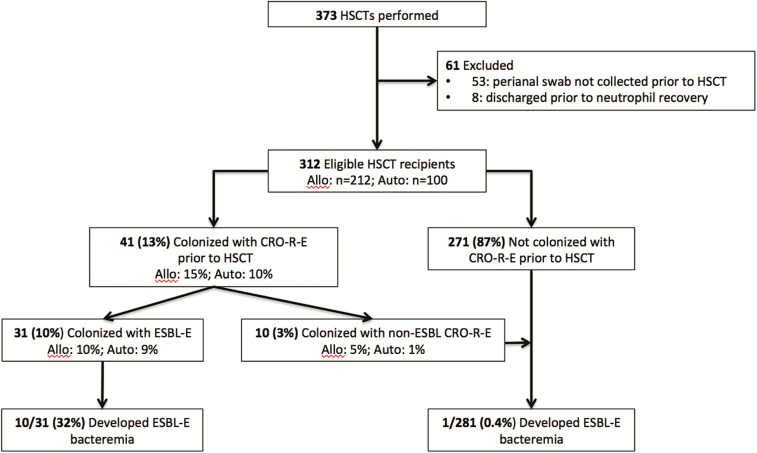
Prior to transplant, 10% of hematopoietic stem cell transplant recipients were colonized with ESBL-producing Enterobacteriaceae (ESBL-E). In the setting of levofloxacin prophylaxis, 32% of colonized patients developed ESBL-E bacteremia during neutropenia and colonizing, and bloodstream ESBL-E were genetically identical.
Keywords: ESBL, neutropenia, transplant, colonization, bacteremia
Abstract
Background
Bacteremia caused by extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae (ESBL-E) is associated with inadequate empirical therapy and substantial mortality in neutropenic patients. Strategies are needed to identify neutropenic patients at high risk of these infections.
Methods
From April 2014 to September 2016, we collected perianal swabs, both at admission and weekly thereafter, from patients undergoing hematopoietic stem cell transplantation (HSCT). Patients received prophylactic levofloxacin while neutropenic. Swabs were plated onto selective agar, colonies were identified and underwent antimicrobial susceptibility testing, and phenotypic ESBL testing and polymerase chain reaction for β-lactamase genes were performed on ceftriaxone-resistant Enterobacteriaceae. We then determined the prevalence of pre-transplant ESBL-E colonization and risk of ESBL-E bacteremia. Colonizing and bloodstream isolates from patients with ESBL-E bacteremia underwent multilocus sequence typing and pulsed-field gel electrophoresis.
Results
We analyzed 312 patients, including 212 allogeneic and 100 autologous HSCT recipients. Ten percent (31/312) of patients had pre-transplant ESBL-E colonization. Susceptibility rates of colonizing ESBL-E were: levofloxacin, 25%; cefepime, 9%; piperacillin-tazobactam, 84%; and meropenem, 97%. Of 31 patients colonized with ESBL-E pre-transplant, 10 (32%) developed ESBL-E bacteremia during their transplant admission, compared to 1 (0.4%) of 281 patients not colonized with ESBL-E (P < .001). All bloodstream ESBL-E were levofloxacin-resistant and colonizing and bloodstream isolates from individual patients had identical genotypic profiles.
Conclusions
HSCT recipients who are colonized with levofloxacin-resistant ESBL-E pre-transplant and receive levofloxacin prophylaxis have high rates of bacteremia from their colonizing strain during neutropenia. Assessing for ESBL-E colonization in neutropenic patients could lead to optimization of empirical antibacterial therapy.
Patients undergoing hematopoietic stem cell transplantation (HSCT) are at high risk of developing bloodstream infections (BSIs) from Gram-negative bacteria because of chemotherapy-induced gastrointestinal mucositis and prolonged neutropenia. Unfortunately, extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae (ESBL-E) are emerging as common causes of bacteremia in these vulnerable patients [1–5]. This development is alarming, because neutropenic patients rely on immediate bactericidal therapy to combat Gram-negative bacteremia and some recommended first-line agents for fever and neutropenia, such as anti-pseudomonal cephalosporins, have limited activity against ESBL-E [6–8]. Thus, neutropenic patients with ESBL-E bacteremia often receive inadequate empirical antimicrobial therapy and have higher mortality rates than those with bacteremia due to ceftriaxone-susceptible Enterobacteriaceae [2, 9–11].
Given the emergence of ESBL-E as causes of bacteremia in HSCT recipients and the increased mortality rates associated with these infections, new strategies are needed to identify HSCT recipients who are at high risk of developing ESBL-E bacteremia. These high-risk patients may then receive optimized empirical therapy with an antibacterial agent that is highly active against ESBL-E, such as a carbapenem, when fever and neutropenia occur. Conversely, patients who are at low risk of developing ESBL-E bacteremia could be empirically treated with an anti-pseudomonal cephalosporin or β-lactam/β-lactamase inhibitor (BL-BLI). We hypothesized that screening the gastrointestinal tract to identify patients colonized with ESBL-E could identify HSCT recipients at high risk of developing ESBL-E bacteremia.
METHODS
Study Population and Sample Collection
This prospective observational study was conducted at an 862-bed New York City hospital and included adult patients (≥18 years old) admitted for autologous or allogeneic HSCT from April 2014 to September 2016. The study was approved by the Institutional Review Board of Weill Cornell Medicine with a waiver of informed consent. All patients were located on a HSCT unit in single-occupancy rooms during their transplant admission. Levofloxacin prophylaxis was initiated 1 day prior to receipt of stem cells. In the setting of fever and neutropenia, at least 2 sets of blood cultures were collected, levofloxacin was discontinued, and an anti-pseudomonal β-lactam agent, most commonly piperacillin-tazobactam, was administered. For each patient, we collected data on patient demographics, malignancy and transplant characteristics, prior antimicrobial exposures, and history of colonization or infection with multidrug-resistant bacteria. We also recorded all episodes of fever and neutropenia and any BSIs associated with these episodes. Any Enterobacteriaceae that were isolated from these patients’ blood cultures were stored at -80°C for subsequent analysis.
