Abstract
INTRODUCTION:
Necitumumab is a monoclonal antibody targeting the epidermal growth factor receptor (EGFR). In the SQUIRE trial, the addition of necitumumab to chemotherapy in squamous cell lung cancer (SCC) significantly improved overall survival (OS) (HR=0.84); in a post-hoc analysis, EGFR copy number gain determined by fluorescence in situ hybridization (FISH) showed a trend towards improved OS (HR=0.70) and progression-free survival (PFS) (HR=0.71) with the addition of necitumumab. We present the analysis of granular EGFR-FISH data from SQUIRE to examine the potential predictive role of high polysomy and gene amplification as both were included in the “FISH-positive” category.
MATERIALS AND METHODS:
Available specimens from SQUIRE underwent FISH analysis in a central laboratory and each sample was evaluated using the Colorado EGFR scoring criteria. The correlation of granular FISH parameters with clinical outcomes was assessed.
RESULTS:
Samples were available for 557 patients (out of 1093); 208 patients (37.3%) were FISH+, including 167 (30.0%) with high polysomy and 41 (7.4%) with gene amplification. In patients with high polysomy, the addition of necitumumab resulted in a statistically significant increase in PFS (6.08 vs. 5.13 months; p=0.044) and nonstatistically significant increase in OS (12.6 vs. 9.5 months; p=0.133); among patients with gene amplification, the addition of necitumumab did not significantly improve PFS (7.4 vs. 5.6 months; p= 0.334), but improved OS (14.8 vs. 7.6 months; p= 0.033).
CONCLUSION:
EGFR copy number gain by FISH might have a role as predictive biomarker for necitumumab in SCC. In our opinion, these data encourage further studies to prospectively evaluate this potential biomarker.
Keywords: non-small cell lung cancer, EGFR, FISH, necitumumab, biomarker
1. INTRODUCTION
Non-small cell lung cancer (NSCLC) is a major healthcare problem and accounts for a high proportion of cancer-related deaths around the world. While cancer research has led to significant improvements in the treatment of this disease, most cases are still diagnosed at advanced stage and the outcomes of such patients are underwhelming [1]. Non-small cell lung cancers are further divided into histo-types, the most common being adenocarcinoma (ADC) and squamous cell carcinoma (SCC). It is widely known that characterizing the correct histo-type is pivotal for the correct treatment approach of advanced disease, as the utilization of several anti-neoplastic treatments and targeted agents are primariliy limited to non-squamous histology [2,3,4].
The expression of the epidermal growth factor receptor (EGFR) was reported to be significantly higher in SCC compared to ADC, suggesting a potential role as therapeutic target that has not been fully exploited yet [5]. Cetuximab, a monoclonal antibody (mAB) targeting EGFR, was employed in combination with first-line chemotherapy in an openlabel, phase III trial involving treatment-naïve patients with advanced NSCLC in presence of EGFR expression [6]. A retrospective, but pre-defined analysis demonstrated an advantage in terms of overall survival (OS) in the patients whose tumor sample expressed higher levels of EGFR as assessed by immunohistochemistry (IHC), defined through a specific histo-score (H-score, which ranges from 0 to 300) [7]. Another subgroup analysis showed a trend for a more pronounced survival advantage in NSCLC patients with squamous histology.
In the open-label, phase III SQUamous NSCLC treatment with the Inhibitor of EGF REceptor (SQUIRE) trial, 1093 patients with advanced squamous NSCLC were randomized to receive first-line chemotherapy of cisplatin-gemcitabine with or without necitumumab, a human mAB directed against EGFR. The combination containing necitumumab achieved superior results in terms of OS (11.5 vs. 9.9 months; hazard ratio [HR]= 0.84; 95% confidence interval [CI]= 0.74–0.96; p= 0.01) as well as progression-free survival (PFS; 5.7 vs. 5.5 months; HR= 0.85; 95% CI= 0.74–0.98; p= 0.02) compared with chemotherapy alone; objective response rate (ORR) did not differ between experimental and control arm (31% vs. 29%; p= 0.40), while disease control rate (DCR) favored the arm containing necitumumab (82% vs. 77%; p= 0.043). The trial included a pre-planned analysis on the basis of low EGFR H-score (< 200) vs. high Hscore (≥ 200). Although the advantage in terms of OS appeared numerically greater with high H-score (HR= 0.75; 95% CI= 0.60–0.94) than with low H-score (HR= 0.90; 95% CI= 0.75–1.07), this difference was not statistically significant (interaction p= 0.235), and an opposite trend was observed for PFS [8]. Notably, another pre-planned analysis suggested that the small sub-group of patients without detectable EGFR protein by IHC, accounting for 5% of the intention-to-treat population (ITT), may not benefit from the addition of necitumumab to cisplatin-gemcitabine [9]. The main findings of SQUIRE have been summarized in supplementary table 1.
