This exploratory recursive partitioning analysis modeled associations between prespecified covariates and survival outcomes using pooled data from 4 randomized clinical trials of adult patients with BRAF V600–mutated metastatic melanoma treated with dacarbazine, vemurafenib, or cobimetinib plus vemurafenib.
Key Points
Question
What are the clinical and genomic factors associated with survival outcomes in patients with BRAF V600–mutated metastatic melanoma treated with BRAF and/or MEK inhibitors?
Findings
In this exploratory recursive partitioning analysis, which modeled associations between prespecified covariates and survival outcomes in 1365 patients, baseline lactate dehydrogenase level, performance status, disease burden, and gene signature were associated with survival outcomes in BRAF V600–mutated metastatic melanoma. Cobimetinib plus vemurafenib provided survival advantages over vemurafenib monotherapy across all prognostic subgroups.
Meaning
The results of this analysis provide a framework that may be used to compare treatment outcomes for patients with metastatic melanoma across trials and regimens and may aid in clinical decision making for individual patients.
Abstract
Importance
Prognostic models may provide insight into clinical trial results and inform the clinical management of patients with BRAF V600–mutated metastatic melanoma.
Objective
To identify subgroups with distinct survival outcomes based on clinical and genomic characteristics and to assess survival in identified prognostic subgroups across cohorts treated with dacarbazine, vemurafenib, or cobimetinib plus vemurafenib.
Design, Setting, and Participants
This retrospective and exploratory recursive partitioning analysis (RPA) modeled associations between prespecified covariates and survival outcomes using pooled data from the BRIM-2, BRIM-3, BRIM-7, and coBRIM studies.
Interventions
Dacarbazine, vemurafenib, or cobimetinib plus vemurafenib.
Main Outcomes and Measures
Progression-free survival (PFS) and overall survival (OS), estimated using the Kaplan-Meier method.
Results
The RPA included 1365 patients (783 men; 57.4%). Of these, 1032 (75.6%) were older than 65 years; 310 received cobimetinib plus vemurafenib; 717, vemurafenib alone; and 338, dacarbazine. Median follow-up was 14.1 months (interquartile range, 6.3-28.3 months). In the RPA that included all patients, baseline lactate dehydrogenase (LDH) level, Eastern Cooperative Oncology Group performance status (ECOG PS), presence or absence of liver metastases, and baseline sum of longest diameters of target lesions (SLDs) were significant prognostic factors for PFS: Median PFS was longest in patients with lower LDH (≤2 × upper limit of normal [ULN]), ECOG PS 0, and shorter SLD (≤44 mm) (11.1 months; 95% CI, 7.0-18.4 months), and shortest in those with elevated LDH (>2 × ULN) (3.5 months; 95% CI, 3.0-3.8 months). The subgroup with normal baseline LDH and no liver metastases had the longest median OS (22.7 months; 95% CI, 20.3-27.2 months). Similar PFS trends were observed when these prognostic subgroups were applied to the cobimetinib plus vemurafenib, vemurafenib alone, and dacarbazine cohorts. Baseline LDH, ECOG PS, and SLD were significant prognostic factors for OS: Median OS was longest in patients with normal LDH and shorter SLD (≤45 mm) (27.2 months; 95% CI, 22.1-32.1 months) and shortest in those with elevated LDH (>2 × ULN) (6.0 months; 95% CI, 5.3-6.8 months). Among patients in the most favorable subgroup (normal LDH and SLD ≤45 mm), 3-year OS rates were 53.3% (95% CI, 39.5%-67.1%) in the cobimetinib plus vemurafenib cohort, 35.6% (95% CI, 27.4%-43.8%) in the vemurafenib cohort, and 35.6% (95% CI, 24.8%-46.4%) in the dacarbazine cohort. Among patients with available gene expression data, RPA identified gene signature as a significant prognostic factor for PFS in those with normal LDH; 3-year PFS rates were 21.9%, (95% CI, 15.4%-28.4%) and 8.8% (95% CI, 3.6%-14.1%) in immune and cell cycle signature, respectively. The RPA for OS did not identify gene signature as a significant factor.
