Abstract
The focus of the cell biology field is now shifting from characterizing cellular activities to organelle and molecular behaviors. This process accompanies the development of new biophysical visualization techniques that offer high spatial and temporal resolutions with ultra-sensitivity and low cell toxicity. They allow the biology research community to observe dynamic behaviors from scales of single molecules, organelles, cells to organoids, and even live animal tissues. In this review, we summarize these biophysical techniques into two major classes: the mechanical nanotools like dynamic force spectroscopy (DFS) and the optical nanotools like single-molecule and super-resolution microscopy. We also discuss their applications in elucidating molecular dynamics and functionally mapping of interactions between inter-cellular networks and intra-cellular components, which is key to understanding cellular processes such as adhesion, trafficking, inheritance, and division.
Keywords: Biophysical nanotools, Dynamic Force Spectroscopy (DFS), Single-molecule micrcoscopy, Super-resolution microscopy
Introduction
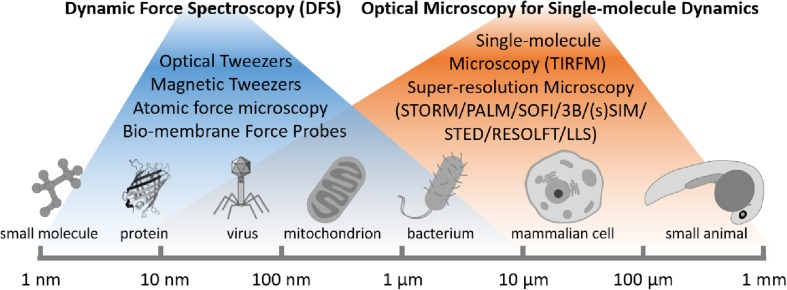
“Seeing is believing”. Most of our current knowledge about the molecular and sub-cellular dynamic processes has been obtained from single-molecule biophysical techniques at single molecule level (Fig. 1). Electron microscopy and crystallography are the most commonly used techniques to visualize conformations and characterize behaviors of purified proteins (Springer and Dustin 2012). However, these approaches only take snapshots of proteins’ stable states and lack real-time details of transient processes. Thus, they cannot be used to investigate the coupling of protein conformational changes with subsequent signaling events on live cells. Over the past two decades, dynamic force spectroscopy (DFS) (Dulin et al. 2015; Liu et al. 2015a; Neuman and Nagy 2008) performed using ultrasensitive force probes (Fig. 2a) such as atomic force microscopy (AFM, Fig. 2b), optical tweezer (OT, Fig. 2c), magnetic tweezer (MT, Fig. 2d), and biomembrane force probe (BFP, Fig. 2e) have provided various biomechanical approaches for manipulation, characterization and visualization of single receptor-ligand interaction and conformational change with controllable force (Liu et al. 2015a). With nanometer spatial, sub-millisecond temporal, and pico-newton force resolutions, these biomechanical approaches can be used to induce, follow and analyze single-molecule behaviors in real time, thus revealing individual molecular details inaccessible by conventional biochemistry methods based on ensemble averaging. As a result, they have elucidated the working mechanisms of many protein nanomachines (Fig. 2f–h) (Liu et al. 2015a). However, most discoveries are obtained from DFS experiments on purified protein systems. How these proteins function in the cellular environment remains largely unknown. Therefore, the live cell DFS technologies are in great demand to move the field forward.
Fig. 1.
The biophysical approaches for single-molecule dynamics
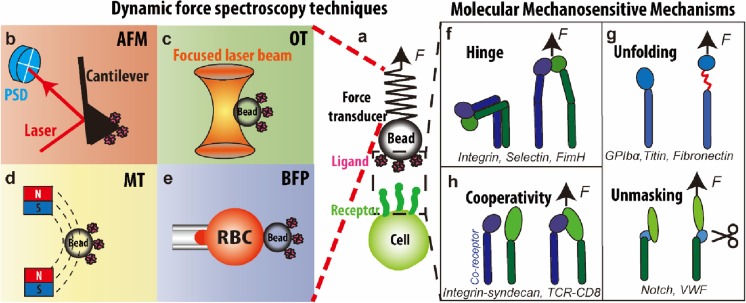
Fig. 2.
a–e Dynamic force spectroscopy techniques. a Generic dynamic force spectroscopy where a force is applied to the receptor–ligand bond spanning between a cell surface and a force transducer. Force is determined respectively by cantilever deflection (b), bead displacement (c), gradient of the magnetic field (d), and RBC deformation (e). b Atomic force microscopy (AFM) where force is applied to individual molecules tethered between a functionalized cantilever and a surface. c Optical tweezers (OT) where a protein-coated bead is held by a laser beam. d Magnetic tweezers (MT) where permanent/electrical magnets are used to manipulate a protein-coated magnetic bead. e Biomembrane force probe (BFP) where a protein-coated bead is attached to the apex of a micropipette-aspirated red blood cell (RBC). f–g Molecular mechanosensitive mechanisms. Mechanosensitive proteins contain a motif or motifs that can change structures in response to mechanical forces, giving rise to hinge movement (f), unfolding and unmasking (g), and multiple receptor cooperativity (h)
The second arm of single-molecule biophysical techniques is the optical imaging (Fig. 1). Cell biologists who study subcellular structures have been dissatisfied by the blurry images. This is due to the diffraction-limited spatial resolution of conventional optical imaging. Addressing this, super-resolution microscopy methods have recently been developed to break the diffraction barrier and push the spatial resolution from 300 to 10 nm. They are particularly being used to visualize structures of molecular machines and characterize their functions underlying important life and disease processes at nanoscale (Huang et al. 2010). Current super-resolution techniques include methods based on point spread function modulation (e.g., stimulated emission depletion (STED), reversible saturable optical linear fluorescence transitions (RESOLFT), structural illumination microscopy (SIM)) and optical stochastic fluctuation (e.g., stochastic optical reconstruction microscopy (STORM), photoactivated localization microscopy (PALM), super-resolution optical fluctuation imaging (SOFI), and Bayesian analysis of blinking and bleaching (3B)). However, most of these techniques require cell samples to be fixed and thin (2D) to achieve the optimal performance. Development of super resolution imaging techniques that are able to work on live cells and even 3D thick samples (> 100 μm) is the focus of the international progress.