The use of fluorescence in situ hybridization (FISH) to predict the efficacy of targeted antineoplastic treatments based on gene copy number (GCN) has a relevant role in clinical practice as well as clinical research for certain solid tumors [10,11]. Available data suggest that EGFR GCN is associated with protein overexpression and might be correlated with survival in NSCLC [12]. Additionally, observations from S0342, a phase II trial conducted by the South West Oncology Group (SWOG), suggested that the outcome of patients treated with an EGFR-mAB might be correlated with EGFR GCN. More specifically, out of 76 patients treated with carboplatin-paclitaxel-cetuximab for advanced NSCLC, 59% were positive for increased EGFR GCN assessed through FISH and achieved improved DCR (81% vs. 55%; p= 0.02), PFS (6 vs. 3 months; p= 0.008), and OS (15 vs. 7 months; p= 0.04) compared to patients with normal GCN [13].
A scheme for classifying NSCLC as EGFR FISH-positive or FISH-negative was developed at the University of Colorado and employed in several clinical studies; these criteria, known as Colorado scoring criteria, are based on granular parameters involving EGFR and chromosome 7 (CEP7) signals [14,15]. The same criteria and laboratory were used to define FISH-positive and FISH-negative patients within the large randomized SWOG 0819 study, which compared chemotherapy with carboplatinpaclitaxel (+/− bevacizumab) with or without cetuximab for advanced NSCLC; with regards to FISH status, while no difference in terms of OS was observed among patients with non-squamous histology, among the 111 FISH-positive patients affected by squamous cell lung carcinoma, the addition of cetuximab to standard chemotherapy (55 patients) compared to standard chemotherapy alone (56 patients) resulted in improved OS (11.8 vs. 6.4 months; HR= 0.56; p= 0.01) [16].
In an exploratory post-hoc analysis of SQUIRE, the correlation between EGFR FISH status (positive vs. negative) and efficacy outcomes of 557 patients (51% of the ITT population from the SQUIRE trial) whose specimens were available for FISH analysis was assessed. Although the treatment-by-marker interaction tests were not statistically significant, a trend for a more favorable HR was observed in the FISH-positive subpopulation. Full details have been reported in supplementary table 2 [9]. Our aim was to retrospectively explore the potential role of FISH granular data beyond the positive/negative status as predictors of benefit from the addition of necitumumab to first-line platinum-based chemotherapy in order to generate hypotheses for future prospective studies.
2. MATERIALS and METHODS
For this study, samples collected from patients enrolled within the SQUIRE trial in agreement with study design (Clinicaltrials.gov NCT: 00981058) [17] and considered suitable for FISH were subsequently analyzed. Of note, tissue availability was an eligibility criterion for SQUIRE with a pre-defined sequence of biomarkers to be evaluated that also included EGFR GCN by FISH. The samples were fixed for 6–48 hours in 10% neutral buffered formalin and stored as formalin-fixed, paraffin embedded blocks; after confirming the presence of tumor, tissue sections of 3–5 μm were mounted on positively charged slides. Dual-target FISH enumeration assay was performed as previously described [15]. Quality assessment for each sample and assay reading were performed by trained personnel. Five tumor areas considered representative of the tumor were selected, each one containing an average of 10 evaluable nuclei (for a total of 50), and the number of EGFR and CEP7 signals in the selected nuclei were counted with single-pass interference filters according to published guidelines [18]. During the enumeration of the signals, doublet and triplet spots that were physically linked were counted as a single signal, while signals in clusters were counted to a maximum of 15 (if signals were >15, the cluster was considered as innumerable and the value was reported as “16”). The EGFR assay was reported as positive if at least 40% of the scored tumor cells displayed four or more copies of the EGFR signal (high polysomy), or in case of gene amplification. Gene amplification was defined as one of the following: A) if EGFR/CEP7 ratio was at least equal to 2 in all scored nuclei and calculated using the mean of EGFR signals per cell divided by the mean of CEP7 signals per cell when mean CEP7 per cell was at least equal to two copies, or B) if gene clusters (at least four spots) were present in at least 10% of neoplastic cells, or else C) if at least 15 copies of the EGFR signals were present in at least 10% of tumor cells. In all other scenarios the EGFR FISH assay was considered negative. According to the Colorado Scoring Criteria summarized in table 1 [18], six categories were identified: disomy (score=1), low trisomy (score=2), high trisomy (score=3), low polysomy (score=4), high polysomy (score=5), and gene amplification (score=6). If a sample met the critieria for both high polysomy and gene amplification, the sample was categorized as score=6.