Conclusions and Relevance
Baseline LDH, ECOG PS, disease burden, and gene signature appear to be key determinants of survival outcomes in patients with BRAF V600–mutated metastatic melanoma treated with BRAF and/or MEK inhibitors. These results are consistent with survival benefits of cobimetinib plus vemurafenib over vemurafenib alone observed in the coBRIM study.
Introduction
The introduction of small-molecule BRAF and MEK inhibitors has improved treatment outcomes in patients with BRAF mutation–positive metastatic melanoma.1,2,3,4,5 In the BRIM-3 study (NCT00949702), BRAF-inhibitor monotherapy with vemurafenib improved overall response rate (ORR), progression-free survival (PFS), and overall survival (OS) compared with dacarbazine in patients with BRAF V600–mutated metastatic melanoma.1,2 Combined BRAF and MEK inhibition with vemurafenib plus cobimetinib demonstrated clinical benefit in the BRIM-7 study (NCT01271803)3 and improved ORR, PFS, and OS compared with vemurafenib monotherapy in the coBRIM study (NCT01689519).4,5 Extended follow-up of studies evaluating BRAF-inhibitor monotherapy or combined BRAF and MEK inhibition show a plateau in survival curves beyond about 3 years, suggestive of a subgroup of patients with prolonged survival.6,7,8,9 Development of prognostic models may provide insight into clinical trial results and inform clinical decision making in the management of patients with metastatic melanoma.
Conventionally accepted prognostic factors for survival in patients with metastatic melanoma include disease stage, baseline lactate dehydrogenase (LDH) levels, and baseline Eastern Cooperative Oncology Group performance status (ECOG PS).10,11 However, these prognostic factors were identified prior to the introduction of BRAF and MEK inhibitors. Recently, Long et al12 identified prognostic subgroups among patients with BRAF V600–mutated metastatic melanoma treated with the BRAF- and MEK-inhibitor combination of dabrafenib plus trametinib.
In patients with BRAF V600–mutated metastatic melanoma treated with vemurafenib alone or in combination with cobimetinib, gene expression profiling previously identified 2 patient subgroups with distinct PFS outcomes: one defined by high baseline expression of genes associated with cell cycle progression and the other characterized by high baseline expression of immune regulatory genes.13 Among vemurafenib-treated patients, the cell cycle signature was associated with shortened PFS compared with the immune signature. The adverse impact of the cell cycle signature on PFS was not observed in patients treated with cobimetinib plus vemurafenib.13 To date, clinical and genomic characteristics have not been evaluated in an integrated model.
The objectives of the present exploratory analysis were (1) to identify patient subgroups defined by baseline prognostic factors with distinct PFS and OS outcomes regardless of treatment; (2) to apply the prognostic subgroups identified to PFS and OS outcomes across the dacarbazine, vemurafenib, and cobimetinib plus vemurafenib cohorts; and (3) to evaluate the association of previously defined gene expression signatures with PFS and OS in the context of already-developed prognostic models in patients with BRAF V600–mutated metastatic melanoma.
Methods
Analysis Population
Data were pooled from the BRIM-2 (NCT00949702), BRIM-3, BRIM-7, and coBRIM studies. Detailed methods have been previously reported. Briefly, BRIM-2 was an open-label, multicenter phase 2 trial of oral vemurafenib 960 mg twice daily14; BRIM-3 was an open-label, multicenter, randomized phase 3 trial of oral vemurafenib, 960 mg twice daily, compared with intravenous dacarbazine, 1000 mg/m2 every 3 weeks1,2; BRIM-7 was an open-label, multicenter phase 1b dose-escalation study of oral cobimetinib, 60, 80, or 100 mg once daily, on a 14 days on/14 days off schedule (14/14), 21 days on/7 days off (21/7), or continuously (28/0) combined with oral vemurafenib, 720 or 960 mg twice daily3; and coBRIM was a randomized, double-blind phase 3 study of oral cobimetinib, 60 mg once daily on a 21/7 schedule, combined with oral vemurafenib, 960 mg twice daily, compared with placebo and vemurafenib.4,5 Key eligibility criteria were similar across trials, including age 18 years or older with unresectable stage IIIC or IV melanoma harboring a BRAF V600 mutation, ECOG PS of 0 or 1, and adequate organ function. BRIM-3 and coBRIM enrolled previously untreated patients only, whereas BRIM-2 enrolled patients who had received prior systemic treatment for advanced disease, and BRIM-7 enrolled both previously treated and untreated patients. All 4 trials allowed enrollment of patients with previously treated, stable brain metastases.