Both mechanical and optical technologies are being evolved to offer the visualization capability in native cell environment. This review will summarize the recent progress of these biophysical techniques. It is worth of noting that most of current knowledge about the sub-cellular dynamic process has come from isolated cells cultured on artificial two-dimensional (2D) surfaces. Furthermore, it has been challenging to study multiple cells in their native and physiological environments, which are three-dimensional (3D) (Rios and Clevers 2018). The barriers mount when cells are hidden in a thick specimen, e.g., a developing metastatic tumor cell in deep tissue, a dividing bacterium in a thick biofilm, a cell circulating in organ-on-a-chip or being 3D printed into an engineered organ. Hereby, this review will also discuss the perspective of the 3D volumetric imaging, which provide untapped spatiotemporal resolutions for subcellular dynamics studies through deep tissue with low background noise and photo-toxicity.
Live cell dynamic force spectroscopy (DFS)
The DFS techniques are commonly used to manipulate and analyze force-induced receptor-ligand unbinding and protein domain unfolding (Fig. 2a). In a typical experiment, automated precise movement brings together ligands and receptors on the respective force probes (i.e., an AFM cantilever in Fig. 2b or a bead in Fig. 2c–e) and targets (i.e., a cell in Fig. 2a bottom). The contact time, area, and force are preset to allow for bond formation under controlled conditions. The following separation of the two surfaces applies a pico-newton (pN) level force to the receptor-ligand bond to induce mechanical changes in the molecules and modulate binding kinetics. These techniques are usually used to perform either force-ramp or force-clamp DFS. Force-ramp DFS exerts linearly increasing forces on the receptor-ligand bond with a range of force application rates, whereas force-clamp DFS exerts a range of constant forces on the bond. However, other modes of operation and manipulation can be used to exert different force waveforms on the molecular bond, for example, jump-and-ramp DFS (Evans et al. 2004) and cyclic force DFS (Kong et al. 2013; Li et al. 2016).
Recently, these DFS techniques have been upgraded with the capability to manipulate single living cells. This enables DFS to examine the live-cell dynamics in response to mechanical (Feng et al. 2017; Sawicka et al. 2017) and biochemical stimulations (Husson et al. 2011; Ju et al. 2017a). When combined with molecular signature analysis (Das et al. 2015; Fiore et al. 2014; Ju et al. 2016), DFS has been used to investigate simultaneous protein conformational or organizational changes in the same native cell context (Chen et al. 2017a). To directly correlate single-molecule dynamics with the triggered cell response requires running DFS analysis with concurrent imaging of intracellular signaling (e.g., calcium flux). We achieved this using a fluorescence BFP (Chen et al. 2015; Ju et al. 2016; Liu et al. 2014a) or micropipette-based adhesion assays (Francis and Heinrich 2018; Pryshchep et al. 2014). Others used fluorescence OT (Alieva et al. 2017; Favre-Bulle et al. 2017; Feng et al. 2017; Kim et al. 2009; Wang et al. 2005), AFM (Chaudhuri et al. 2009; Hu and Butte 2016), and MT (Matthews et al. 2006). This ability to manipulate protein dynamics and concurrently analyze the resulting receptor–ligand unbinding, receptor mechanical events, and intracellular signaling enables one to examine how cells interpret the mechanical stimulation then respond differently. Furthermore, a novel upgrade of BFP to a dual BFP allows one to analyze dual receptor crosstalk by quantifying the spatiotemporal requirements and functional consequences of the up- and down-stream signaling events (Ju et al. 2017b). In contrast to conventional biochemistry and cell biology methods that are usually population-averaged and not imaged in real-time, the dual BFP provides precise control and quantitative readouts in both mechanical and chemical terms, which is particularly suited for juxtacrine signaling and mechanosensing studies.
Overall, the live cell DFS led to many discoveries on inner-workings of many protein nanomachines in the context of mechanobiology including T cell receptors (Liu et al. 2014a), integrin (Chen et al. 2012; Ju et al. 2018; Passam et al. 2018; Xu et al. 2018), syndecan (Fiore et al. 2014), notch (Luca et al. 2017), glycoprotein Ibα (Deng et al. 2016; Ju et al. 2016; Ju et al. 2015), and von Willebrand factor (Butera et al. 2018; Ju et al. 2013). Their dynamic molecular mechanisms can be classified into three categories:
-
(i)
Hinge movement (Fig. 2f). For proteins or their subunits that consist of two or more distinct globular domains connected by a hinge region, such as integrin (Luo and Springer 2006), selectin (Somers et al. 2000), and FimH (Le Trong et al. 2010), force can relieve the conformational constraints and allosterically promote hinge opening. Take the integrin as an example, numerous studies have demonstrated that force applied through ligand binding facilitates the conformational switch from a bent to an extended conformation (Chen et al. 2017b; Springer and Dustin 2012).
-
(ii)
Unfolding and unmasking (Fig. 2g). Force can unfold and unmask a specific protein domain to expose cryptic cleaving, binding, or enzymatic sites. For example, several recent studies have demonstrated that force can unfold distinct domains of GPIb to mediate signal transduction (Deng et al. 2016; Ju et al. 2016; Zhang et al. 2015). Force-induced exposure of a binding site has been shown for intracellular adaptor proteins such as talin (del Rio et al. 2009) as well as extracellular proteins such as fibronectin (Kong et al. 2009; Smith et al. 2007) and VWF (Fu et al. 2017; Ju et al. 2013). Force-induced exposure of an enzymatic cleavage site has also been shown for Notch activation (Stephenson and Avis 2012) and VWF proteolysis (Wu et al. 2010; Zhang et al. 2009).