Table 1.
Colorado Scoring Criteria for EGFR FISH. The table includes the number of eligible patients from the SQUIRE trial, divided by scoring category and treatment arm.
| Score | Observation | N# of patients (%) | EGFR signal features | FISH result Patients (%) | ||
|---|---|---|---|---|---|---|
| Arm A | Arm B | Overall | ||||
| 1 | Disomy | 11 (3.9%) | 13 (4.7%) | 24 (4.3%) | < 40% of cells displaying ≥ 4 copies of the EGFR signal AND EGFR/CEP7 ratio < 2 AND absence of gene clusters AND < 10% of cells displaying ≥ 15 copies of EGFR |
Negative
349 (62.7%) |
| 2 | Low trisomy | 61 (21.6%) | 56 (20.4%) | 117 (21.0%) | ||
| 3 | High trisomy | 10 (3.5%) | 13 (4.7%) | 23 (4.1%) | ||
| 4 | Low polysomy | 89 (31.6%) | 96 (34.9%) | 185 (33.2%) | ||
| 5 | High polysomy | 89 (31.6%) | 78 (28.4%) | 167 (30.0%) | ≥ 40% of cells displaying ≥ 4 copies of the EGFR signal* |
Positive
208 (37.3%) |
| 6 | Gene amplification | 22 (7.8%) | 19 (6.9%) | 41 (7.4%) | EGFR/CEP7 ratio ≥ 2 OR presence of gene clusters (≥ 4 spots) OR ≥ 10% of cells displaying ≥ 15 copies of EGFR* |
|
| TOTAL | 282 (50.6%) | 275 (49.4%) | 557 (100%) | |||
Arm A: cisplatin-gemcitabine-necitumumab; Arm B: cisplatin-gemcitabine.
If a sample met the critieria for both high polysomy and gene amplification, the sample was categorized as score=6.
Hence, the clinical data from the SQUIRE patients corresponding to the available samples, including age, gender, smoking history, Eastern Cooperative Oncology Group performance status, ethnicity, and efficacy outcomes (PFS and OS) were compared with the FISH assay results, in order to determine any possible correlation. Our study includes the comparison between patients with high polysomy and patients with gene amplification within the FISH positive population; additionally, the comparison included patients with ≥ 40% vs. < 40% of cells with ≥ 4 EGFR copies, patients with ≥ 10% vs. < 10% of cells with ≥ 15 EGFR copies, and patients with EGFR/CEP7 ratio ≥ 2 vs. < 2.
3. STATISTICAL ANALYSIS
Baseline demographic and disease characteristics between trial arms or between the subset of patients were summarized to examine if substantial imbalance existed. For exploratory efficacy analyses, we presented quantiles of PFS and OS within each subset, and used the Cox proportional hazards model for time-to-event endpoints (PFS or OS). Interaction model was used where an efficacy endpoint was modeled as a function of treatment, EGFR FISH class and their interaction. Hazard ratio estimates from a Cox regression model, 95% CI and log-rank p-values were calculated. Because of the relatively small sample size and exploratory nature, we only considered unstratified model in all analyses without multiplicity adjustment.
4. RESULTS
Among the patients enrolled in SQUIRE, 557 had tissue samples suitable for FISH analysis and provided valid results, corresponding to 51% of the ITT population. The comparative survival data for the ITT population and for the FISH analysis population are shown in table 2. Among the patients subjected to FISH analysis, median PFS in the experimental arm and in the standard arm was 5.75 and 5.49 months, respectively (HR= 0.901; p= 0.2768), while OS in the experimental arm and in the standard arm was 11.73 and 9.99 months, respectively (HR= 0.892; p= 0.2309).
Table 2.