Perianal swabs were collected on admission for transplant and weekly thereafter until hospital discharge. Perianal flocked BD Eswabs (Becton Dickinson, Franklin Lakes, NJ) were collected by either the patient or the nursing staff. Patients who did not have a swab collected before their transplant or who were discharged prior to neutrophil recovery were excluded. Any ceftriaxone-resistant Enterobacteriaceae (CRO-R-E) that were isolated from these swabs were reported in the medical record to the patient care team. Patients found to be colonized with CRO-R-E were placed on contact precautions with gown and gloves.
Microbiologic Analysis
Perianal swabs were vortexed and 100 μL of Amies liquid was inoculated onto HardyCHROM ESBL or carbapenem-resistant enterobacteriaceae chromogenic agar plates (Hardy Diagnostics, Santa Maria, CA). These plates were incubated at 37°C for 48 and 24 hours, respectively. Colonies identified after incubation on either plate were subcultured onto BD Trypticase soy agar II plates (Becton Dickinson, Franklin Lakes, NJ) and incubated overnight. The next day, colonies from these plates were identified and underwent antimicrobial susceptibility testing using the Microscan Walkaway plus System (Panel NM42, Beckman Coulter, Inc., Brea, CA). Etest strips (bioMérieux, Durham, NC) were used on all CRO-R-E to obtain susceptibilities to ceftolozane-tazobactam and ceftazidime-avibactam. Interpretive criteria of the Clinical and Laboratory Standards Institute were applied [12].
All CRO-R-E isolated from perianal swabs or blood cultures underwent phenotypic testing for ESBL production, using cefotaxime and ceftazidime discs, with and without clavulanate [12]. Furthermore, these isolates underwent testing by molecular beacon probes in a real-time PCR assay for the following β-lactamase genes, followed by Sanger sequencing: blaKPC, blaOXA-48, blaVIM, blaIMP, blaNDM, blaCTX-M (groups 1, 2, 9, 8/25), blaCMY1/MOX, blaCMY2/LAT/CEF, blaACT/MIR, blaDHA, blaTEM (wildtype and mutants 104K, 164S, 164H, 164C, 238S, 240K), and blaSHV (wildtype and mutants 156D, 238S, 240K) [13].
For patients who had a perianal swab and a blood culture that grew CRO-R-E, their swab and bloodstream isolates underwent multilocus sequence typing (MLST) to assess the genetic relatedness of these paired strains [14]. Pulsed-field gel electrophoresis (PFGE) was also performed on these isolates using the Xba1 restriction enzyme [15]. Salmonella enterica subsp. enterica serovar Braenderup (ATCC BAA-664) was used as a DNA size marker.
Definitions
ESBL producers were defined as CRO-R-E that displayed ESBL production phenotypically and did not harbor a carbapenemase gene. ESBL acquisition was defined as any patient who did not have a swab that yielded ESBL-E prior to HSCT, but had a subsequent swab that yielded ESBL-E after transplantation. Fever and neutropenia (FN) was defined as a temperature ≥38°C with a concurrent absolute neutrophil count ≤500 cells/mm3. Coagulase-negative staphylococci and other common skin commensals were only considered causes of BSI if isolated from 2 separate sets of blood cultures.
Statistical Analyses
We first determined the proportion of patients who were colonized with CRO-R-E and ESBL-E prior to HSCT. Factors associated with pre-transplant ESBL-E colonization were assessed by comparing characteristics of patients colonized with ESBL-E to those not colonized with CRO-R-E, using Fisher’s exact or chi-square tests for categorical variables and the Wilcoxon rank-sum test for continuous variables. We then compared the cumulative incidence of developing ESBL-E bacteremia during the transplant admission, as well as the incidence and etiologies of FN and BSI, initial empirical therapies for FN, and transplant outcomes in patients with and without pre-transplant ESBL-E colonization. Finally, we assessed the risk of ESBL-E acquisition in patients who were not initially colonized with ESBL-E during their transplant admission. For this acquisition analysis, we only included patients who had a subsequent swab collected within 1 week of neutrophil recovery. STATA, version 12.0 (StataCorp, College Station, TX) was used for statistical analyses and P values ≤0.05 (2-tailed) were considered statistically significant.
RESULTS
Patients and Pre-transplant Colonization Status
Of 373 patients who received a HSCT during the study period, 312 (84%) were eligible for analysis, including 212 allogeneic and 100 autologous HSCT recipients (Figure 1). The median patient age was 58 years and the most common underlying malignancies were acute myeloid leukemia (36%), multiple myeloma (19%), and non-Hodgkin’s lymphoma (19%; Table 1). The median duration of neutropenia was 10 days (interquartile range: 8–14 days). Most (92%) pre-transplant perianal swabs were collected prior to the initiation of levofloxacin prophylaxis. Prior to transplant, 41 (13%) HSCT recipients were colonized with CRO-R-E (allogeneic: 15%; autologous: 10%) and 31 (10%) were colonized with ESBL-E (allogeneic: 10%; autologous: 9%; Figure 1).
Figure 1.
Flow diagram of patients included in the study and their risk of ESBL-E bacteremia, stratified by colonization status. Abbreviations: CRO-R-E, ceftriaxone-resistant Enterobacteriaceae; ESBL-E, extended-spectrum β-lactamase-producing Enterobacteriaceae; HSCT, hematopoietic stem cell transplantation.
Table 1.
