SUMMARY
Parkinson’s disease is characterized by the progressive loss of midbrain dopamine neurons. Dopamine replacement therapy with levodopa alleviates parkinsonian motor symptoms but is complicated by the development of involuntary movements, termed levodopa-induced dyskinesia (LID). Aberrant activity in the striatum has been hypothesized to cause LID. Here, to establish a direct link between striatal activity and dyskinesia, we combine optogenetics and a method to manipulate dyskinesia-associated neurons, targeted recombination in active populations (TRAP). We find that TRAPed cells are a stable subset of sensorimotor striatal neurons, predominantly from the direct pathway, and that reactivation of TRAPed striatal neurons causes dyskinesia in the absence of levodopa. Inhibition of TRAPed cells, but not a nonspecific subset of direct pathway neurons, ameliorates LID. These results establish that a distinct subset of striatal neurons is causally involved in LID and indicate that successful therapeutic strategies for treating LID may require targeting functionally selective neuronal subtypes.
In Brief
Girasole et al. use the FosTRAP system to capture and manipulate populations associated with levodopa-induced dyskinesia (LID) brain-wide. They show that a subset of striatal neurons is necessary and sufficient in the production of LID.
INTRODUCTION
In Parkinson’s disease (PD), degeneration of midbrain dopamine neurons leads to slowing of movement (bradykinesia), rigidity, and tremor. Levodopa, the gold standard dopamine replacement therapy, alleviates motor symptoms but eventually triggers abnormal involuntary movements, termed levodopa-induced dyskinesia (LID). Pharmacological treatment for LID is limited; better understanding of the relationship of specific brain areas and cell types to dyskinesia would accelerate therapeutic development.
LID-associated aberrant activity has been observed in several brain regions, but which area and/or cell types cause dyskinesia is unknown. One hypothesis is that by increasing dopamine release, levodopa evokes abnormal activity in the striatum, the primary input nucleus of the basal ganglia, and a major target of midbrain dopamine neurons. Compelling pharmacological, biochemical, and electrophysiological data from rodent and nonhuman primate models of LID indirectly support this hypothesis: local striatal infusion of levodopa triggers dyskinesia in parkinsonian rats (Buck et al., 2010), striatal firing rate and pattern change markedly in parkinsonian monkeys with LID (Liang et al., 2008; Singh et al., 2015), and biochemical changes are seen in parkinsonian rodents treated with levodopa. Immediate-early genes (IEGs) such as c-Fos, FosB, and ΔFosB are consistently upregulated in the striatum of animals with LID (Jenner, 2008). However, other brain regions have also been implicated, including primary motor cortex (M1) (Halje et al., 2012; Lindenbach and Bishop, 2013; Swann et al., 2016) and primary somatosensory cortex (S1) (Alam et al., 2017). Interconnections between these brain regions add uncertainty regarding the origin of aberrant activity and heighten the need to look brain-wide to find the specific cell types and patterns of activity that cause LID.
Striatal IEG expression correlates with dyskinesia severity in parkinsonian primates (Berton et al., 2009), but these findings do not indicate whether striatal activation is causal. To investigate which brain regions and cell types cause LID, we used a transgenic mouse tool, targeted recombination in active populations (TRAP) (Guenthner et al., 2013), in combination with a well-established mouse model of LID (Cenci and Lundblad, 2007). Based on Fos-driven CreER, TRAP allows identification and subsequent manipulation of activated neurons, captured during a time window defined by tamoxifen administration. Using TRAP, we find LID-associated neurons in several brain regions, including M1, S1, and striatum. Optical reactivation of LID-associated striatal neurons, but not LID-associated neurons in other brain regions, triggered dyskinesia in the absence of levodopa. Furthermore, inhibiting striatal LID-associated neurons, but not nonspecifically labeled neurons in the same region, reduced dyskinesia, indicating that a distinct subpopulation of striatal neurons mediates LID.
RESULTS
Targeted Recombination in Active Populations Captures Levodopa-Induced Dyskinesia-Associated Neurons
We first validated the mouse model of LID in wild-type (WT) mice, examining behavior and IEG expression (Figure S1A). To control for the effects of parkinsonism and levodopa treatment itself, we divided mice (N = 34) into four experimental groups, based on parkinsonian state (unilateral intrastriatal injection of saline or the neurotoxin 6-OHDA, the latter to deplete dopamine neurons, Figure S1B) and systemic treatment (daily intraperitoneal saline or levodopa/benserazide). During behavioral sessions, a blinded experimenter measured open field locomotor behavior (drug-induced rotations) or scored dyskinesia in response to saline or levodopa injection. While parkinsonian animals rotate ipsilateral to the dopamine-depleted side, levodopa evokes contralateral rotations (6-OHDA/levodopa group, Figure S1E). Dyskinesia was quantified using a validated rating scale for abnormal involuntary movements (AIMs) (Cenci and Lundblad, 2007). Neither nonparkinsonian animals treated with systemic saline (saline/saline, N = 6) or levodopa (saline/levodopa, N = 6), nor parkinsonian animals treated with systemic saline (6-OHDA/saline, N = 6) developed dyskinesia (Figure S1C). However, parkinsonian mice receiving levodopa (6-OHDA/levodopa, N = 16) developed dyskinesia, peaking 20–40 min after injection (Figure S1C). LID was reliably evoked over 3 weeks of treatment (Figure S1D). To evaluate IEG expression in LID, we sacrificed animals 2 hr after saline or levodopa injection and immunostained for c-Fos (Figures S1F–S1I). As expected, the sensorimotor, or dorsolateral striatum (DLS) of 6-OHDA/levodopa-treated mice contained numerous c-Fos-positive nuclei (Figure S1I). However, striatal expression of c-Fos was not increased in other groups (Figure S1J), consistent with similar studies (Pavón et al., 2006).
We repeated these experiments in FosTRAP (FosCreER/+ R26Ai14/+) mice (N = 39), again in four experimental groups based on intrastriatal injection (Figure 1A) and systemic treatment (Figure 1B). Postmortem staining for tyrosine hydroxylase (TH) was used to confirm dopamine depletion (Figure 1C). As in WT mice, FosTRAP mice treated with 6-OHDA and levodopa (N = 11) showed contralateral rotations (Figure 1F) and dyskinesia (Figures 1D, 1E, and 1G), while the three control groups did not (N = 10 saline/saline, N = 9 saline/levodopa, and N = 9 6-OHDA/saline mice). We then used FosTRAP to capture neurons activated during LID by administering the short-acting tamoxifen metabolite 4-hydroxytamoxifen (4-OHT) during a single levodopa or saline session. In FosTRAP mice, the pairing of 4-OHT and a behavioral state that activates c-Fos, such as dyskinesia, leads to Cre-dependent expression of tdTomato (Guenthner et al., 2013). We captured activated cells 1 week into daily saline or levodopa treatment (Figure 1B) and sacrificed animals 2 weeks later to quantify tdTomato expression (henceforth called “TRAPed cells”) brain-wide (Figure 1H). We focused on three candidate brain areas: the striatum, primary somatosensory cortex (S1), and primary motor cortex (M1). TRAPed cells were found in each region (Figure S1K) but differed in abundance. As with c-Fos immunostaining, we found a marked increase in TRAPed cells in the DLS of 6OHDA/levodopa-treated mice (Figure 1H, right). The density and number of TRAPed cells was increased in the striatum of these mice as compared to the three control groups (Figures 1I and S1L, p < 0.01, N = 10 animals). TRAPed cells were also observed in S1 and M1 but were not significantly enriched in 6-OHDA/levodopa-treated mice over control groups (Figures 1I and S1L, N = 10 animals each). These results are consistent with previous studies showing strong striatal IEG activation in rodents with LID (Jenner, 2008).
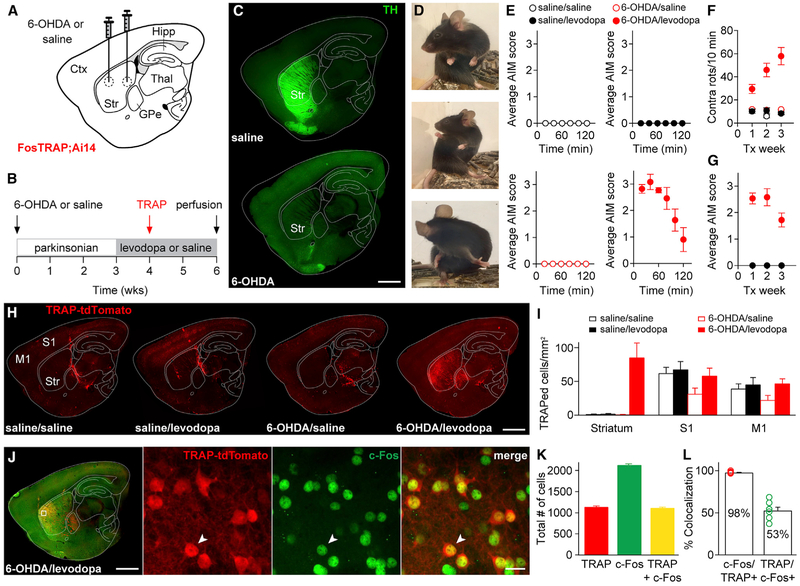
Figure 1. FosTRAP Captures Levodopa-Induced Dyskinesia-Associated Striatal Cells.
FosTRAP mice were separated into four groups based on intracerebral injections (saline or 6-OHDA) and chronic drug treatment (saline or levodopa).
(A) Sagittal schematic showing 6-OHDA or saline injections.
(B) Experimental timeline.
(C) Sagittal sections from mice injected with saline (top) or 6-OHDA (bottom) stained with anti-tyrosine hydroxylase (TH).
(D) Still images showing levodopa-induced dyskinesia in a parkinsonian FosTRAP mouse.
(E) Average abnormal involuntary movement (AIM) score measured after levodopa or saline injection in all groups.
(F) Contralateral rotations over 3 weeks of treatment.
(G) Average composite AIM score over 3 weeks of treatment (note: all four groups are displayed, but three groups showed no AIMS).