Survival data for the ITT population and for the FISH analysis population of SQUIRE.
| ITT POPULATION | FISH ANALYSIS POPULATION | ||||||
|---|---|---|---|---|---|---|---|
| PARAMETER | ARM A (N=545) | ARM B (N=548) | ARM A (N=282) | ARM B (N=275) | |||
| OS | Number of Events, n (%) | 418 (76.70) | 442 (80.66) | 218 (77.30) | 221 (80.36) | ||
| Number of Censored, n (%) | 127 (23.30) | 106 (19.34) | 64 (22.70) | 54 (19.64) | |||
| Observed range (min, max) (month) | (0.03,36.99) | (0.03,38.54) | (0.03,36.99) | (0.07,37.39) | |||
| 25th percentile (month) (95% CI) | 6.77 (6.05,7.75) | 5.68 (5.13,6.14) | 7.49 (6.11,8.48) | 5.68 (4.99,6.31) | |||
| Median OS (month) (95% CI) | 11.50 (10.45,12.58) | 9.86 (8.87,11.10) | 11.73 (10.61,12.98) | 9.99 (8.77,11.86) | |||
| 75th percentile (month) (95% CI) | 20.27 (17.91,22.90) | 17.74(16.39,19.91) | 20.24 (17.51,22.90) | 18.00 (16.16,21.06) | |||
| Unstratified HR (95% CI) (a) | 0.850 (0.744,0.972) | 0.892 (0.739,1.076) | |||||
| Unstratified Log-rank p-value (b) | 0.0176 | 0.2309 | |||||
| PFS | Number of Events, n (%) | 431 (79.08) | 417(76.09) | 227(80.50) | 207(75.27) | ||
| Number of Censored, n (%) | 114 (20.92) | 131(23.91) | 55(19.50) | 68(24.73) | |||
| Observed range (min, max) (month) | (0.03,33.31) | (0.03,30.49) | (0.03,27.56) | (0.03,30.49) | |||
| 25th percentile (month) (95% CI) | 4.07 (3.06,4.21) | 2.86 (2.76,3.22) | 4.14 (3.12,4.27) | 2.99 (2.76,4.11) | |||
| Median PFS (month) (95% CI) | 5.68 (5.59,5.98) | 5.45 (4.83,5.55) | 5.75 (5.59,6.77) | 5.49 (4.70,5.62) | |||
| 75th percentile (month) (95% CI) | 8.34 (8.21,9.66) | 7.26 (6.93,8.21) | 8.87 (7.98,9.76) | 7.29 (6.90,8.38) | |||
| Unstratified HR (95% CI) (a) | 0.852 (0.745,0.976) | 0.901 (0.745,1.088) | |||||
| Unstratified Log-rank p-value (b) | 0.0196 | 0.2768 | |||||
Arm A: cisplatin-gemcitabine-necitumumab; Arm B: cisplatin-gemcitabine.
Hazard ratio and 95% confidence interval obtained using a unstratified Cox proportional hazards model.
Log-rank p-value (2-sided). P-value is not adjusted for multiple testing.
CI: confidence interval; HR: hazard ratio; ITT: intention-to-treat; OS: overall survival; PFS: progression-free survival.
Arm A: cisplatin-gemcitabine-necitumumab; Arm B: cisplatin-gemcitabine.
CEP7: chromosome 7; CI= confidence interval; EGFR: epidermal growth factor receptor (gene) HR= hazard ratio; OS: overall survival; PFS: progression-free survival.
The categorization of the evaluable patients according to the Colorado Scoring Criteria is summarized in table 1, while their demographic characteristics are summarized in supplementary table 3. Among the EGFR FISH positive patients, 167 (80.3% of the FISH positive patients; 30.0% of the FISH analysis population) had high polysomy, while 41 (18.8% of the FISH positive patients; 7.4% of the FISH analysis population) had gene amplification. The demographic characteristics of positive vs. negative patients are reported in supplementary table 4. With regards to FISH positive subclassifications, all but one FISH positive patient had at least 40% of cells with at least four EGFR copies, therefore results for this categorization are essentially identical to positive FISH; all but two gene-amplified patients (Colorado Score=6) had at least 10% of cells with at least 15 copies, therefore these categorizations are essentially identical for the purpose of the analysis.
The EGFR/CEP7 ratio was ≥ 2 in 34 patients (6.10%) and < 2 in 523 patients (93.90%), respectively. Among the patients with EGFR/CEP7 ratio ≥ 2, median OS in the experimental and in the standard arm was 12.1 and 7.6 months, respectively (HR=0.63; p=0.245), while median PFS was 7.4 and 5.6 months, respectively (HR=0.80; p=0.581). Among the patients with EGFR/CEP7 ratio < 2, median OS in the experimental arm and in the standard arm was 11.7 and 10.0 months, respectively (HR=0.91; p=0.355), while median PFS was 5.7 and 5.5 months, respectively (HR=0.91; p=0.345). The survival analysis according to EGFR/CEP7 ratio is reported in table 3.
