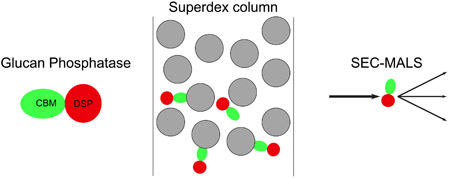
Abstract
Glucan phosphatases are a unique subset of the phosphatase family that bind to and dephosphorylate carbohydrate substrates. Family members are found in diverse organisms ranging from single-cell red algae to humans. The nature of their functional oligomerization has been a source of considerable debate. We demonstrate that the human laforin protein behaves aberrantly when subjected to Size Exclusion Chromotography (SEC) analysis due to interaction with the carbohydrate-based matrix. This interaction complicates the analysis of laforin human disease mutations. Herein, we show that SEC with Multi-Angle static Light Scattering (SEC-MALS) provides a method to robustly define the oligomerization state of laforin and laforin variants. We further analyzed glucan phosphatases from photosynthetic organisms to define if this interaction was characteristic of all glucan phosphatases. Starch EXcess-four (SEX4) from green plants was found to lack significant interaction with the matrix and instead exists as a monomer. Conversely, Cm-laforin, from red algae, exists as a monomer in solution while still exhibiting significant interaction with the matrix. These data demonstrate a range of oligomerization behaviors of members of the glucan phosphatase family, and establish SEC-MALS as a robust methodology to quantify and compare oligomerization states between different proteins and protein variants in this family.
Keywords: SEC-MALS, Glucan Phosphatase, Laforin, Oligomerization
Graphical Abstract
Introduction
Glucan phosphatases are members of the Dual Specificity Phosphatase (DSP) family [1]. Their substrates are phosphorylated carbohydrates, and they function as key regulators of carbohydrate metabolism. Glucan phosphatases from various kingdoms of life have been identified based on the presence of both DSP and Carbohydrate Binding Module (CBM) domains [2]. The human glucan phosphatase laforin is a critical regulator of glycogen metabolism, and mutations in the laforin gene result in Lafora’s Disease (LD), a devastating, fatal, autosomal recessive juvenile epilepsy [3]. Plant glucan phosphatases are critical for starch metabolism, and loss results in a Starch EXcess phenotype [4].
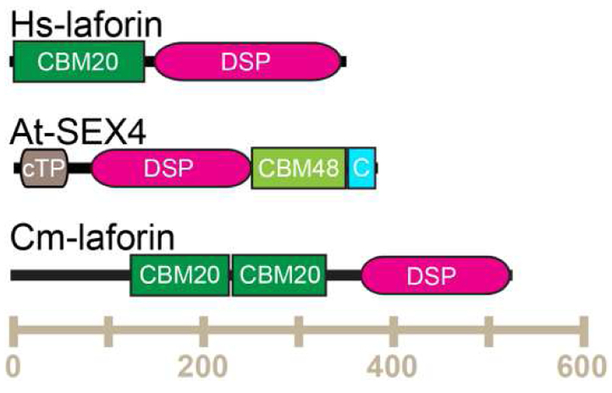
Glucan phosphatases from different kingdoms act on different carbohydrate substrates, and possess varied domain architecture (Fig. 1). Vertebrate laforin, typified by the human protein, Hs-laforin possesses an N-terminal CBM20 carbohydrate binding domain and C-terminal DSP domain. Green plant glucan phosphatases, typified by the Arabidopsis thaliana (At) SEX4, possess an N-terminal chloroplast Targeting Peptide (cTP), DSP, CBM48, and plant-specific C-terminus (CT) [5]. Green plants additionally contain two other glucan phosphatases, Like-SEX4 (LSF)1 and LSF2 [6]. Red algae, typified by the Cyanidioschyzon merolae (Cm)-laforin, contain a variable N-terminal region that is predicted to be intrinsically disordered, followed by two CBM20, and a C-terminal DSP domain. Because of the domain identity and organization, it has been hypothesized that vertebrate and red algae proteins are most similar [2]. However, since they act on different substrates and different domain combinations and structure, the similarity and difference between the function of glucan phosphatases from different kingdoms remains to be determined.
