthetic reducing system, a DNA-based sensor would likely lose binding sensitivity within this pH range.
Cobalt
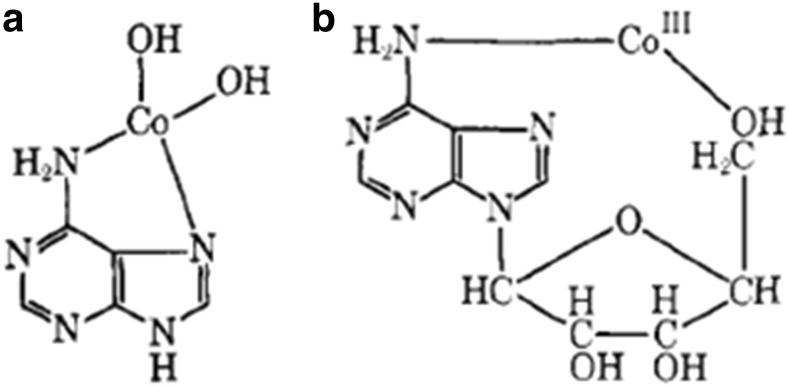
The four main species of cobalt that interact with DNA are metallic cobalt (Co), tungsten carbide in a Co matrix (WC–Co), Co(II) (as CoCl2) (Izatt et al. 1971; Lison and Lauwerys 1994), and Co(III). WC–Co and Co predominantly damage DNA via reactive oxygen species. Co(II)-adenine complexes involve the C6”2 and N7 groups with two additional OH groups bound to Co(II). As shown in Fig. 2, Co(II) exclusively and directly coordinates to N-7 moieties of guanine in an octahedral geometry, and also incomplete hydration shells (Gao et al. 1993; Izatt et al. 1971). Such interactions may induce significant conformational changes in either β-DNA or α-DNA. This highlights the importance of having DNA probes in sensors with initial conformation that are carefully designed and controlled.
Fig. 2.
Co-adenine complexes for (a) Co(II) and (b) Co(III). Copyright (2018) American Chemical Society
Whilst crystallographic studies examine DNA in a different state compared to DNA in solution, some of the observed binding behaviour might be extrapolated into DNA-metal interactions that occur in water. Such behaviour might be used as the logical starting point for the design of selective DNA probes. One notable study examined Mg forms of ‘native’ CGCGCG and CGCGTG Z-DNA oligocrystals (Gao et al. 1993). The stronger DNA affinity of Co(II) to G-N7 groups in the CGCGTG oligo causes Co(II) to displace Mg(II) and Na(I) ions (that were initially bound to the oligo). During this displacement interaction, two Co(II) ions bind to separate G bases (G4 and G8), while a third Co(II) ion binds simultaneously to two other G bases (G10 and G12). In the CGCGCG oligocrystal, three Co(II) ions coordinate with G4, G6 and G10 bases, with some incomplete hydration shells. The hydration shell of Co(II) invokes conformational changes in both α- and β-DNA because of van der Walls clashes and steric interference at the Co(II) binding pocket). In α-DNA, the preferred G-N7 groups are inaccessible to Co(II) (and other small metal ions such as Ni(II)) due to the N7 group being buried very deeply in the narrow major groove of DNA. Coordination of these ions (surrounded by their hydration shells) would create unfavourable contacts and significantly destabilise the DNA helix (Jia and Marzilli 1991), whilst larger metal cation such as Ba(II) or Pb(II) may bind to G-N7 or G-06 groups without significantly affecting the DNA structure (Gao et al. 1995).
Compared to Co(II), Cu(II) assumes a trigonal planar coordination geometry with three water molecules whilst simultaneously binding to G12-N7 and G10-N7 groups in the CGCGTC oligo. The third hydrated coordination site of Cu(II) links the G12–O6 and G10–O6 groups (similar to Ba(II)), which suggests Cu(II) is capable of pulling G away from the helix axis in the CGCGTC crystal lattice without destabilising the crystal. Such clear differences in DNA-binding behaviours provide a basis for the logical construction of highly selective DNA sensor probes that are able to differentiate the atomically similar Co and Cu ions.
Copper
G-N7 is typically the most reactive base interaction site for cations that bind both phosphate and base DNA sites (Izatt et al. 1971; Shin and Eichhorn 1968). Amongst metals that binds both nucleotide bases and phosphates, Cu(II) has the strongest affinity for bases (Duguid et al. 1993; Shin and Eichhorn 1968). This affinity can destabilise DNA regions that are rich in GC more so than AT-rich regions (Zimmer et al. 1971). The high affinity can also cause dsDNA helix unwinding in the presence of other salts (Izatt et al. 1971). Cu(II) binds to bases in the order G > A > C > T (Fiskin and Beer 2002). Cu(II) has also been seen to stabilise the single helical form depending on the relative amount of electrolytes and non-complementary C-A base pairs (Sponer et al. 1999). Therefore, minimising the accessibility of G-N7 groups and maximising (GC)n-rich regions in the DNA probe will likely optimise the specificity for Cu(II) in DNA-based sensors.
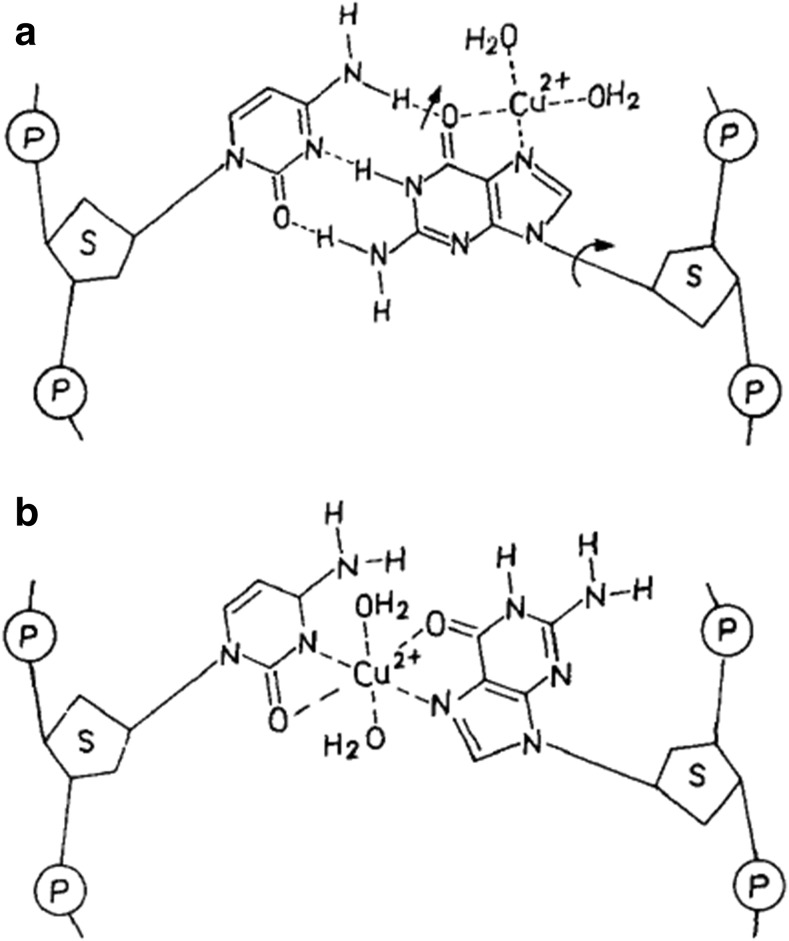
Two main models describe Cu(II)-dsDNA complex structures. The first (Fig. 3) suggests Cu(II) intercalates between two G-C BPs and two adjacent guanine residues in the same strand (Fig. 3a) with subsequent breaking of hydrogen bonds, loosening of base-base interactions and tilting of the plane of the bases (Fig. 3b) (Zimmer et al. 1971). The second model suggests Cu(II) binds between G-N7 and the closest phosphate group of the same strand (Duguid et al. 1993; Richard et al. 1973) and that β-DNA is not structurally distorted after Cu(II) is bound to two adjacent G residues due to interactions between ligands of the adjacent complexes (Geierstanger et al. 1991). It is therefore likely that introducing dsDNA GC produces a mismatch in DNA-based sensor leading to a decrease in the stabilisation of Cu(II) binding. Alternatively, the DNA may be deliberately destabilised via pH, temperature or salt changes in solutions suspected of containing Cu(II). DNA-based sensors in this case may be interpreted as an increased DNA stability, i.e. a ‘positive reading’ for the presence of Cu(II).
Fig. 3.