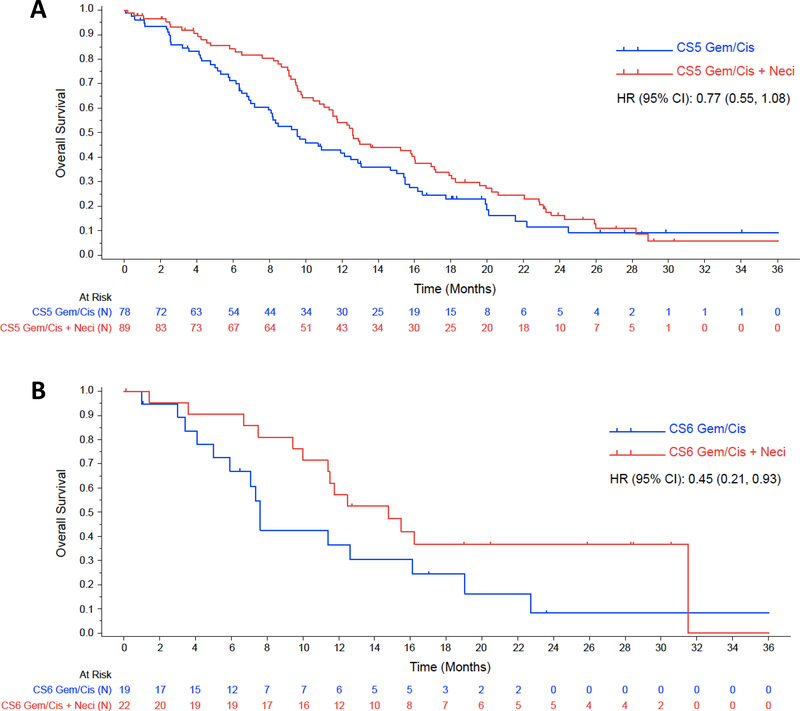
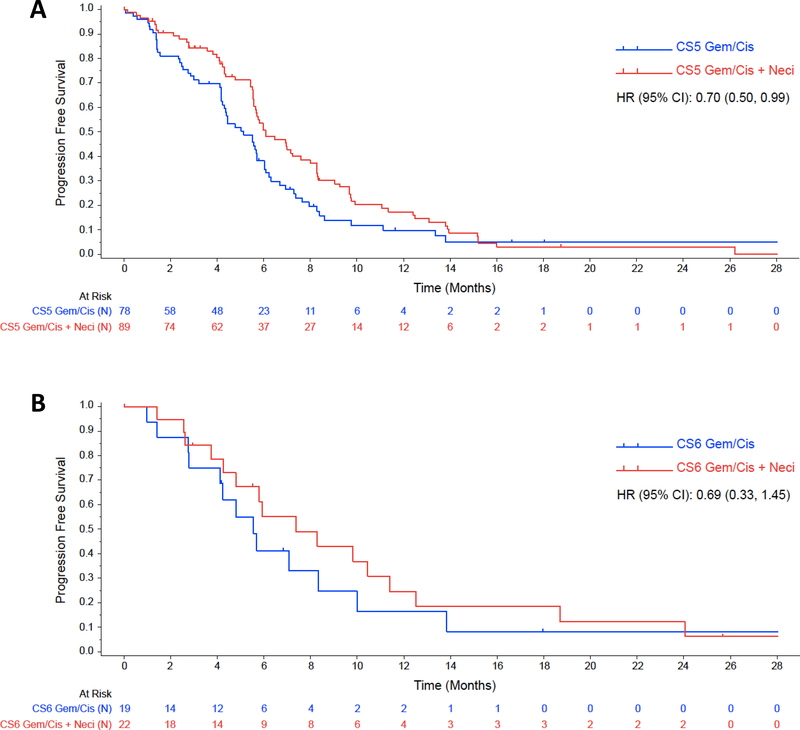
When the analysis was limited to FISH-positive patients, the addition of necitumumab to chemotherapy, compared to chemotherapy alone, in high polysomy (Colorado Score=5) patients, resulted in a significant increase in terms of PFS (6.1 vs. 5.1 months; p=0.044), while the OS effect was not statistically significant (12.6 vs. 9.5 months; p=0.133). On the contrary, in the gene amplified patients (Colorado Score=6), the addition of necitumumab did not improve PFS significantly (7.4 vs. 5.6 months; p= 0.334), but improved OS (14.8 vs. 7.6 months; p= 0.033). However, neither the OS nor the PFS interactions were significant. These results are presented in table 4. Analysis conclusions are limited by sample number. The Kaplan-Meier estimates for OS and PFS for this analysis are shown in figure 1 and figure 2, respectively.