(H) Representative sagittal sections from the four experimental groups. Increased striatal expression of TRAP-tdTomato was only observed in 6-OHDA/levodopa-treated animals.
(I) Density of TRAPed cells measured in the striatum, S1, and M1.
(J-L) FosTRAP mice were administered levodopa 2 hr prior to perfusion. TRAPed cells reflect those activated during a levodopa session approximately 2 weeks prior, while c-Fos-positive neurons reflect cells activated during the terminal levodopa session. (J) Left: sagittal section showing TRAP-tdTomato and immunostained for c-Fos. Inset: confocal images showing colocalization (arrowhead) of TRAP-tdTomato (left), c-Fos (middle), and merged image (right). (K) Total number of TRAP-tdTomato, c-Fos, and colocalized cells. (L) Percent colocalization of c-Fos with TRAP-tdTomato (left) and TRAP with c-Fos (right).
Scale bar represents 1 mm in (C), (H), and (J) (left) and 20 μm in (J) (inset). Data are displayed as average ± SEM. See also Figure S1.
As FosTRAP requires time for Cre-dependent fluorophore expression, and c-Fos labeling reflects activation just prior to sacrifice, it is not possible to confirm whether FosTRAP and c-Fos label the same cells in a given levodopa session. However, we examined whether FosTRAP and c-Fos capture a similar group of cells across two sessions 2–3 weeks apart, by c-Fos immunostaining tissue from FosTRAP mice administered levodopa 2 hr prior to sacrifice. Remarkably, we saw extensive overlap between striatal TRAPed and c-Fos-positive cells (Figures 1J–1L, 98.1% ± 0.4%, N = 7 animals). We also examined the expression of two other LID-associated IEGs, ΔFosB and Erg-1, in TRAPed cells (Figures S1M and S1O). As with c-Fos, we saw extensive overlap between TRAPed cells and these cellular markers (Figures S1N and S1P, ΔFosB, 83.3% ± 1.9%, Erg-1, 83.4% ± 1.7%, N = 3 animals each). Together, these results suggested that TRAP and IEGs capture overlapping populations of neurons, but further, that LID-associated striatal cells represent a consistent, nonrandom population session to session (Figure S1Q).
TRAPed Cells Are Primarily Direct Pathway Medium Spiny Neurons
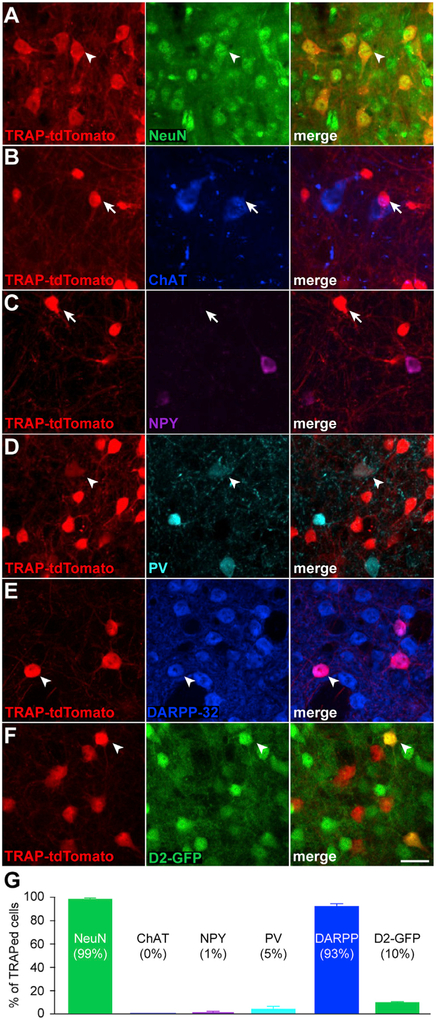
We next investigated which striatal cell types were TRAPed during LID, using confocal imaging to quantify colocalization of tdTomato and neuronal markers in postmortem tissue. Not surprisingly, we found that 99.0% ± 0.10% of activated cells were neurons, by colocalization with the marker NeuN (Figures 2A and 2G, N = 4 animals). Next, we examined whether TRAPed cells were striatal projection neurons (medium spiny neurons [MSNs]), cholinergic interneurons, or GABAergic interneurons. We hypothesized that levodopa-evoked striatal dopamine release activates neurons via “excitatory” D1-like receptors (Surmeier et al., 2011), which are expressed on a subset of MSNs and parvalbumin (PV)-positive GABAergic interneurons. Other cell types express “inhibitory” D2-like receptors and would be less likely to be activated by levodopa. Cholinergic interneurons express both D5 (Bergson et al., 1995) and D2 receptors, so it was difficult to predict whether they would be activated by levodopa. Immunohistochemistry revealed that none of the TRAPed cells expressed choline acetyltransferase (ChAT, Figures 2B and 2G, N = 3 animals), 1.0% ± 1.0% expressed Neuropeptide Y (NPY, Figures 2C and 2G, N = 3 animals), and 5.0% ± 2.0% expressed PV (Figures 2D and 2G, N = 3 animals). From these results, we reasoned that most TRAPed cells are MSNs. To confirm this hypothesis, we examined colocalization of the MSN marker DARPP-32 (dopamine- and cAMP-regulated phos-phoprotein Mr~32,000) (Figure 2E, N = 4 animals). 93.0% ±2.0% of all TRAPed cells were DARPP-32 positive (Figure 2G), confirming the vast majority of TRAPed neurons are MSNs.
Figure 2. TRAPed Cells Are Primarily Direct Pathway Medium Spiny Neurons.
(A-F) Representative 40x confocal images from the striatum of FosTRAP mice treated with 6-OHDA and levodopa, immunostained for different cellular markers. Left columns show TRAP-tdTomato, middle columns show antibody staining for NeuN (A), Choline Acetyltransferase (ChAT, B), Neuropeptide Y (NPY, C), Parvalbumin (PV, D), DARPP-32 (E), D2-GFP (F), and right columns show merged images. White arrowheads denote colocalization, while white arrows show non-colocalized cells.
(G) Percent of TRAPed striatal cells positive for each cellular marker.
Scale bar represents 20 mm in (A)-(F). Data are displayed as average ± SEM. See also Figure S2.
Based on their expression of D1 receptors (Gerfen etal., 1990), many have hypothesized that in parkinsonian animals, levodopa treatment leads to increased activity of D1-bearing direct pathway MSNs (dMSNs), and excessive activity in the case of dyskinesia. Physiological recordings in downstream basal ganglia nuclei are consistent with, but cannot directly confirm this hypothesis (Boraud et al., 2001; Filion et al., 1991; Levy et al., 2001; Lozano et al., 2000; Papa et al., 1999). It is unclear how D2-bearing indirect pathway MSNs (iMSNs) are involved. To determine whether activated neurons are dMSNs, we repeated similar experiments in two additional cohorts of mice: FosTRAP;Ai14;D2-GFP mice, in which GFP is expressed in iMSNs, and in Drd1a-tdTomato mice, in which tdTomato is expressed in dMSNs. In the former cohort, we found that 10.4% ± 0.1% of TRAPed neurons (Figures 2F and 2G, N = 3 animals) and 3.7% ± 0.5% of c-Fos-positive neurons (Figures S2F and S2H) were D2-GFP positive, suggesting that the majority of TRAPed cells are dMSNs. We supported this hypothesis in the second cohort, by colocalization of D1-tdTomato with c-Fos (Figures S2A and S2B, N = 18 animals). As in previous experiments, 6-OHDA/levodopa-treated Drd1a-tdTomato mice showed drug-induced rotations and dyskinesia (Figures S2C–S2E). In postmortem tissue from animals with LID, approximately 70.0% ± 4.0% of c-Fos-positive nuclei showed colocalization with the dMSN reporter (Figures S2F and S2G, N = 5 animals). Though all dMSNs express D1 receptors, only a subset may be activated by a dopaminergic manipulation. Differential synaptic inputs or sensitivity to dopamine may lead to heterogeneous dMSN responses. We found that only 20.0% ± 2.0% of all striatal neurons were TRAPed. Indeed, even among dMSNs, only 53.0% ± 4.0% show c-Fos expression (Figure S2G). Together, these findings suggest that LID recruits a stable subset of predominantly dMSNs.
Optogenetic Reactivation of TRAPed Striatal Cells, but Not TRAPed S1 or M1 Cortical Cells, Causes Dyskinesia in the Absence of Levodopa
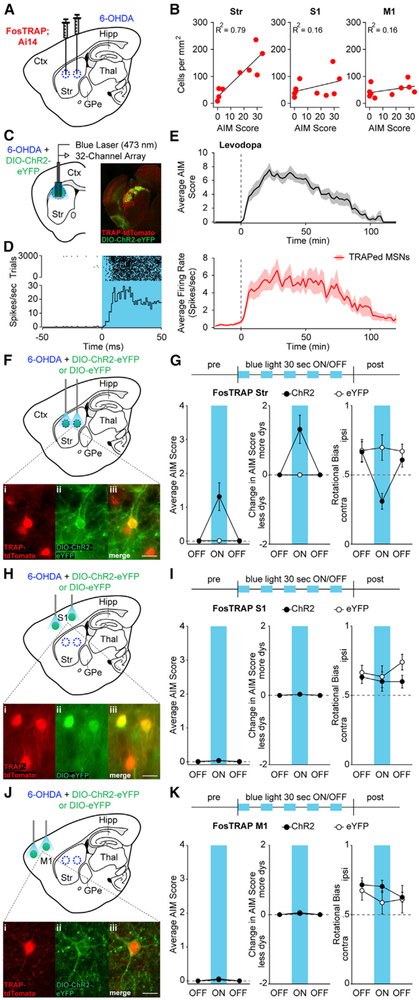
TRAPed striatal neurons may be the cause or the effect of dyskinesia, or they may be activated by levodopa, but unrelated to dyskinesia. To investigate the relationship between TRAPed cells and dyskinesia, we asked whether the number or density of activated neurons correlated with dyskinesia severity across animals (Figures 3A and S3A). The density and number of TRAPed cells correlated more strongly with AIM score in the striatum than in S1 and M1 (Figures 3B and S3B; N = 10 animals per area), consistent with, but not proving, the hypothesis that TRAPed cells cause dyskinesia.