Each trial was conducted in accordance with the Declaration of Helsinki and the principles of Good Clinical and Laboratory Practice and with the approval of appropriate ethics committees. All patients provided written informed consent.
Gene Expression Signatures
Identification and validation of the cell cycle and immune gene expression signatures (eTable 1 in the Supplement) were previously described.13 Gene expression was measured using the nCOUNTER platform (NanoString Technologies) in archival formalin-fixed, paraffin-embedded tumor samples. The tumor samples were from either primary lesions or metastases. Patients with gene expression data were classified into the cell cycle or immune subgroup as previously described.13 The most common reason for exclusion of patients from the gene signature analysis was the nonavailability of archival tumor tissue.
Recursive Partitioning Analysis
This exploratory and retrospective recursive partitioning analysis (RPA) was performed using the pooled data set to model associations between prespecified covariates and PFS or OS. Additional RPAs using data from the subset of patients with available gene expression signature data were conducted to model associations between prespecified covariates, gene expression signatures, and PFS or OS.
Included covariates were prespecified and/or previously identified as potential prognostic factors in peer-reviewed literature. These included age (<65 vs ≥65 years), sex (male vs female), race (white vs nonwhite), geographic region (North America, Europe, or Australia/New Zealand/other), baseline ECOG PS (0, 1, or 2), baseline LDH (normal vs elevated), disease stage at enrollment (unresectable IIIC/M1a/M1b vs M1c), liver metastases (present vs absent), and baseline sum of longest diameters (SLDs) of target lesions. A small proportion of patients (≤10% in the overall populations in the PFS, OS, and gene signature analysis) were excluded because of missing covariate data.
For the RPAs, we used a unified framework for conditional inference (permutation tests) using censored response variables to avoid bias in the selection of covariates and minimize overfitting of the data.15 This approach ensured a right-sized tree with no need for pruning or cross-validation. The global null hypothesis of independence between the prespecified input variables and survival was tested. If the null hypothesis could not be rejected, the analysis was stopped; otherwise, the covariate most strongly associated with survival was identified based on univariate P values, and a binary split was implemented. These steps were repeated until no further covariates with a significant association with survival could be distinguished. The stop criterion was based on either multiplicity-adjusted univariate P values or a prespecified threshold value of the test statistic (both criteria were maximized using 1 − P value).
Statistical Analysis
The end points for this exploratory analysis were PFS and OS, estimated using the Kaplan-Meier method. The independent association of gene expression signatures with PFS and OS outcomes was tested by multivariate Cox proportional hazards modeling. All analyses were conducted using data cutoff dates of February 1, 2012, for BRIM-2; July 8, 2015, for BRIM-3; April 25, 2016, for BRIM-7; and June 20, 2016, for coBRIM. All eligible patients, as defined by each study protocol, were included in the analysis regardless of treatment assignment.
Results
Patient Population
Baseline characteristics of patients in the pooled population and pooled treatment cohorts and were comparable across cohorts (eTable 2 in the Supplement). Gene expression signature data were available for 608 patients across pooled studies; 312 patients (51.3%) had the cell cycle signature, and 296 patients (48.7%) had the immune signature. The distribution of baseline characteristics for patients with available gene signatures was similar to that of the overall pooled population and comparable across cohorts (eTable 3 in the Supplement).
The median follow-up duration for the pooled population was 14.1 months (interquartile range [IQR], 6.3-28.3 months). Median (IQR) follow-up durations were 21.7 (10.2-32.1) months in the cobimetinib plus vemurafenib cohort, 13.9 (6.7-25.2) months in the vemurafenib cohort, and 9.2 (3.0-21.0) months in the dacarbazine cohort.