-
(iii)
Cooperativity (Fig. 2h). External force facilitates the cooperative binding of two receptors to one ligand. In a dual receptor system, for example, integrin and syndecan bind to their mutual ligand Thy-1 (Fiore et al. 2014). The trimolecular bond displays a unique bond-strengthening phenomenon, in which increasing force abruptly stiffens the complex by bridging both receptors through one ligand. A similar mechanism is also implied in the TCR/CD8 co-receptor system (Jiang et al. 2011).
Optical microscopy for single-molecule dynamics
Optical microscopy has been widely utilized to observe the protein and membrane dynamics on single cells. With the advance of fluorescent labeling techniques, such as genetically expressed fluorescent proteins, tag/ligand labeling system, and specific organelle-targeted organic dyes (Valm et al. 2017), cell behavior within a millisecond timeframe can be described at single-molecule level. Remarked by the 2014 Nobel Prize in Chemistry, the development of single-molecule and super-resolution microscopy enables the visualization of subcellular structures at the resolutions down to tens of nanometers scale and the dynamic information at milliseconds level (Liu et al. 2014b; Valm et al. 2017). Here, we summarize these single-molecule and super-resolution imaging methods.
Single-molecule microscopy
Thanks to its high signal-to-noise ratio and localization accuracy, the total internal reflection fluorescence (TIRF) microscopy is widely used to observe dynamic processes of individual molecules and their assemblies. Using the TIRF, fluorophores within a short axial distance above surface (100~200 nm) can be selectively excited to achieve higher signal-to-noise ratio, lower photo-toxicity, and shorter exposure time than conventional fluorescence microscopies. In the biophysics field, TIRF is an effective approach for tracking molecular motors (myosin, kinesin and dynein) while they move along the cytoskeleton with < 1.5 nm precision at video rates (Sun et al. 2007; Yildiz et al. 2003). Multiple single-molecule in vitro assays have been developed around TIRF to characterize motor conformations and interactions with cytoskeleton filaments under various biochemical conditions (Guan et al. 2017; Shen et al. 2016).
Moreover, TIRF has recently been used to investigate the movements of single molecules in cells. The single-particle tracking, single-molecule FRET, and the correlation-based approaches have been developed. For example, the TIRF has been used to demonstrate motor driven intracellular membrane dynamics (such as vesicles transport, deformation, tubulation and reformation) and their important roles for cargo transportation and organelle biogenesis (Su et al. 2016). In recent studies, it reveals the essential role of kinesin1 motor driven nanotube dynamics in the autophagic lysosome reformation during late-stage autophagy (Du et al. 2016) and mitochondrial network remodeling in the subzone of the cell plasma (Wang et al. 2015). Notably, a novel synthetic biology method has been developed to reconstitute purified organelles with different genetic modification, artificial liposomes with different lipid component, recombinant motor proteins, and polymerized microtubule filaments (Chen et al. 2018b). The big advantage for such in vitro reconstitution system is the feasibility of manipulating small molecules like ATP, GTP, and ions which are impossible in cultured cell line systems. Combined with TIRF imaging, we can investigate the molecular mechanisms of membrane and protein dynamics from in situ cell environment to in vitro systems reconstituted from purified organelles and artificial liposomes/vesicles.
Super-resolution microscopy
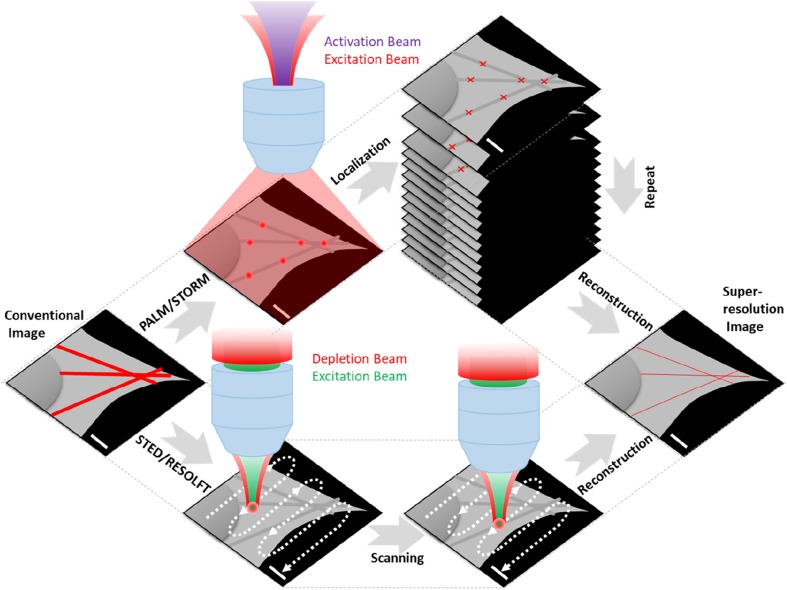
Advances in super-resolution fluorescence imaging have enabled unprecedented insights on the inner life of a cell under the diffraction-limit (200~300 nm) resolution (Huang et al. 2010). To improve lateral (and more recently axial) resolution, several localization-based methods, such as PALM, STORM, and SOFI, reconstruct fluorescent images from large datasets consisting of stochastically sparse and localized centroids, to achieve the typical lateral resolutions of 20~40 nm (Sahl et al. 2017) (Fig. 3, top). Another set of super-resolution microscopies is based on the point spread function modification, e.g., STED, RESOLFT, SIM, resulting a lateral resolution of 50–100 nm (Sahl et al. 2017) (Fig. 3, bottom). These super-resolution microscopies are sufficient to visualize precise subcellular structures and membrane protein interactions at the single molecule level.
Fig. 3.