Table 4.
Survival data for sub-populations of FISH positive patients.
| HIGH POLYSOMY | GENE AMPLIFICATION | |||
|---|---|---|---|---|
| Arm A | Arm B | Arm A | Arm B | |
| N | 89 | 78 | 22 | 19 |
| Median OS (95% CI) | 12.58 (11.04–16.00) | 9.53 (7.16–12.48) | 14.78 (10.02–31.51) | 7.62 (4.99–16.10) |
| HR within subgroup (Interaction model) | 0.77 (0.55–1.08) p = 0.133 | 0.45 (0.21–0.93) p = 0.033 | ||
| Interaction p-value | 0.189 | |||
| Median PFS (95% CI) | 6.08 (5.59–7.59) | 5.13 (4.24–5.72) | 7.36 (4.27–11.40) | 5.55 (2.79–8.34) |
| HR within subgroup (Interaction model) | 0.70 (0.50–0.99) p = 0.044 | 0.69 (0.33–1.45) p = 0.334 | ||
| Interaction p-value | 0.980 | |||
Arm A: cisplatin-gemcitabine-necitumumab; arm B: cisplatin-gemcitabine.
CI: confidence interval; HR: hazard ratio; OS: overall survival; PFS: progression-free survival.
FIGURE 1.
Kaplan-Meier estimates for OS according to treatment arm among EGFR FISH-positive patients; figure 1A shows estimates for patients with Colorado score 5, while figure 1B shows estimates for patients with Colorado score 6.
CS5: Colorado Score 5; CS6: Colorado score 6; Gem/Cis: gemcitabine plus cisplatin; Gem/Cis + Neci: Gemcitabine plus cisplatin plus necitumumab.
FIGURE 2.
Kaplan-Meier estimates for PFS according to treatment arm among EGFR FISH-positive patients; figure 2A shows estimates for patients with Colorado score 5, while figure 2B shows estimates for patients with Colorado score 6.
CS5: Colorado Score 5; CS6: Colorado score 6; Gem/Cis: gemcitabine plus cisplatin; Gem/Cis + Neci: Gemcitabine plus cisplatin plus necitumumab.
6. DISCUSSION
The addition of necitumumab to first-line chemotherapy with cisplatin-gemcitabine resulted in a significant survival advantage in patients with squamous NSCLC in the randomized, phase III SQUIRE trial; however, to date, no strong predictor of benefit has been identified. Since a post-hoc analysis of SQUIRE EGFR FISH status has suggested a trend towards significance in terms of outcome prediction, our hypothesis was that granular FISH data might provide potential predictive biomarkers and therefore allow identifying patients who are more likely to benefit from the addition of necitumumab. The FISH analysis population included 51% of the patients from ITT population. Within the FISH analysis population, the addition of necitumumab resulted in an improvement of PFS and OS that was numerically similar to what was observed within the ITT population; the improvements were not statistically significant within the FISH analysis population, but this observation may be related to the smaller number of patients compared to the ITT analysis. When FISH sub-populations were taken into account, the addition of necitumumab to chemotherapy resulted in an increase in OS (12.1 vs. 7.6 months) among the patients harboring an EGFR/CEP7 ratio ≥ 2; although not statistically significant, the HR for this difference was 0.63. Notably, the number of patients with increased EGFR/CEP7 ratio was limited; therefore, definitive conclusions cannot be drawn. Finally, among the FISH positive patients, a significant advantage in terms of PFS was observed in the patients with high polysomy receiving chemotherapy plus necitumumab, while a significant advantage in terms of OS was observed in patients with gene amplification; while the interaction p-values were not significant (likely due to the limited sample sizes), the HR are not negligible, with particular mention to the HR for OS in gene-amplified patients (0.45).
There are limitations associated with this analysis, mostly deriving from the granular nature of the data. First, while in this study a relatively large number of patients with SCC had samples available for FISH analysis, the number of patients for each subcategory is limited, influencing the statistical test results; this is especially true for patients with gene amplification, which represent only 7.4% of the evaluable patients. Additionally, since this is a post-hoc analysis, only a proportion (albeit generally representative) of the ITT population was eligible for this analysis and the patients had not been stratified for FISH status, potentially leading to some imbalances among demographic characteristics. Finally, while a statistically significant advantage in terms of PFS was observed in the high polysomy sub-category, but not among the patients with gene amplification, it should be noted that this specific end-point might be influenced by different parameters and variables, especially considering the lack of preplanned stratification for granular FISH data and the number of patients for each subcategory.