Figure 3. Optogenetic Reactivation of TRAPed Striatal Neurons, but Not TRAPed S1 or M1 Cortical Neurons, Causes Dyskinesia in the Absence of Levodopa.
(A) FosTRAP mice were injected intrastriatally with 6-OHDA.
(B) Correlation between the density of TRAPed cells and total AIM score.
(C) Coronal schematic of optrode recording configuration (left) and representative postmortem section from a dopamine-depleted FosTRAP mouse injected with DIO-ChR2-eYFP (right).
(D) Representative optogenetically identified unit, showing short-latency spiking to blue light pulses. Top: rasterized firing during 3,000 trials. Bottom: peristimulus time histogram.
(E) Top: average AIM score from sessions in which a TRAPed neuron was identified. Levodopa was administered at time zero (dotted line). Bottom: average firing rate of optogenetically identified TRAPed MSNs in response to levodopa.
(F–K) FosTRAP mice were injected with intrastriatal 6-OHDA and DIOChR2-eYFP or DIO-eYFP in either the striatum (Str, F), primary somatosensory cortex (S1, H), or primary motor cortex (M1, J), after which behavioral testing commenced in the absence of levodopa (G, I, and K). (F, H, and J) Sagittal schematics showing sites of injection and light activation. Insets: postmortem histology showing TRAP-tdTomato (i), ChR2-eYFP (ii), and merge (iii) in the striatum, S1, and M1. (G, I, K) Top: optical activation protocol (1 mW) in Str, S1, and M1. Average (left) and change in (middle) AIM scores before, during, and after blue light in FosTRAP-ChR2 or eYFP mice. Right: rotational bias.
Scale bar represents 20 μm in (F), (H), and (J). Data are displayed as average ± SEM. See also Figure S3.
We next wondered how levodopa altered firing of TRAPed striatal neurons, hypothesizing that levodopa would increase the firing rate of TRAPed neurons. To characterize this change in firing rate of TRAPed neurons, we performed optrode recordings in freely moving mice. We expressed Cre-dependent Channelrhodopsin (ChR2) in the DLS of dopamine-depleted FosTRAP mice, who were subsequently implanted with 32-channel optrode arrays (Figure 3C). Animals developed levodopa-evoked contralateral rotations and dyskinesia as in other cohorts of FosTRAP mice (data not shown), and single-unit recordings commenced 1–2 weeks following 4-OHT, to allow for ChR2 expression. During each session, we recorded striatal activity before and after levodopa administration, and at the end of the session, delivered a series of blue light pulses to determine whether the unit expressed ChR2 (and thus was TRAPed) (Kravitz et al., 2013). Those units with short-latency responses to blue light pulses (Figure 3D) were classified as optically identified TRAPed neurons. As expected, all optically labeled TRAPed putative MSNs showed a significant levodopa-evoked increase in firing rate (Figure 3E, baseline: 0.27 ± 0.1 spikes/s, levodopa: 4.51 ± 1.3 spikes/s, n = 8 cells, N = 4 animals). These results suggested the majority of TRAPed neurons were positively modulated by dopamine and that increased activity of such neurons paralleled LID.
Given both the increase in TRAPed striatal neuron firing rate during dyskinesia and the correlation between the number of TRAPed striatal neurons and dyskinesia severity, we next determined whether reactivating LID-associated neuronal ensembles in the striatum, in the absence of levodopa, causes dyskinesia. We expressed ChR2 or eYFP in the DLS of FosTRAP mice and implanted optical fibers (Figures 3F and S3D). Animals showed levodopa-evoked contralateral rotations and dyskinesia over 3 weeks of levodopa treatment as in previous cohorts (data not shown). ChR2-eYFP expression in TRAPed cells was confirmed by postmortem histology (Figures 3F and S3E) and current-clamp recordings showing light-evoked spiking (Figure S3F). After 4-OHT administration and Cre-dependent opsin expression, two blinded raters scored behavior during optical stimulation (1 mW blue light). In optrode recordings, we found that 30 s, 1 mW light stimulation transiently evoked firing rates of 3.9 ± 1.6 spikes/s (Figure S3C) in TRAPed striatal neurons, similar to peak levodopa-evoked firing rates in TRAPed striatal neurons (Figure 3E). In FosTRAP-ChR2 mice, blue light qualitatively reproduced the motor effects of levodopa, including contralateral rotations and dyskinesia (Figure 3G, left; Figure S3G and Movie S1). Light triggered dyskinesia in FosTRAP-ChR2 mice (N = 14 mice; p = 0.001), but not in FosTRAP-eYFP animals (N = 14 mice; p = 1.0). The light-evoked change in both dyskinesia and rotations was larger in FosTRAP-ChR2 mice than in eYFP animals (Figure 3G, middle: p = 0.0002; Figure 3G, right: p = 0.01). These results suggest that reactivation of striatal LID-associated neurons is sufficient to cause dyskinesia in the absence of levodopa.
In contrast, we found that reactivation of TRAPed neurons in S1 and M1 did not evoke dyskinesia. Using a similar approach, we expressed ChR2 or eYFP in S1 or M1 TRAPed cortical cells (Figures 3H, 3J, S3H, S3I, S3L, and S3M), which showed light-evoked spiking (Figures S3J and S3N). We then administered, in separate sessions, either continuous or pulsatile (10 Hz) blue light, to activate S1 or M1 TRAPed neurons (p values for each condition are noted). In FosTRAP-ChR2 mice, optical stimulation in S1 (N = 10), with continuous or pulsatile blue light, failed to increase rotational behavior (Figure 3I, right; S1: p = 0.77 and 0.89, respectively) or dyskinesia (Figure 3I, left; p = 0.25 and 0.31, respectively). In FosTRAP-eYFP mice (N = 8), S1 light also failed to change rotations (p = 0.52 and 0.91, respectively) or dyskinesia (p = 0.25 and 0.25, respectively). No differences were noted in light-evoked rotations (p = 0.81 and 0.11) or dyskinesia (Figure 3I, middle; Figure S3K; p = 0.86 and 0.83) between FosTRAP-ChR2 and eYFP cohorts.
Optical stimulation of M1, with continuous or 10 Hz pulsatile stimulation, also failed to evoke rotations (Figure 3K, right; p = 0.89 and p = 0.96) or dyskinesia (Figure 3K, left; Figure S3O; p = 1.0 and p = 1.0) in FosTRAP-ChR2 mice (N = 6). As predicted, in M1 FosTRAP-eYFP mice (N = 5), light administration also did not evoke changes in rotations (p = 0.55 and p = 0.65) or dyskinesia (p = 0.5 and p = 0.25). Finally, there were no differences in the light-evoked change in rotations (p = 0.34 and p = 0.53) or dyskinesia between the M1 ChR2 and eYFP groups (p = 0.85 and p = 0.12). We did, however, see a significant increase in velocity and total distance traveled with M1 activation in FosTRAP-ChR2 animals (distance 679.2 ± 236.8 cm to 846.7 ± 257.2 cm, p = 0.009; velocity 5.5 ±1.9 cm/s to 6.9 ± 2.1 cm/s, p = 0.009), as expected for M1 stimulation. These results confirmed that reactivation of TRAPed striatal cells, but not S1 or M1 cortical cells, is sufficient for dyskinesia in the absence of levodopa.
Optogenetic Inhibition of TRAPed Striatal Neurons, but Not Nonspecific Direct Pathway Neurons, Ameliorates LID
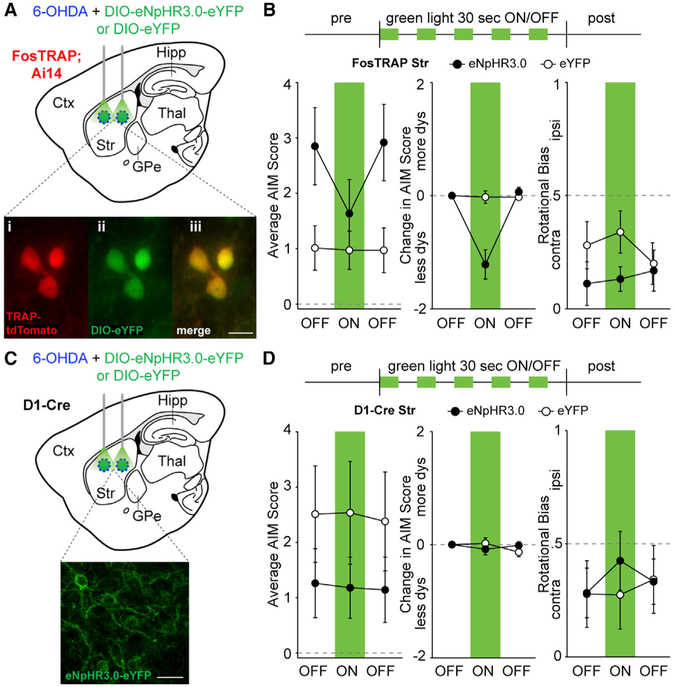
Dyskinesia triggered by optical reactivation of striatal TRAPed cells suggests sufficiency, but does not definitively show that TRAPed neurons cause LID under normal conditions. To test whether activity of TRAPed striatal neurons is required for LID, we expressed the inhibitory opsin eNpHR3.0 or eYFP (Figures 4A and S4A) in the DLS of parkinsonian FosTRAP mice treated with daily levodopa. We confirmed eNpHR3.0 expression with postmortem histology (Figures 4A and S4B) and slice physiology showing light-evoked inhibition of spiking (Figure S4C). To determine whether inhibition of TRAPed neurons reduces LID, we delivered green light during peak LID. Green light did not change rotational behavior in eNpHR3.0 (N = 12) or eYFP-expressing (N = 15) animals (Figure 4B right, p = 0.55 and 0.52, respectively). However, in FosTRAP-eNpHR3.0 mice, light reduced dyskinesia severity (Figures 4B, left, and S4D; Movie S2, p = 0.0005); in eYFP controls, it did not (p = 0.81). The light-evoked reduction in dyskinesia was greater in eNpHR3.0 animals as compared to eYFP controls (Figure 4B middle, p = 0.0002). These results suggested TRAPed striatal cells are also necessary for dyskinesia, though it remains unclear how they contribute to rotational behavior.