Characteristics of Patients Colonized With Extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBL- E); Colonized With Non-ESBL, Ceftriaxone-resistant Enterobacteriaceae (CRO-R-E); and Not Colonized With CRO-R-E Prior to Hematopoietic Stem Cell Transplantation
| Patient Characteristic | Colonized With ESBL-Ea (n = 31) |
Colonized With Non-ESBL CRO-R-E (n = 10) | Not Colonized With CRO-R-E (n = 271) | P Valueb |
|---|---|---|---|---|
| Demographics | ||||
| Age, years | 59 (43–65) | 50 (42–63) | 58 (48–65) | 0.76 |
| Female gender | 12 (39) | 5 (50) | 123 (45) | 0.48 |
| Ethnicity/Race | ||||
| Hispanic, White | 6 (19) | 1 (10) | 26 (10) | 0.12 |
| Hispanic, Black | 0 | 0 | 2 (1) | 1.00 |
| Non-Hispanic | ||||
| White | 15 (48) | 6 (60) | 174 (64) | 0.085 |
| Black | 3 (10) | 1 (10) | 31 (11) | 1.00 |
| Asian | 2 (6) | 2 (20) | 22 (8) | 1.00 |
| Middle Eastern | 5 (16) | 0 | 16 (6) | 0.051 |
| Residence outside of the United States | 3 (10) | 0 | 9 (3) | 0.11 |
| Underlying malignancy | ||||
| AML | 7 (23) | 5 (50) | 101 (37) | 0.11 |
| ALL | 4 (13) | 0 | 16 (6) | 0.14 |
| CML | 1 (3) | 0 | 6 (2) | 0.72 |
| CLL | 0 | 0 | 2 (1) | 1.00 |
| Non-Hodgkin’s lymphoma | 4 (13) | 3 (30) | 52 (19) | 0.39 |
| Hodgkin’s lymphoma | 2 (6) | 0 | 12 (4) | 0.64 |
| Multiple myeloma | 7 (23) | 1 (10) | 51 (19) | 0.62 |
| MDS or MPD | 6 (19) | 0 | 18 (7) | 0.025 |
| Others | 0 | 1 (10) | 13 (5) | 0.38 |
| ASBMT RFI risk classification | ||||
| Low risk | 13 (42) | 4 (40) | 103 (38) | 0.67 |
| Intermediate risk | 6 (19) | 3 (30) | 52 (19) | 0.98 |
| High risk | 10 (32) | 2 (20) | 107 (39) | 0.43 |
| N/A | 2 (7) | 1 (10) | 9 (3) | 0.38 |
| Prior transplant | 3 (10) | 0 | 19 (7) | 0.59 |
| Type of transplant | ||||
| Allogeneic | 22 (71) | 9 (90) | 181 (67) | 0.88 |
| Autologous | 9 (29) | 1 (10) | 90 (33) | |
| Conditioning regimen | ||||
| Fludarabine-melphalan | 16 (52) | 8 (80) | 145 (54) | 0.84 |
| BEAMc | 5 (16) | 1 (10) | 48 (18) | 0.83 |
| Melphalan | 7 (23) | 0 | 38 (14) | 0.19 |
| Bendamustine-melphalan | 0 | 0 | 9 (3) | 0.61 |
| Other | 3 (10) | 1 (10) | 31 (11) | 1.00 |
| Adjunctive conditioning therapies | ||||
| Use of rituximab | 3 (10) | 1 (10) | 29 (11) | 1.00 |
| Use of total body irradiation | 9 (29) | 1 (10) | 55 (20) | 0.26 |
| Anti-T cell therapies for GVHD prophylaxis | ||||
| Anti-thymocyte globulin | 7 (23) | 4 (40) | 71 (26) | 0.66 |
| Alemtuzumab | 12 (38) | 5 (50) | 104 (38) | 0.97 |
| Hospitalization within previous 90 days | 18 (58) | 5 (50) | 153 (56) | 0.86 |
| Antibacterials within previous 90 days | ||||
| Beta-lactams | 14 (45) | 4 (40) | 113 (42) | 0.71 |
| Fluoroquinolones | 5 (16) | 1 (10) | 58 (21) | 0.49 |
| Vancomycin | 4 (13) | 3 (30) | 66 (24) | 0.15 |
| History of MDR bacteria colonization or infectiond | ||||
| CRO-R-E | 2 (6) | 0 | 6 (2) | 0.18 |
| VRE | 0 | 2 (20) | 29 (10) | 0.09 |
| Clostridium difficile | 5 (16) | 1 (10) | 28 (10) | 0.35 |
| Duration of neutropenia, days | 10 (8–15) | 7 (6–10) | 10 (8–14) | 0.60 |
Values are expressed as number (% of total) or median (interquartile range).
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; ASBMT RFI, American Society of Bone Marrow Transplant Request for Information; CLL, chronic lymphocytic leukemia; CML, chronic myeloid leukemia; CRO-R-E, ceftriaxone-resistant Enterobacteriaceae; ESBL, extended-spectrum β-lactamase; ESBL-E, ESBL-producing Enterobacteriaceae; GVHD, graft-versus-host disease; MDR, multidrug-resistant; MDS, myelodysplastic syndrome; MPD, myeloproliferative disorder; N/A, not applicable; VRE, vancomycin-resistant enterococci.
aWe defined an organism as an ESBL producer if the addition of clavulanate to cefotaxime or ceftazidime led to a ≥5 mm increase in disc diffusion zone diameter and no carbapenemase genes were detected [12].
bComparison of characteristics of patients colonized with ESBL-E to those of patients not colonized with CRO-R-E.
cBEAM refers to carmustine, etoposide, cytarabine, and melphalan.
dPatients were not routinely screened for CRO-R-E or VRE prior to their admission for HSCT.