Progression-Free Survival
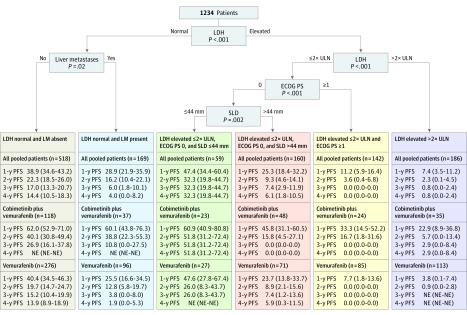
In the RPA that included all patients (n = 1365), baseline LDH, ECOG PS, presence or absence of liver metastases, and baseline SLD were significant prognostic factors for PFS, producing 6 subgroups with distinct outcomes; supporting data can be found in the Figure. Baseline LDH was the strongest determinant of PFS; outcomes were further distinguished by presence or absence of liver metastases among patients with normal LDH, and by degree of LDH elevation (≤2 × vs >2 × upper limit of normal [ULN]) among those with elevated LDH. For those with elevated LDH less than or equal to 2 × ULN, outcomes were further distinguished by baseline ECOG PS (0 vs ≥1) and SLD (≤44 vs >44 mm). Median PFS was longest in patients with elevated LDH less than or equal to 2 × ULN, ECOG PS 0, and SLD less than or equal to 44 mm (11.1 months; 95% CI, 7.0-18.4 months), and shortest in those with elevated LDH greater than 2 × ULN (3.5 months; 95% CI, 3.0-3.8 months).
Figure. Recursive Partitioning Decision Tree and Progression-Free Survival (PFS) Outcomes by Identified Prognostic Subgroups in All Pooled Patients.
Unless otherwise indicated, data are reported as mean PFS (95% CI) measured in months. A total of 131 patients (9.5%) were excluded from the PFS analysis. ECOG PS indicates Eastern Cooperative Oncology Group performance status; LDH, lactate dehydrogenase level; LM, lentigo maligna; NE, not estimable; PFS, progression-free survival; SLD, sum of longest diameters of target lesions; ULN, upper limit of normal.
Similar trends in PFS were observed when these prognostic subgroups were applied to the cobimetinib plus vemurafenib, vemurafenib monotherapy, and dacarbazine cohorts (Figure and eFigure 1 in the Supplement). Within and across prognostic subgroups, PFS outcomes were improved in the cobimetinib plus vemurafenib cohort compared with the vemurafenib monotherapy cohort. Among patients with elevated LDH less than or equal to 2 × ULN, ECOG PS 0, and SLD less than or equal to 44 mm (the most favorable subgroup), 3-year PFS rates were 51.8% (95% CI, 31.2%-72.4%) in the cobimetinib plus vemurafenib cohort, 26.0% (95% CI, 8.3%-43.7%) in the vemurafenib cohort, and 0.0% (95% CI, 0.0-0.0) in the dacarbazine cohort.
Overall Survival
In the RPA that included all patients (n = 1365), baseline LDH, ECOG PS, and SLD were significant prognostic factors for OS, producing 5 subgroups with distinct outcomes (supporting data provided in eFigure 2 in the Supplement). Baseline LDH was the strongest prognostic factor for OS outcomes. Among patients with normal LDH, OS outcomes were further determined by SLD (≤45 vs >45 mm). In patients with elevated LDH, distinct OS outcomes were observed according to degree of LDH elevation (≤2 × ULN vs >2 × ULN): among those with modest LDH elevations (≤2 × ULN), outcomes were further distinguished by ECOG PS (0 vs ≥1). Median OS was longest in patients with normal LDH and SLD less than or equal to 45 mm (27.2 months; 95% CI, 22.1-32.1 months) and shortest in those with LDH greater than 2 × ULN (6.0 months; 95% CI, 5.3-6.8 months). Patients with modest LDH elevations (≤2 × ULN) and ECOG PS 0 had intermediate outcomes (median OS, 16.0 months; 95% CI, 13.3-18.5 months), and some patients experienced prolonged OS, with 3- and 4-year OS rates of 22% and 20%, respectively.
Similar trends in OS were observed when these prognostic subgroups identified were applied to the cobimetinib plus vemurafenib, vemurafenib monotherapy, and dacarbazine cohorts (eFigure 2 and eFigure 3 in the Supplement). Within and across prognostic subgroups, OS outcomes were improved in the cobimetinib plus vemurafenib cohort compared with the vemurafenib monotherapy cohort. Among patients with normal LDH levels and SLD less than or equal to 45 mm (the most favorable subgroup), 3-year OS rates were 53.3% (95% CI, 39.5%-67.1%) in the cobimetinib plus vemurafenib cohort, 35.6% (95% CI, 27.4%-43.8%) in the vemurafenib cohort, and 35.6% (95% CI, 24.8%-46.4%) in the dacarbazine cohort.