Schematic representation of a complex binding model of Cu(II) to a GC BP in DNA. a Most probable first attachment site. b Complexing of Cu(II) between guanine and cytosine moieties. Copyright (2018) American Chemical Society
Cu(II) assumes a trigonal planar coordination geometry with three water molecules when simultaneously bound to the N-7 sites of both G-10 and G-12 G bases of a Z-DNA CGCGTC oligonucleotide (Gao et al. 1993). The third hydrated coordination site of Cu(II) bridged the O-6 atoms of both G bases (similarly to Ba[II]), which suggests Cu(II) is capable of pulling G away from the helix axis in the CGCGTC crystal lattice without destabilising the crystal.
Aluminium
Al typically exists as either Al(III) or insoluble Al (Macdonald and Martin 1988). Al(III) irreversibly unwinds DNA (Rao and Divakar 1993), by interacting more strongly with phosphate groups stronger than most metal ions in acidic solutions and forms strong bis-complexes that promote BP stacking (Kiss 1995; Kiss et al. 1991). In addition to phosphate groups, Al(III) also interacts with the alcoholic hydroxyl group(s) of ribose or deoxyribose, or to carbonyl O and/or N ring donors in the nucleotide bases (Harris et al. 1996; Kiss 1995).
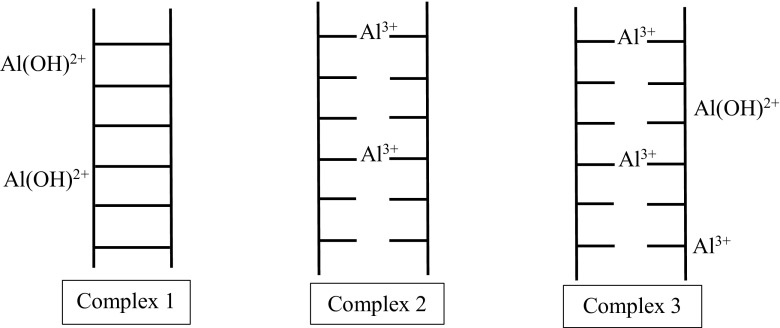
In aqueous solutions, Al(III) or [A1(H2O)6]3+ exists in a complex equilibrium between various hydroxylated species that are highly dependent on the concentration of Al and the pH of the solution. For example, when Al hydrolyses at ~ 10 μM into the following five water coordinated water molecules: Al(OH)2+, A1(OH)2+, Al(OH)3 and Al(OH)4− and a single isopolycation A11304(OH)247+ (Karlik et al. 1980). Three of these hydroxylated states are shown in Fig. 4 (Karlik et al. 1980). Clearly, Al ions that constitute complex 1 are more hydroxylated than those seen in complex 2. Not only does the stability constant for the formation of Al-phosphate exceed most metal ions at pH ≈ 4 (including Cu[III]), but Al(III) also forms strong bis-complexes (Kiss et al. 1991). At low millimolar concentrations, bis-complex formation neutralises the repulsive charges in minimally stacked BPs in DNA. Subsequently, this increases the density of BP stacking to promote bis-complex formation (Kiss 1995).
Fig. 4.
Three pH-dependant Al complexes with DNA (Karlik et al. 1980). High pH and the formation of A1(OH)2+ is required for the formation of complex 1, which stabilises the dsDNA. In contrast, low pH values (~pH 3.5–5.5) promote Al(III) formation and binding to BPs (complex 2) which irreversibly destabilises, cross-links, and unwinds DNA in a manner similar to Hg(II) (Karlik et al. 1980; Macdonald and Martin 1988; Wu et al. 2005)
Complex 3 is a hybrid of complexes 1 and 2 with both Al(III) and Al(OH)2+ simultaneously binding to the DNA. Although complex 3 occurs at all pH values, it is most frequently seen at intermediate pH values and is characterised by BP cross-linking at pH < 6 (Karlik et al. 1980). Between pH 6.0 to 7.0, Al normally exists as an insoluble hydroxide complex that completely precipitates DNA without binding to the DNA molecule (Wu et al. 2005). The neutral and negative species Al(OH)3 and Al(OH)4− have no electrostatic attraction for DNA phosphate groups, while the large and highly charged A11304(OH)247+ ion destabilises, twists and/or folds dsDNA via electrostatic and steric interactions. Therefore, Al(OH)3, Al(OH)4− and A11304(OH)247+ probably do not contribute to the formation of complex 1. Al(OH)2+ does not form complex 1 according to measured Tm values of complex 1 (Yamane and Davidson 1962). It is likely that an Al DNA-based sensor will require the selection of at least three different DNA probes to compensate for these three modes of DNA binding. Additionally, the field sample may need to have their pH adjusted from neutral pH values to avoid Al-induced DNA precipitation.
Scandium, yttrium and selected lanthanides
La(III) and selected lanthanides (Ce(III), Tb(III) and Dy(III)) bind to DNA oligo nucleotides preferably at GC-rich and at phosphate oxygen groups (Rossetto and Nieboer 1994). Such binding resulted in biphasic transitions of the conformation of the DNA with strengths that are weaker than those observed for Ag(I), Hg(II), Pd(II), Pt(II) and Ru(III), Cd(II), Cu(II), Co(II), Mn(II), Ni(II), Pb(II), Zn(II), Fe(III) or In(III) complexes. Other lanthanides (Pr, Eu, Gd, Tb, Lu and Sm) bind to DNA and yeast phenylalanine transfer ribonucleic acid (YP-tRNA) phosphate oxygen groups, rather than forming direct coordination complexes to bases like the other lanthanides (Kim et al. 1985). Dimeric forms of Y, and Nd, participate in active dsDNA-catalysed DNA degradation, whilst monomeric complexes displayed no activity (Bligh et al. 2001).
La has similar chemical properties to the alkaline earth elements with (Das et al. 1988). La(II) precipitates DNA by binding to phosphate groups of adjacent DNA chains (Das et al. 1988; Stern and Steinberg 1953). It possesses multiple DNA-binding sites, interacts with DNA in a dose-dependent manner (Das et al. 1988) and binds DNA in a 1:9 ratio (similar to Nd) (Kerner and Anderson 1955; Rosoff and Spencer 1979).
The actinide, Sc, almost exclusively exists as Sc(III) but can form species with various valences (Corbett 1981; Dudis et al. 1986; Takahashi et al. 1995). Due to its smaller ionic size to the lanthanide series, Sc ions display lower coordination numbers than larger lanthanides. The ionic radii of six-coordinate Sc(III), La(III) and Lu(III) are 0.745 A, 1.032 Å and 0.861 Å respectively (Mioduski 1993).
Interestingly, some rare earth cation-DNA complexes have characteristic colours. For example, Nd-DNA solutions are pale pink, Pr-DNA is pale green, while Y, Sm and La DNA complexes were reported to be colourless (Stern and Steinberg 1953). Additionally, strong absorption bands in the visible spectrum for Nd (510–512. 520–525 and 570–590 nm) and Pr (440–450, 465–470 and 475–480 nm) were observed, whilst the addition of both Nd and Pr to the DNA produced different band patterns (440–450, 465–470, 520–525 and 570–575 nm). When dried, the DNA preparations were white and appeared similar to asbestos fibres. This suggests that spectral DNA-based sensors are not possible unless algorithms can distinguish between the many overlapping spectral bands observed in mixtures of the rare earth lanthanides. The influence of other metals and non-metals would also need to be anticipated and evaluated before they are incorporated into a DNA-based sensor.
Platinum
Importantly, Pt(IV) exclusively binds to YP-tRNA where there are two adjacent “C-A” BPs to form three possible types of complexes YP-tRNA (Kim et al. 1985). In the first binding mode of Pt(IV) to YP-tRNA, Pt(IV) binds to N-4 of cytosine and N-7 of adenine with hydroxylation of the remaining coordination sites. The second mode of Pt(IV)-YP-tRNA binding involves binding directly to N-4 of cytosine and N-7 of adenine. A third coordination site of Pt(IV) between the terminal O-3′ (or O-2′) of adenine (YP-tRNA residues 76) and the adenine of YP-tRNA residue 36 of an adjacent molecule.
Despite the obvious different sugar moieties (ribose in RNA and deoxyribose in DNA), the above evidence warrants a thorough study of Pt-DNA interaction, which focus on the AC motifs, and A-A mismatches. Emphasis should focus on the initial conformation of DNA probes due to the effect of β- and Z-DNA conformation on cis-Pt(II) coordination (Rossetto and Nieboer 1994). Pt binding to β-DNA induces intra-strand cross-linkage between two G residues separated by a C BP, to convert the DNA into a distorted Z-helix. By comparison, the same cis-Pt(II) ion induces a mono-dentate Z-type adduct if the DNA was originally in the Z-helix conformation. This behaviour is especially problematic to DNA-based sensors if non-target molecules in test samples affect the DNA conformation of the original DNA. To further complicate matters, the stoichiometry and reactivity of Pt to DNA is significantly affected by the number of reactive ligands within its coordination sphere (Macquet and Theophanides 1975). Interestingly, intra- or intermolecular condensation increase Pt(II) binding to DNA (Macquet and Theophanides 1975). It has also been suggested that Pt(II), and the chemically similar Pd(II), initiate intra-strand and inter-strand cross-linkages in (GC)n oligos (Lippard 1982).