Figure 1:
Domain schematic of glucan phosphatases. Different domain type and organization are characteristic of the enzymes from different kingdoms, amino acid number indicated along the horizontal axis. Domain types include: Carbohydrate Binding Module (CBM), Dual Specificity Phosphatase (DSP), chloroplast Targeting Peptide (cTP), and C-terminus (CT).
Laforin is a multi-domain and multi-functional protein. Previous studies have identified key aspects of the functionality of laforin, including carbohydrate binding, protein interactions, catalytic activity, and dimerization. There has been debate regarding the equilibrium, regulation, and function of different laforin oligomers [7-9] Recent work demonstrated the structural basis for stable dimerization in solution, mediated by the DSP domain, and showed that human disease mutations were found at the dimer interface and are deleterious to protein oligomerization [10]. This result motivated studies of laforin dimerization, and development of methods to reliably determine the oligomerization state of patient mutations. We demonstrate that SEC-MALS allows reliable determination of laforin dimerization and is well suited for assessing the oligomerization of human patient mutations, critically decoupling carbohydrate binding from oligomerization. The oligomerization of glucan phosphatases from other kingdoms of life was also analyzed, and revealed that glucan phosphatases from other kingdoms exist in monomeric form. These data establish a reliable method to assess the differing oligomerization behaviors of glucan phosphatases and variants from different kingdoms.
Materials and Methods
Protein expression and purification
Wild type (WT) and mutant human laforin [10], SEX4 [5], and SEX4ΔCBM [11] were expressed and purified as previously reported. Cm-laforin (domains 123, residues 157-532 C480S) and sub-domain constructs, CBM linker (domains 12l, residues 157-379), CBM (domains 12, residues 157-356), and DSP (domains 3, residues 379-532 C480S) were expressed from pET28b in E. coli strain Rosetta-2 (DE3) (Millipore Sigma, Burlington, MA). Cells were grown in Terrific Broth to an OD600 1.5, cold-shocked for 15 minutes, induced with 1mM IPTG, and grown overnight at 16°C. Cell pellets were harvested and frozen at −20°C. Cells were lysed using lysozyme and sonication. Initial purification was accomplished by Immobilized Metal Affinity Chromatography (IMAC) with HIS-Select HF resin (Millipore Sigma, St. Louis, MO). Protein was loaded and washed in 20mM Tris, 100mM NaCl, 5mM 2-mercaptoethanol (BME), pH 8.0 and eluted with 300 mM Imidazole, 100 mM NaCl, 5mM BME, pH 8.0. Subsequently, proteins were purified by preparative size exclusion using an AKTA pure with Superdex75 HiLoad 16/60 column (GE Healthcare Life Sciences, Marlborough, MA) equilibrated and run in SEC buffer [20 mM Tris, 100 mM NaCl, 2mM Dithiothreitol (DTT), pH 7.5]. Additional SEC analysis was performed using a silica-based Bio-Select SEC 125-5 column (Bio-Rad, Hercules, CA) run in SEC buffer.
SEC-MALS
Proteins were run on an AKTA pure with inline Superdex 75 Increase 10/300 column (GE Healthcare Life Sciences), miniDAWN TREOS, and Optilab T-rEX (Wyatt Technologies, Santa Barbara, CA). The column was equilibrated and run in SEC buffer, 500μL of 1-2mg/mL protein was loaded, and the column run at 0.5mL/min. Addition of 100mM maltose in the running buffer was used to determine the effect of laforin retention. The apparent molecular weight from SEC analysis was calculated by fitting the elution volume of the protein standards Bovine albumin, Ovalbumin, Carbonic Anhydrase and Lysozme. Light scattering data were processed using ASTRA (Wyatt Technologies). Molecular weight was determined by analyzing peaks at half height using Refractive Index (RI) as the concentration source. Further analysis and graphics were prepared using Prism (Graphpad Software, La Jolla, CA).
