Abstract
The objective of this study was to perform genetic diversity analysis of 13 strains isolated from South Korean foods by multilocus sequence typing (MLST). For typing, seven housekeeping loci (atpA, dnaA, dnaK, gyrB, pheS, pyrG, and rpoA) were selected, amplified and analyzed. Fifty-one polymorphic sites varying from 1 to 22 in each species were identified. Thirteen sequence types were generated with allele numbers ranged from 2 to 10. The overall relationship between strains was assessed by unweighted pair group method with arithmetic mean dendrogram and minimum spanning tree. In addition, combined spits tree analysis revealed intragenic recombination. No clear relationship was observed between the isolation sources and strains. The developed MLST scheme enhanced our knowledge of the population diversity of Leu. citreum strains and will be used further for the selection of industrially important strain.
Keywords: Lactic acid bacteria, Leuconostoc citreum, MLST, Sequence type, Alleles
Introduction
The genus Leuconostoc consisted of 24 different species characterized as Gram-positive, non-motile, catalase-negative and facultative anaerobes (Kot et al., 2014). This genus has been found to be associated with dairy products, meat, fish and poultry and various other plant-originated materials (Björkroth and Holzapfel, 2006). It has also been used as starter cultures in fermented milk and vegetables. The important species of this genus is Leuconostoc citreum (formerly Leu. amelibiosum) considered as one of the most predominant lactic acid bacteria (LAB) which helps in the production of kimchi (Korean traditional fermented cabbages), the top-known Korean traditional dish (Chang and Chang, 2010). It is a heterofermentative lactic acid bacterium which is an inhabitant of various fermented foods of plant and dairy origins, such as Pozol (a fermented corn Beverage-Mexican), cassava starch, sourdough, and recently used for the direct fermentation of semolina (Björkroth and Holzapfel, 2006).
Commonly, typing methods of bacteria include phenotyping and genotyping. Phenotypic methods are traditional (biotypes, serotypes, phage-types, and antibiograms), which often lead to an uncertain identification of the LAB (Dan et al., 2014). Therefore, genotypic methods are playing a significant role in phylogenetic classification and identification of the bacterial species. Till date, many molecular methods have been used for the typing of Leuconostoc genus like restriction fragment length polymorphism (RFLP), random amplified polymorphic DNA (RAPD)-PCR, amplified fragment length polymorphism (AFLP), pulsed-field gel electrophoresis (PFGE), repetitive element palindromic PCR (rep-PCR) using different primers, and multilocus sequence typing (MLST) (Sharma et al., 2018). MLST is based on the principles of multi-locus enzyme electrophoresis (MLEE) (Maiden et al., 1998). For characterization of bacteria on the basis of alleles, this technique utilizes the polymorphisms in the sequences of the candidate housekeeping genes. Compared to other methods, this typing tool is showing clear results and the sequence data can be transferred worldwide. It gives valuable information about the species regarding their population organization and evolution. Initially, the technique was described in 1998 for the identification of virulent strains of Neisseria meningitides (Maiden et al., 1998). Continuously, several MLST schemes have been created for bacterial species including the LAB (Sharma et al., 2018). Identification of industrially important bacterial strain is essential in both basic and applied research. MLST may help to differentiate strains at the genetic level and also to identify an industrially important strain. By considering the importance of the technique, the present study was carried out to develop an MLST scheme to determine the genetic diversity of Leu. citreum strains at species level from South Korea.
Materials and methods
Bacterial strains and DNA isolation
Bacterial strains were isolated from six food sources of Korea i.e., kimchi (6), young radish kimchi (1), salted oyster (1), salted fish (1), and pear (1) from Seongnam and salted small octopus (3) from Sunchang and Seongnam. The strains were grown in MRS broth (BD, Franklin Lakes, NJ, USA) at 37 °C overnight. Total DNA was extracted from 3.0 mL of fresh cultures using the DNA isolation kit (Bioneer, AccuPrep Genomic DNA Extraction Kit, Daejeon, Korea), as described in the previous study (Kaur et al., 2017a). The good quality DNA was obtained; estimated in ethidium bromide stained 0.8% (w/v) agarose (Seakem, Lonza Ltd., Basel, Switzerland) gel.