Gene Expression Signature
In multivariate analyses, the immune signature was associated with improved PFS (adjusted hazard ratio [HR], 0.75; 95% CI, 0.62-0.89; P = .002) and OS (adjusted HR, 0.74; 95% CI, 0.61-0.90; P = .002) compared with the cell cycle signature (Table). The association of gene expression signatures with PFS and OS was statistically significant in the all-pooled-patients and the vemurafenib cohorts, but not in the cobimetinib plus vemurafenib cohort. Compared with vemurafenib monotherapy, cobimetinib plus vemurafenib improved both PFS and OS regardless of gene expression signature.
Table. Survival Outcomes According to Gene Expression Signatures Across Treatment Cohorts in Patients With Gene Expression Dataa.
| Treatment Group | Progression-Free Survival | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|---|
| Immune | Cell Cycle | HR (95% CI)b | P Value | Immune | Cell Cycle | HR (95% CI)b | P Value | |
| All pooled patients (n = 608) | 7.39 (6.70-8.97) | 5.42 (4.44-6.54) | 0.75 (0.62-0.89) | .002 | 18.83 (15.61-23.16) | 13.31 (11.30-15.24) | 0.74 (0.61-0.90) | .002 |
| Cobimetinib plus vemurafenib (n = 143) | 12.94 (9.69-20.34) | 10.51 (7.92-13.50) | 0.76 (0.50-1.17) | .21 | 27.96 (21.49-NE) | 21.55 (17.54-31.08) | 0.71 (0.44-1.14) | .16 |
| Vemurafenib (n = 320) | 7.84 (6.97-9.56) | 5.55 (4.80-6.74) | 0.70 (0.54-0.90) | .01 | 18.76 (15.51-25.56) | 12.29 (10.05-14.09) | 0.74 (0.56-0.98) | .04 |
| Dacarbazine (n = 145) | 1.74 (1.48-3.12) | 1.63 (1.48-2.10) | 0.72 (0.51-1.03) | .07 | 11.14 (7.66-17.84) | 9.63 (7.26-13.31) | 0.70 (0.48-1.03) | .07 |
Abbreviation: HR, hazard ratio.
Unless otherwise indicated, data are reported as median (95% CI) survival time measured in months.
Adjusted HR for immune vs cell cycle.
In the RPA that included all patients with gene expression data (n = 608), baseline LDH, SLD, and gene expression signature were identified as significant prognostic factors for PFS, yielding 4 subgroups with distinct outcomes (eFigure 4 in the Supplement). Baseline LDH was the strongest prognostic factor for PFS; outcomes were further determined by gene expression signature (immune vs cell cycle) among patients with normal LDH and by SLD (≤60 vs >60 mm) among those with elevated LDH. The subgroup with normal baseline LDH and immune signature had the longest median PFS (9.0 months; 95% CI, 7.1-11.2 months), whereas the subgroup with elevated LDH and SLD greater than 60 mm had the shortest median PFS (4.0 months; 95% CI, 3.5-4.7 months).
Gene expression signature was not identified as a prognostic factor for OS in the RPA that included all patients with gene expression data (n = 608). For all pooled patients with gene expression signature data, baseline LDH, ECOG PS, and liver metastasis were identified as prognostic factors for OS, yielding 5 groups with distinct OS outcomes (eFigure 5 in the Supplement). The subgroup with normal baseline LDH and no liver metastases had the longest median OS (22.7 months; 95% CI, 20.3-27.2 months), whereas the subgroup with elevated baseline LDH greater than 2 × ULN had the shortest median OS (6.0 months; 95% CI, 5.1-8.0 months).
The immune signature was significantly more prevalent in favorable prognostic groups based on clinical variables identified for PFS and OS in the RPA in the all-pooled-patients population (both P < .001 by 1-sided Cochrane Armitage trend test) (eFigure 6 in the Supplement). The trend was significant in the cobimetinib plus vemurafenib and vemurafenib monotherapy cohorts, but not in the dacarbazine cohort (eTables 4 and 5 in the Supplement).