Results and Discussion
Analysis of human laforin
One of the key features of laforin is its ability to oligomerize, but there is considerable debate as to the nature and effect of patient mutations on oligomerization. We first analyzed WT human laforin using SEC-MALS. We found that laforin elutes as a single species. When calibrated versus proteins of known molecular weight, SEC analysis indicates that it elutes at a volume consistent with monomeric protein. However, SEC-MALS indicates that it exists as a dimer in solution (Fig. 2A, Table 1), consistent with our previous determination by analytical ultracentrifugation and Small Angle X-ray Scattering (SAXS). To test whether the divergent retention of wild-type laforin was due to residual matrix interactions, since Superdex is a carbohydrate-based size exclusion matrix, we tested the behavior of wild-type laforin in the presence and absence of maltose. Indeed, we find that there is a significant decrease in elution volume in the presence of maltose and corresponding increase in molecular weight (Fig. 2A, Table 1). This suggests that previous assessments of the oligomerization of laforin by SEC were ambiguous due to combined effects from both oligomerization and laforin interactions with the carbohydrate-based resin, providing an explanation for previous discrepant findings.
Figure 2:
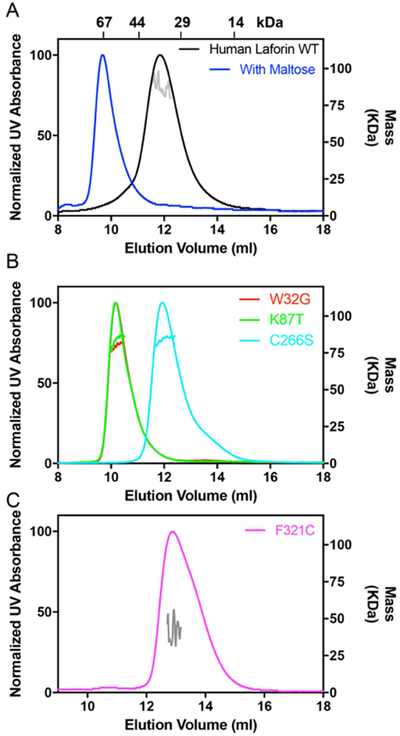
Behavior of the human glucan phosphatase laforin. A) WT laforin without and with (blue) added maltose. The right Y-axis represents the MALS-measured mass, with individual fits determined from the elution range indicated in each panel. The elution volume of SEC standards is indicated at the top of the panel. B) Laforin W32G, K87T, and C266S C) Laforin F321C.
Table 1:
Determination of glucan phosphatase oligomerization
| Protein | Theoretical Monomer M.W. (KDa) |
Apparent M.W. SEC (KDa) |
SEC-MALS M.W. (KDa) |
Ratio SEC /Theoretical |
Ratio MALS /Theoretical |
|---|---|---|---|---|---|
| Hs-laforin WT | 39.1 | 39.9 | 78.2 (±5.4%) | 1.0 | 2.0 |
| Hs-laforin WT + Maltose | 39.4 | 62.0 | 95.2 (±3.7%) | 1.6 | 2.4 |
| Hs-laforin W32G | 39.0 | 56.8 | 80.1 (±1.2%) | 1.5 | 2.1 |
| Hs-laforin K87T | 39.1 | 57.0 | 76.9 (±3.2%) | 1.5 | 2.0 |
| Hs-laforin C266S | 39.1 | 39.0 | 77.4 (±1.6%) | 1.0 | 2.0 |
| Hs-laforin F321C | 39.1 | 29.4 | 41.6 (±29.6%) | 0.8 | 1.1 |
| At-SEX4 | 33.0 | 39.6 | 34.6 (±1.4%) | 1.2 | 1.0 |
| At-SEX4 ΔCBM | 25.9 | 32.5 | 25.8 (±1.5%) | 1.3 | 1.0 |
| Cm-laforin 123 | 44.9 | 33.3 | 44.3 (±2.0%) | 0.7 | 1.0 |
| Cm-laforin 12l-CBM | 27.8 | −73.8 | 28.3 (±2.3%) | −2.7 | 1.0 |
| Cm-laforin 3-DSP | 19.5 | 23.4 | 19.2 (±0.8%) | 1.2 | 1.0 |
These results indicate that SEC-MALS is a reliable method to directly determine the oligomerization state of laforin patient mutations, without effects from other functionality of the protein. We next analyzed two patient mutations, W32G and K87T, both located at the carbohydrate binding interface of the CBM domain which have dramatically reduced carbohydrate binding. In both cases the proteins eluted as a single peak, but at different volume than wild-type protein strongly suggesting that residual interaction of the CBM domain with the column matrix is responsible for the aberrant elution behavior (Fig. 2B). SEC analysis suggested an apparent molecular weight between monomer and dimer. However, analysis by SEC-MALS indicated that the proteins exist as stable dimers indicating a robust ability to decouple changes in oligomerization and carbohydrate binding (Table 1). We next assessed the C266S mutation, located in the laforin DSP-domain active site, which is catalytically inactive. In this case, the mutant protein behaved as wild-type protein and SEC-MALS analysis demonstrated the protein exists as a stable dimer (Fig. 2B, Table 1). These data indicate that laforin oligomerization and carbohydrate binding, but not catalytic activity, affects the apparent molecular weight of the protein.