Principle of single-molecule localization and point-spread-function modification based super-resolution microscopies. (Top) A weak violet light activates only a few fluorescent molecules whose distance is much longer than the diffraction limit of 200 nm. A strong red light is used to turn them on and off frame-by-frame until photobleaching; (bottom) A high-power laser (red) that could quench the fluorophores is used to modify the excitation laser (green) beam to a pretty tiny volume < 200 nm and then scan the sample point-by-point
However, to see fine structural detail with high spatial resolution together with fast dynamic biological tracking with high temporal resolution is mutually contradicted as temporal resolution is typically sacrificed to achieve higher spatial resolution and vice versa. Developed by Betzig et al., the lattice light sheet (LLS) microscope uses a light sheet generated by a 2D optical lattice for confined illumination. This is an extraordinary example of super-resolution imaging method that can resolve both dynamic processes (90 ms) and subcellular structures (~ 400 nm) (Chen et al. 2014). With the LLS microscope, for the first time, scientists are able to pinpoint the subcellular organelle interactome dynamics in the living cells (Valm et al. 2017) and even in the living animals (Liu et al. 2018).
Recently, new imaging techniques, such as super-resolution by polarization demodulation (SPoD) (Hafi et al. 2014) and super-resolution dipole orientation mapping (SDOM) (Zhanghao et al. 2016) that use information on fluorophore dipole orientation, have been employed to overcome the diffraction limit and produce super-resolution images with the information of molecular orientation on the membrane. Notably, SDOM can map F-actin orientations and membrane proteins on different organelles juxtaposed on super resolution cell images. This new technique may potentially be used to characterize individual protein conformational changes with high spatial and temporal resolutions at the same time.
Next-generation imaging methods to visualize cells in native state
New nanoprobes for super-resolution functional imaging
The development of super resolution imaging methods has created a need for robust probes that can track single molecules and image subcellular structures in real time. Rapid advances in material sciences provide a large collection of near-infrared (NIR) luminescent probes, such as upconversion nanocrystals (Lu et al. 2013; Zhao et al. 2013), fluorescent nanodiamonds (Reineck et al. 2018), and fluorescent proteins (Ghodke et al. 2016). They are super bright, produce non-bleaching, non-blinking emissions, and can be precisely tuned to emit different lifetime signatures (take turns to shine) and to show distinct colors at NIR-IR range. These properties are ideal for simultaneous long-term imaging of multiple analytes deep within tissues and under background-free conditions. These developments proved the concept of single-molecule imaging with deep tissue penetration, free of optical alignment of multiple laser beams. In addition, time-gated microscopy has also been developed to overcome the autofluorescence of conventional in vivo imaging (Miao et al. 2017). Considering their time-gated luminescence properties (Lu et al. 2013), the upconversion nanocrystals based NIR-IR super resolution microscopy have a promising future that can solve the critical bottlenecks of deep tissue imaging as one stone two birds. The new library of Super Dots®, τ-dots, and Hyper Dots provide multi-photon luminescent probes, through the so-called “transparent biology window” (650–1350 nm), which is vitally important to obtain maximum penetration depth for deep tissue imaging (Chen et al. 2018a).
New modalities for super-resolution imaging in three dimensions (3D)
Most of our current knowledge about sub-cellular dynamic processes has been obtained from imaging isolated cells, cultured on a two-dimensional (2D) slide or petri dish. An ultimate challenge for cell biology research is to study molecular and cellular behavior inside their 3D natural environments. Current super-resolution imaging methods like STORM and PALM are relatively limited to thin samples (< 50 μm) as a reliable separation against a diffusive background is needed. To address this, several leading groups have been working on integrating various optical facilities to enable super resolution cell imaging in thick samples (> 100 μm) or “Volumetric Imaging.” In this regard, Betzig et al. have developed Bessel beam LLS microscope and integrated adaptive optics in both excitation and emission paths so as to correct the sample-induced aberrations (Chen et al. 2014). This exciting new LLS microscope for the first time enables video-rate super resolution imaging in 3D physiological environment (Liu et al. 2018). In addition, Hell et al. have developed the parallel fast super-resolution microscopy to decode the principle underlying vital biological processes (Balzarotti et al. 2017; Chmyrov et al. 2013). In the conventional microscopy domain, Zong et al. designed a miniaturized two-photon microscope with improved speed (40 Hz) and spatial resolution (0.64 μm), and utilized it to resolve activity at single dendritic spines in freely-moving animals (Zong et al. 2017; Zong et al. 2015). Chen et al. developed a new Hessian SIM for fast, long-term, super-resolved rapid imaging of moving vesicles or loops in the endoplasmic reticulum with a spatiotemporal resolution of 88 nm and 188 Hz (Huang et al. 2018).
Discussion
This review summarizes the latest biophysical technologies for biologists at levels of single molecules, cells, and tissues. The mechanical techniques DFS have strength in manipulating molecular behaviors whereas the optical techniques super resolution microscopies harness the power of directly visualizing biological processes. Currently, biophysicists are trying to combine the two approaches to investigate molecular biology with both mechanical control and optical information (Heller et al. 2013). Researchers have also made efforts to apply DFS in vivo. The IR optical tweezers have been developed to trap and manipulate red blood cells in capillaries of living mice (Zhong et al. 2013a; Zhong et al. 2013b), and more recently trap 55 μm otoliths in larval zebrafish that causes tail movements (Favre-Bulle et al. 2017). The magnetic tweezers have been combined with the magnetic nanoparticles for the control of cerebral ischemia and stroke investigation in living mice (Jia et al. 2017). Furthermore, a series of optomechanical and photothermal actuator nanoparticles become available (Guo et al. 2018; Liu et al. 2015b), providing new avenues of cell manipulation when combined with DFS in future. Taken together, we could foresee the DFS techniques will push towards native state molecular and cellular analysis.
On the imaging side, super resolution microscopies are being combined with multifunctional imaging probes, i.e., Super Dots towards a future vision of a complete, progressive “street view” of intra-cellular traffic. Importantly, it will enable functional imaging of system-level organelle interactions on a single cell. On one hand, it will monitor the interactions between at least five membrane-bound organelles, e.g., cell-membrane bound integrins, endoplasmic reticulum, Golgi complex, mitochondria, and lysosomes (Valm et al. 2017). On the other hand, it will simultaneously visualize cellular responses to the local environment changes in mechanical shear stress, pH value, viscosity, temperature, and membrane potential (ref). Taken together, these optical approaches will be used to examine subcellular processes such as energy metabolism, trafficking, inheritance, autophagy, and cell division. In the near future, scientists could generate a “street-view” version of functional maps upon how membrane receptors and organelles respond under different nutrient conditions in living cells. By using spectral barcoding and/or combinatorial labeling, it may also be possible to study 3D interactions of a number of proteins simultaneously in the same cell and to understand how they coordinate with each other to achieve complex biological functions.