The current study of gene copy number alterations by FISH is performed in the same laboratory and using the same criteria as the previously mentioned SWOG 0819 study comparing chemotherapy with- or without cetuximab in patients with advanced NSCLC, in which also in the SCC group of patients EGFR FISH seemed to predict better outcome with the addition of cetuximab [19]. Thus, the potential predictive role of the EGFR FISH assay with regard to EGFR antibodies seems to be consistent in the two studies where inter-laboratory variability as a confounding factor is eliminated. Eventually, a future meta-analysis of the results from these two studies might give a statistically stronger evidence regarding the predictive role of EGFR gene copy number and the FISH assay’s predictive role for EGFR antibody therapy in patients with NSCLC. Additionally, further studies might allow the evaluation of a greater number of patients with gene amplification, resulting in more robust data and eventually confirming the consistence of our findings.
In conclusion, while our results are not robust enough for drawing definitive conclusions with immediate clinical application, they suggest that EGFR copy number gain by FISH might have a potential role as a predictive biomarker of clinical outcomes for EGFR mABs in SCC, especially in the case of gene amplification. By consequence, we consider these data encouraging and advise the development of future studies designed to prospectively address the potential role of EGFR FISH in this setting.
Supplementary Material
Table 3.
Survival data according to EGFR/CEP7 ratio (≥ 2 vs. < 2).
| EGFR/CEP7 RATIO | ||||
|---|---|---|---|---|
| ≥ 2 | < 2 | |||
| ARM A | ARM B | ARM A | ARM B | |
| N | 18 | 16 | 264 | 259 |
| Median OS (95% CI) | 12.11 (9.43, 31.51) | 7.62 (4.99, 19.06) | 11.73 (10.51, 13.60) | 9.99 (8.90, 11.93) |
| HR within subgroup (interaction model) | 0.63 (0.29, 1.37) p = 0.245 | 0.91 (0.75, 1.11) p = 0.355 | ||
| Interaction p-value | 0.365 | |||
| Median PFS (95% CI) | 7.36 (3.75, 11.40) | 5.55 (2.76, 9.99) | 5.72 (5.55, 6.47) | 5.49 (4.60, 5.62) |
| HR within subgroup (interaction model) | 0.80 (0.36, 1.78) p = 0.581 | 0.91 (0.75, 1.11) p = 0.345 | ||
| Interaction p-value | 0.752 | |||
Arm A: cisplatin-gemcitabine-necitumumab; Arm B: cisplatin-gemcitabine.
CEP7: chromosome 7; CI= confidence interval; EGFR: epidermal growth factor receptor (gene) HR= hazard ratio; OS: overall survival; PFS: progression-free survival.
Acknowledgments
ACKNOWLEDGMENTS: none
7. FUNDING
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
DISCLAIMERS: none
DISCLOSURES:
Carlo Genova: honoraria from Astra Zeneca, Boehringer-Ingelheim and Bristol-Myers Squibb.
Mark A. Socinski: none
Rebecca R. Hozak: employee and stockholder of Eli Lilly and Company.
Gu Mi: employee and stockholder of Eli Lilly and Company.
Raffael Kurek: employee and stockholder of Eli Lilly and Company.
Javad Shahidi: former employee of Eli Lilly and Company; employee of Daiichi Snakyo Inc.
Luis Paz-Ares: medical advisory fees from Astra Zeneca, Boehringer-Ingelheim, Bristol-Myers Squibb, Merck Sharp & Dohme, Roche, Novartis, Eli Lilly and Company, Pfizer.
Nick Thatcher: speaker and board fees from Eli Lilly and Company.
Christopher J. Rivard: none
Marileila Varella- Garcia: patent related to use of EGFR as a biomarker for selecting therapy in lung cancer licensed to Abbott Molecular.