Silver
Unlike Hg(II), low concentrations of Ag(I) do not alter β-DNA conformation (Nordén et al. 1986). At higher Ag(I)-DNA ratios, BP orientations are significantly tilted/twisted to fold and alter the flexibility of the DNA (Nordén et al. 1986; Wilhelm and Daune 1969). Similar to Cu(II), Ag(I) induces DNA folding and accompanying decreases in native DNA rigidity (Wilhelm and Daune 1969). When binding to DNA, Ag(I) does so reversibly and with a preference for both GC-rich regions and denatured DNA by participating in at least three different cation-DNA complexes (Jensen and Davidson 1966). The first Ag complex (complex I) occurs in a GC-dependent manner where the ratio of Ag/nucleotide being less than 2 and is characterised by a GC-Ag(I)–base “sandwich” (Sissoeff et al. 1976). Ag(I) has a high affinity and stabilises C-C miss-matched BPs in dsDNA to produce C-Ag(I)-C BPs (Izatt et al. 1971) by binding to N-7 or N-3 of alternate BPs (Yamane and Davidson 1962).
The formation of a second Ag complex (complex 2) (Fig. 5) is conditional both upon the ratio of Ag/nucleotide being between 0.2 and 0.5 and also conditional upon no bases are affected by complex I formation. As shown in Fig. 5, Ag(I) binds between A-N1 and T-N3 groups within AT base sequences, and also binds between G-N7 or G-N3 groups in GC base sequences (Izatt et al. 1971; Yamane and Davidson 1962), forming a stabilised reverse Hoogsteen CA BP (Sponer et al. 1999). Complex 2 forms weaker bonds than complex 1, and also displays different binding characteristics depending on whether the DNA is natural or synthetic oligos such as (AT)n (Yamane and Davidson 1962). Complex III (and higher complexes) are formed at pH > 7 higher densities of metal within the DNA and are characterised by the formation of DNA precipitation (Izatt et al. 1971). These complexes can be perhaps incorporated into the logical construction of DNA probes for DNA-based sensors.
Fig. 5.
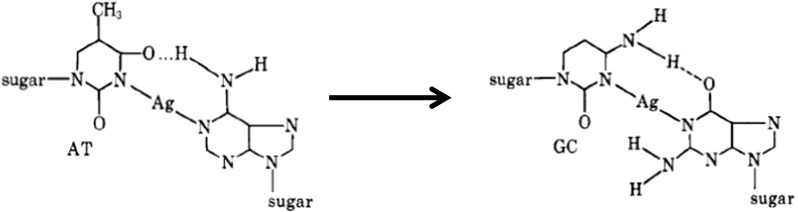
Proposed complex II formation of Ag(I) to DNA whereby N-H~N hydrogen bonds of complementary BPs are converted into N–Ag–N bonds. In addition, such binding occurs independent of complex 1 formation. Copyright (2018) American Chemical Society
Tin
Sn is a potent DNA-DNA and DNA-protein cross-linking agent because it binds to two or more ligand groups and because it can extend its linking range via Sn-Sn or Sn-O bridges (Veith and Recktenwald 1982). Different organic Sn(IV) complexes form interesting complexes with DNA (Barbieri et al. 1999; Casini et al. 2001; Silvestri et al. 2000). Sn(IV), without cleaving the dsDNA, directly coordinates to oxygen atoms of the negatively-charged DNA phosphate groups (Li et al. 1996) via electrostatic interactions (Arjmand and Jamsheera 2011). Additionally, Sn(IV) can effectively neutralise the negative charges of the phosphate sugar backbone (Tabassum and Pettinari 2006). DNA-based probes for Sn(IV) should therefore capitalise on the long Sn-Sn or Sn-O bridges, by immobilising very short oligos onto suitable substrates. Additionally, Sn(IV) binding to these oligonucleotides can delocalise enough electrons/charges on the immobilisation surface so that Sn binding events can be detected using voltammetry or another technique to produce a sensor with exceptional Sn selectivity and sensitivity.
Uranium
Although uranyl, UO(II), is the most soluble species, U exists in many species in aqueous solutions (Lee et al. 2008). Uranyl prefers to break ssDNA and form adducts in DsDNA (Yellowhair et al. 2018) and link phosphate groups on opposite strands of the minor groove of DNA (Nielsen et al. 1992). A direct catalysed hydrolysis of DNA by UO2(II) occurs upon coordination with adenosine BP and α-phosphate groups of the DNA backbone (Yazzie et al. 2003). This only occurs however, when both these groups are part of the same molecule and are geometrically aligned in a favourable manner for chelation (Izatt et al. 1971). Therefore, a sensor for Uranium would need to provide individual binding pockets cater for the various DNA-binding characteristics of UO2+ and the many other U aqueous species.
Lithium
Amongst the alkaline metals, DNA binding-affinity is approximately in the sequence order: Cs(I) > Li(I) > Na(I) > K(I) (Lyubartsev and Laaksonen 1999; Ross and Scruggs 1964). Li(I) typically coordinates with water and phosphate groups in the minor grooves of DNA helices in differing ways in a BP-dependent manner (Lyubartsev and Laaksonen 1999). Of the three Li(I)-Adenine complexes, the first and most stable complex is a five-membered ring that links A-N6 and A-N7 (Lee 2002). The second Li(I)-Adenine complex links A-N1 and A-N6 groups, while the third Li(I)-Adenine complex forms an open bond to the N3 group of A (Lee 2002). Two Li(I)-G complexes exist. The first links G-N2 and G-N3, whilst the second complex links G-O6 and G-N7. The two Li(I)-cystosine complexes link C-O2 and C-N3 or C-N3 and C-N4. The two Li(I)-Thymine complexes bind to either T-O2 or T-O4 of the carbonyl groups (Lee 2002). Based on the above evidence, sensitive and selective sensor for Li(I) must recognise and provide optimal binding pockets for each type of Li-BP complex.
Rubidium and caesium
Rb(I) and Cs(I) interact with T-O4 and T-O2 groups with a slight preference for the T-O4 group due to the close proximity of the sugar moiety to the T-O2 binding site (Marino et al. 2010). These two ions also display nucleobases-specific behaviours. For example, Rb(I)-C and Cs(I)-C binding results in four stable complexes, with the most stable complex being a mono-coordinated C-O2 complex (Marino et al. 2010). In addition, six stable Rb(I)-G and Cs(I)-G complexes have been found, with Rb(I)-G12 and Cs(I)-G12 being the most stable. Overall, Rb(I) and Cs(I) display the highest affinity for G, with BP affinities in the order of G > C > T > U > A (Marino et al. 2010). This trend is typical amongst alkaline earth metals, along with a general decrease in BP affinity with increasing atomic number (Zhu et al. 2004).
Cs(I) binds directly to the O18 atoms of DNA sugars in the minor groove (Lyubartsev and Laaksonen 1999). Cs(I) at high concentrations does not break hydrogen bonds of BPs in (dA-dT)n oligonucleotides nor does it cause altered overlap geometries (e.g. altered dsDNA structures) (Patel et al. 1981). Importantly, one study reported Cs forms a unique G-pentaplex when complexed to isoguanine-rich DNA (Lin et al. 2016). Isoguanine is an analogy of G, whereby the carbonyl and amino groups are swapped.
By carefully introducing sugar moieties into synthetic nucleotides, a specific Rb(I)/Cs(I) sensor might be devised. Differentiating Rb and Cs using DNA probes, given their similar DNA-binding properties, will likely rely upon the Cs-unique G-pentaplex formed with isoguanine-rich DNA. Complicating the differentiation between Cs and Rb ions is the preferential binding of both ions to minor grooves of dsDNA that contain An tracts (Stellwagen et al. 2005). Care will be needed when considering the stability of Rb and Cs complexes relative to other metals, and that reiterative selection may prove useful in finding specific DNA probes for these two metals.