PCR amplification and sequencing
The strains were already identified using 16S rRNA gene sequencing as described in the previous publication (Kaur et al., 2017b). For MLST gene amplification, the primers were designed using the primer blast tool from the NCBI website (www.ncbi.nlm.nih.gov/) (Table 2). The PCR amplifications of the MLST loci, i.e., ATP synthase subunit alpha (atpA), chromosomal replication initiation protein (dnaA), chaperone hsp70 (dnaK), gyrase subunit B (gyrB), phenylalanyl-tRNA synthetase subunit α (pheS), CTP synthase (pyrG), and RNA polymerase (rpoA) were performed in 20 μL PCR reaction mixture premix kit (Bioneer, Daejeon, Korea) containing 1 µL of genomic DNA, 1 µL of each (forward and reverse) primer (10 pmol/µL) and 17 µL of distilled water. Amplification was performed with the automatic thermal cycler (MyCycler™, BioRad, Hercules, CA, USA) following conditions: 94 °C for 2 min, 35 cycles of denaturation at 95 °C for 20 s, annealing temperature different for each locus, i.e., atpA, 51.1 °C; dnaA, 54.2 °C; dnaK, 56.8 °C; gyrB, 54.6 °C; pheS, 46.6 °C; pyrG, 58.0 °C; rpoA, 56.0 °C for 30 s, extension at 72 °C for 30 s, followed by a final elongation at 72 °C for 7 min. The amplified products were purified using a Wizard SV Gel and PCR Cleanup system kit (Promega, Madison, WI, USA) and sequenced by Macrogen-Humanizing Genomics (Seoul, Korea) and Bioneer (Bioneer, Daejeon, Korea).
Table 2.
Primers used and nucleotide diversity observed within the Leu. citreum strains
| Locus | Primer sequence (5′–3′) | Length (bp) | Sequence used (bp) | Polymorphic sites | No. of alleles | G + C content | π Value | Tajima’s D value | dN/dS ratio |
|---|---|---|---|---|---|---|---|---|---|
| atpA | F-GTTTTCGAGCCATTACAA R-GTGATTGAGATAACGTTTG |
590 | 567 | 7 | 5 | 40.74 | 0.00529 | − 0.746820 | 0 |
| dnaA | F-CCAATTACAAAAGAGGAACTA R-TTATCTTGTTGGTTGCGTG |
1261 | 1100 | 3 | 4 | 37.79 | 0.00151 | 0.167656 | 0 |
| dnaK | F-GAAGGTGGCGAACCAAAAA R-AGCCAATGCTTCTGTCTTAG |
1647 | 806 | 22 | 10 | 41.28 | 0.00896 | − 0.337078 | 1.1951 |
| gyrB | F-ATAAAGTCTCTGGTGGATT R-TGGTAGGATAGCTTGTGTTA |
1022 | 800 | 1 | 2 | 41.62 | 0.00067 | 1.475424 | 0 |
| pheS | F-CACAAACTTCACCTGT R-ACGTCTACTTCAACTG |
286 | 264 | 5 | 8 | 41.71 | 0.00757 | 0.168432 | 0.3874 |
| pyrG | F-ACCGTGGCTTAAAATTGGC R-TCACTGCCAAGATCTGATTTCAT |
415 | 398 | 11 | 9 | 39.06 | 0.01060 | 0.203116 | 0.1152 |
| rpoA | F-CGTTGTTGAAGATGTCACAC R-GCCAACACGCCGATAG |
291 | 271 | 2 | 3 | 42.9 | 0.00113 | − 1.468006 | 0.2459 |
π, nucleotide diversity per site; dN, number of nonsynonymous changes per nonsynonymous site; dS, number of synonymous changes per synonymous site
Data analysis
For MLST analysis, forward and reverse sequences were cropped and examined with Bioedit Sequence Alignment Editor ver. 7.2.5 for every gene (Hall, 1999) and aligned with the ClustalX ver. 1.83 software. The GC content was calculated with DnaSP ver. 5.10 (Librado and Rozas, 2009) and the polymorphic sites, Tajima’s D value and nucleotide diversity per site (π) by MEGA software ver. 7 (Kumar et al., 2016). The ratios of the synonymous substitutions (dS) to non-synonymous substitutions (dN) were analyzed online by the SNAP tool (www.hiv.lanl.gov) (Korber, 2000). Generation of allelic profiles, sequence types (STs), dendrogram and minimum spanning tree (MST) were done using Bionumerics ver. 7.6 (Applied-Maths, Sint Maartens-Latem, Belgium) software. Split decomposition analysis was completed with SplitsTree ver. 4.14 software (Huson and Bryant, 2006).