Middle Eastern ethnicity (P = .05) and myelodysplastic syndrome or myeloproliferative disorder (P = .03) were the only variables associated with pre-transplant ESBL-E colonization (Table 1). Of the 31 patients colonized with ESBL-E prior to transplantation, only 2 (6%) had a previously positive culture for CRO-R-E.
Microbiologic Characteristics of Pre-transplant Colonizing CRO-R-E
There were 32 ESBL-E among the 31 patients colonized prior to transplantation, of which 28 were Escherichia coli and 4 were Klebsiella pneumoniae (Table 2). Of these 32, 29 (91%) ESBL-E were CTX-M-producers, of which CTX-M-15 was most common (n = 18). Co-production of non-ESBL β-lactamases was present in 16 of the ESBL-E (50%), with TEM-1 being the most common. Of the 10 non-ESBL CRO-R-E, 6 were AmpC-producers, 3 produced K. pneumoniae carbapenemase (KPC), and 1 did not have a CRO resistance mechanism identified.
Table 2.
Species and Resistance Mechanisms of 42 Colonizing Ceftriaxone-resistant Enterobacteriaceaea
| Organism Type | No. |
|---|---|
| ESBL-producers | 32 |
| Escherichia coli | 28 |
| Klebsiella pneumoniae | 4 |
| CTX-M-producer | 29/32 (91%) |
| CTX-M-15 | 18 |
| CTX-M-14 | 5 |
| CTX-M-27 | 4 |
| Othersb | 2 |
| SHV-β-lactamasesc | 3/32 (9%) |
| Co-harbored other β-lactamasesd | 16/32 (50%) |
| AmpC-producerse | 6 |
| CMY2/LAT/CEF | 4 |
| Othersf | 2 |
| Carbapenemase-producers (all KPC)g | 3 |
| Unknown ceftriaxone resistance mechanism | 1 |
Abbreviations: ESBL, extended-spectrum β-lactamase; KPC, Klebsiella pneumoniae carbapenemase.
aIn 1 patient, two different ceftriaxone-resistant Enterobacteriaceae colonized.
bThe other CTX-M enzymes were CTX-M-3 and CTX-M-130 (n = 1, for each).
cThe other SHV ESBLs were SHV-12 and variants of SHV-1 and SHV-11 (n = 1, for each).
dOther non-ESBL β-lactamases were TEM-1 (n = 12), TEM-1 and SHV-11 (n = 2), and CMY-6 and CMY-70 (n = 1, for each).
eThe AmpC-producing isolates were Escherichia coli (n = 4), Citrobacter freundii, and Enterobacter cloacae (n = 1, for each).
fThe other AmpC β-lactamases were ACT/MIR and DHA (n = 1, for each).
gThe 3 KPC-producing isolates were Enterobacter cloacae, Escherichia coli, and Klebsiella pneumoniae (n = 1, for each).
The antimicrobial susceptibilities of the 32 colonizing ESBL-E to recommended agents for fever and neutropenia were as follows (Table 3): cefepime, 9%; ceftazidime, 13%; piperacillin-tazobactam, 84%; meropenem, 97%; ceftolozane-tazobactam, 88%; and ceftazidime-avibactam, 100%. Only one-fourth of the ESBL-E were levofloxacin-susceptible.
Table 3.
Antimicrobial Susceptibilities of Colonizing Ceftriaxone-resistant Enterobacteriaceae on Admission for Hematopoietic Stem Cell Transplantation
| Antimicrobial Agent | ESBL-E | Non-ESBL CRO-R-Ea |
|---|---|---|
| %Susceptible (n = 32) | %Susceptible (n = 10) | |
| Amikacin | 97% | 80% |
| Ampicillin | 0% | 0% |
| Ampicillin-sulbactam | 25% | 0% |
| Amoxicillin-clavulanate | 44% | 10% |
| Aztreonam | 25% | 10% |
| Cefepime | 9% | 60% |
| Ceftazidime | 13% | 10% |
| Ceftolozane-tazobactam | 88% | 70% |
| Ceftazidime-avibactam | 100% | 100% |
| Ciprofloxacin | 25% | 70% |
| Ertapenem | 94% | 40% |
| Imipenem | 97% | 70% |
| Gentamicin | 59% | 90% |
| Levofloxacin | 25% | 70% |
| Meropenem | 97% | 70% |
| Piperacillin-tazobactam | 84% | 40% |
| Tigecycline | 100% | 100% |
| TMP-SMX | 16% | 40% |
| Tobramycin | 47% | 60% |
Abbreviations: CRO-R-E, ceftriaxone-resistant Enterobacteriaceae; ESBL, extended-spectrum β-lactamase; ESBL-E, ESBL-producing Enterobacteriaceae; TMP-SMX, trimethoprim-sulfamethoxazole.
aThis group includes AmpC- and carbapenemase-producing Enterobacteriaceae.