Nickel
Ni in its common speciation Ni(II) interacts essentially the same as Mn(II) and Co(II), with the affinity of Ni(II) for base sites being similar to Co(II) (Shin and Eichhorn 1968). Co(II) and Ni(II) also have octahedral solvation shell dimensions similar to Mg(II) (Abrescia et al. 1999). Both Co(II) and Ni(II) have a strong affinity for the N7 atom of guanine (Abrescia et al. 1999). Such interactions cannot occur within standard α- or β- dsDNA, but instead occur only at terminal or extra-helical G bases (Gao et al. 1993). The resulting complex is characterised by G-N7-Ni-N7-G bridges, which may at times have an extra cation to stabilise the interaction with the dsDNA (Abrescia et al. 1999). Ni has been recently shown to intercalate between BPs in DNA (Wu et al. 2012). Destabilisation of dsDNA by Ni may be used to dislodge reporter molecules (such as fluorescent dyes (Foreman et al. 2011)) into solution from within the DNA molecule to provide a measurable change in quantum yield/signalling. The question to ask is whether DNA conformation changes are significantly varied between metals to facilitate differentiation between Ni(II), Mn(II) and Co(II) and other metals in solution.
Manganese
Mn(II) probably binds to DNA bases and phosphates at high and low concentrations (respectively) (Shin and Eichhorn 1968) and forms chelates between G-N7 and a phosphate as indicated by the decreased helix stability of Mn(II)-DNA complexes (Richard et al. 1973). It has been reported that below pH 6.4, Mn(II) induces C-DNA conformation changes most likely without direct chelation (Polyanichko et al. 2004). Mn(II)-DNA complexes open DNA secondary structures and disturb local conformational changes in close proximities of GC pairs via G-N7 binding (Sissoeff et al. 1976). Like Cu(II) and Zn(II), Mn(II) stabilises random coil and DNA helix structures due to its strong affinity for base interactions that hold complementary strands in close proximity as well as strong phosphate binding affinities. This conserves large proportions of dsDNA structure during unwinding associated with heating and cooling cycles (Shin and Eichhorn 1968). Conformational transitions from β-DNA to C-DNA have been documented in solution at the molar ratio Mn(II): DNA-phosphates between 0.1 and 1.5 (Polyanichko et al. 2004).
Strong similarities between Mn(II) and Zn(II) are observed following the NMR spectral analysis of Mn(II) binding specificity to the Dickerson dodecamer via G-N7 interactions (G4 > > G2, G10, G12) (Montrel et al. 1998). Mn(II) prefers binding to the minor groove of the 5′-most ApA step in a TnAn sequence element such as the AATT sequence in Dickerson dodecamer (Hud and Feigon 1997, 2002).
Zinc
Zn(II) binds to G-N7, but probably not as a G-Zn(II)-G sandwich or GN7-ZN(II)-GN7 cross-linking complex (Jia and Marzilli 1991). N7-Zn(II)-N7 chelate and G-Zn-G sandwich structures are not observed in 1H NMR studies, and nor are there any suggestions of phosphate groups or A residue binding by Zn(II) (Jia and Marzilli 1991). Additionally, correlations of Tm values to DNA GC support Zn(II) binding to bases (but with lower base-specificity compared to Cu(II). Zn(II) can stabilise random coil or single helical forms (Sissoeff et al. 1976) and can reversibly unwind and rewind DNA upon heating and cooling respectively (Izatt et al. 1971). DNA melting profiles in solution suggest that Zn(II) holds the strands of the dsDNA in close proximity so as to permit helix formation on cooling (Shin and Eichhorn 1968). Weaker base binding of Zn(II) results in rewinding without needing to add concentrated electrolytes (Izatt et al. 1971).
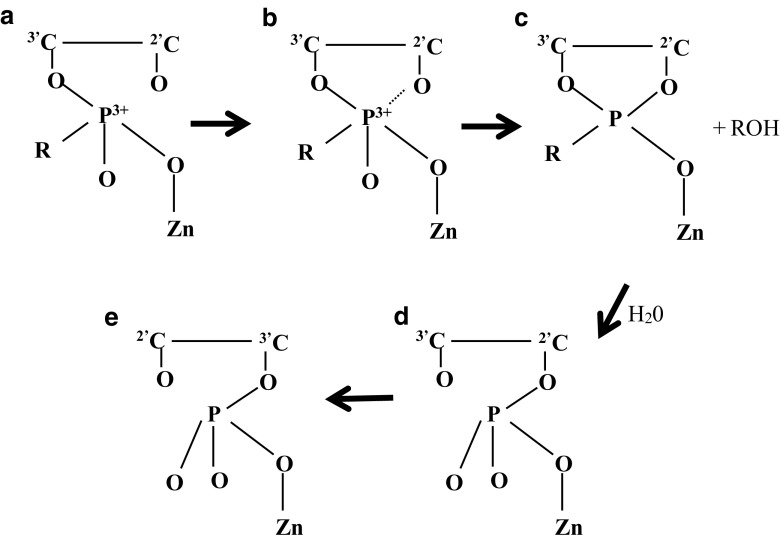
Zn(II) displays incredibly high binding specificity to d(CGCGAATTCGCG)2 (with a G selectivity of G4 > > G2, G10 and G12) (Montrel et al. 1998). Zn(II) also binds to d(ATGGGTACCCAT)2 with a specificity in the order of G3,G4 > > G5,A (Jia and Marzilli 1991). G in 5′-(G-Pu) steps is most favourable for Zn(II) binding (Montrel et al. 1998). Binding mechanisms have been previously proposed as illustrated in Fig. 6 (Izatt et al. 1971).
Fig. 6.
Zn(II) cations bind phosphate groups which polarises the PO linkage to produce a positive dipole on the phosphorus atom. The formation of a ring with a 2′-hydroxyl group leads to cleavage of the phosphodiester linkage. It is postulated that other metal cations (e.g. Mn2+, Co2+, Ni2+, Cu2+, La3+, Ce3+ and Lu3+) that depolymerize RNA can also bind via similar mechanisms. Copyright (2018) American Chemical Society
Cadmium
A recent review has suggested that there is a general consensus ion of Cd(II) ion is a human carcinogen (Hartwig 2010), which mediates its carcinogenicity predominantly via multiple distinct protein-based mechanisms. The review suggests the presence of several categories of targets that are influenced by Cd, such as anti-oxidative pathways, DNA repair processes, and acting as a tumour suppressor and in the signal transduction of proteins. However, some studies suggest that Cd(II) covalently binds to N-7 moieties of A and G and possibly forms intra-strand bi-functional AT adducts (Hossain and Huq 2002). Similarly to Fe(II), Cd(II) chelates to DNA between G-N7 and a phosphate group (Sissoeff et al. 1976). Cd(II) promotes DNA renaturation by forming sandwich complexes between two adjacent G bases (Richard et al. 1973). Cd(II) also forms Cd(II)-base bonds that are strong enough to remain intact (contrary to Zn[II]) and subsequently prevent regeneration of the double helix upon heating-cooling experimental protocols(Shin and Eichhorn 1968).
Iron
At low pH, Fe(III) forms an octahedral arrangement of oxygen atoms when complexed to AMP, ADP, or ATP (the formation constant increases in this sequence) which indicates significant binding only to the phosphate moieties (Izatt et al. 1971). Interestingly, at pH 7, one or more ring nitrogen atoms have replaced oxygen donor atoms on the Fe(III) (Izatt et al. 1971). Although Fe(II) binds to phosphates rather than the interior DNA base sites, it remains unclear whether Fe(III) binds to the interior base or exterior phosphate sites (Izatt et al. 1971). The difficulty in identifying the exact mechanism(s) of Fe(II) and Fe(III) complex formation to DNA or RNA probably is due to the responsiveness of Fe complex formation to experimental conditions such as pH, temperature and presence of other cations (Izatt et al. 1971)
Lead
Pb(II) creates double bonds between DNA BPs. It also removes protons from the T methyl groups and from each of the C–H bonds of 2-deoxyribose (Cooke et al. 2003). This leads to single- or double-strand breaks, purine, pyrimidine or deoxyribose modifications, and DNA cross-links (Klaunig and Kamendulis 2004; Wozniak and Blasiak 2003).
Pb(II) weakly interacts with G (N7 and C6 O atoms) and C (N3 and C2 O atoms), followed by the phosphate moiety (Da Costa and Sigel 2000; Knobloch et al. 2005). Furthermore, Pb(II) interacts simultaneously with G and the phosphate group to form macro chelates (Da Costa and Sigel 2000). It also interacts strongly with two neighbouring phosphates in solution (Knobloch et al. 2005). Pb(II) also has a well-documented propensity to form G-quartets, which are self-assembled G nucleoside square co-planar arrays of four G bases, in which each base is hydrogen bonded to its neighbours (Liu et al. 2009). Stacking of multiple G-quartets in DNA has been observed to produce interesting G-quadruplex structures (Simonsson 2001). Pb(II) coordinated with eight oxygen atoms (of the C6 = O groups) displays geometries intermediate between cubic and a square ant-prisms, which resemble potassium complexes in nucleic acid structures (Kotch et al. 2000). The potential for competitive analogue replacement of K with Pb may play a pivotal role in the genotoxicity of Pb. Additionally, coordination of the Pb(II) ion and the thrombin-binding aptamer, ‘TBA’, d(G2T2G2TGTG2T2G2) revealed that the cation binds between the two quartets in the Pb(II)-TBA complex and coordinates with the eight surrounding G O6 atoms.