Results and discussion
The present study describes the MLST scheme of 13 bacterial strains isolated from South Korea. Isolated strains were examined for their genetic diversity using comparative sequence analysis of selected seven housekeeping genes. Gene sequences of the seven loci have been deposited in GenBank under the accession numbers KX286339-KX286348, KX286350-KX286352 for 16S rRNA gene and KY763993-KY764057 and KY767960-KY767985 for MLST loci.
MLST typing and locus variation
Seven target genes were amplified, and the length of genes varied from 286 bp for pheS to 1647 bp for dnaK. The MLST scheme defined alleles between 264 (pheS) and 1100 bp (dnaA). The number of alleles per gene varied between 2 for gyrB and 10 for dnaK. From the gene sequence data of the seven loci, 13 different sequence types (STs) were obtained. Each ST was represented by a single strain of Leu. citreum, indicating about the genetic diversity in selected strains. The genetic variations of the Leu. citreum strains for the different loci have been shown in Table 1. All STs were differed at various loci, whereas ST-2, ST-4 and ST-6, ST-7 were having 5 alleles identical out of 7, depicting around 71% similarity. In addition, it can be observed that the allelic frequency was dominated for alleles 1 and 2 compared to other alleles among the selected few gene loci for MLST analysis. For example, dominance in frequencies was observed in allele 1 for atpA, dnaA, and gyrB loci and in allele 2 for gyrB and rpoA loci (Table 1). The number of polymorphic sites varied from 1 for gyrB to 22 for the dnaK, the most polymorphic locus, therefore, a total of 51 SNPs has been detected (Table 2). Out of 7, three loci namely gyrB, rpoA, and dnaA had low polymorphism showing 1, 2, and 3 polymorphic sites respectively. The apparent low levels of biodiversity in gyrB, rpoA, and dnaA suggested that the partial gene sequences of the loci were somewhat conserved amongst 13 strains, and have the lower discriminatory ability than the other housekeeping loci used in the study. The remaining four loci namely atpA, dnaK, pheS, and pyrG had more polymorphic sites (between 5 and 22), suggesting their good discriminatory ability. Fifty-one polymorphic sites were detected among 4206 nucleotides giving polymorphism rate of 1.21%. This value was higher than that observed for Leu. lactis, which showed 47 SNPs (0.88%) amongst 5325 nucleotides (Dan et al., 2014). Comparatively low number of polymorphic sites among seven housekeeping loci suggested that these are conserved amongst the 13 Leu. citreum isolates. The Tajima’s D value (Table 2) ranged between − 1.46 to 1.47, which supported the complete fitting of the population in the model system (Andreani et al., 2014) and since the values are between − 2 and 2 the evolution of these genes was likely driven by neutral selection. The mean GC content of the MLST gene fragments varied from the 37.79% for dnaA to 42.9% for rpoA (Table 2), which was compared with already reported GC content (38–44%) for the genus Leuconostoc (De Bruyne et al., 2007). The nucleotide diversity index (π) ranging from the 0.00067 for gyrB to 0.01060 for pyrG (Table 2). It has been reported that the higher value of π is related to high intragenic nucleotide polymorphism (Cai et al., 2007). Similarly, in this study, the loci with high π value were represented with more polymorphic sites compared to the others. Analysis of synonymous substitutions to non-synonymous substitutions in the allele sequence of a locus can be used to determine whether the genes are under positive selection, therefore a ratio greater than 1 implies selection for amino acid changes. In this genetic analysis, six housekeeping loci had dN/dS ratio lower than 1. Even for three loci, i.e., atpA, dnaA, and gyrB the ratio was found equal to zero, where substitution was synonymous indicating that the amino acid composition of these genes did not change (Table 2). However, one exception was observed for locus dnaK, which had a dN/dS ratio, 1.1951 > 1, this is not typically observed for housekeeping genes. In addition, a similar exception was also reported earlier in a study of Leu. mesenteroides using 9 housekeeping loci, dN/dS ratio varied from 0.0219 to 2.4379 and was found to be > 1 only for one locus, i.e., murC (2.4379) (Zhang et al., 2015).
Table 1.