Figure 4. Optogenetic Inhibition of TRAPed Striatal Neurons, but Not All Direct Pathway Neurons, Ameliorates Levodopa-Induced Dyskinesia.
(A–D) FosTRAP (A and B) or D1-Cre mice (C and D) were injected with intrastriatal 6-OHDA and DIO-eNpHR3.0-eYFP or DIO-eYFP. Behavioral testing was performed after the injection of levodopa. (A and C) Sagittal schematics showing sites of injection and light stimulation in FosTRAP (A) and D1-Cre (C) mice. (A) Inset: postmortem histology showing TRAP-tdTomato (i), eYFP (ii), and merge (iii). (B and D). Top: optogenetic inhibition protocol (5 mW). Average (left) and change in (middle) AIM scores before, during, and after striatal green light in eNpHR3.0 or eYFP mice. Right: rotational bias. (C) Inset: postmortem histology showing eNpHR3.0-eYFP positive striatal neurons. Right: rotational bias.
Scale bar represents 20 μm in (A) and (C). Data are displayed as average ± SEM. See also Figure S4
Although LID-associated TRAPed cells are predominantly dMSNs, they represent only a subset of all dMSNs. To determine whether this specific subset is necessary for LID, we tested whether inhibiting a random subset of dMSNs (as opposed to TRAPed neurons) could reduce LID, by optically inhibiting DLS dMSNs in parkinsonian D1-Cre animals treated with levodopa. We confirmed striatal eNpHR3.0 expression with histology and slice physiology (Figures 4C and S4E–S4G). Optical inhibition of a comparable number of random dMSNs did not alter rotations or dyskinesia in D1-Cre-eNpHR3.0 mice (Figure 4D, left and right, S4H; p = 0.48 and 0.75, respectively; N = 10 mice). As expected, green light did not alter rotations or dyskinesia in D1-Cre-eYFP mice (p = 1.0 and 1.0, respectively, N = 9 mice). No differences in light-evoked dyskinesia was observed between the eNpHR3.0 and eYFP groups (Figure 4D, middle, p = 0.88). It was somewhat surprising that dyskinesia was not reduced in D1-eNpHR3.0 mice, despite the fact that we likely manipulated significantly more neurons in D1-eNpHR3.0 mice than FosTRAP-eNpHR3.0 mice (Figure S4I). Taken together, our results suggest that TRAPed cells may represent a specific subset of striatal direct pathway neurons necessary for levodopa-induced dyskinesia (Figure S4J).
DISCUSSION
We used optogenetics and TRAP to investigate the neural populations that cause levodopa-induced dyskinesia (LID). Although LID-associated neurons were found brain-wide, the number of activated neurons in the striatum correlated most strongly with dyskinesia severity, and optogenetic reactivation of these neurons caused dyskinesia in the absence of levodopa. Inhibition of TRAPed striatal neurons, but not a random set of dMSNs, ameliorated LID. While these results corroborate many studies implicating the striatum in LID (Andersson et al., 1999; Berton et al., 2009; Buck et al., 2010; Cao et al., 2010; Engeln et al., 2016; Westin et al., 2001), our study is the first to establish that a specific and stable subset of striatal neurons mediates LID.
Many suspect that LID is the result of excessive dMSN activity, and previous studies have treated dMSNs as a single group modulated during LID. Our results combining FosTRAP with c-Fos staining showed that dyskinetic attacks triggered weeks apart activated a highly overlapping population of striatal neurons. This surprising finding suggests that the dMSNs activated during LID are a selective and stable subgroup. This subpopulation contained many dMSNs, as suggested in previous studies. We also found that TRAPed MSNs showed elevated firing rates in response to levodopa in vivo. Our experiments also showed LID recruits a handful of PV-positive interneurons, and somewhat surprisingly, a small number of iMSNs. Taken together, these results suggest LID is mediated by a varied, but specialized, ensemble within the striatum.
Optical stimulation may impose non-physiological levels or patterns of activity on brain circuits. Recent studies demonstrated that optical or chemogenetic stimulation of sensorimotor MSNs in parkinsonian animals can evoke dyskinesia (Alcacer et al., 2017; Hernández et al., 2017; Perez et al., 2017); dyskinesias have also been reported with dMSN stimulation in healthy animals (Rothwell et al., 2015). These observations suggest synchronous activation of many dMSNs in the sensorimotor striatum may be sufficient to cause dyskinesia but do not indicate which neurons are normally engaged in LID. Optical reactivation of LID-associated neurons in our study has many of the same methodological caveats, though single-unit recordings of TRAPed striatal neurons suggest light-evoked firing in TRAPed neurons approximated rates achieved by TRAPed neurons during LID. In addition, inhibiting LID-associated striatal neurons reduced LID, whereas inhibiting a comparable number of dMSNs did not. This observation suggests (1) TRAPed striatal neurons are necessary and sufficient to produce dyskinesia in parkinsonian animals, and (2) TRAPed neurons represent a more LID-specific subset of neurons than dMSNs overall.
Why is inhibiting TRAPed striatal neurons more effective than inhibiting dMSNs to reduce LID? One explanation may be that circuit reorganization, triggered by parkinsonism itself, or by chronic levodopa treatment, is heterogeneous within the striatum. For example, TRAPed neurons may be particularly vulnerable to aberrant corticostriatal plasticity, which has been reported in rodent models of LID (Bagetta et al., 2012; Fieblinger et al., 2014; Picconi et al., 2003; Shen et al., 2015). Alternatively, TRAPed cells may differ from neighboring dMSNs in local inhibitory connections (Gittis et al., 2011a), synaptic output (Borgkvist et al., 2015), or sensitivity to dopamine (Heiman et al., 2014), which in turn may drive differences in behavior evoked by manipulating TRAPed versus random dMSNs. Finally, the TRAPed population included PV-positive striatal neurons and iMSNs, which may contribute to circuit dysfunction and dyskinesia. Though several studies have implicated PV-positive interneurons in dyskinesia (Alberico et al., 2017; Gernert et al., 2000; Gittis et al., 2011b), at this point it is unclear how these interneurons contribute to dyskinesia without cell-type-specific experiments directly testing causality.
Our results suggest that LID is caused by a distinct and stable group of striatal neurons. To take full advantage of this observation, the activity patterns, behavioral correlates, and cellular and synaptic properties of these neurons will require further study. Together, these results could help identify new pharmacological targets for the prevention and management of LID.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alexandra Nelson (alexandra.nelson@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Six different types of transgenic mice, of either sex, aged 3-6 months, all on a C57BL/6 background, were used in this study. Hemizygous FosTRAP mice (Liqun Luo, Stanford) were bred to either wild-type C57BL/6 mice (WT, Jackson Labs) or homozygous Ai14 mice (Jackson Labs) to yield FosTRAP;WT or FosTRAP;Ai14 mice. Hemizygous Drd1a-tdTomato mice (Shuen etal., 2008) were bred against WT mice to produce Drd1a-tdTomato;WT animals. Hemizygous D2-GFP mice (Gong et al., 2003) were bred against WT mice to produce D2-GFP;WT animals. To look for colocalization of FosTRAP and D2-GFP in the striatum, hemizygous FosTRAP;Ai14 mice were bred to hemizygous D2-GFP mice to yield FosTRAP;Ai14;D2-GFP mice. To manipulate direct pathway neurons, hemizygous Drd1-Cre (D1-Cre-217) mice were crossed to WT animals to yield D1-Cre;WT mice. Animals were housed 1-5 per cage on a 12-hour light/dark cycle with ad libitum access to rodent chow and water. All behavioral manipulations were performed during the light phase. We complied with local and national ethical and legal regulations regarding the use of mice in research. All experimental protocols were approved by the UC San Francisco Institutional Animal Care and Use Committee.
METHOD DETAILS
Surgical Procedures
All surgical procedures were performed at 3-6 months of age. Anesthesia was induced with intraperitoneal (IP) injection of ketamine/xylazine and maintained with 0.5%–1.0% inhaled isoflurane. Mice were placed in a stereotaxic frame and a mounted drill was used to create two small holes over the left dorsolateral striatum. The left dorsolateral striatum (DLS, ± 1.0 AP, + 2.4 ML, − 3.0 mm DV) was injected at two sites using a 33-gauge needle with 2-2.5 μL per site 6-Hydroxydopamine (6-OHDA)-bromide (to render mice parkinsonian) or normal saline (for control animals). In some experiments, AAV5-DIO-ChR2-eYFP (UPenn Vector Core, 1 μL of 1:1 diluted virus per site), AAV5-DIO-eYFP (UNC Vector Core, 1 μL per site), or AAV5-DIO-eNpHR3.0 (UNC Vector Core, 1 μL per site) was also injected in the DLS (±1.0 AP, + 2.4 ML, − 3.0 mm DV). 6-OHDA, saline, and virus were injected at a rate of 0.15 μl/min, after which the injection cannula was left in place for 10-15 min prior to being withdrawn and the scalp being sutured. For experiments targeting cortical regions, AAV5-DIO-ChR2-eYFP or AAV5-DIO-eYFP was injected at two sites in primary somatosensory cortex (S1, +0.9 AP, +3.0 ML, −0.5 mm DV and −1.1 AP, +3.0 ML, −0.4 mm DV) or primary motor cortex (M1, +1.9 AP, +1.9 ML, −0.75 mm DV and −0.1 AP, +1.15 ML, −0.6 mm DV). For subsequent optical manipulations, the scalp was reopened 4-5 weeks after initial surgeries to implant optical fiber-ferrule assemblies in the DLS, S1, or M1 sites: DLS (±1.0 AP, + 2.4 ML, − 2.8 mm DV), S1 (+0.9 AP, +3.0 ML, −0.2 mm DV and −1.1 AP, +3.0 ML, −0.2 mm DV), and M1 (+1.9 AP, +1.9 ML, −0.55 mm DV and −0.1 AP, +1.15 ML, −0.55 mm DV). Ferrules were secured in place with dental cement (Metabond) and dental acrylic (Ortho-Jet). The scalp was then sutured and the mouse was allowed to recover from anesthesia.