Barium and strontium
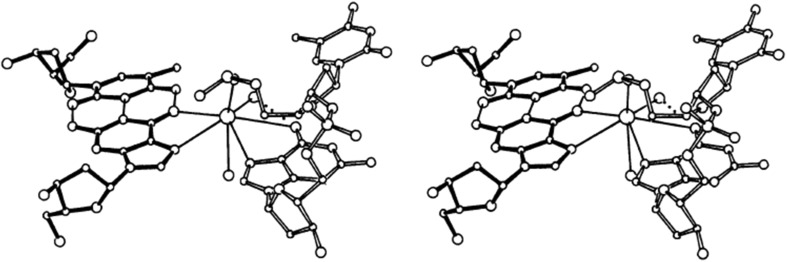
One study examining Z-DNA hexamers revealed Ba(II) ions possess eight coordination sites, such that a single cation coordinates to four water molecules, bridges two adjacent helices in the crystalline DNA and simultaneously coordinates with the O6 and N7 atoms of G bases (Gao et al. 1993, 1995; Jean et al. 1993). Ba(II) also bis-coordinates with aminohexyl-CGCGCG (m5CGm5CGTG), a Ba(II)-dependent crystal structure, without disrupting the Z-DNA lattice (Jean et al. 1993) (see Fig. 7). Both Sr(II) and Ba(II) cations complex with the O2 sites of both U and T (Marino et al. 2010). This same study found C1-Sr(II) and C1-Ba(II) complexes are the most stable and therefore most likely to bind these complexes to C mono-coordinated ions with O-M(II)-bond lengths of 2.302 and 2.442 A.
Fig. 7.
Stereoscopic view of Ba(II) ion bis-coordination between G10 and G12 in crystals m5CGm5CGTG grown in the presence of Ba(II). G12 belongs to a symmetry-related helix and is shown in filled bonds. Copyright (2018) American Chemical Society
Vanadium
It is possible that A BPs stabilises the V(III) oxidation state and with VCl3 forms weak monomeric and dimeric complexes in the 1.8–4.5 pH range, depending on the concentration of V (Bukietynska and Krot-Lacina 2001). VO(II) binds to DNA through G and A N-7 atoms and via the backbone PO2 moieties. VO3− exhibits weaker binding through T, A and G BPs and essentially no interaction with the backbone phosphate group. The same study reported partial β- to α-DNA transitions occurring upon V-O complex formation, while DNA remains in the β-family structure in the VO3− complexes (Ouameur et al. 2006).
Antimony
Sb displays both acute and chronic toxicity, with suggestions that Sb(III) is 10× more toxic than Sb(V) (Filella et al. 2009). This is unsurprising since Sb(V) species do not interact with DNA (Li et al. 2011). Sb(III) binds to the S atoms of C BP residues (Yan et al. 2000)
Discussion
This review has so far outlined and described the chemical properties of heavy metals with DNA. The observations argue that thymus genomic DNA is an effective and simple-to-use biological sensor to test for heavy metal ion toxicity in actual environmental water samples. This publication provided empirical evidence that genomic DNA could be used to detect toxicity of quite complex mixtures of chemical species. The chemical summaries presented in this review help to explain why some metal ions are more toxic than others. Foreman et al. showed that it is possible to demonstrate that simple quantification of the loss of fluorescence of acridine orange bound to calf thymus DNA. This loss is due to the toxic chemicals binding to the DNA and altering its structure. The drop-in fluorescence is approximately proportional to the total toxicity of the sample. The fact that this change in fluorescence can be detected in real time in the field using hand-held fluorimeters can be used to track down the source(s) of chemical toxicity in the field.
Author contributions
All the authors wrote, revised, and approved the final manuscript.
Conflict of interest
Cris dos Remedios declares that he/she has no conflict of interest. Vangelis George Kanellis declares that he/she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Contributor Information
Vangelis George Kanellis, Email: vangeliskanellis@gmail.com.
Cristobal G. dos Remedios, Email: cris.dosremedios@sydney.edu.au
References
- Abrescia NGA, Malinina L, Fernandez LG, Subirana JA, Huynh-Dinh T, Neidle S. Structure of the oligonucleotide d(CGTATATACG) as a site-specific complex with nickel ions. Nucleic Acids Res. 1999;27(7):1593–1599. doi: 10.1093/nar/27.7.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjmand F, Jamsheera A. DNA binding studies of new valine derived chiral complexes of tin(IV) and zirconium(IV) Spectrochim Acta A Mol Biomol Spectrosc. 2011;78(1):45–51. doi: 10.1016/j.saa.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Barbieri R, Huber F, Silvestri A, Ruisi G, Rossi M, Barone G, Paulsen AB. The interaction of S,N-coordinated dimethyltin(IV) derivatives with deoxyribonucleic acid: structure and dynamics by 119Sn Mössbauer spectroscopy. Appl Organomet Chem. 1999;13:595–603. doi: 10.1002/(SICI)1099-0739(199908)13:8<595::AID-AOC894>3.0.CO;2-0. [DOI] [Google Scholar]
- Batley GE, Apte SC, Stauber JL. Speciation and bioavailability of trace metals in water: progress since 1982. Aust J Chem. 2004;57:903–919. doi: 10.1071/CH04095. [DOI] [Google Scholar]
- Bligh SWA, Choi N, Evagorou EG, McPartlin M, White KN. Dimeric yttrium(iii) and neodymium(iii) macrocyclic complexes: potential catalysts for hydrolysis of double-stranded DNA. J Chem Soc Dalton Trans. 2001;0:3169–3172. doi: 10.1039/b102533n. [DOI] [Google Scholar]
- Bukietynska K, Krot-Lacina K. Vanadium(III) interaction with adenine. Polyhedron. 2001;20(18):2353–2361. doi: 10.1016/s0277-5387(01)00840-3. [DOI] [Google Scholar]
- Casini A, Messori L, Orioli P, Gielen M, Kemmer M, Willem R. Interactions of two cytotoxic organotin(IV) compounds with calf thymus DNA. J Inorg Biochem. 2001;85(4):297–300. doi: 10.1016/s0162-0134(01)00215-x. [DOI] [PubMed] [Google Scholar]
- Cohen MD, Kargacin B, Klein CB, Costa M. Mechanisms of chromium carcinogenicity and toxicity. Crit Rev Toxicol. 1993;23(3):255–281. doi: 10.3109/10408449309105012. [DOI] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17(10):1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- Corbett JD. Extended metal-metal bonding in halides of the early transition metals. Acc Chem Res. 1981;14(8):239–246. doi: 10.1021/ar00068a003. [DOI] [Google Scholar]
- Corner EDS, Sparrow BW. The modes of action of toxic agents I. Observations on the poisoning of certain crustaceans by copper and mercury. J Mar Biol Assoc U K. 1956;35(3):531–548. doi: 10.1017/S0025315400010390. [DOI] [Google Scholar]
- Da Costa CP, Sigel H. Lead(II)-binding properties of the 5‘-monophosphates of adenosine (AMP2-), inosine (IMP2-), and guanosine (GMP2-) in aqueous solution. Evidence for nucleobase−lead(II) interactions. Inorg Chem. 2000;39(26):5985–5993. doi: 10.1021/ic0007207. [DOI] [PubMed] [Google Scholar]
- Das T, Sharma A, Talukder G. Effects of lanthanum in cellular systems. Biol Trace Elem Res. 1988;18(1):201–228. doi: 10.1007/bf02917504. [DOI] [PubMed] [Google Scholar]
- Destouni Georgia, Jarsjö Jerker. Zones of untreatable water pollution call for better appreciation of mitigation limits and opportunities. Wiley Interdisciplinary Reviews: Water. 2018;5(6):e1312. doi: 10.1002/wat2.1312. [DOI] [Google Scholar]
- Dudis DS, Corbett JD, Hwu SJ. Synthesis, characterization, and crystal structures of two scandium cluster carbides and a boride, Sc7X12C (X = I, Br) and Sc7I12B. Inorg Chem. 1986;25(19):3434–3438. doi: 10.1021/ic00239a023. [DOI] [Google Scholar]