Source and typing data of Leu. citreum strains analyzed in this study
| Strain | KCCP number | Source | MLST (sequence type) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ST | atpA | dnaA | dnaK | gyrB | pheS | pyrG | rpoA | |||
| SC53 | 11037 | Salted small octopus | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| YKC002 | 11076 | Kimchi | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 |
| YKC003 | 11077 | Kimchi | 3 | 2 | 1 | 3 | 1 | 2 | 2 | 3 |
| YKC019 | 11082 | Kimchi | 4 | 2 | 1 | 2 | 1 | 1 | 3 | 2 |
| PEAR008 | 11093 | Pear | 5 | 1 | 3 | 4 | 1 | 3 | 3 | 2 |
| NJ1 | 11382 | Salted small octopus | 6 | 1 | 1 | 4 | 2 | 4 | 4 | 2 |
| CMK2 | 11388 | Young reddish kimchi | 7 | 3 | 1 | 4 | 2 | 4 | 2 | 2 |
| NJG3-1 | 11391 | Salted small octopus | 8 | 1 | 4 | 5 | 2 | 5 | 5 | 2 |
| KCF001 | 11413 | Kimchi | 9 | 1 | 1 | 6 | 2 | 5 | 6 | 2 |
| KCF002 | 11414 | Kimchi | 10 | 1 | 1 | 7 | 2 | 2 | 7 | 2 |
| KCG001 | 11422 | Kimchi | 11 | 1 | 1 | 8 | 1 | 6 | 7 | 2 |
| OJ1 | 11432 | Salted oyster | 12 | 4 | 4 | 9 | 1 | 7 | 8 | 2 |
| SF4 | 11447 | Salted fish | 13 | 5 | 2 | 10 | 2 | 8 | 9 | 2 |
KCCP Korean Culture Collection of Probiotics
Splits tree analysis
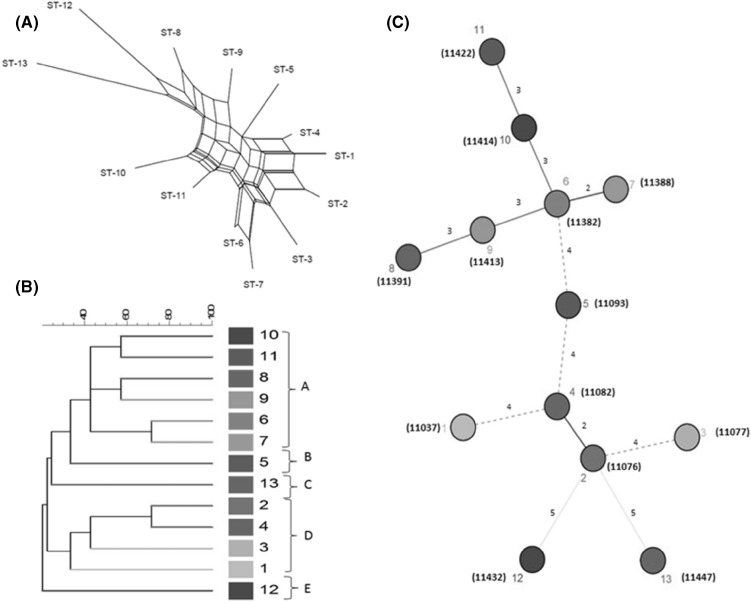
In order to further analyze the population structure of Leu. citreum we used split decomposition analysis. A tree-like structure is created when the descent is clonal, but an interconnecting network will appear when recombination plays a role in the evolutionary history of Leu. citreum genes. A combined split graph based on all alleles in the seven MLST loci displayed a network-like structure with different rays of length [Fig. 1(A)]. Parallelogram-shaped groupings were observed suggesting their descents were originated from interspecies recombination events. It has been reported in the previous literature that usually in the genus Leuconostoc recombination could occur due to various mobile elements (transposable elements, genomic islands, and bacteriophages) in their genome sequences (Meslier et al., 2012). In addition, plasmid has also been identified from Leu. citreum (Chang and Chang, 2009). Furthermore, it can be observed from the figure that all the STs were showing some degree of relationship, except ST-12 and ST-13, which were distantly related to remaining 11 isolates.
Fig. 1.