Fred R. Hirsch: patent related to use of EGFR as a biomarker for selecting therapy in lung cancer licensed to Abbott Molecular.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2012, National Cancer Institute; Bethesda, MD, http://seer.cancer.gov/csr/1975_2012/ based on November 2014 SEER data submission, posted to the SEER web site, April 2015 [Google Scholar]
- [2].Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res. 2015. February;4(1):36–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Thomas A, Liu SV, Subramaniam DS, Giaccone G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol. 2015. September;12(9):511–26 [DOI] [PubMed] [Google Scholar]
- [4].Ang YL, Tan HL, Soo RA. Best practice in the treatment of advanced squamous cell lung cancer. Ther Adv Respir Dis. 2015. April 22 pii: 1753465815581147. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- [5].López-Malpartida AV, Ludeña MD, Varela G, García Pichel J. Differential ErbB receptor expression and intracellular signaling activity in lung adenocarcinomas and squamous cell carcinomas. Lung Cancer. 2009. July;65(1):25–33 [DOI] [PubMed] [Google Scholar]
- [6].Pirker R, Pereira JR, Szczesna A, et al. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX): an open-label randomised phase III trial. Lancet. 2009. May 2;373(9674):1525–31 [DOI] [PubMed] [Google Scholar]
- [7].Pirker R, Pereira JR, von Pawel J, et al. EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Oncol. 2012. January;13(1):33–42 [DOI] [PubMed] [Google Scholar]
- [8].Thatcher N, Hirsch FR, Luft AV, et al. Necitumumab plus gemcitabine and cisplatin versus gemcitabine and cisplatin alone as first-line therapy in patients with stage IV squamous non-small-cell lung cancer (SQUIRE): an open-label, randomised, controlled phase 3 trial. Lancet Oncol. 2015. July;16(7):763–74 [DOI] [PubMed] [Google Scholar]
- [9].Paz-Ares L, Socinski MA, Shahidi J, et al. Correlation of EGFR-expression with safety and efficacy outcomes in SQUIRE: a randomized, multicenter, open-label, phase III study of gemcitabine-cisplatin plus necitumumab versus gemcitabine-cisplatin alone in the first-line treatment of patients with stage IV squamous non-small-cell lung cancer. Ann Oncol. 2016; 27(8):1573–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Del Mastro L, Lambertini M, Bighin C, et al. Trastuzumab as first-line therapy in HER2-positive metastatic breast cancer patients. Expert Rev Anticancer Ther. 2012. November;12(11):1391–405 [DOI] [PubMed] [Google Scholar]
- [11].Cappuzzo F, Finocchiaro G, Grossi F, et al. Phase II study of afatinib, an irreversible ErbB family blocker, in EGFR FISH-positive non-small-cell lung cancer. J Thorac Oncol. 2015. April;10(4):665–72 [DOI] [PubMed] [Google Scholar]
- [12].Hirsch FR, Varella-Garcia M, Bunn PA Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003. October 15;21(20):3798–807 [DOI] [PubMed] [Google Scholar]
- [13].Hirsch FR, Herbst RS, Olsen C, et al. Increased EGFR gene copy number detected by fluorescent in situ hybridization predicts outcome in non-small-cell lung cancer patients treated with cetuximab and chemotherapy. J Clin Oncol. 2008. July 10;26(20):3351–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Varella-Garcia M Stratification of non-small cell lung cancer patients for therapy with epidermal growth factor receptor inhibitors: the EGFR fluorescence in situ hybridization assay. Diagn Pathol. 2006. August 15;1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst. 2005. May 4;97(9):643–55 [DOI] [PubMed] [Google Scholar]
- [16].Hirsch FR, Redman MW, Herbst RS, et al. Biomarker-enriched efficacy of cetuximab-based therapy: Squamous subset analysis from S0819, a phase III trial of chemotherapy with or without cetuximab in advanced NSCLC. J Clin Oncol 34, 2016. (suppl; abstr 9090). Presented at the 2016 ASCO Annual Meeting; Chicago (IL), USA, May 30- June 4 [Google Scholar]
- [17].https://clinicaltrials.gov/ct2/show/NCT00981058?term=nct00981058&rank=1https://clinicaltrials.gov/ct2/show/NCT00981058?term=nct00981058&rank=1 [accessed on April 12 2017]
- [18].Varella-Garcia M, Diebold J, Eberhard DA, et al. EGFR fluorescence in situ hybridisation assay: guidelines for application to non-small-cell lung cancer. J Clin Pathol. 2009. November;62(11):970–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Herbst R, Redman MW, Kim ES, et al. : A randomized, phase III study comparing carboplatin/paclitaxel or carboplatin/paclitaxel/bevacizumab with or without concurrent cetuximab in patients with advanced non-small cell lung cancer (NSCLC): SWOG S0819. Presented at the 16th World Conference on Lung Cancer; Denver (CO), USA, September 6–9, 2015 (Abstract PLEN04.01) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.