- Duguid J, Bloomfield VA, Benevides J, Thomas GJ., Jr Raman spectroscopy of DNA-metal complexes. I. Interactions and conformational effects of the divalent cations: Mg, Ca, Sr, Ba, Mn, Co, Ni, Cu, Pd, and Cd. Biophys J. 1993;65(5):1916–1928. doi: 10.1016/S0006-3495(93)81263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filella M, Williams PA, Belzile N. Antimony in the environment: knowns and unknowns. Environ Chem. 2009;6(2):95–105. doi: 10.1071/EN09007. [DOI] [Google Scholar]
- Fiskin AM, Beer M. Determination of base sequence in nucleic acids with the electron microscope. IV. Nucleoside complexes with certain metal ions. Biochemistry. 2002;4(7):1289–1294. doi: 10.1021/bi00883a012. [DOI] [PubMed] [Google Scholar]
- Foreman AL, Phillips L, Kanellis VG, Hammoudeh D, Naumann C, Wong H, Chisari R, Hibbert DB, Lee GCH, Patra R, Julli M, Chapman J, Cooke AR, dos Remedios CG. A DNA-based assay for toxic chemicals in wastewater. Environ Toxicol Chem. 2011;30(8):1810–1818. doi: 10.1002/etc.568. [DOI] [PubMed] [Google Scholar]
- Gao Y-G, Sriram M, Wang AH-J. Crystallographic studies of metal ion - DNA interactions: different binding modes of cobalt(II), copper(II) and barium(II) to N7of guanines in Z-DNA and a drug-DNA complex. Nucleic Acids Res. 1993;21(17):4093–4101. doi: 10.1093/nar/21.17.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YG, Robinson H, van Boom JH, Wang AH. Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA. Biophys J. 1995;69(2):559–568. doi: 10.1016/S0006-3495(95)79929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geierstanger BH, Kagawa TF, Chen SL, Quigley GJ, Ho PS. Base-specific binding of copper(II) to Z-DNA. The 1.3-A single crystal structure of d(m5CGUAm5CG) in the presence of CuCl2. J Biol Chem. 1991;266(30):20185–20191. doi: 10.2210/pdb1d40/pdb. [DOI] [PubMed] [Google Scholar]
- Gruber D. Biological monitoring and our water resources. Endeavour. 1989;13(3):135–140. doi: 10.1016/0160-9327(89)90088-4. [DOI] [PubMed] [Google Scholar]
- Gruenwedel DW, Cruikshank MK. Mercury-induced DNA polymorphism: probing the conformation of Hg(II)-DNA via staphylococcal nuclease digestion and circular dichroism measurements. Biochemistry. 1990;29(8):2110–2116. doi: 10.1021/bi00460a021. [DOI] [PubMed] [Google Scholar]
- Haight GP, Jr, Huang TJ, Shakhashiri BZ. Reactions of chromium(IV) J Inorg Nucl Chem. 1971;33(7):2169–2175. doi: 10.1016/0022-1902(71)80578-x. [DOI] [Google Scholar]
- Harris WR, Berthon G, Day JP, Exley C, Flaten TP, Forbes WF, Zatta PF. Speciation of aluminum in biological systems. J Toxicol Environ Health. 1996;48(6):543–568. doi: 10.1080/009841096161069. [DOI] [PubMed] [Google Scholar]
- Hartwig A. Mechanisms in cadmium-induced carcinogenicity: recent insights. Biometals. 2010;23(5):951–960. doi: 10.1007/s10534-010-9330-4. [DOI] [PubMed] [Google Scholar]
- Hossain Z, Huq F. Studies on the interaction between Cd2+ ions and DNA. J Inorg Biochem. 2002;90(3–4):85–96. doi: 10.1016/s0162-0134(02)00412-9. [DOI] [PubMed] [Google Scholar]
- Hud NV, Feigon J. Localization of divalent metal ions in the minor groove of DNA A-tracts. J Am Chem Soc. 1997;119(24):5756–5757. doi: 10.1021/ja9704085. [DOI] [Google Scholar]
- Hud NV, Feigon J. Characterization of divalent Cation localization in the minor groove of the AnTn and TnAn DNA sequence elements by 1H NMR spectroscopy and manganese(II)†. Biochemistry. 2002;41(31):9900–9910. doi: 10.1021/bi020159j. [DOI] [PubMed] [Google Scholar]
- Hyne RV, Pablo F, Julli M, Markich SJ. Influence of water chemistry on the acute toxicity of copper and zinc to the cladoceran Ceriodaphnia cf dubia. Environ Toxicol Chem. 2005;24:1667–1675. doi: 10.1897/04-497R.1. [DOI] [PubMed] [Google Scholar]
- Izatt RM, Christensen JJ, Rytting JH. Sites and thermodynamic quantities associated with proton and metal ion interaction with ribonucleic acid, deoxyribonucleic acid, and their constituent bases, nucleosides, and nucleotides. Chem Rev. 1971;71(5):439–481. doi: 10.1021/cr60273a002. [DOI] [PubMed] [Google Scholar]
- Jean YC, Gao YG, Wang AH. Z-DNA structure of a modified DNA hexamer at 1.4—a resolution: aminohexyl-5ʼ-d(pCpGp[br5C]pGpCpG) Biochemistry. 1993;32(1):381–388. doi: 10.1021/bi00052a047. [DOI] [PubMed] [Google Scholar]
- Jensen RH, Davidson N. Spectrophotometric, potentiometric, and density gradient ultracentrifugation studies of the binding of silver ion by DNA. Biopolymers. 1966;4:17–32. doi: 10.1002/bip.1966.360040104. [DOI] [Google Scholar]
- Jia X, Marzilli LG. Zinc ion-DNA polymer interactions. Biopolymers. 1991;31(1):23–44. doi: 10.1002/bip.360310104. [DOI] [PubMed] [Google Scholar]
- Karlik SJ, Eichhorn GL, Lewis PN, Crapper DR. Interaction of aluminum species with deoxyribonucleic acid. Biochemistry. 1980;19(26):5991–5998. doi: 10.1021/bi00567a008. [DOI] [PubMed] [Google Scholar]
- Katz S. The reversible reaction of Hg (II) and double-stranded polynucleotides. A step-function theory and its significance. Biochim Biophys Acta. 1963;68:240–253. doi: 10.1016/0926-6550(63)90435-3. [DOI] [PubMed] [Google Scholar]
- Kerner MW, Anderson HH. The uptake of yttrium and cerium144 by viable Entamoeba histolytica in culture. Exp Parasitol. 1955;4(6):564–568. doi: 10.1016/0014-4894(55)90044-8. [DOI] [PubMed] [Google Scholar]
- Kim SH, Shin WC, Warrant RW. Heavy metal ion-nucleic acid interaction. Methods Enzymol. 1985;114:156–167. doi: 10.1016/0076-6879(85)14016-4. [DOI] [PubMed] [Google Scholar]
- Kiss T. Interaction of aluminum with biomolecules—any relevance to Alzheimer's disease? Arch Gerontol Geriatr. 1995;21(1):99–112. doi: 10.1016/0167-4943(95)00642-X. [DOI] [PubMed] [Google Scholar]
- Kiss T, Sovago I, Martin RB. Aluminum(3+) binding by adenosine 5′-phosphates: AMP, ADP, and ATP. Inorg Chem. 1991;30(9):2130–2132. doi: 10.1021/ic00009a032. [DOI] [Google Scholar]
- Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annu Rev Pharmacol Toxicol. 2004;44(1):239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- Knobloch B, Linert W, Sigel H, Gray HB. Metal ion-binding properties of (N3)-deprotonated uridine, thymidine, and related pyrimidine nucleosides in aqueous solution. Proc Natl Acad Sci U S A. 2005;102(21):7459–7464. doi: 10.1073/pnas.0501446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltover I, Wagner K, Safinya CR. DNA condensation in two dimensions. Proc Natl Acad Sci. 2000;97(26):14046–14051. doi: 10.1073/pnas.97.26.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortenkamp A, Casadevall M, Da Cruz Fresco P. The reductive conversion of the carcinogen chromium (VI) and its role in the formation of DNA lesions. Ann Clin Lab Sci. 1996;26(2):160–175. [PubMed] [Google Scholar]
- Kortenkamp A, Casadevall M, Faux SP, Jenner A, Shayer RO, Woodbridge N, O'Brien P. A role for molecular oxygen in the formation of DNA damage during the reduction of the carcinogen chromium (VI) by glutathione. Arch Biochem Biophys. 1996;329(2):199–207. doi: 10.1006/abbi.1996.0209. [DOI] [PubMed] [Google Scholar]
- Kotch FW, Fettinger JC, Davis JT. A lead-filled G-Quadruplex: insight into the G-Quartet’s selectivity for Pb2+ over K+ Org Lett. 2000;2(21):3277–3280. doi: 10.1021/ol0065120. [DOI] [PubMed] [Google Scholar]