In preparation for in vivo single-unit recordings, FosTRAP;Ai14 mice were injected with 6-OHDA and ChR2, as described above, and optrode arrays were implanted in a subsequent surgical procedure. After the skull was reopened, a large craniectomy (1.5 × 1 mm) was created over the DLS, and two additional holes were drilled for placement of a skull screw (Fine Scientific Tools, FST) and ground wire, both over the contralateral hemisphere. A fixed multichannel electrode array (32 tungsten microwires, Innovative Neurophysiology) coupled to a 200 mm optical fiber (Thorlabs) was slowly lowered through the craniectomy into the DLS. The final location of the array was targeted 100-200 μm above the previous 6-OHDA and ChR2 injection (−2.7-2.8mmDV). The array was covered and secured into place with dental cement and acrylic, as above. In 2 of 4 mice used for in vivo unit recordings, a medial forebrain bundle (MFB) dopamine depletion was used instead of an intrastriatal depletion. In this case, 1 μL of 6-OHDA was injected unilaterally into the MFB (−1.0 AP, +1.0 ML, −4.9 mm DV).
All animals were given buprenorphine (IP, 0.05 mg/kg) and ketoprofen (subcutaneous injection, 5 mg/kg) for postoperative analgesia. Parkinsonian animals were monitored closely for 1 week following surgery: mouse cages were kept on a heating pad, animals received daily saline injections and were fed nutritional supplements (Diet-Gel Recovery Packs and forage/trail mix).
Behavior
Postoperatively, parkinsonian mice were monitored in the open field 1-2 times per week for 10 min per session. All mice were habituated to the open field (clear acrylic cylinders, 25 cm diameter) for 30 min 1-2 days prior to behavioral sessions. The mice were monitored via two cameras, one directly above (to capture overall movement) and one in front of the chamber (to capture fine motor behaviors). Video-tracking software (Noldus Ethovision) was used to quantify locomotor activity, including rotations (full 360° contralateral or ipsilateral turns), distance traveled, and velocity. After a three-week baseline period, daily injections of levodopa commenced. Levodopa-induced dyskinesia (LID) was scored during weekly sessions in which mice were injected, then placed in a clean, clear cage for visualization. Dyskinesia was quantified using a standard scoring method (Cenci and Lundblad, 2007), which takes into account abnormal involuntary movements (AIMs) in axial, limb, and orofacial (ALO) body segments. Briefly, dyskinesia was quantified every 20 min, over a 2-hr period, using a scale of 0-4. A score of 0 indicates no abnormal movement, and a score of 4 describes continuous and uninterruptable dyskinetic movements; 12 (4 × 3 body segments) is the maximum score possible for a given time point. For regular weekly dyskinesia scoring, 1-2 blinded experimenters rated AIMs. For optical reactivation or inhibition experiments, 2 blinded raters scored mice.
Pharmacology
6-OHDA (Sigma Aldrich) for intrastriatal dopamine depletions was prepared at 2.5 μg/μL in normal saline solution. 6-OHDA (Sigma Aldrich) for MFB dopamine depletions was prepared at 5 μg/μL in normal saline solution. Levodopa (Sigma Aldrich) was always co-administered with benserazide (Sigma Aldrich) and prepared in normal saline solution. For cell counts and cell density experiments, (Figures 1 and S1) WT and FosTRAP;Ai14 animals received 20 mg/kg levodopa and 10 mg/kg benserazide. For optical reactivation experiments (Figures 3, S3, and Movie S1), 10 mg/kg levodopa and 5 mg/kg benserazide were used. For optical inhibition experiments (Figures 4, S4, and Movie S2), 5-10 mg/kg levodopa and 2.5-5 mg/kg benserazide were administered. Levodopa was given via IP injection 5-7 days per week. On the 7th day of levodopa treatment for FosTRAP;Ai14 mice, animals were given 4-hydroxytamoxifen (4-OHT, 50 mg/kg in Chen oil, IP) exactly 1 hr post-levodopa injection to capture dyskinesia-associated neurons (Figure 1B). 4-OHT was prepared as previously described (Guenthner et al., 2013). Briefly, to prepare a 20 mg/mL stock of 4-OHT, 4-OHT was added to 200 proof ethanol, vortexed, and placed on a horizontal shaker at 37°C for 30 min or until the 4-OHT dissolved. The stock solution was kept covered in foil to minimize light exposure. Next, to prepare a 10 mg/mL working solution in oil, the 4-OHT/ethanol mixture was combined with Chen Oil (a mixture of 4 parts sunflower seed oil and 1 part castor oil) and placed into 1.5 mL Eppendorf tubes. The tubes were vigorously mixed, wrapped in foil, and left on a nutator for 45 min at room temperature, vortexing, and shaking periodically. The tubes were then placed in a speed-vac for 2-3 hr to evaporate the ethanol. If necessary, the final volume was adjusted with Chen Oil to 1mL to reach a final concentration of 10 mg/mL. Both levodopa and 4-OHT were injected in a quiet, familiar environment, and animals were returned to their home cages, to minimize additional stimuli. Daily levodopa injections continued for 2-6 weeks to allow expression of Cre-dependent constructs. For in vitro experiments, picrotoxin (Sigma) was dissolved in warm water to prepare a 5 mM stock solution, which was subsequently diluted in ACSF for a final concentration of 50 μM.
Optogenetic Manipulations
Prior to optical stimulation experiments, animals were habituated to tethering with lightweight patch cables (components: PrecisionFiber Products and ThorLabs) coupled to an optical commutator (Doric Lenses) in the open field for 30 min, over 1-2 days. Optical stimulation experiments consisted of a 2-min baseline followed by 30 s light on/30 s light off (repeated 5-10x) and then a 2-min post period. TTL-controlled blue (488 nm, 1 mW, Shanghai Laser and Optics Century) or green laser light (593 nm, 5 mW, Shanghai Laser and Optics Century) was delivered continuously for all striatal experiments and either continuously or in pulse trains (5 ms, 10Hz, controlled with Master8, A.M.P.I.)for S1 and M1 experiments. Animals were manually scored for dyskinetic behavior by raters blinded to the manipulation (eg ChR2 versus eYFP) during the baseline, light on/off, and post period. Videotracking software was used to measure movement. For optical reactivation experiments, animals had not received levodopa for at least the previous 24 hr. For optical inhibition experiments, levodopa was administered 30 min before testing, to capture maximal dyskinesia. At the end of experimental procedures, animals were returned to their home cages.
In Vivo Electrophysiology
Two weeks after optrode array implantation, mice were habituated to tethering and the recording chamber for 1-2 days. After habituation, experimental sessions occurred 3-5 times per week for 2-6 weeks. During each session, electrical signals (single-unit and LFP data from each of 32 channels) were collected using a multiplexed 32 channel headstage (Triangle Biosystems), an electrical commutator equipped with a fluid bore (Dragonfly), filtered, amplified, and recorded on a MAP system, using RASPUTIN 2.4 HLK3 acquisition software (Plexon). Spike waveforms were filtered at 154-8,800 Hz and digitized at 40 kHz. The experimenter manually set a threshold for storage of electrical events.
During recording sessions, after a baseline period of 30 min, levodopa (5-10 mg/kg) was injected IP. After a period of 2-3 hr of recording spontaneous activity in the open field, an optogenetic cell identification protocol was applied (Kravitz et al., 2013), consisting of 100 ms blue light pulses, given at 1 Hz. At each of 4 light powers (0.5, 1, 2, and 4 mW), 1000 light pulses were delivered via a lightweight patch cable (Doric Lenses) connected to a blue laser (Shanghai Laser and Optics Century), via an optical commutator (Doric Lenses), and controlled by TTL pulses from a behavioral monitoring system (Noldus Ethovision). In a subset of experiments, an additional cell identification protocol was applied, which consisted of continuous 30 s light on/off (repeated 5-10x) blue light pulses given at each light power (0.5, 1, 2, and 4 mW). At the end of all cell identification protocols, animals were detached from the electrical and optical cables and returned to their home cages.
Single units were identified offline by manual sorting using Offline Sorter 3.3.5 (Plexon) and principle components analysis (PCA). Clusters were considered to represent a single unit if (1) the unit’s waveforms were statistically different from multiunit activity and any other single units on the same wire, in 3D PCA space, (2) no interspike interval <1 ms was observed. Single units were then classified as putative medium spiny neurons (MSNs) as previously described (Berke et al., 2004; Gage et al., 2010; Harris et al., 2000) using features of the spike waveform (peak to valley and peak width), as well as inter-spike interval distribution. Only putative MSNs were included in subsequent analyses.
After single units had been selected for further study, their firing activity was analyzed using NeuroExplorer 4.133 (Nex Technologies). To determine if a unit was optogenetically identified, a peristimulus time histogram was constructed around the onset of laser pulses. To be considered optogenetically identified, a unit had to fulfill 3 criteria: (1) the unit had to increase firing rate above the 95% confidence interval of the baseline within 15 ms of laser onset; (2) the unit’s firing was above this threshold for at least 15 ms; (3) the unit’s laser-activated waveforms were not statistically distinguishable from spontaneous waveforms.
In Vitro Electrophysiology
To prepare ex vivo slices for whole-cell recordings, mice were deeply anesthetized with an IP ketamine-xylazine injection, transcardially perfused with ice-cold glycerol-based slicing solution, decapitated, and the brain was removed. Glycerol-based slicing solution contained (in mM): 250 glycerol, 2.5 KCl, 1.2 NaH2PO4, 10 HEPES, 21 NaHCO3, 5 glucose, 2 MgCl2, 2 CaCl2. The brain was mounted on a submerged chuck, and sequential 250-300 μm coronal slices were cut on a vibrating microtome (Leica), transferred to a chamber of warm (34°C) carbogenated ACSF containing (in mM) 125 NaCl, 26 NaHCO3, 2.5 KCl, 1 MgCl2, 2 CaCl2, 1.25 NaH2PO4, 12.5 glucose for 30-60 min, then stored in carbogenated ACSF at room temperature. Each slice was then submerged in a chamber super-fused with carbogenated ACSF at 31°C-33°C for recordings.