Risk of ESBL-E Bacteremia Stratified by Pre-transplant Colonization Status
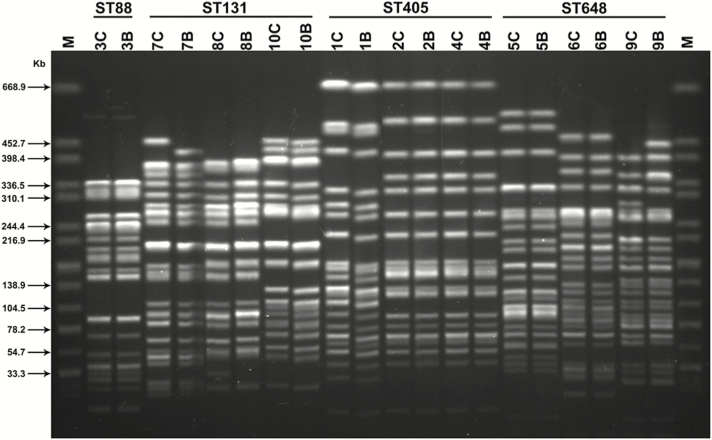
Of 31 patients colonized with ESBL-E pre-transplant, 10 (32%) developed ESBL-E bacteremia while neutropenic during their transplant admission, compared to 1 (0.4%) of 281 patients without pre-transplant ESBL-E colonization (P < .001; Figure 1). ESBL-E bacteremia developed in 8 (36%) of the 22 colonized allogeneic HSCT recipients and 2 (22%) of the 9 colonized autologous HSCT recipients. Among patients with pre-transplant ESBL-E colonization, all ESBL-E bacteremias occurred in patients colonized with levofloxacin-resistant CTX-M-producing E. coli. In fact, the risk of ESBL-E bacteremia was 42% (10/24) in such patients. The antimicrobial susceptibility profiles of each of the 10 pairs of colonizing and bloodstream ESBL-E were nearly identical, with identical susceptibility interpretations for at least 24 of 25 tested antimicrobial agents for all pairs. The genetic analysis demonstrated that all 10 pairs had identical MLST patterns and ESBL genes (Figure 2). Furthermore, >95% genetic similarity was observed by PFGE (Figure 2). Conversely, there was significant genetic heterogeneity of ESBL-E among the majority of patients, arguing against a clonal origin of these bacteria (Figure 2).
Figure 2.
Pulsed-field gel electrophoresis profiles of XbaI digested DNA of paired colonizing and bloodstream ESBL-producing Enterobacteriaceae, stratified by multilocus sequence type (ST). Each number refers to a unique patient, C refers to their colonizing strain, and B refers to their bloodstream strain. Strains from patients 1, 2, 4, 5, and 10 harbored blaCTX-M-15; 3, 6, and 9 harbored blaCTX-M-14; and 7 and 8 harbored blaCTX-M-27. Strains from patients 3, 5, 6, and 9 co-harbored blaTEM-1. Lane M: DNA size maker Salmonella enterica subsp. enterica serovar Braenderup (ATCC BA-664).
The patient who was not initially colonized with ESBL-E but developed ESBL-E bacteremia had a subsequent swab collected 2 days after transplantation that yielded ESBL-E and became bacteremic 4 days later. One patient who had pre-transplant colonization with an ESBL-producing E. coli developed a BSI caused by KPC-producing K. pneumoniae. Of the 6 patients colonized with AmpC-producing Enterobacteriaceae pre-transplant, none developed a subsequent BSI due to these organisms. One of the 3 patients colonized with KPC-producing K. pneumoniae developed subsequent BSI due to this organism.
Incidence of Fever and Neutropenia and Etiologies of Bloodstream Infections Stratified by Pre-transplant Colonization Status
Fever and neutropenia (FN) occurred in 22 (71%) of 31 patients colonized with ESBL-E pre-transplant, compared to 150 (53%) of 281 patients not colonized with ESBL-E pre-transplant (P = .06; Table 4). Patients colonized with ESBL-E were more likely to receive a carbapenem as initial empirical therapy for FN than patients not colonized with ESBL-E. The first episode of FN was associated with BSI in 55% of patients colonized with ESBL-E, compared to 28% in patients not colonized with ESBL-E (P = .01). Of the 12 BSIs associated with the first FN episode in patients with pre-transplant ESBL-E colonization, 9 were caused by their colonizing ESBL-E strain, 2 were caused by CRO-susceptible E. coli, 1 was caused by KPC-producing K. pneumoniae, and 0 were caused by Gram-positive bacteria. Gram-negative bloodstream isolates from patients colonized with ESBL-E were less likely to be susceptible to aztreonam, cefepime, ceftazidime, ceftriaxone, gentamicin, and tobramycin than those from patients without ESBL-E colonization (Supplementary Table). There was no significant difference in time to administration of an antimicrobial agent to which the bloodstream isolate was susceptible in patients colonized and not colonized with ESBL-E.
Table 4.
Comparison of the Incidence of Fever and Neutropenia, Initial Empirical Therapies, and Bloodstream Infections During the First Febrile Neutropenic Episode in Patients Who Were and Were Not Colonized With Extended-spectrum β-lactamase producing Enterobacteriaceae Prior to Transplant
| Colonized With ESBL-E (n = 31) |
Not Colonized With ESBL-E (n = 281) | P Value | |
|---|---|---|---|
| FN | 22 (71%) | 150 (53%) | .062 |
| Initial Gram-negative empirical therapy for FN | |||
| Piperacillin-tazobactam | 6/22 (27%) | 111/150 (74%) | <.001 |
| Meropenem | 14/22 (64%) | 35/150 (23%) | <.001 |
| Other | 2/22 (9%) | 4/150 (3%) | .16 |
| Etiology of FNa | |||
| Bloodstream infection | 12/22 (55%) | 42/150 (28%) | .012 |
| Gram-positive bacteremia | 0 | 26 (17%) | .034 |
| Viridans group streptococcib | 0 | 15 (10%) | .12 |
| Vancomycin-resistant Enterococcus faecium | 0 | 5 (3%) | 1.00 |
| Coagulase-negative staphylococcic | 0 | 3 (2%) | 1.00 |
| Otherd | 0 | 3 (2%) | 1.00 |
| Gram-negative bacteremia | 12 (55%) | 13 (9%) | <.001 |
| Escherichia coli | 11 (50%) | 11 (7%) | <.001 |
| Ceftriaxone-resistant | 9 (41%)e | 1 (1%) | <.001 |
| Ceftriaxone-susceptible | 2 (9%) | 9 (6%) | .64 |
| Klebsiella pneumoniae | 1 (5%) | 1 (1%) | 1.00 |
| Fusobacterium nucleatum | 0 | 1 (1%) | 1.00 |
| Polymicrobial | 0 | 2 (1%) | 1.00 |
| Candidemia | 0 | 1 (1%) | 1.00 |
Values expressed are number (% of total).