However, advances in instrumentation alone are not sufficient to enable studies in native physiological environments. This is largely owing to the gap between real animal tissues and cell lines grown in laboratories that are widely used for years (Vlachogiannis et al. 2018). It is worth of noting that these cell lines often do not mimic the tissues they originally come from. The results from animals and humans are also not always comparable. In this regard, several alternative in vitro methods have been developed. The first is the organ-on-a-chip that grows tissues on a microfluidic chip to simulate the microarchitectures and functions of living organs like the kidney, lungs, bone morrow, intestine, and others (Bhatia and Ingber 2014; Takebe et al. 2017). The other one is the organoids, announced as the Method of the Year 2017 by Nature Methods (Eisenstein 2018). These clusters of cells organize themselves into mini versions of our organs, including the liver, lungs, and even brain cells. The organoids are usually grown in a 3D hydrogel that allows them to develop three dimensionally; thereafter, they fill the gap between 3D animal tissues and 2D cultured cell lines by overcoming the current limitation in life sciences and clinic applications. Taken together, these powerful approaches allow scientists to better study the development and treatment for human diseases at organ levels, and could be implemented in personalized medicine programs (Vlachogiannis et al. 2018).
In our view, the time is ripe to combine the biophysical techniques summarized in this review with these 3D human-mimicked models (Takebe et al. 2017). It will certainly facilitate the study of cell functions and heterogeneity in an ideal multicellular microenvironment that mimics human-specific physiology and pathophysiology and helps to identify therapeutic targets. In the meantime, the demand of biophysical techniques for physiological imaging of subcellular dynamics in native physiological environments, through deep tissue or over a large population of cellular networks in 3D in vitro models will definitely accelerate the design, fabrication, characterization and validation of new luminescent probes, photonics imaging techniques and organ-on-chip devices. This would also facilitate the translation into a single, practical point-of-care device to aid disease diagnostics and improve healthcare outcomes.
Acknowledgments
We thank Shaun P. Jackson, Dayong Jin, and their labs for helpful discussion.
Funding
This work was supported by National Heart Foundation of Australia Postdoctoral Fellowship 101285 (LJ), Sydney Local Health District - Annual Health Research Infrastructure Award (LJ), The Royal College of Pathologists of Australasia Kanematsu Research Award (LJ).
Author contributions
Q.P.S. and L.A.J. contributed equally, prepared figures, and wrote and edited the manuscript together.
Conflict of interest
Qian Peter Su declares that he has no conflict of interest. Lining Arnold Ju declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Contributor Information
Qian Peter Su, Email: qian.su@uts.edu.au.
Lining Arnold Ju, Email: arnold.ju@sydney.edu.au.
References
- Alieva, N.O., Efremov, A.K., Hu, S., Oh, D., Chen, Z., Natarajan, M., Ong, H.T., Jegou, A., Romet-Lemonne, G., Groves, J.T., et al. (2017). Force dependence of filopodia adhesion: involvement of myosin II and formins. bioRxiv [DOI] [PMC free article] [PubMed]
- Balzarotti F, Eilers Y, Gwosch KC, Gynna AH, Westphal V, Stefani FD, Elf J, Hell SW. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science. 2017;355:606–612. doi: 10.1126/science.aak9913. [DOI] [PubMed] [Google Scholar]
- Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Butera Diego, Passam Freda, Ju Lining, Cook Kristina M., Woon Heng, Aponte-Santamaría Camilo, Gardiner Elizabeth, Davis Amanda K., Murphy Deirdre A., Bronowska Agnieszka, Luken Brenda M., Baldauf Carsten, Jackson Shaun, Andrews Robert, Gräter Frauke, Hogg Philip J. Autoregulation of von Willebrand factor function by a disulfide bond switch. Science Advances. 2018;4(2):eaaq1477. doi: 10.1126/sciadv.aaq1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O, Parekh SH, Lam WA, Fletcher DA. Combined atomic force microscopy and side-view optical imaging for mechanical studies of cells. Nat Meth. 2009;6:383–387. doi: 10.1038/nmeth.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Lou J, Evans EA, Zhu C. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J Cell Biol. 2012;199:497–512. doi: 10.1083/jcb.201201091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, 3rd, Liu Z, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu B, Ju L, Hong J, Ji Q, Chen W, Zhu C (2015) Fluorescence biomembrane force probe: concurrent quantitation of receptor-ligand kinetics and binding-induced intracellular signaling on a single cell. J Vis Exp. 10.3791/52975 [DOI] [PMC free article] [PubMed]
- Chen Y, Ju L, Rushdi M, Ge C, Zhu C. Receptor-mediated cell mechanosensing. Mol Biol Cell. 2017;28:3134–3155. doi: 10.1091/mbc.e17-04-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lee H, Tong H, Schwartz M, Zhu C. Force regulated conformational change of integrin alphaVbeta3. Matrix Biol. 2017;60-61:70–85. doi: 10.1016/j.matbio.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Wang F, Wen S, Su PQ, Wu MCL, Liu Y, Wang B, Li D, Shan X, Kianinia M, Aharonovich I, Toth M, Jackson SP, Xi P, Jin D (2018a) Multi-photon near-infrared emission saturation nanoscopy using upconversion nanoparticles. Nat Commun. 10.1038/s41467-018-05842-w [DOI] [PMC free article] [PubMed]