(A) Combined split-decomposition analysis based on concatenated sequences of the seven MLST loci. The numbering in the figure refers to ST. (B) UPGMA dendrogram showing the genetic relationship between 13 STs belonging to Leu. citreum. The numbering in the figure refers to ST. (C) Minimum-spanning tree analysis of the 13 Leu. citreum strains from South Korea. Each circle relates to a sequence type (ST) and a line between isolates indicates the strength of the genetic relationship between these isolates (bold, strong relationship; solid dashed, intermediate relationship; very small dotted line, weak relationship)
UPGMA tree analysis and minimum spanning tree
Genetic relatedness among Leu. citreum strains were investigated by constructing a UPGMA (Unweighted Pair Group Method with Arithmetic Mean) dendrogram [Fig. 1(B)]. As it can be observed from the figure, strains were showing the low degree of similarity. As shown in the figure, there were two major groups of the strains on the basis of similarity in their allelic profiles. Larger group A consisted of six strains (ST-6 to ST-11). Groups B and C were represented by ST-5 and ST-13, whereas group D consists of strains (ST-1 to ST-4). ST-6 and ST-7 from group A and ST-2 and ST-4 from group D were showing maximum similarity (71.4%) amongst 13 strains on the basis of 5 common alleles out of seven. Earlier, the use of other genotypic methods namely rep-PCR (Kaur et al., 2017c) and RAPD-PCR (Kaur et al., 2017b) have also been reported for the analysis of Leu. citreum using REP, ERIC, (GTG)5 and 239, Kay3 primers respectively. In addition, minimum spanning tree (MST), an algorithm was also used to perform a phylogenetic analysis of the strains based on the allelic profiles. Usually, in MST, the size of the circle is proportional to the number of isolates with that unique profile. Nevertheless, as represented in the figure, the size of each circle is similar, as there were 13 circles of different color, representing one isolate each. Different circles were connected to each other by different lines representing the degree of relationship between Leu. citreum strains [Fig. 1(C)]. The numbers on the lines were depicting the difference in the alleles between two isolates. On the top of the MST, bold lines between strains (11391, 11382, 11388, 11413, 11414, and 11422) indicating a comparatively strong relationship. Among these strains, 2 (11382 and 11388) or 3 (11391, 11413, 11414, and 11422) alleles were found to be different from each other. These strains were isolated from three different sources namely, kimchi, salted small octopus, and young radish kimchi. Similarly, strains 11082 and 11076 were connected with each other with a bold line, both were isolated from kimchi, and showing a difference for two alleles out of seven. Moreover, the strains 11037, 11093, and 11077 were connected with dashed lines to other strains showing they have 4 alleles different and isolated from salted small octopus, pear and kimchi respectively. Lastly, 11432, and 11447 strains isolated from salted oyster and salted fish were connected with dotted lines, showing a very weak relationship on the basis of alleles (maximum allelic difference). The strains were not clustered in the common circle, indicating variability at the genetic level. Comparable results were obtained with MST, UPGMA tree and concatenated splits tree analysis. However, despite the same isolation source for some of the STs, no strong relationship was observed, as can be seen from MST as well as allelic profiles also [Fig. 1(C)]. It can be observed from tree analysis that no appropriate association was found between ST and the source from which the bacteria were isolated (Table 1). Comparable results have been reported in Leu. lactis, where no significant associations between STs and the sources of the isolates could be found (Picozzi et al., 2010). The absence of such an association in Leu. citreum strains may be because of the genetic diversity of individual Leu. citreum strains. The variation in MLST data from one study to another could be because of different bacterial species (habitats), isolation sources and housekeeping genes selected for individual analysis.
In summary, the presented MLST scheme was found to be a useful tool for the typing of Leu. citreum strains and showed a sufficient discriminatory power to determine the genetic variability. The protocol was followed for the structural analysis of 13 bacterial species, which were directed into thirteen sequence types. It was found that the evolution of different Leuconostoc strains was not related to respective food sources. For a better understanding of Leu. citreum from South Korea, a large number of strains must need to be evaluated which will further contribute details on the evolution and population genetics of the species. As MLST allows precise identification, and easy assessment, the future application of this molecular method could be useful for the identification of valuable Leu. citreum strains and nonpathogenic food production bacteria for their use in the food industry with a valuable application.
Acknowledgements
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through High Value-added Food Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (Grant Number: 316051-03-2-HD020).