Striatal or cortical neurons were targeted for recordings using differential interference contrast (DIC) optics in FosTRAP;Ai14 or D1-Cre mice on a Olympus BX 51 WIF microscope. In FosTRAP;Ai14 mice, TRAPed neurons were identified by their td-Tomato positive somata and D1 positive neurons were identified by eYFP fluorescence from DIO-eYFP or DIO-eNpHR3.0-eYFP. Neurons were patched in the whole-cell configuration using borosilicate glass electrodes (3-5 MΩ) filled with potassium methanesulfonate-based internal solution containing (in mM): 130 KMeSO3, 10 NaCl, 2 MgCl2, 0.16 CaCl2, 0.5 EGTA, 10 HEPES, 2 MgATP, 0.3 NaGTP, pH 7.3. Picrotoxin was added to all external solutions.
Whole-cell current-clamp recordings were made using a MultiClamp 700B amplifier (Molecular Devices) and digitized with an ITC-18 A/D board (HEKA). Data were acquired using Igor Pro 6.0 software (Wavemetrics) and custom acquisition routines (mafPC, courtesy of M. A. Xu-Friedman). Current-clamp recordings were filtered at 5 kHz and digitized at 10 kHz. To validate ChR2 or eNpHR3.0 function in slice, light pulses were delivered to the slice by a TTL-controlled LED (Olympus), passed through a GFP (473 nm) or TxRed (562 nm) filter (Chroma) and the 40X immersion objective. LED intensity was adjusted to yield an output of approximately 1-10 mW at the slice. Light pulses were given at 5-2,000 ms in duration. For optogenetic inhibition experiments, cells were given current injections (200-500 pA) to produce stable spiking at roughly 10 Hz. After 500 ms, a 500-1,000 ms light pulse was delivered to assess the ability of eNpHR3.0 to inhibit spiking. Holding current and input resistance were continuously monitored as proxies of recording stability.
Histology, Microscopy, and Cell Counting
After optogenetic or behavioral experiments, mice were deeply anesthetized with IP ketamine-xylazine and transcardially perfused with 4% paraformaldehyde in PBS. After perfusion, the brain was dissected from the skull and post-fixed overnight in 4% paraformaldehyde, then placed in 30% sucrose at 4°C for cryoprotection. The brain was then cut into 35 μm coronal or sagittal sections on a freezing microtome (Leica) and then mounted in Vectashield Mounting Medium onto glass slides for imaging. For immunohistochemistry, the tissue was blocked with 3% normal donkey serum (NDS) and permeabilized with 0.1% Triton X-100 for 2 hr at room temperature on a shaker. A subset of stains used 5% normal donkey serum with 1.0% Triton X-100 (anti-ChAT). Primary antibodies were added to 3% NDS and incubated overnight at 4°C on a shaker. Primary antibodies used: Rabbit anti-TH (Pel-Freez, 1:1,000), Chicken anti-TH (Sigma, 1:1,000), Rabbit anti-c-Fos (Cell Signalizing Technology, 1:1,000), Rabbit anti-c-Fos (Santa Cruz, 1:1,000), Goat anti-ChAT (Millipore, 1:500), Rabbit anti-DARPP-32 (Cell Signaling Technology, 1:1,000), Mouse anti-RFP (Rockland Immunochemicals, 1:500), Rabbit anti-NeuN (Millipore, 1:1,000), Rabbit anti-NPY (Cell Signaling Technology, 1:1,000), Rabbit anti-PV (Swant, 1:2,000), Rabbit anti-Erg-1 (Cell Signaling Technology, 1:500), or Goat anti-ΔFosB (Santa Cruz Biotechnology, 1:500). Slices were then incubated in secondary antibodies (donkey anti-rabbit, mouse, goat, or chicken Alexa Fluor 488, 593, or 647, 1:500, JacksonImmuno Research) for 2-4 hr at 4°C on a shaker, washed, and mounted onto slides for imaging. For a subset of animals (WT LID cohort, Figures S1A–S1J), development with diaminobenzidine (DAB) was used to visualize c-Fos. After 2-4 hr of incubation with secondary antibodies, the tissue was washed and then incubated for 60 min in Avidin-Biotin Complex (ABCVector Elite). After washing in PBS, DAB was prepared and added to each well. The color reaction was monitored under a dissecting microscope. Slices were then washed and mounted onto slides for imaging. 4 or 10x images were acquired on a Nikon 6D conventional widefield microscope.
QUANTIFICATION AND STATISTICAL ANALYSIS
Behavior
Average AIM score per session (Figures 1E and S1C) is an average ALO score per 20 min of scoring. Average AIM score per week (Figures 1G and S1D) is an average ALO score over the entire 120-min scoring session. Levodopa-induced contralateral rotations were measured weekly using video tracking (Figures 1F and S1E). Mice were placed in the open field 20-30 min post-levodopa injection and monitored for 10 min in the open field.
Optogenetic Experiments
For each stimulation experiment, we calculated all behavioral measurements (both AIMs and rotations) during three periods, termed ‘OFF (1)’, ‘ON’, and ‘OFF (2)’. The ‘OFF (1)’ period was the 25 s preceding the laser. The ‘ON’ period was the period starting 5 s after the illumination of the laser and lasting until the end of laser illumination (25 s long). The ‘OFF (2)’ period was the period between 5 and 30 s after the end of the laser illumination. The two-minute baseline preceding stimulation (‘pre’), and 2-min baseline after stimulation (‘post’) was not included in calculations. Rotation data are presented as a rotation ratio (total ipsilateral rotations)/(total contralateral + total ipsilateral rotations). As dyskinesia intensity varied from animal to animal, data were also presented with the baseline subtracted (Figure 3G, 3I,3K, middle panels; Figure 4B, 4D, middle panels). The Wilcoxon rank-sum test was used to compare animals expressing eYFP versus an opsin (ChR2 or eNpHR3.0). To compare ‘OFF’ (1) and ‘ON’ periods within an animal, the Wilcoxon sign-rank test was employed. If postmortem studies showed evidence of inadequate dopamine depletion, viral expression, or no TRAPed cells (based on user error), its results were excluded from further analysis (N = 2 animals excluded from the entire study, 1 based on inadequate viral expression (1 ChR2) and 1 based on improper injection of 4-OHT).
In Vivo Electrophysiology
For display and analysis purposes, the firing rate of single units was averaged in one-minute bins. For analyses of firing rate before and during levodopa administration, the average firing rate from 0-30 min prior to injection and 35-65 min after injection were calculated. To determine whether a unit’s firing rate significantly changed after levodopa injection, the firing rates during the baseline period were compared with the firing rates between 35-65 min after levodopa injection, using a Wilcoxon Signed Rank test.
Histology, Microscopy, and Cell Counting
To quantify colocalization between TRAPed cells or c-Fos positive nuclei or for high-magnification images, confocal imaging was implemented using a Nikon Spinning Disk confocal microscope with a 40x objective. Exposure times were matched between images of the same type.
To quantify the number of TRAPed cells in the striatum, S1, or M1 (Figure S1L) or c-Fos positive nuclei in the striatum (Figure S1J), individual images were stitched together to produce an entire coronal or sagittal image. We consulted the Allen Brain Atlas (http://www.brain-map.org/) and the Mouse Brain in Stereotaxic Coordinates (hard copy, 4th edition, by George Paxinos) for anatomy and chose 5-7 representative coronal or sagittal sections (at specified coordinates from bregma) for each brain region. To quantify the number of striatal c-Fos positive nuclei we used the coronal coordinates (+1) - (−1) for quantification. To quantify the number or density of TRAPed cells in the striatum, S1, or M1 we used the following sagittal coordinates for quantification, striatum: 1.725-3.60 ML, S1: 1.08-3.72 ML, and M1: 1.0-3.1 ML. We used FIJI/ImageJ software to count cells, using both auto-threshold detection and manual counting. Slices were discarded if there was tissue damage to the designated area. One-way ANOVA was used to assess any differences in cell numbers for c-Fos positive nuclei in the striatum (Figure S1J) or TRAPed cell or cell density in the striatum, S1, and M1 (Figures 1I and S1L). For all cohorts, the post hoc Tukey HSD test was used if the analysis of variance yielded a significant F-ratio. For the WT LID cohort (Figure S1J), we saw no statistical difference in cell numbers between the control groups (saline/saline, saline/levodopa, and 6-OHDA/saline), and pooled the data together to compare to the 6-OHDA/levodopa treated group. To determine correlations between behavior and TRAPed neurons (Figures 3B and S3B), the average number or density of TRAPed cells was calculated by averaging cell counts or density over the defined coordinates for each brain area. The AIM score was calculated by summing all AIM scores over the entire 120 min scoring session closest to the day of 4-OHT administration.
For immunohistochemical experiments to investigate colocalization between TRAPed cells and markers of striatal cell types (Figures 2 and S2), 5-7 representative coronal or sagittal slices per animal were chosen, focusing next on 5-7 specific fields of view (1×1 screen size) in the DLS. Confocal z stacks in each field of view were acquired, matching exposure time and laser power for each fluorophore in all images captured. To assess colocalization between TRAPed cells and different neuronal markers, maximum z-projections were made and merged to produce an overlay with TRAPed cells and the corresponding cellular marker. For each animal, a minimum of 100 cells of each type (TRAP-tdTomato, cellular marker, or c-Fos) were manually counted using custom scripts in FIJI/ImageJ. For a cell to be counted as colocalized, it needed to appear on both individual and merged channels in the z stack and maximum z-projection showing identical overlap with the other cell. Any cell showing ambiguous overlap was not counted as colocalized. The same procedure was used to assess colocalization of c-Fos and Drd1a-tdTomato (Figures S2F and S2G) and c-Fos and D2-GFP (Figures S2F and S2H).