- Chen Y, Su QP, Sun Y, Yu L. Visualizing autophagic lysosome reformation in cells using in vitro reconstitution systems. Current Protocols Cell Biology. 2018;78:11.24.11–11.24.15. doi: 10.1002/cpcb.44. [DOI] [PubMed] [Google Scholar]
- Chmyrov A, Keller J, Grotjohann T, Ratz M, d'Este E, Jakobs S, Eggeling C, Hell SW. Nanoscopy with more than 100,000 'doughnuts'. Nat Methods. 2013;10:737–740. doi: 10.1038/nmeth.2556. [DOI] [PubMed] [Google Scholar]
- Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, Brady SK, Wang JH, Wagner G, Reinherz EL, et al. Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci U S A. 2015;112:1517–1522. doi: 10.1073/pnas.1424829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Xu Y, Chen W, Paul DS, Syed AK, Dragovich MA, Liang X, Zakas P, Berndt MC, Di Paola J, et al. Platelet clearance via shear-induced unfolding of a membrane mechanoreceptor. Nat Commun. 2016;7:12863. doi: 10.1038/ncomms12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Su QP, Chen Y, Zhu Y, Jiang D, Rong Y, Zhang S, Zhang Y, Ren H, Zhang C, et al. Kinesin 1 drives autolysosome tubulation. Dev Cell. 2016;37:326–336. doi: 10.1016/j.devcel.2016.04.014. [DOI] [PubMed] [Google Scholar]
- Dulin D, Berghuis BA, Depken M, Dekker NH. Untangling reaction pathways through modern approaches to high-throughput single-molecule force-spectroscopy experiments. Curr Opin Struct Biol. 2015;34:116–122. doi: 10.1016/j.sbi.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Eisenstein M. Organoids: the body builders. Nat Methods. 2018;15:19. doi: 10.1038/nmeth.4538. [DOI] [Google Scholar]
- Evans E, Leung A, Heinrich V, Zhu C. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc Natl Acad Sci. 2004;101:11281–11286. doi: 10.1073/pnas.0401870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre-Bulle IA, Stilgoe AB, Rubinsztein-Dunlop H, Scott EK. Optical trapping of otoliths drives vestibular behaviours in larval zebrafish. Nat Commun. 2017;8:630. doi: 10.1038/s41467-017-00713-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Brazin KN, Kobayashi E, Mallis RJ, Reinherz EL, Lang MJ. Mechanosensing drives acuity of alphabeta T-cell recognition. Proc Natl Acad Sci U S A. 2017;114:E8204–E8213. doi: 10.1073/pnas.1703559114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore VF, Ju L, Chen Y, Zhu C, Barker TH. Dynamic catch of a Thy-1–α5β1+syndecan-4 trimolecular complex. Nat Commun. 2014;5:4886. doi: 10.1038/ncomms5886. [DOI] [PubMed] [Google Scholar]
- Francis Emmet A., Heinrich Volkmar. Extension of chemotactic pseudopods by nonadherent human neutrophils does not require or cause calcium bursts. Science Signaling. 2018;11(521):eaal4289. doi: 10.1126/scisignal.aal4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Jiang Y, Yang D, Scheiflinger F, Wong WP, Springer TA. Flow-induced elongation of von Willebrand factor precedes tension-dependent activation. Nat Commun. 2017;8:324. doi: 10.1038/s41467-017-00230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodke H, Caldas VE, Punter CM, van Oijen AM, Robinson A. Single-molecule specific mislocalization of red fluorescent proteins in live Escherichia coli. Biophys J. 2016;111:25–27. doi: 10.1016/j.bpj.2016.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Zhang L, Su QP, Mickolajczyk KJ, Chen GY, Hancock WO, Sun Y, Zhao Y, Chen Z. Crystal structure of Zen4 in the apo state reveals a missing conformation of kinesin. Nat Commun. 2017;8:14951. doi: 10.1038/ncomms14951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Bishop CJ, Meyer RA, Wilson DR, Olasov L, Schlesinger DE, Mather PT, Spicer JB, Elisseeff JH, Green JJ. Entanglement-based thermoplastic shape memory polymeric particles with photothermal actuation for biomedical applications. ACS Appl Mater Interfaces. 2018;10:13333–13341. doi: 10.1021/acsami.8b01582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafi N, Grunwald M, van den Heuvel LS, Aspelmeier T, Chen JH, Zagrebelsky M, Schutte OM, Steinem C, Korte M, Munk A, et al. Fluorescence nanoscopy by polarization modulation and polarization angle narrowing. Nat Methods. 2014;11:579–584. doi: 10.1038/nmeth.2919. [DOI] [PubMed] [Google Scholar]
- Heller I, Sitters G, Broekmans OD, Farge G, Menges C, Wende W, Hell SW, Peterman EJ, Wuite GJ. STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nat Methods. 2013;10:910–916. doi: 10.1038/nmeth.2599. [DOI] [PubMed] [Google Scholar]
- Hu KH, Butte MJ. T cell activation requires force generation. J Cell Biol. 2016;213:535–542. doi: 10.1083/jcb.201511053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Xiaoshuai, Fan Junchao, Li Liuju, Liu Haosen, Wu Runlong, Wu Yi, Wei Lisi, Mao Heng, Lal Amit, Xi Peng, Tang Liqiang, Zhang Yunfeng, Liu Yanmei, Tan Shan, Chen Liangyi. Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nature Biotechnology. 2018;36(5):451–459. doi: 10.1038/nbt.4115. [DOI] [PubMed] [Google Scholar]