To further test this method, we analyzed the novel F321C mutation, a recently identified patient mutation located in the dimer interface. F321C also eluted as a single peak, but with different volume than wild-type protein (Fig. 2C). In this case, SEC and SEC-MALS analysis indicated that the mutant protein exists as a monomer, further supporting a DSP-mediated dimeric form of the enzyme and providing a basis for understanding its role in disease (Table 1). Taken together, these data demonstrate that SEC-MALS is a valuable tool in ongoing efforts focused on defining the biochemical basis for the dysfunction of laforin mutations.
Analysis of glucan phosphatases from photosynthetic organisms
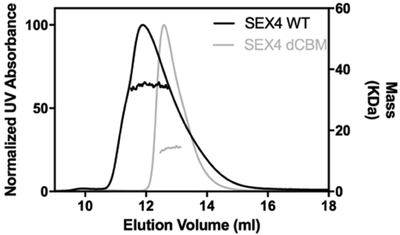
We next assessed glucan phosphatases from diverse photosynthetic organisms. We first examined the behavior of the glucan phosphatases SEX4, which is the conserved glucan phosphatase found in green plants. SEX4 dephosphorylates starch and binds to linear oligosaccharides using an integrated CBM/DSP interface [12]. When analyzed by SEC-MALS, SEX4 was found to exist as a monomer in solution (Figure 3, Table 1), consistent with previous structural studies [5]. In contrast to laforin, SEX4 eluted from the gel filtration at the expected volume. Further deletion of the SEX4 CBM had little effect on the apparent mobility or oligomerization state of SEX4 (Figure 3, Table 1). While both SEX4 and laforin proteins have CBM domains, they belong to different families. SEX4 has a CBM48 domain while laforin has a CBM20 domain [13]. The different CBM families have different binding properties for carbohydrates [14], potentially explaining the difference in observed matrix interactions.
Figure 3:
Behavior of the green plant glucan phosphatase SEX4. SEC-MALS analysis SEX4 WT and dCBM.
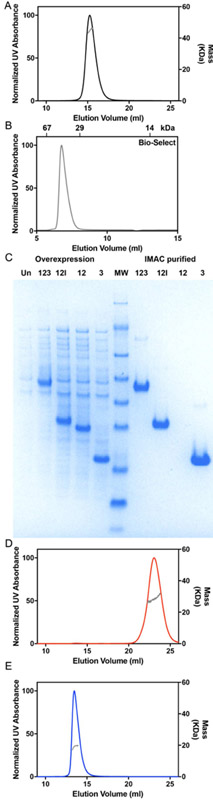
We next assessed Cm-laforin, to determine the behavior of the glucan phosphatase from red algae that has two CBM20 domains and a DSP domain (Fig. 1). Cm-laforin is well behaved on gel filtration eluting as a single species (Fig. 4A). However, the apparent mobility of the protein produces an estimate for molecular weight 25% lower than that of monomeric protein, which is not physically plausible (Table 1). However, SEC-MALS analysis demonstrates that the protein exists as a monomer in solution (Table 1). Based on the data from Hs-laforin, we hypothesized that the CBM domains may be interacting with the matrix, resulting in the observed anomalous mobility. To test this hypothesis, we first performed SEC analysis using a silica-based Bio-Silect column. Strikingly, the apparent molecular weight of Cm-laforin (46 kDa) corresponds to a monomeric species, consistent with the oligomeric state determined from MALS (Fig. 4B). This result further supports the finding that residual matrix interactions are the basis for anomalous elution from the Superdex column. To define the contribution of different domains, we designed sub-domain constructs that expressed either CBM domains or the DSP domain with and without the CBM-DSP linker. Intriguingly, we find that the linker is required for CBM domain expression, but dispensable for the DSP domain (Fig. 4C). We next assessed the behavior of separate CBM and DSP containing constructs. Strikingly, the CBM-containing construct displayed very anomalous retention on gel filtration. This construct actually eluted after the total column volume, resulting in a physically impossible estimate of molecular weight by SEC (Fig. 4D, Table 1). In contrast, the DSP domain alone elutes at the expected volume and, in fact, prior to the elution volume of the much larger full protein (Fig. 4E). Importantly, SEC-MALS analysis demonstrates that both constructs are well behaved and monomeric, consistent with the full three domain construct (Table 1).