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- Andreani NA, Martino ME, Fasolato L, Carraro L, Montemurro F, Mioni R, Bordin P, Cardazzo B. Tracking the blue: A MLST approach to characterize the Pseudomonas fluorescens group. Food Microbiol. 2014;39:116–126. doi: 10.1016/j.fm.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Björkroth J, Holzapfel W. Genera Leuconostoc, Oenococcus and Weissella. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes: A Handbook on the Biology of Bacteria: Firmicutes, Cyanobacteria. New York, NY, USA: Springer; 2006. pp. 267–319. [Google Scholar]
- Cai H, Rodriguez BT, Zhang W, Broadbent JR, Steele JL. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology. 2007;153:2655–2665. doi: 10.1099/mic.0.2007/006452-0. [DOI] [PubMed] [Google Scholar]
- Chang JY, Chang HC. Identification of a replicon from pCC3, a cryptic plasmid from Leuconostoc citreum C4 derived from kimchi, and development of a new host vector system. Biotechnol. Lett. 2009;31:685–696. doi: 10.1007/s10529-009-9912-9. [DOI] [PubMed] [Google Scholar]
- Chang JY, Chang HC. Improvements in the quality and shelf-life of kimchi by fermentation with the induced bacteriocin-producing strain, Leuconostoc citreum GJ7, as a starter. J. Food Sci. 2010;75:103–110. doi: 10.1111/j.1750-3841.2009.01486.x. [DOI] [PubMed] [Google Scholar]
- Dan T, Liu W, Sun Z, Lv Q, Xu H, Song Y, Zhang H. A novel multi-locus sequence typing (MLST) protocol for Leuconostoc lactis isolates from traditional dairy products in China and Mongolia. BMC Microbiol. 2014;14:150. doi: 10.1186/1471-2180-14-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruyne K, Schillinger U, Caroline L, Boehringer B, Cleenwerck I, Vancanneyt M, De Vuyst L, Franz CM, Vandamme P. Leuconostoc holzapfelii sp. nov., isolated from Ethiopian coffee fermentation and assessment of sequence analysis of housekeeping genes for delineation of Leuconostoc species. Int. J. Syst. Evol. Microbiol. 2007;57:2952–2959. doi: 10.1099/ijs.0.65292-0. [DOI] [PubMed] [Google Scholar]
- Hall TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Kaur J, Lee S, Park YS, Sharma A. RAPD analysis of Leuconostoc mesenteroides strains associated with vegetables and food products from Korea. LWT–Food. Sci. Technol. 2017;77:383–388. [Google Scholar]
- Kaur J, Lee S, Sharma A, Park YS. Molecular typing of Leuconostoc citreum strains isolated from korean fermented foods using a random amplified polymorphic DNA Marker. Food Eng. Prog. 2017;21:174–179. doi: 10.13050/foodengprog.2017.21.2.174. [DOI] [Google Scholar]
- Kaur J, Sharma A, Lee S, Park YS. DNA profiling of Leuconostoc citreum strains in fermented foods by repetitive element polymerase chain reaction. J. Microbiol. Biotechnol. 2017;27(10):1778–1782. doi: 10.4014/jmb.1705.05022. [DOI] [PubMed] [Google Scholar]
- Korber B. HIV signature and sequence variation analysis. In: Rodrigo AG, Learn GH, editors. Computational Analysis of HIV Molecular Sequences. Dordrecht: Dordrecht, Kluwer Academic Publishers; 2000. pp. 55–72. [Google Scholar]
- Kot W, Neve H, Heller KJ, Vogensen FK. Bacteriophages of Leuconostoc, Oenococcus, and Weissella. Front. Microbiol. 2014;5:186. doi: 10.3389/fmicb.2014.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA: Molecular Evolutionary Genetic analysis 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSPv5: A software for comprehensive analysis of DNA polymorphisms data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meslier V, Loux V, Renault P. Genome sequence of Leuconostoc pseudomesenteroides strain 4882, isolated from a dairy starter culture. J. Bacteriol. 2012;194:696–712. doi: 10.1128/JB.01696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picozzi C, Bonacina G, Vigentini I, Foschino R. Genetic diversity in Italian Lactobacillus sanfranciscensis strains assessed by Multilocus sequence typing and pulsed-field gel electrophoresis analyses. Microbiology. 2010;156:2035–2045. doi: 10.1099/mic.0.037341-0. [DOI] [PubMed] [Google Scholar]
- Sharma A, Kaur J, Lee S, Park Y-S. Genetic diversity analysis of Leuconostoc mesenteroides from Korean vegetables and food products by multilocus sequence typing. Appl. Microbiol. Biotechnol. 2018 doi: 10.1007/s00253-018-8942-4. [DOI] [PubMed] [Google Scholar]
- Zhang W, Liu W, Song Y, Xu H, Menghe B, Zhang H, Sun Z. Multilocus sequence typing of a dairy-associated Leuconostoc mesenteroides population reveals clonal structure with intragenic homologous recombination. J. Dairy Sci. 2015;98:1–10. doi: 10.3168/jds.2014-8433. [DOI] [PubMed] [Google Scholar]