- Kuklenyik Z, Marzilli LG. Mercury(II) site-selective binding to a DNA hairpin. Relationship of sequence-dependent intra- and interstrand cross-linking to the hairpin−duplex conformational transition. Inorg Chem. 1996;35(19):5654–5662. doi: 10.1021/ic960260a. [DOI] [PubMed] [Google Scholar]
- Lee GY. DFT studies of the lithium complexes of DNA bases. Bull Kor Chem Soc. 2002;23(7):1023–1026. doi: 10.5012/bkcs.2002.23.7.1023. [DOI] [Google Scholar]
- Lee JH, Wang Z, Liu J, Lu Y. Highly sensitive and selective colorimetric sensors for uranyl (UO22+): development and comparison of labeled and label-free DNAzyme-gold nanoparticle systems. J Am Chem Soc. 2008;130(43):14217–14226. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescrinier E, Froeyen M, Herdewijn P. Difference in conformational diversity between nucleic acids with a six-membered ‘sugar’ unit and natural ‘furanose’ nucleic acids. Nucleic Acids Res. 2003;31(12):2975–2989. doi: 10.1093/nar/gkg407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Yang P, Wang H, Guo M. Diorganotin(IV) antitumor agent. (C2H5)2SnCl2 (phen)/nucleotides aqueous and solid-state coordination chemistry and its DNA binding studies. J Inorg Biochem. 1996;64(3):181–195. doi: 10.1016/0162-0134(96)00039-6. [DOI] [PubMed] [Google Scholar]
- Li Y, Liu J-M, Han F, Jiang Y, Yan X-P. Probing interactions of antimony species with DNA by short column capillary electrophoresis coupled with inductively coupled plasma mass spectrometry. J Anal At Spectrom. 2011;26:94–99. doi: 10.1039/C0JA00079E. [DOI] [Google Scholar]
- Lin S, Yang C, Mao Z, He B, Wang Y-T, Leung C-H, Ma D-L. A G-pentaplex-based assay for Cs+ ions in aqueous solution using a luminescent Ir(III) complex. Biosens Bioelectron. 2016;77:609–612. doi: 10.1016/j.bios.2015.10.026. [DOI] [PubMed] [Google Scholar]
- Lippard SJ. New chemistry of an old molecule: cis-[Pt(NH3)2Cl2] Science. 1982;218(4577):1075–1082. doi: 10.1126/science.6890712. [DOI] [PubMed] [Google Scholar]
- Lison D, Lauwerys R. Cobalt bioavailability from hard metal particles. Arch Toxicol. 1994;68(8):528–531. doi: 10.1007/s002040050108. [DOI] [PubMed] [Google Scholar]
- Liu C-W, Huang C-C, Chang H-T. Highly selective DNA-based sensor for lead(II) and mercury(II) ions. Anal Chem. 2009;81(6):2383–2387. doi: 10.1021/ac8022185. [DOI] [PubMed] [Google Scholar]
- Lyubartsev AP, Laaksonen A. Effective potentials for ion–DNA interactions. J Chem Phys. 1999;111(24):11207–11215. doi: 10.1063/1.480476. [DOI] [Google Scholar]
- Macdonald TL, Martin RB. Aluminum ion in biological systems. Trends Biochem Sci. 1988;13(1):15–19. doi: 10.1016/0968-0004(88)90012-6. [DOI] [PubMed] [Google Scholar]
- Macquet JP, Theophanides T. Specificity of the interaction of DNA-platinum, amount of platinum, and pH measurement. Biopolymers. 1975;14(4):781–799. doi: 10.1002/bip.1975.360140409. [DOI] [PubMed] [Google Scholar]
- Manning GS. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978;11(02):179. doi: 10.1017/S0033583500002031. [DOI] [PubMed] [Google Scholar]
- Marino T, Mazzuca D, Russo N, Toscano M, Grand A. On the interaction of rubidium and cesium mono-, strontium and barium bi-cations with DNA and RNA bases. A theoretical study. Int J Quantum Chem. 2010;110:138–147. doi: 10.1002/qua.22076. [DOI] [Google Scholar]
- Matsuda M, Takeuchi E. The interaction of Hg++ with deoxyribonucleic acid. J Biochem. 1967;61(4):523–526. doi: 10.1093/oxfordjournals.jbchem.a128580. [DOI] [PubMed] [Google Scholar]
- Mioduski T. Quasilanthanide(III) behavior of scandium(III) and coordination number. Comments Inorg Chem. 1993;14(5):263–281. doi: 10.1080/02603599308048664. [DOI] [Google Scholar]
- Miyake Y, et al. MercuryII-mediated formation of thymine−HgII−thymine base pairs in DNA duplexes. J Am Chem Soc. 2006;128(7):2172–2173. doi: 10.1021/ja056354d. [DOI] [PubMed] [Google Scholar]
- Moldrheim E, Andersen B, Frøystein NA, Sletten E. Interaction of manganese(II), cobalt(II) and nickel(II) with DNA oligomers studied by 1H NMR spectroscopy. Inorg Chim Acta. 1998;273(1–2):41–46. doi: 10.1016/s0020-1693(97)06010-6. [DOI] [Google Scholar]
- Montrel M, Chuprina VP, Poltev VI, Nerdal W, Sletten E. Sequence-dependent binding of metal ion to DNA oligomeres. A comparison of molecular electrostatic potentials with NMR data. J Biomol Struct Dyn. 1998;16(3):631–637. doi: 10.1080/07391102.1998.10508275. [DOI] [PubMed] [Google Scholar]
- Nandi US, Wang JC, Davidson N. Separation of deoxyribonucleic acids by hg(II) binding and Cs2SO4 density-gradient centrifugation. Biochemistry. 1965;4(9):1687–1696. doi: 10.1021/bi00885a001. [DOI] [Google Scholar]
- Nielsen PE, Hiort C, Soennischsen SO, Buchardt O, Dahl O, Norden B. DNA binding and photocleavage by Uranyl VI salts. J Am Chem Soc. 1992;114(13):4967–4975. doi: 10.1021/ja00039a003. [DOI] [Google Scholar]
- Nordén B, Matsuoka Y, Kurucsev T. Nucleic acid–metal interactions: V. the effect of silver(I) on the structures of A- and B-DNA forms. Biopolymers. 1986;25(8):1531–1545. doi: 10.1002/bip.360250811. [DOI] [PubMed] [Google Scholar]
- Onyido I, Norris AR, Buncel E. Biomolecule—mercury interactions: modalities of DNA base--mercury binding mechanisms. Remediation strategies. Chem Rev. 2004;104(12):5911–5929. doi: 10.1021/cr030443w. [DOI] [PubMed] [Google Scholar]
- Ouameur AA, Arakawa H, Tajmir-Riahi HA. Binding of oxovanadium ions to the major and minor grooves of DNA duplex: stability and structural models. Biochem Cell Biol. 2006;84(5):677–683. doi: 10.1139/o06-043. [DOI] [PubMed] [Google Scholar]
- Pack GR. Theory of ion effects on DNA conformation: a high-salt model. Int J Quantum Chem. 1982;22:81–101. doi: 10.1002/qua.560220710. [DOI] [Google Scholar]
- Passow H, Rothstein A, Clarkson TW. The general pharmacology of the heavy metals. Pharm Rev. 1961;13:185–224. [PubMed] [Google Scholar]
- Patel DJ, Sharon AK, Suggs JW, Cox SD. Right-handed alternating DNA conformation: poly(dA-dT) adopts the same dinucleotide repeat with cesium, tetraalkylammonium, and 3α ,5β ,17β -dipyrrolidinium steroid dimethiodide cations in aqueous solution. Proc Natl Acad Sci U S A. 1981;78(7):4063–4067. doi: 10.1073/pnas.78.7.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyanichko AM, Andrushchenko VV, Chikhirzhina EV, Vorob’ev VI, Wieser H. The effect of manganese(II) on DNA structure: electronic and vibrational circular dichroism studies. Nucleic Acids Res. 2004;32(3):989–996. doi: 10.1093/nar/gkh242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao KS, Divakar S. Spectroscopic studies on the effects of aluminum ion on calf-thymus DNA. Bull Environ Contam Toxicol. 1993;50(1):92–99. doi: 10.1007/BF00196546. [DOI] [PubMed] [Google Scholar]
- Richard H, Schreiber JP, Daune M. Interactions of metallic ions with DNA. V. DNA renaturation mechanism in the presence of Cu 2+ Biopolymers. 1973;12(1):1–10. doi: 10.1002/bip.1973.360120102. [DOI] [PubMed] [Google Scholar]
- Rosoff B, Spencer H. Binding of rare earths to serum proteins and DNA. Clin Chim Acta. 1979;93(3):311–319. doi: 10.1016/0009-8981(79)90280-8. [DOI] [PubMed] [Google Scholar]