Figure 4:
Behavior of the red algae glucan phosphatase Cm-laforin. A) SEC-MALS analysis of the red algae glucan phosphatase Cm-laforin using a Superdex column. B) SEC analysis of Cm-laforin using a Bio-Silect SEC column. The elution volume of SEC standards is indicated at the top of the panel. C) Overexpression/purification gels of Cm-laforin construct containing the full two CBM and DSP domains (123), both CBMs with linker (12l), both CBMs (12), and DSP (3). The linker is required for the CBM domains, since expression and purification of 12l but not 12 results in soluble and folded protein. D) SEC-MALS analysis of isolated CBM-domain containing (12l) and E) DSP-domain (3) constructs.
These data are intriguing, since Cm-laforin oligomerization is different from vertebrate laforin and more similar to plant SEX4. While bioinformatics analysis of the glucan phosphatase from Cyanidioschyzon merolae indicated it is more closely related to the vertebrate glucan phosphatase laforin, and thus it was named Cm-laforin, its functional relationship remains to be determined. In terms of oligomerization, these data indicate that Cm-laforin is biochemically more similar to SEX4 from land plants rather than vertebrate laforin. This functional conservation may be due to differences between the carbohydrate substrate with vertebrates possessing soluble glycogen while both plants and algae produce insoluble starch. Indeed, recent studies have demonstrated that activity of different glucan phosphatases is highly dependent on the nature of the glucan substrate utilized [11].
Conclusions
We report the use of SEC-MALS as a reliable method to assess the oligomeric state of glucan phosphatases and protein variants without a contribution from carbohydrate binding. These results highlight that using SEC alone to estimate the molecular weight of proteins that contain carbohydrate binding modules can be problematic. Because SEC-MALS allows rapid assessment of glucan phosphatase enyzmes and variants and direct determination of oligomeric state, it is the preferred method for determination of oligomerization of glucan phosphatases and likely other families of carbohydrate-binding proteins as well.
This method allows determination of differences in the oligomeric state of novel human patient mutations identified in LD patients. Also, due to its recessive inheritance, a significant number of potential carrier mutations have been identified from large scale population-based sequencing efforts. This approach will assist ongoing studies to reveal the biochemical features of both novel disease mutations and potential carrier mutations, towards the goal of patient-specific diagnosis and treatment.
Further, we demonstrate that glucan phosphatases from different kingdoms display a range of oligomerization behaviors. Interestingly, glucan phosphatases from very divergent photosynthetic organisms, land plants and red algae, are both found to be monomeric. Future studies will define the nature of shared and unique aspects of glucan phosphatase function in different kingdoms of life.
Highlights.
Glucan phosphatases dephosphorylate carbohydrate substrates.
Different oligomeric states are important for their correct biological function.
Laforin shows interaction with the size-exclusion column carbohydrate-based matrix.
Glucan phosphatases from photosynthetic organisms exist as monomers.
SEC-MALS allows determination and comparison of glucan phosphatase oligomerization.
Acknowledgements
We thank Dr. Emilia Galperin, Ms. Katy Brewer, and members of the Vander Kooi and Gentry laboratories for helpful discussions and assistance.
Funding
This work was supported by the National Science Foundation (CHE-1808304 to C.W.V.K and MCB-1252345 to M.S.G.) and National Institutes of Health (P20GM103486, core support; R01NS070899 to M.S.G.; and P01NS097197 to M.S.G.).