Abbreviations: ESBL-E, extended-spectrum β-lactamase-producing Enterobacteriaceae; FN, fever and neutropenia.
aThe denominator for bloodstream infection analyses are the number of patients with FN.
bViridans group streptococci (VGS) included S. mitis (n = 8), S. oralis (n = 4), S. salivarius (n = 1) and 2 other VGS that were not identified to the species level.
cCoagulase-negative staphylococci were only considered causes of bloodstream infection if recovered from ≥2 sets of blood cultures.
dOther Gram-positive bloodstream isolates were Clostridium innocuum, Peptostreptococcus micros, and methicillin-resistant Staphylococcus aureus (n = 1, for each).
eAll of the isolates were ESBL-E.
Risk of Acquisition of ESBL-E During the Transplant Admission and Transplant Outcomes
Of the 281 patients who were not initially colonized with ESBL-E, 248 (88%) were eligible for the acquisition analysis. Of those, 8 (3.2%) acquired ESBL-E during their transplant admission; the acquisition rates were similar in recipients of allogeneic (6/172; 3.5%) and autologous transplants (2/76; 2.6%). All but 1 of the acquired ESBL-E were levofloxacin-resistant. In patients with and without pre-transplant ESBL-E colonization, there were no significant differences in mortality rate during the transplant admission (6% vs. 2%, P = .15), 100-day mortality rate (10% vs. 7%, P = .55), or the proportion of allogeneic HSCT recipients who developed graft-versus-host disease (47% vs. 36%, P = .30).
DISCUSSION
In this single-center study of 312 HSCT recipients, we found that 10% of patients were colonized with ESBL-E prior to their transplant. Nearly one-third of patients with pre-transplant ESBL-E colonization developed subsequent ESBL-E bacteremia while neutropenic after their transplant, compared to <1% of patients who were not initially colonized with ESBL-E. Furthermore, the bloodstream and gastrointestinal ESBL-E had identical MLST and PFGE profiles in all cases, suggesting that these patients developed bacteremia from their colonizing isolates. All patients in this study received levofloxacin prophylaxis and, importantly, ESBL-E bacteremia only occurred in patients colonized with levofloxacin-resistant ESBL-E.
The rate of ESBL-E bacteremia found in this study in HSCT recipients colonized with ESBL-E (32%) is greater than the risk reported in prior studies that evaluated patients with hematologic malignancies. Analyses of patients with hematologic malignancies from 3 European studies reported ESBL-E bacteremia rates of 2–7% in colonized patients [16–18] and 1 study from Mexico reported a 22% risk of ESBL-E bacteremia [19]. In addition to only evaluating HSCT recipients, a critical difference between our study and these prior reports is that our patients received levofloxacin prophylaxis, whereas antibacterial prophylaxis was not routinely administered in these other studies. We found that only patients colonized with levofloxacin-resistant ESBL-E developed subsequent ESBL-E bacteremia, and that ESBL-E represented the vast majority of BSIs in these patients. We hypothesize that levofloxacin prophylaxis eradicated other enteric bacteria, which led to intestinal domination by levofloxacin-resistant ESBL-E, and that this state led to translocation of ESBL-E into the bloodstream during chemotherapy-induced neutropenia and mucositis. This model has been demonstrated with vancomycin-resistant enterococci in HSCT recipients [20]. We believe that our findings warrant additional investigations to evaluate the role of fluoroquinolone prophylaxis in neutropenic patients who are colonized with fluoroquinolone-resistant ESBL-E and to identify optimal infection prevention strategies in such patients.
The high rate of ESBL-E bacteremia in colonized patients is clinically important, because almost all of these organisms were resistant to anti-pseudomonal cephalosporins, which are recommended empirical therapies for fever and neutropenia [6]. Colonized neutropenic patients who develop cephalosporin-resistant ESBL-E bacteremia and are treated empirically with cephalosporins while awaiting culture results are likely to have poor clinical outcomes. In contrast to cephalosporins, 84% of ESBL-E were susceptible to piperacillin-tazobactam, another recommended agent for fever and neutropenia [6]. Despite this high rate of in vitro susceptibility to piperacillin-tazobactam, carbapenems have been considered the treatments of choice for ESBL-E bacteremia in neutropenic patients [6]. In a study of hospitalized patients, empirical therapy with piperacillin-tazobactam for ESBL-E bacteremia was associated with a 2-fold increased risk of death compared to empirical therapy with a carbapenem [21]. Although a recent study demonstrated similar outcomes in neutropenic patients treated with BL-BLIs compared to carbapenems, relatively few patients received BL-BLIs in this study, and thus it had limited statistical power to detect a difference between therapies [22]. Thus, we believe that carbapenems remain the optimal initial therapy for fever and neutropenia in neutropenic patients who are at high risk of ESBL-E bacteremia, such as patients colonized with ESBL-E in our study.
We are not aware of any prior studies that reported the prevalence of ESBL-E colonization prior to HSCT. However, the initial ESBL-E colonization rate of 10% in this study is similar to the 11–17% admission ESBL-E colonization rates that have been reported from Europe in patients with hematologic malignancies [16–18]. We did not find any association between ESBL-E colonization on admission for HSCT and recent antibacterial exposures, recent hospitalizations, or prior infection with multidrug-resistant bacteria. In fact, almost all patients found to be colonized with ESBL-E did not have a prior positive culture for ESBL-E. These data suggest a relatively high rate of ESBL-E colonization in the ambulatory setting and are consistent with reports of the rising incidence of ESBL-E in community-acquired infections [23, 24]. The only variables associated with ESBL-E colonization were Middle Eastern ethnicity and underlying myelodysplastic syndrome or myeloproliferative disorder, and these characteristics were uncommon. Thus, our data suggest that risk stratification for colonization with ESBL-E based on clinical criteria is challenging, and all HSCT recipients would need to be screened for gastrointestinal carriage to reliably identify patients colonized with ESBL-E.