To determine the average number of FosTRAP-eNpHR3.0 versus D1-eNpHR3.0 per field, confocal images were acquired from at least 2 sagittal or coronal sections where the optical ferrule could be seen. We focused on 5-7 specific fields of view near the ferrule mark and acquired images over a strict depth of 20 μm for each animal. Cells were manually counted, using the outline of the cell body and cell processes as a guide, in FIJI/ImageJ using both the maximum z-projection and individual images within the stack. At least two blinded experimenters validated the presence or absence of an eNpHR3.0 positive cell.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-TH | Pel-Freez Biologicals | Cat# P40101-150, RRID: AB_2617184 |
| Chicken anti-TH | Milipore | Cat# AB9702, RRID: AB_570923, Lot #2586825 |
| Rabbit anti-c-Fos (9F6) | Cell Signaling Technology | Cat# 2250 also 2250S, 2250P, RRID: AB_2247211, Lot #8 |
| Rabbit anti-c-Fos | Santa Cruz Biotechnology | Cat# sc-52, RRID: AB_2106783 |
| Goat anti-ChAT | Millipore | Cat# AB144P, RRID: AB_2079751 |
| Rabbit anti-DARPP-32 (19A3) | Cell Signaling Technology | Cat# 2306S, RRID: AB_823479, Lot #6 |
| Mouse anti-RFP | Rockland | Cat# 200-301-379, RRID: AB_2611063 |
| Rabbit anti-NeuN | Millipore | Cat# MAB377, RRID: AB_2298772 |
| Rabbit anti-NPY | Cell Signaling Technology | Cat# 11976, RRID: AB_2716286, Lot #1 |
| Rabbit anti-PV | Swant | Cat# PV27, RRID: AB_2631173 |
| Rabbit anti- Erg-1 | Cell Signaling Technology | Cat# 4153 RRID: AB_2097038 |
| Goat anti- ΔFosB | Santa Cruz Biotechnology | Cat# sc-48-G, RRID: AB_631516, Lot #C0716 |
| Alexa Fluor 488- Donkey Anti-Rabbit IgG | Jackson ImmunoResearch Labs | Cat# 711-546-152, RRID: AB_2340619, Lot #128227 |
| Alexa Fluor 488- Donkey Anti-Goat IgG | Jackson ImmunoResearch Labs | Cat# 705-545-147, RRID: AB_2336933, Lot #128271 |
| Alexa Fluor 594- Donkey Anti-Mouse IgG | Jackson ImmunoResearch Labs | Cat# 715-586-150, RRID: AB_2340857, Lot #122479 |
| Alexa Fluor 647- Donkey Anti-Rabbit IgG | Jackson ImmunoResearch Labs | Cat# 711-606-152, RRID: AB_2340625, Lot #118661 |
| Alexa Fluor 647- Anti-Goat IgG | Jackson ImmunoResearch Labs | Cat# 705-605-147, RRID: AB_2340437, Lot #128808 |
| Alexa Fluor 647- Donkey Anti-Chicken IgY (IgG) | Jackson ImmunoResearch Labs | Cat# 703-606-155, RRID: AB_2340380, Lot #123720 |
| Alexa Fluor 647- Donkey Anti-Mouse IgG | Jackson ImmunoResearch Labs | Cat# 715-606-150, RRID: AB_2340865, Lot #120518 |
|
Bacterial and Virus Strains | ||
| AAV5-EF1a-DIO-eYFP | UNC Vector Core | Lot #AV4310g |
| AAV5-EF1a-DIO-hCHR2(H134R)-eYFP-wpre-hGH | Penn Vector Core | AV-5-20298P, Lot #CS0384 |
| rAAV5/EF1a-DIO-eNpHR3.0-eYFP | UNC Vector Core | Lot #AV4806e |
|
Chemicals, Peptides, and Recombinant Proteins | ||
| Picrotoxin | Sigma-Aldrich | P1675 |
| Lidocaine N-ethyl chloride | Sigma-Aldrich | L1663 |
| 6-Hydroxydopamine hydrobromide | Sigma-Aldrich | H116 |
| 4-Hydroxytamoxifen | Sigma-Aldrich | H6278 |
| Potassium methanesulfonate | Sigma-Aldrich | 83000 |
| Guanosine 5′-triphosphate sodium salt hydrate | Sigma-Aldrich | G8877 |
| Adenosine 5′-triphosphate magnesium salt | Sigma-Aldrich | A9187 |
| Desipramine hydrochloride | Sigma-Aldrich | D3900 |
|
Critical Commercial Assays | ||
| VECTASHIELD Antifade Mounting Medium | Vector Laboratories | Cat# H-1000, RRID: AB_2336789 |
| VECTASTAIN Elite ABC-Peroxidase Kit antibody | Vector Laboratories | Cat# PK-6100, RRID: AB_2336819 |
|
Experimental Models: Organisms/Strains | ||
| Mouse: WT: C57BL/6J | The Jackson Laboratory | RRID: IMSR_JAX:000664 |
| Mouse: B6.129(Cg)-Fostm1.1(cre/ERT2)Luo/J | The Jackson Laboratory | RRID: IMSR_JAX:021882 |
| Mouse: B6.Cg-Tg(Drd1a-tdTomato)6Calak/J Mus musculus |
The Jackson Laboratory | RRID: IMSR_JAX:016204 |
| Mouse: STOCK Tg(Drd2-EGFP)S118Gsat/ Mmnc Mus musculus |
MMRRC | RRID: MMRRC_000230-UNC |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J |
The Jackson Laboratory | RRID: IMSR_JAX:007914 |
| Mouse: B6.FVB(Cg)-Tg(Drd1-cre)EY217Gsat/ Mmucd B6.FVB(Cg)-Tg(Drd1-cre)EY217Gsat/Mmucd |
MMRRC | RRID: MMRRC_034258-UCD |
|
Software and Algorithms | ||
| Igor Pro | Wavemetrics | http://www.wavemetrics.com/products/igorpro/igorpro.htm; RRID: SCR_000325 |
| MATLAB R2015a | MathWorks | https://www.mathworks.com/products/matlab/; RRID: SCR_001622 |
| ImageJ | NIH | https://imagej.nih.gov/ij/; RRID: SCR_003070 |
| EthoVision XT | Noldus | http://www.noldus.com/animal-behavior-research/products/ethovision-xt; RRID: SCR_000441 |
| Adobe Illustrator CS5 | Adobe | https://www.adobe.com/products/illustrator.html; RRID: SCR_014198 |
| MAP Software | Plexon | http://plexon.com/products/map-software; RRID: SCR_003170 |
| Offline Sorter | Plexon | http://plexon.com/products/offline-sorter; RRID: SCR_000012 |
| NeuroExplorer | NeuroExplorer | http://www.neuroexplorer.com/; RRID: SCR_001818 |
|
Other | ||
| 32-channel fixed optrode array | Innovative Neurophysiology | http://www.inphysiology.com/optogenetic-applications/ |
| 200 μm Core TECS-Clad Multimode Optical Fiber, 0.39 NA | Thorlabs | Catalog # FT200UMT |
| 1.25 mm Multimode LC/PC Ceramic Ferrule, 230 μm Bore Size | Thorlabs | Catalog # CFLC230-10 |
| Ceramic Split Mating Sleeve for 1.25 mm (LC/PC) Ferrules | Thorlabs | Catalog # ADAL1 |
| 150mW DPSS 473nm Blue Laser | Shanghai Laser & Optics Century | BL473T8-150 + ADR-800A |
| 200mW Fiber-coupled DPSS Green laser, 532nm | Shanghai Laser & Optics Century | GL532T3-200 + ADR-700A |
| Master-8 | A.M.P.I. | http://www.ampi.co.il/master8cp.html |
Highlights.