- Husson J, Chemin K, Bohineust A, Hivroz C, Henry N. Force generation upon T cell receptor engagement. PLoS One. 2011;6:e19680. doi: 10.1371/journal.pone.0019680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia JM, Chowdary PD, Gao X, Ci B, Li W, Mulgaonkar A, Plautz EJ, Hassan G, Kumar A, Stowe AM, et al. Control of cerebral ischemia with magnetic nanoparticles. Nat Methods. 2017;14:160–166. doi: 10.1038/nmeth.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Huang J, Edwards LJ, Liu B, Zhang Y, Beal CD, Evavold BD, Zhu C. Two-stage cooperative T cell receptor-peptide major histocompatibility complex-CD8 trimolecular interactions amplify antigen discrimination. Immunity. 2011;34:13–23. doi: 10.1016/j.immuni.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Dong J-F, Cruz MA, Zhu C. The N-terminal flanking region of the A1 domain regulates the force-dependent binding of von Willebrand factor to platelet glycoprotein Ibα. J Biol Chem. 2013;288:32289–32301. doi: 10.1074/jbc.M113.504001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Lou J, Chen Y, Li Z, Zhu C. Force-induced unfolding of leucine-rich repeats of glycoprotein Ibα strengthens ligand interaction. Biophys J. 2015;109:1781–1784. doi: 10.1016/j.bpj.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Chen Y, Xue L, Du X, Zhu C (2016) Cooperative unfolding of distinctive mechanoreceptor domains transduces force into signals. ELife 5:e15447 [DOI] [PMC free article] [PubMed]
- Ju L, Chen Y, Li K, Yuan Z, Liu B, Jackson SP, Zhu C. Dual biomembrane force probe enables single-cell mechanical analysis of signal crosstalk between multiple molecular species. Sci Rep. 2017;7:14185. doi: 10.1038/s41598-017-13793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Lining, Chen Yunfeng, Rushdi Muaz Nik, Chen Wei, Zhu Cheng. The Immune Synapse. New York, NY: Springer New York; 2017. Two-Dimensional Analysis of Cross-Junctional Molecular Interaction by Force Probes; pp. 231–258. [DOI] [PubMed] [Google Scholar]
- Ju L, McFadyen JD, Al-Daher S, Alwis I, Chen Y, Tonnesen LL, Maiocchi S, Coulter B, Calkin AC, Felner EI, et al. Compression force sensing regulates integrin alphaIIbbeta3 adhesive function on diabetic platelets. Nat Commun. 2018;9:1087. doi: 10.1038/s41467-018-03430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Takeuchi K, Sun ZY, Touma M, Castro CE, Fahmy A, Lang MJ, Wagner G, Reinherz EL. The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem. 2009;284:31028–31037. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, F., Li, Z., Parks, W.M., Dumbauld, D.W., García, A.J., Mould, A.P., Humphries, M.J., and Zhu, C. (2013). Cyclic mechanical reinforcement of integrin-ligand interactions. Mol Cell [DOI] [PMC free article] [PubMed]
- Le Trong I, Aprikian P, Kidd BA, Forero-Shelton M, Tchesnokova V, Rajagopal P, Rodriguez V, Interlandi G, Klevit R, Vogel V, et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like β sheet twisting. Cell. 2010;141:645–655. doi: 10.1016/j.cell.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kong F, Zhu C. A model for cyclic mechanical reinforcement. Sci Rep. 2016;6:35954. doi: 10.1038/srep35954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell. 2014;157:357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xing D, Su QP, Zhu Y, Zhang J, Kong X, Xue B, Wang S, Sun H, Tao Y, Sun Y (2014b) Super-resolution imaging and tracking of protein–protein interactions in sub-diffraction cellular space. Nat Commun 5:4443 [DOI] [PMC free article] [PubMed]
- Liu B, Chen W, Zhu C. Molecular force spectroscopy on cells. Annu Rev Phys Chem. 2015;66:427–451. doi: 10.1146/annurev-physchem-040214-121742. [DOI] [PubMed] [Google Scholar]
- Liu Z, Liu Y, Chang Y, Seyf HR, Henry A, Mattheyses AL, Yehl K, Zhang Y, Huang Z, Salaita K (2015b) Nanoscale optomechanical actuators for controlling mechanotransduction in living cells. Nat Meth:1–8 [DOI] [PMC free article] [PubMed]
- Liu TL, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J et al (2018) Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science 360(6386):eaaq1392 [DOI] [PMC free article] [PubMed]
- Lu Y, Zhao J, Zhang R, Liu Y, Liu D, Goldys EM, Yang X, Xi P, Sunna A, Lu J, et al. Tunable lifetime multiplexing using luminescent nanocrystals. Nat Photonics. 2013;8:32–36. doi: 10.1038/nphoton.2013.322. [DOI] [Google Scholar]
- Luca, V.C., Kim, B.C., Ge, C., Kakuda, S., Wu, D., Roein-Peikar, M., Haltiwanger, R.S., Zhu, C., Ha, T., and Garcia, K.C. (2017). Notch-jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science [DOI] [PMC free article] [PubMed]
- Luo BH, Springer TA. Integrin structures and conformational signaling. Curr Opin Cell Biol. 2006;18:579–586. doi: 10.1016/j.ceb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BD, Overby DR, Mannix R, Ingber DE. Cellular adaptation to mechanical stress: role of integrins, Rho, cytoskeletal tension and mechanosensitive ion channels. J Cell Sci. 2006;119:508–518. doi: 10.1242/jcs.02760. [DOI] [PubMed] [Google Scholar]
- Miao Q, Xie C, Zhen X, Lyu Y, Duan H, Liu X, Jokerst JV, Pu K. Molecular afterglow imaging with bright, biodegradable polymer nanoparticles. Nat Biotechnol. 2017;35:1102–1110. doi: 10.1038/nbt.3987. [DOI] [PubMed] [Google Scholar]
- Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Meth. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passam F, Chiu J, Ju L, Pijning A, Jahan Z, Mor-Cohen R, Yeheskel A, Kolsek K, Tharichen L, Aponte-Santamaria C et al (2018) Mechano-redox control of integrin de-adhesion. eLife 7:e34843 [DOI] [PMC free article] [PubMed]