ESBL-E acquisition during the transplant admission occurred in only 3% of patients. Studies of patients in hematology and intensive care units have also demonstrated low rates of inpatient ESBL-E acquisition [18, 25]. Our low rate of ESBL-E acquisition should be interpreted in the context of 2 factors: (1) patients were located in single-occupancy rooms on a dedicated HSCT unit, and (2) patients found to be colonized with ESBL-E were placed on contact precautions. In addition to low rates of ESBL-E acquisition during the transplant admission, the marked genetic heterogeneity among patients who were initially colonized with ESBL-E suggests that these patients did not typically acquire ESBL-E strains from each other during previous hospitalizations.
Our study has several limitations. We do not know if the high rate of ESBL-E bacteremia in colonized patients in this single-center study would apply to other neutropenic patient populations who do not receive a HSCT or to other transplant centers, particularly centers that do not administer fluoroquinolone prophylaxis. Second, the results of the perianal swab cultures were reported to the clinical staff, and thus many patients colonized with ESBL-E were treated empirically with carbapenems for FN, instead of being treated with our center’s typical first-line agent, piperacillin-tazobactam. Thus, nearly all ESBL-E bacteremia episodes occurred during the first episode of fever and neutropenia, prior to the administration of a carbapenem. Third, we did not perform a broth enrichment step when isolating CRO-R-E from perianal swabs, a method that has been found to increase the yield of detecting ESBL-E [26–29]. Thus, it is possible that the ESBL-E colonization rates were underestimated. However, the direct plating methods used in this study had sufficient sensitivity to distinguish patients who were at high risk of ESBL-E bacteremia from those at low risk, presumably because the additional yield of broth enrichment would have only identified patients with a low burden of organisms. Fourth, few patients were colonized with AmpC- and carbapenemase-producing Enterobacteriaceae, and thus we had limited ability to estimate the risk of bacteremia in patients colonized with these organisms. Finally, we have not demonstrated in this study that screening for ESBL-E carriage and initiating empirical carbapenem therapy for FN leads to improved clinical outcomes. This potential benefit must be balanced against the potential risks of future selection of carbapenem resistance. However, we believe that our data provide a compelling rationale to initiate a multicenter, interventional trial designed to assess the benefits and risks of a strategy of screening for carriage of levofloxacin-resistant ESBL-E and of individualizing empirical therapy.
Our findings have implications for antibacterial prophylaxis and empirical therapy at HSCT centers where ESBL-E are prevalent bloodstream pathogens. We demonstrated that, in the setting of levofloxacin prophylaxis, screening the gastrointestinal tract for colonization with levofloxacin-resistant ESBL-E identifies patients who are at high risk of developing ESBL-E bacteremia during their first episode of FN.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (grant numbers K23 AI114994 to M. J. S. and R01 AI090155 to B. N. K.) and the New York Community Trust (grant number P13-000934 to B. N. K.).
Potential conflicts of interest. M. J. S, A. N. S., and T. J. W. have received research grants through Weill Cornell Medicine from Allergan and Merck. M. J. S. has also consulted for Achaogen. S. G. J. reports grants from Merck, outside the submitted work. L. F. W. reports grants from Accelerate Diagnostics, outside the submitted work. L. C. reports grants from National Institutes of Allergy and Infectious Diseases, outside the submitted work. T. B. reports other income from Janssen Research & Development, outside the submitted work. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Mikulska M, Del Bono V, Raiola AM et al. Blood stream infections in allogeneic hematopoietic stem cell transplant recipients: reemergence of Gram-negative rods and increasing antibiotic resistance. Biol Blood Marrow Transplant 2009; 15:47–53. [DOI] [PubMed] [Google Scholar]
- 2. Gudiol C, Calatayud L, Garcia-Vidal C et al. Bacteraemia due to extended-spectrum beta-lactamase-producing Escherichia coli (ESBL-EC) in cancer patients: clinical features, risk factors, molecular epidemiology and outcome. J Antimicrob Chemother 2010; 65:333–41. [DOI] [PubMed] [Google Scholar]
- 3. Ha YE, Kang CI, Cha MK et al. Epidemiology and clinical outcomes of bloodstream infections caused by extended-spectrum β-lactamase-producing Escherichia coli in patients with cancer. Int J Antimicrob Agents 2013; 42:403–9. [DOI] [PubMed] [Google Scholar]
- 4. Kim SH, Kwon JC, Choi SM et al. Escherichia coli and Klebsiella pneumoniae bacteremia in patients with neutropenic fever: factors associated with extended-spectrum β-lactamase production and its impact on outcome. Ann Hematol 2013; 92:533–41. [DOI] [PubMed] [Google Scholar]
- 5. Yemişen M, Balkan II, Salihoğlu A et al. The changing epidemiology of bloodstream infections and resistance in hematopoietic stem cell transplantation recipients. Turk J Hematol 2016; 33:216–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Freifeld AG, Bow EJ, Sepkowitz KA et al. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 2011; 52:e56–93. [DOI] [PubMed] [Google Scholar]
- 7. Lee NY, Lee CC, Huang WH, Tsui KC, Hsueh PR, Ko WC. Cefepime therapy for monomicrobial bacteremia caused by cefepime-susceptible extended-spectrum beta-lactamase-producing Enterobacteriaceae: MIC matters. Clin Infect Dis 2013; 56:488–95. [DOI] [PubMed] [Google Scholar]