- Ross PD, Scruggs RL. Electrophoresis of DNA. II. Specific interactions of univalent and divalent cations with DNA. Biopolymers. 1964;2:79–89. doi: 10.1002/bip.1964.360020111. [DOI] [Google Scholar]
- Rossetto FE, Nieboer E. The interaction of metal ions with synthetic DNA: induction of conformational and structural transitions. J Inorg Biochem. 1994;54(3):167–186. doi: 10.1016/0162-0134(94)80011-1. [DOI] [PubMed] [Google Scholar]
- Savelyev A, MacKerell AD. Differential deformability of the DNA minor groove and altered BI/BII backbone conformational equilibrium by the monovalent ions Li(+), Na(+), K(+) and Rb(+) via water-mediated hydrogen bonding. J Chem Theory Comput. 2015;11(9):4473–4485. doi: 10.1021/acs.jctc.5b00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savreux-Lenglet G, et al. Protein recognition in drug-induced DNA alkylation: when the moonlight protein GAPDH meets S23906-1/DNA minor groove adducts. Int J Mol Sci. 2015;16(11):26555–26581. doi: 10.3390/ijms161125971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YA, Eichhorn GL. Interactions of metal ions with polynucleotides and related compounds. XI. The reversible unwinding and rewinding of deoxyribonucleic acid by zinc (II) ions through temperature manipulation. Biochemistry. 1968;7(3):1026–1032. doi: 10.1021/bi00843a022. [DOI] [PubMed] [Google Scholar]
- Silvestri A, Ruisi G, Barbieri R. The interaction of DNA with organotin(IV) salts and complexes. Hyperfine Interact. 2000;126(1):43–46. doi: 10.1023/a:1012676224584. [DOI] [Google Scholar]
- Simonsson T. G-Quadruplex DNA structures variations on a theme. J Biol Chem. 2001;382(4):621–628. doi: 10.1515/bc.2001.073. [DOI] [PubMed] [Google Scholar]
- Sissoeff I, Grisvard J, Guille E. Studies on metal ions-DNA interactions: specific behaviour of reiterative DNA sequences. Prog Biophys Mol Biol. 1976;31(2):165–199. doi: 10.1016/0079-6107(78)90008-1. [DOI] [PubMed] [Google Scholar]
- Sponer J, Sabat M, Burda JV, Leszczynski J, Hobza P, Lippert B. Metal ions in non-complementary DNA base pairs: an ab initio study of Cu(I), Ag(I), and Au(I) complexes with the cytosine-adenine base pair. J Biol Inorg Chem. 1999;4(5):537–545. doi: 10.1007/s007750050376. [DOI] [PubMed] [Google Scholar]
- Šponer J, Riley KE, Hobza P. Nature and magnitude of aromatic stacking of nucleic acid bases. Phys Chem Chem Phys. 2008;10(19):2595–2610. doi: 10.1039/B719370J. [DOI] [PubMed] [Google Scholar]
- Stellwagen E, Dong Q, Stellwagen NC. Monovalent cations affect the free solution mobility of DNA by perturbing the hydrogen-bonded structure of water. Biopolymers. 2005;78(2):62–68. doi: 10.1002/bip.20260. [DOI] [PubMed] [Google Scholar]
- Stern KG, Steinberg MA. Desoxyribonucleic acid complexes of rare earths. Biochim Biophys Acta. 1953;11:553–558. doi: 10.1016/0006-3002(53)90095-x. [DOI] [PubMed] [Google Scholar]
- Sugden KD. Formation of modified cleavage termini from the reaction of chromium(V) with DNA. J Inorg Biochem. 1999;77(3–4):177–183. doi: 10.1016/s0162-0134(99)00189-0. [DOI] [PubMed] [Google Scholar]
- Tabassum S, Pettinari C. Chemical and biotechnological developments in organotin cancer chemotherapy. J Organomet Chem. 2006;691(8):1761–1766. doi: 10.1016/j.jorganchem.2005.12.033. [DOI] [Google Scholar]
- Takahashi T, Ito A, Inakuma M, Shinohara H. Divalent scandium atoms in the cage of C84. Phys Rev B Condens Matter. 1995;52(19):13812–13814. doi: 10.1103/PhysRevB.52.13812. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Oda S, Yamaguchi H, Kondo Y, Kojima C, Ono A. 15N−15N J-coupling across HgII: direct observation of HgII-mediated T−T base pairs in a DNA duplex. J Am Chem Soc. 2007;129(2):244–245. doi: 10.1021/ja065552h. [DOI] [PubMed] [Google Scholar]
- Tchounwou PB, Yedjou CG, Patlolla AK, Sutton DJ. Heavy metals toxicity and the environment. EXS. 2012;101:133–164. doi: 10.1007/978-3-7643-8340-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsapakos MJ, Wetterhahn KE. The interaction of chromium with nucleic acids. Chem Biol Interact. 1983;46(2):265–277. doi: 10.1016/0009-2797(83)90034-0. [DOI] [PubMed] [Google Scholar]
- Veith M, Recktenwald O. Structure and reactivity of monomeric, molecular Tin(II) compounds. Organotin Compounds. 1982;104:1–55. doi: 10.1007/BFb0012243. [DOI] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Wilhelm FX, Daune M. Interactions des ions métalliques avec le DNA. III. Stabilité et configuration des complexes Ag–DNA. Biopolymers. 1969;8(1):121–137. doi: 10.1002/bip.1969.360080110. [DOI] [Google Scholar]
- Wozniak K, Blasiak J. In vitro genotoxicity of lead acetate: induction of single and double DNA strand breaks and DNA-protein cross-links. Mutat Res Genet Toxicol Environ Mutagen. 2003;535(2):127–139. doi: 10.1016/s1383-5718(02)00295-4. [DOI] [PubMed] [Google Scholar]
- Wu J, Du F, Zhang P, Khan IA, Chen J, Liang Y. Thermodynamics of the interaction of aluminum ions with DNA: implications for the biological function of aluminum. J Inorg Biochem. 2005;99(5):1145–1154. doi: 10.1016/j.jinorgbio.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Wu H, Sun T, Li K, Liu B, Kou F, Jia F, Bai Y. Synthesis, crystal structure, and DNA-binding studies of a nickel(II) complex with the Bis(2-benzimidazolymethyl)amine ligand. Bioinorg Chem Appl. 2012;2012:609796. doi: 10.1155/2012/609796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane T, Davidson N. On the complexing of deoxyribonucleic acid by silver(I) Biochim Biophys Acta. 1962;55(5):609–621. doi: 10.1016/0006-3002(62)90839-9. [DOI] [PubMed] [Google Scholar]
- Yan S, Ding K, Zhang L, Sun H. Complexation of antimony(III) by trypanothione. Angew Chem Int Ed. 2000;39:4260–4262. doi: 10.1002/1521-3773(20001201)39:23<4260::AID-ANIE4260>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Yazzie M, Gamble SL, Civitello ER, Stearns DM. Uranyl acetate causes DNA single Strand breaks in vitro in the presence of Ascorbate (vitamin C) Chem Res Toxicol. 2003;16(4):524–530. doi: 10.1021/tx025685q. [DOI] [PubMed] [Google Scholar]
- Yellowhair M, Romanotto MR, Stearns DM, Clark Lantz R. Uranyl acetate induced DNA single strand breaks and AP sites in Chinese hamster ovary cells. Toxicol Appl Pharmacol. 2018;349:29–38. doi: 10.1016/j.taap.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhitkovich A, Voitkun V, Costa M. Formation of the amino acid-DNA complexes by hexavalent and trivalent chromium in vitro: importance of trivalent chromium and the phosphate group. Biochemistry. 1996;35(22):7275–7282. doi: 10.1021/bi960147wbi960147w. [DOI] [PubMed] [Google Scholar]
- Zhu W, Luo X, Puah CM, Tan X, Shen J, Gu J, Jiang H. The multiplicity, strength, and nature of the interaction of nucleobases with alkaline and alkaline earth metal cations: a density functional theory investigation. J Phys Chem A. 2004;108(18):4008–4018. doi: 10.1021/jp036911n. [DOI] [Google Scholar]
- Zimmer C, Luck G, Fritzsche H, Triebel H. DNA–copper(II) complex and the DNA conformation. Biopolymers. 1971;10(3):441–463. doi: 10.1002/bip.360100303. [DOI] [PubMed] [Google Scholar]