- Pryshchep S, Zarnitsyna VI, Hong J, Evavold BD, Zhu C. Accumulation of serial forces on TCR and CD8 frequently applied by agonist antigenic peptides embedded in MHC molecules triggers calcium in T cells. J Immunol. 2014;193:68–76. doi: 10.4049/jimmunol.1303436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineck P, Lau DWM, Wilson ER, Nunn N, Shenderova OA, Gibson BC. Visible to near-IR fluorescence from single-digit detonation nanodiamonds: excitation wavelength and pH dependence. Sci Rep. 2018;8:2478. doi: 10.1038/s41598-018-20905-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios AC, Clevers H. Imaging organoids: a bright future ahead. Nat Methods. 2018;15:24. doi: 10.1038/nmeth.4537. [DOI] [PubMed] [Google Scholar]
- Sahl SJ, Hell SW, Jakobs S. Fluorescence nanoscopy in cell biology. Nat Rev Mol Cell Biol. 2017;18:685. doi: 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- Sawicka A, Babataheri A, Dogniaux S, Barakat AI, Gonzalez-Rodriguez D, Hivroz C, Husson J. Micropipette force probe to quantify single-cell force generation: application to T-cell activation. Mol Biol Cell. 2017;28:3229–3239. doi: 10.1091/mbc.e17-06-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Zhang N, Zheng S, Zhang WB, Zhang HM, Lu Z, Su QP, Sun Y, Ye K, Li XD. Calmodulin in complex with the first IQ motif of myosin-5a functions as an intact calcium sensor. Proc Natl Acad Sci U S A. 2016;113:E5812–e5820. doi: 10.1073/pnas.1607702113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers WS, Tang J, Shaw GD, Camphausen RT. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell. 2000;103:467–479. doi: 10.1016/S0092-8674(00)00138-0. [DOI] [PubMed] [Google Scholar]
- Springer TA, Dustin ML. Integrin inside-out signaling and the immunological synapse. Curr Opin Cell Biol. 2012;24:107–115. doi: 10.1016/j.ceb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson NL, Avis JM. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proc Natl Acad Sci U S A. 2012;109:E2757–E2765. doi: 10.1073/pnas.1205788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su QP, Du W, Ji Q, Xue B, Jiang D, Zhu Y, Lou J, Yu L, Sun Y. Vesicle size regulates nanotube formation in the cell. Sci Rep. 2016;6:24002. doi: 10.1038/srep24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Schroeder HW, 3rd, Beausang JF, Homma K, Ikebe M, Goldman YE. Myosin VI walks “wiggly” on actin with large and variable tilting. Mol Cell. 2007;28:954–964. doi: 10.1016/j.molcel.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe T, Zhang B, Radisic M. Synergistic engineering: organoids meet organs-on-a-Chip. Cell Stem Cell. 2017;21:297–300. doi: 10.1016/j.stem.2017.08.016. [DOI] [PubMed] [Google Scholar]
- Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott-Schwartz J. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546:162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Du W, Su QP, Zhu M, Feng P, Li Y, Zhou Y, Mi N, Zhu Y, Jiang D, et al. Dynamic tubulation of mitochondria drives mitochondrial network formation. Cell Res. 2015;25:1108–1120. doi: 10.1038/cr.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Botvinick E, Zhao Y, Berns M, Usami S, Tsien R, Chien S. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- Wu T, Lin J, Cruz MA, Dong JF, Zhu C. Force-induced cleavage of single VWFA1A2A3 tridomains by ADAMTS-13. Blood. 2010;115:370–378. doi: 10.1182/blood-2009-03-210369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XR, Wang Y, Adili R, Ju L, Spring, CM, Jin, JW, Yang H, Neves MAD, Chen P, Yang Y, Lei X, Chen Y, Gallant RC, Xu M, Zhang H, Song J, Ke P, Zhang D, Carrim N, Yu SY, Zhu G, She YM, Cyr T, Fu W, Liu G, Connelly PW, Rand ML, Adeli K, Freedman J, Lee JE, Tso P, Marchese P, Davidson WS, Jackson SP, Zhu C, Ruggeri ZM, Ni H (2018) Apolipoprotein A-IV binds aIIbß3 integrin and inhibits thrombosis. Nat Commun. 10.1038/s41467-018-05806-0 [DOI] [PMC free article] [PubMed]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- Zhang W, Deng W, Zhou L, Xu Y, Yang W, Liang X, Wang Y, Kulman JD, Zhang XF, Li R. Identification of a juxtamembrane mechanosensitive domain in the platelet mechanosensor glycoprotein Ib-IX complex. Blood. 2015;125:562–569. doi: 10.1182/blood-2014-07-589507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanghao K, Chen L, Yang X-S, Wang M-Y, Jing Z-L, Han H-B, Zhang MQ, Jin D, Gao J-T, Xi P. Super-resolution dipole orientation mapping via polarization demodulation. Light: Sci Applications. 2016;5:e16166–e16166. doi: 10.1038/lsa.2016.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Jin D, Schartner EP, Lu Y, Liu Y, Zvyagin AV, Zhang L, Dawes JM, Xi P, Piper JA, et al. Single-nanocrystal sensitivity achieved by enhanced upconversion luminescence. Nat Nanotechnol. 2013;8:729–734. doi: 10.1038/nnano.2013.171. [DOI] [PubMed] [Google Scholar]
- Zhong MC, Gong L, Zhou JH, Wang ZQ, Li YM. Optical trapping of red blood cells in living animals with a water immersion objective. Opt Lett. 2013;38:5134–5137. doi: 10.1364/OL.38.005134. [DOI] [PubMed] [Google Scholar]
- Zhong MC, Wei XB, Zhou JH, Wang ZQ, Li YM. Trapping red blood cells in living animals using optical tweezers. Nat Commun. 2013;4:1768. doi: 10.1038/ncomms2786. [DOI] [PubMed] [Google Scholar]
- Zong W, Wu R, Li M, Hu Y, Li Y, Li J, Rong H, Wu H, Xu Y, Lu Y, et al. Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nat Methods. 2017;14:713–719. doi: 10.1038/nmeth.4305. [DOI] [PubMed] [Google Scholar]
- Zong W, Zhao J, Chen X, Lin Y, Ren H, Zhang Y, Fan M, Zhou Z, Cheng H, Sun Y, et al. Large-field high-resolution two-photon digital scanned light-sheet microscopy. Cell Res. 2015;25:254–257. doi: 10.1038/cr.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]