- 8. Wang R, Cosgrove SE, Tschudin-Sutter S et al. Cefepime therapy for cefepime-susceptible extended-spectrum β-lactamase-producing Enterobacteriaceae bacteremia. Open Forum Infect Dis 2016; 3:ofw132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Trecarichi EM, Pagano L, Candoni A et al. ; HeMABIS Registry—SEIFEM Group, Italy Current epidemiology and antimicrobial resistance data for bacterial bloodstream infections in patients with hematologic malignancies: an Italian multicentre prospective survey. Clin Microbiol Infect 2015; 21:337–43. [DOI] [PubMed] [Google Scholar]
- 10. Cornejo-Juárez P, Pérez-Jiménez C, Silva-Sánchez J et al. Molecular analysis and risk factors for Escherichia coli producing extended-spectrum β-lactamase bloodstream infection in hematological malignancies. PLoS One 2012; 7:e35780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kang CI, Chung DR, Ko KS, Peck KR, Song JH; Korean Network for Study of Infectious Diseases Risk factors for infection and treatment outcome of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae bacteremia in patients with hematologic malignancy. Ann Hematol 2012; 91:115–21. [DOI] [PubMed] [Google Scholar]
- 12. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 26th informational supplement. CLSI M100-S26. Wayne, PA: CLSI; 2016. [Google Scholar]
- 13. Chavda KD, Satlin MJ, Chen L et al. Evaluation of a multiplex PCR assay to rapidly detect enterobacteriaceae with a broad range of β-lactamases directly from perianal swabs. Antimicrob Agents Chemother 2016; 60:6957–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wirth T, Falush D, Lan R et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 2006; 60:1136–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Durmaz R, Otlu B, Koksal F et al. The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella spp. Jpn J Infect Dis 2009; 62:372–7. [PubMed] [Google Scholar]
- 16. Arnan M, Gudiol C, Calatayud L et al. Risk factors for, and clinical relevance of, faecal extended-spectrum β-lactamase producing Escherichia coli (ESBL-EC) carriage in neutropenic patients with haematological malignancies. Eur J Clin Microbiol Infect Dis 2011; 30:355–60. [DOI] [PubMed] [Google Scholar]
- 17. Liss BJ, Vehreschild JJ, Cornely OA et al. Intestinal colonisation and blood stream infections due to vancomycin-resistant enterococci (VRE) and extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBLE) in patients with haematological and oncological malignancies. Infection 2012; 40:613–9. [DOI] [PubMed] [Google Scholar]
- 18. Vehreschild MJ, Hamprecht A, Peterson L et al. A multicentre cohort study on colonization and infection with ESBL-producing Enterobacteriaceae in high-risk patients with haematological malignancies. J Antimicrob Chemother 2014; 69:3387–92. [DOI] [PubMed] [Google Scholar]
- 19. Cornejo-Juárez P, Suárez-Cuenca JA, Volkow-Fernández P et al. Fecal ESBL Escherichia coli carriage as a risk factor for bacteremia in patients with hematological malignancies. Support Care Cancer 2016; 24:253–9. [DOI] [PubMed] [Google Scholar]
- 20. Taur Y, Xavier JB, Lipuma L et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012; 55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tamma PD, Han JH, Rock C et al. ; Antibacterial Resistance Leadership Group Carbapenem therapy is associated with improved survival compared with piperacillin-tazobactam for patients with extended-spectrum β-lactamase bacteremia. Clin Infect Dis 2015; 60:1319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gudiol C, Royo-Cebrecos C, Abdala E et al. Efficacy of β-lactam/β-lactamase inhibitor combinations for the treatment of bloodstream infection due to extended-spectrum β-lactamase-producing Enterobacteriaceae in hematological patients with neutropenia. Antimicrob Agents Chemother 2017; 61:e00164–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karanika S, Karantanos T, Arvanitis M, Grigoras C, Mylonakis E. Fecal colonization with extended-spectrum beta-lactamase-producing Enterobacteriaceae and risk factors among healthy individuals: a systematic review and metaanalysis. Clin Infect Dis 2016; 63:310–8. [DOI] [PubMed] [Google Scholar]
- 24. Anesi JA, Lautenbach E, Nachamkin I et al. Clinical and molecular characterization of community-onset urinary tract infections due to extended-spectrum cephalosporin-resistant Enterobacteriaceae. Infect Control Hosp Epidemiol 2016; 37:1433–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tschudin-Sutter S, Frei R, Dangel M, Stranden A, Widmer AF. Rate of transmission of extended-spectrum beta-lactamase-producing Enterobacteriaceae without contact isolation. Clin Infect Dis 2012; 55:1505–11. [DOI] [PubMed] [Google Scholar]
- 26. Murk JL, Heddema ER, Hess DL, Bogaards JA, Vandenbroucke-Grauls CM, Debets-Ossenkopp YJ. Enrichment broth improved detection of extended-spectrum-beta-lactamase-producing bacteria in throat and rectal surveillance cultures of samples from patients in intensive care units. J Clin Microbiol 2009; 47:1885–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Diederen B, Chang C, Euser S, Stuart JC. Evaluation of four screening protocols for detection of extended-spectrum β-lactamase-producing members of the Enterobacteriaceae. J Med Microbiol 2012; 61:452–3. [DOI] [PubMed] [Google Scholar]
- 28. Kluytmans-van den Bergh MF, Verhulst C, Willemsen LE, Verkade E, Bonten MJ, Kluytmans JA. Rectal carriage of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in hospitalized patients: selective preenrichment increases yield of screening. J Clin Microbiol 2015; 53:2709–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jazmati N, Hein R, Hamprecht A. Use of an enrichment broth improves detection of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in clinical stool samples. J Clin Microbiol 2016; 54:467–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.