FosTRAP captures neurons activated in levodopa-induced dyskinesia (LID) brain-wide
Primarily striatal direct pathway neurons (dMSNs) are activated during dyskinesia
Reactivation of TRAPed striatal neurons produces dyskinesia in the absence of levodopa
Inhibition of TRAPed striatal neurons, but not nonspecific dMSNs, reduces LID
ACKNOWLEDGMENTS
We thank Viktor Kharazia, Scott Wegner (UCSF ACTG), and DeLaine Larsen (UCSF Nikon Imaging Center) for excellent histological and microscopy assistance, and Kevin Bender, Phillip Starr, Massimo Scanziani, Michael Ryan, and the Nelson and Kreitzer laboratories for valuable feedback on this manuscript. This work was supported by the Brain Research Foundation (A.B.N.), NINDS (K08 NS081001, A.B.N.), NSF Graduate Research Fellowship (A.E.G.), and UCSF Discovery Fellows Program (A.E.G.). A.B.N. is the Richard and Shirley Cahill Endowed Chair in Parkinson’s Disease Research.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and two movies and can be found with this article online athttps://doi.org/10.1016/j.neuron.2018.01.017
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alam M, Rumpel R, Jin X, von Wrangel C, Tschirner SK, Krauss JK, Grothe C, Ratzka A, and Schwabe K (2017). Altered somatosensory cortex neuronal activity in a rat model of Parkinson’s disease and levodopa-induced dyskinesias. Exp. Neurol 294, 19–31. [DOI] [PubMed] [Google Scholar]
- Alberico SL, Kim YC, Lence T, and Narayanan NS (2017). Axial levodopa-induced dyskinesias and neuronal activity in the dorsal striatum. Neuroscience 343, 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcacer C, Andreoli L, Sebastianutto I, Jakobsson J, Fieblinger T, and Cenci MA (2017). Chemogenetic stimulation of striatal projection neurons modulates responses to Parkinson’s disease therapy. J. Clin. Invest 127, 720–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M, Hilbertson A, and Cenci MA (1999). Striatal fosB expression is causally linked with l-DOPA-induced abnormal involuntary movements and the associated upregulation of striatal prodynorphin mRNA in a rat model of Parkinson’s disease. Neurobiol. Dis 6, 461–474. [DOI] [PubMed] [Google Scholar]
- Bagetta V, Sgobio C, Pendolino V, Del Papa G, Tozzi A, Ghiglieri V, Giampà C, Zianni E, Gardoni F, Calabresi P, and Picconi B (2012). Rebalance of striatal NMDA/AMPA receptor ratio underlies the reduced emergence of dyskinesia during D2-like dopamine agonist treatment in experimental Parkinson’s disease. J. Neurosci 32, 17921–17931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergson C, Mrzljak L, Smiley JF, Pappy M, Levenson R, and Goldman-Rakic PS (1995). Regional, cellular, and subcellular variations in the distribution of D1 and D5 dopamine receptors in primate brain. J. Neurosci 15, 7821–7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke JD, Okatan M, Skurski J, and Eichenbaum HB (2004). Oscillatory entrainment of striatal neurons in freely moving rats. Neuron 43, 883–896. [DOI] [PubMed] [Google Scholar]
- Berton O, Guigoni C, Li Q, Bioulac BH, Aubert I, Gross CE, Dileone RJ, Nestler EJ, and Bezard E (2009). Striatal overexpression of DeltaJunD resets L-DOPA-induced dyskinesia in a primate model of Parkinson disease. Biol. Psychiatry 66, 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraud T, Bezard E, Bioulac B, and Gross CE (2001). Dopamine agonist-induced dyskinesias are correlated to both firing pattern and frequency alterations of pallidal neurones in the MPTP-treated monkey. Brain 124, 546–557. [DOI] [PubMed] [Google Scholar]
- Borgkvist A, Avegno EM, Wong MY, Kheirbek MA, Sonders MS, Hen R, and Sulzer D (2015). Loss of Striatonigral GABAergic Presynaptic Inhibition Enables Motor Sensitization in Parkinsonian Mice. Neuron 87, 976–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck K, Voehringer P, and Ferger B (2010). Site-specific action of L-3,4-dihydroxyphenylalanine in the striatum but not globus pallidus and substantia nigra pars reticulata evokes dyskinetic movements in chronic L-3,4-dihydroxyphenylalanine-treated 6-hydroxydopamine-lesioned rats. Neuroscience 166, 355–358. [DOI] [PubMed] [Google Scholar]
- Cao X, Yasuda T, Uthayathas S, Watts RL, Mouradian MM, Mochizuki H, and Papa SM (2010). Striatal overexpression of DeltaFosB reproduces chronic levodopa-induced involuntary movements. J. Neurosci 30, 7335–7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci MA, and Lundblad M (2007). Ratings of L-DOPA-induced dyskinesia in the unilateral 6-OHDA lesion model of Parkinson’s disease in rats and mice. Curr. Protoc. Neurosci. Chapter 9, 25. [DOI] [PubMed] [Google Scholar]
- Engeln M, Bastide MF, Toulmé E, Dehay B, Bourdenx M, Doudnikoff E, Li Q, Gross CE, Boué-Grabot E, Pisani A, et al. (2016). Selective inactivation of striatal FosB/ΔFosB-expressing neurons alleviates L-Dopa-induced Dyskinesia. Biol. Psychiatry 79, 354–361. [DOI] [PubMed] [Google Scholar]
- Fieblinger T, Graves SM, Sebel LE, Alcacer C, Plotkin JL, Gertler TS, Chan CS, Heiman M, Greengard P, Cenci MA, and Surmeier DJ (2014). Cell type-specific plasticity of striatal projection neurons in parkinsonism and L-DOPA-induced dyskinesia. Nat. Commun 5, 5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion M, Tremblay L, and Bédard PJ (1991). Effects of dopamine agonists on the spontaneous activity of globus pallidus neurons in monkeys with MPTP-induced parkinsonism. Brain Res 547, 152–161. [PubMed] [Google Scholar]
- Gage GJ, Stoetzner CR, Wiltschko AB, and Berke JD (2010). Selective activation of striatal fast-spiking interneurons during choice execution. Neuron 67, 466–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Engber TM, Mahan LC, Susel Z, Chase TN, Monsma FJ Jr., and Sibley DR (1990). D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science 250, 1429–1432. [DOI] [PubMed] [Google Scholar]
- Gernert M, Hamann M, Bennay M, Löscher W, and Richter A (2000). Deficit of striatal parvalbumin-reactive GABAergic interneurons and decreased basal ganglia output in a genetic rodent model of idiopathic paroxysmal dystonia. J. Neurosci 20, 7052–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittis AH, Hang GB, LaDow ES, Shoenfeld LR, Atallah BV, Finkbeiner S, and Kreitzer AC (2011a). Rapid target-specific remodeling of fast-spiking inhibitory circuits after loss of dopamine. Neuron 71, 858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittis AH, Leventhal DK, Fensterheim BA, Pettibone JR, Berke JD, and Kreitzer AC (2011b). Selective inhibition of striatal fast-spiking interneurons causes dyskinesias. J. Neurosci 31, 15727–15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, and Heintz N (2003). A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925. [DOI] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, and Luo L (2013). Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halje P, Tamtè M, Richter U, Mohammed M, Cenci MA, and Petersson P (2012). Levodopa-induced dyskinesia is strongly associated with resonant cortical oscillations. J. Neurosci 32, 16541–16551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Henze DA, Csicsvari J, Hirase H, and Buzsáki G (2000). Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. J. Neurophysiol 84, 401–414. [DOI] [PubMed] [Google Scholar]
- Heiman M, Heilbut A, Francardo V, Kulicke R, Fenster RJ, Kolaczyk ED, Mesirov JP, Surmeier DJ, Cenci MA, and Greengard P (2014). Molecular adaptations of striatal spiny projection neurons during levodopa-induced dyskinesia. Proc. Natl. Acad. Sci. USA 111, 4578–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández LF, Castela I, Ruiz-DeDiego I, Obeso JA, and Moratalla R (2017). Striatal activation by optogenetics induces dyskinesias in the 6-hydroxydopamine rat model of Parkinson disease. Mov. Disord 32, 530–537. [DOI] [PubMed] [Google Scholar]
- Jenner P (2008). Molecular mechanisms of L-DOPA-induced dyskinesia. Nat. Rev. Neurosci 9, 665–677. [DOI] [PubMed] [Google Scholar]
- Kravitz AV, Owen SF, and Kreitzer AC (2013). Optogenetic identification of striatal projection neuron subtypes during in vivo recordings. Brain Res 1511,21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R, Dostrovsky JO, Lang AE, Sime E, Hutchison WD, and Lozano AM (2001). Effects of apomorphine on subthalamic nucleus and globus pallidus internus neurons in patients with Parkinson’s disease. J. Neurophysiol 86, 249–260. [DOI] [PubMed] [Google Scholar]
- Liang L, DeLong MR, and Papa SM (2008). Inversion of dopamine responses in striatal medium spiny neurons and involuntary movements. J. Neurosci 28, 7537–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach D, and Bishop C (2013). Critical involvement of the motor cortex in the pathophysiology and treatment of Parkinson’s disease. Neurosci. Biobehav. Rev 37, 2737–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano AM, Lang AE, Levy R, Hutchison W, and Dostrovsky J (2000). Neuronal recordings in Parkinson’s disease patients with dyskinesias induced by apomorphine. Ann. Neurol 47 (4, Suppl 1), S141–S146. [PubMed] [Google Scholar]
- Papa SM, Desimone R, Fiorani M, and Oldfield EH (1999). Internal globus pallidus discharge is nearly suppressed during levodopa-induced dyskinesias. Ann. Neurol 46, 732–738. [DOI] [PubMed] [Google Scholar]
- Pavón N, Martín AB, Mendialdua A, and Moratalla R (2006). ERK phosphorylation and FosB expression are associated with L-DOPA-induced dyskinesia in hemiparkinsonian mice. Biol. Psychiatry 59, 64–74. [DOI] [PubMed] [Google Scholar]
- Perez XA, Zhang D, Bordia T, and Quik M (2017). Striatal D1 medium spiny neuron activation induces dyskinesias in parkinsonian mice. Mov. Disord 32, 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picconi B, Centonze D, Håkansson K, Bernardi G, Greengard P, Fisone G, Cenci MA, and Calabresi P (2003). Loss of bidirectional striatal synaptic plasticity in L-DOPA-induced dyskinesia. Nat. Neurosci 6, 501–506. [DOI] [PubMed] [Google Scholar]
- Rothwell PE, Hayton SJ, Sun GL, Fuccillo MV, Lim BK, and Malenka RC (2015). Input- and Output-Specific Regulation of Serial Order Performance by Corticostriatal Circuits. Neuron 88, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Plotkin JL, Francardo V, Ko WK, Xie Z, Li Q, Fieblinger T, Wess J, Neubig RR, Lindsley CW, et al. (2015). M4 Muscarinic Receptor Signaling Ameliorates Striatal Plasticity Deficits in Models of L-DOPA-Induced Dyskinesia. Neuron 88, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuen JA, Chen M, Gloss B, and Calakos N (2008). Drd1a-tdTomato BAC transgenic mice for simultaneous visualization of medium spiny neurons in the direct and indirect pathways of the basal ganglia. J. Neurosci 28, 2681–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Liang L, Kaneoke Y, Cao X, and Papa SM (2015). Dopamine regulates distinctively the activity patterns of striatal output neurons in advanced parkinsonian primates. J. Neurophysiol 113, 1533–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier DJ, Carrillo-Reid L, and Bargas J (2011). Dopaminergic modulation of striatal neurons, circuits, and assemblies. Neuroscience 198, 3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann NC, de Hemptinne C, Miocinovic S, Qasim S, Wang SS, Ziman N, Ostrem JL, San Luciano M, Galifianakis NB, and Starr PA (2016). Gamma Oscillations in the Hyperkinetic State Detected with Chronic Human Brain Recordings in Parkinson’s Disease. J. Neurosci 36, 6445–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin JE, Andersson M, Lundblad M, and Cenci MA (2001). Persistent changes in striatal gene expression induced by long-term L-DOPA treatment in a rat model of Parkinson’s disease. Eur. J. Neurosci 14, 1171–1176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.