Abbreviations
- SEC
Size Exclusion Chromotography
- SEC-MALS
Size Exclusion Chromatography with Multi-Angle static Light Scattering
- SEX4
Starch EXcess-four
- DSP
Dual Specificity Phosphatase
- CBM
Carbohydrate Binding Module
- LD
Lafora’s Disease
- At
Arabidopsis thaliana
- cTP
chloroplast Targeting Peptide
- CT
C-terminus
- LSF
Like-SEX4
- Cm
Cyanidioschyzon merolae
- WT
Wild type
- IMAC
Immobilized Metal Affinity Chromatography
- BME
2-mercaptoethanol
- DTT
Dithiothreitol
- RI
Refractive Index
- SAXS
Small Angle X-ray Scattering
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: MSG and CWVK are founders of OptiMol Enzymes LLC.
References
- [1].Gentry MS, Brewer MK, Vander Kooi CW, Structural biology of glucan phosphatases from humans to plants, Curr Opin Struct Biol, 40 (2016) 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gentry MS, Dowen RH 3rd, Worby CA, Mattoo S, Ecker JR, Dixon JE, The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease, J Cell Biol, 178 (2007) 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gentry MS, Guinovart JJ, Minassian BA, Roach PJ, Serratosa J, Lafora disease offers a unique window into neuronal glycogen metabolism, J Biol Chem, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kotting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M, Ritte G, Zeeman SC, STARCH-EXCESS4 is a laforin-like Phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana, Plant Cell, 21 (2009) 334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vander Kooi CW, Taylor AO, Pace RM, Meekins DA, Guo HF, Kim Y, Gentry MS, Structural basis for the glucan phosphatase activity of Starch Excess4, Proc Natl Acad Sci U S A, 107 (2010) 15379–15384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Meekins DA, Vander Kooi CW, Gentry MS, Structural mechanisms of plant glucan phosphatases in starch metabolism, FEBS J, 283 (2016) 2427–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu Y, Wang Y, Wu C, Liu Y, Zheng P, Dimerization of Laforin is required for its optimal phosphatase activity, regulation of GSK3beta phosphorylation, and Wnt signaling, J Biol Chem, 281 (2006) 34768–34774. [DOI] [PubMed] [Google Scholar]
- [8].Dukhande VV, Rogers DM, Roma-Mateo C, Donderis J, Marina A, Taylor AO, Sanz P, Gentry MS, Laforin, a dual specificity phosphatase involved in Lafora disease, is present mainly as monomeric form with full phosphatase activity, PLoS One, 6 (2011) e24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sankhala RS, Koksal AC, Ho L, Nitschke F, Minassian BA, Cingolani G, Dimeric quaternary structure of human laforin, J Biol Chem, 290 (2015) 4552–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Raththagala M, Brewer MK, Parker MW, Sherwood AR, Wong BK, Hsu S, Bridges TM, Paasch BC, Hellman LM, Husodo S, Meekins DA, Taylor AO, Turner BD, Auger KD, Dukhande VV, Chakravarthy S, Sanz P, Woods VL Jr., Li S, Vander Kooi CW, Gentry MS, Structural mechanism of laforin function in glycogen dephosphorylation and lafora disease, Mol Cell, 57 (2015) 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Meekins DA, Raththagala M, Auger KD, Turner BD, Santelia D, Kotting O, Gentry MS, Vander Kooi CW, Mechanistic Insights into Glucan Phosphatase Activity against Polyglucan Substrates, J Biol Chem, 290 (2015) 23361–23370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Meekins DA, Raththagala M, Husodo S, White CJ, Guo HF, Kotting O, Vander Kooi CW, Gentry MS, Phosphoglucan-bound structure of starch phosphatase Starch Excess4 reveals the mechanism for C6 specificity, Proc Natl Acad Sci U S A, 111 (2014) 7272–7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kuchtova A, Gentry MS, Janecek S, The unique evolution of the carbohydrate-binding module CBM20 in laforin, FEBS Lett, 592 (2018) 586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wilkens C, Auger KD, Anderson NT, Meekins DA, Raththagala M, Abou Hachem M, Payne CM, Gentry MS, Svensson B, Plant alpha-glucan phosphatases SEX4 and LSF2 display different affinity for amylopectin and amylose, FEBS Lett, 590 (2016) 118–128. [DOI] [PubMed] [Google Scholar]