Objective Response
Within and across prognostic groups, ORRs were improved with cobimetinib plus vemurafenib (range, 54%-78%) compared with vemurafenib monotherapy (range, 33%-67%) (eTable 6 in the Supplement). Similar results were also observed for duration of response (eTable 7 in the Supplement).
Complete responses (CRs) occurred predominantly in the most favorable prognostic subgroups (eFigure 7 in the Supplement). In analyses that included all patients, CR rates were 39%, 19%, and 11% in the cobimetinib plus vemurafenib, vemurafenib monotherapy, and dacarbazine cohorts, respectively, in the most favorable prognostic subgroup for PFS (elevated LDH ≤2 × ULN, ECOG PS 0, and SLD ≤44 mm) and 36%, 17%, and 4%, respectively, in the most favorable prognostic subgroup for OS (normal LDH levels and SLD ≤45 mm). In comparison, CR rates were 0% to 3% across treatment cohorts in the least favorable subgroup (LDH >2 × ULN). In patients with available gene expression data, CR rates were 28%, 17%, and 2% in the cobimetinib plus vemurafenib, vemurafenib monotherapy, and dacarbazine cohorts, respectively, in the most favorable prognostic subgroup for PFS (normal LDH and immune gene signature), and 6%, 2%, and 0%, respectively, in the least favorable subgroup (elevated LDH and SLD >60 mm).
Discussion
In this exploratory pooled analysis of 4 key trials of vemurafenib or cobimetinib plus vemurafenib incorporating demographic, clinical, and genomic data, LDH level, presence or absence of liver metastases, SLD, and ECOG PS emerged as key prognostic factors defining groups with distinct PFS and OS outcomes in patients with BRAF V600–mutated melanoma. Gene expression signatures demonstrated independent prognostic associations in an integrated RPA model for PFS but not OS outcomes. Regardless of the prognostic model used, PFS and OS outcomes were favorable for cobimetinib plus vemurafenib compared with vemurafenib monotherapy across all prognostic groups.
Identification of LDH level and ECOG PS as prognostic factors is consistent with previous findings in patients with metastatic melanoma,10,11 including those with BRAF mutations treated with BRAF and/or MEK inhibitors.12,16 Despite differences in statistical methodology and treatment, these findings are remarkably consistent with those recently reported by Long et al12 and Schadendorf et al,16 who conducted pooled analyses of patients with BRAF V600–mutated metastatic melanoma treated with dabrafenib + trametinib. The LDH level, number of metastatic sites, ECOG PS, and SLD were identified as key prognostic factors for PFS and/or OS in these analyses.12,16 Although number of metastatic sites was not identified as a significant prognostic factor in our models, all analyses identified 1 or more measures of tumor burden (presence or absence of liver metastasis, SLD, or number of metastatic sites) as having an independent prognostic association.
Baseline LDH was identified as the strongest prognostic factor across all analyses in the present study and in previously reported analyses, including those conducted by Long et al12 and Schadendorf et al.16 All analyses further discriminated outcomes between patients with modest (≤2 × ULN) vs more substantial (>2 × ULN) elevations of LDH, suggesting that additional stratification beyond normal vs elevated LDH may be useful. A unique finding of our analysis was identification of a subgroup of patients with elevated LDH (LDH ≤2 × ULN, ECOG PS 0, and SLD ≤44 mm) who exhibited survival outcomes comparable to those with normal LDH. In contrast, analyses of patients treated with dabrafenib plus trametinib did not identify a similar elevated LDH subset with favorable survival outcomes.
Another common finding across analyses was clustering of CRs in favorable prognostic subgroups and a notable absence of CR in the least favorable prognostic subgroup (consistently defined as LDH ≥2 × ULN).12,16 These observations appear to extend to immunotherapy using checkpoint inhibitors. A recent pooled analysis of patients with advanced melanoma receiving nivolumab plus ipilimumab or nivolumab alone found that patients who did not achieve a CR had worse prognostic features (elevated LDH and higher tumor burden based on disease stage and SLD).17 Additional analyses are ongoing to better understand these populations within our data set.
Among patients with available gene expression data, PFS and OS were significantly improved for those with the immune signature compared with those with the cell cycle signature, as previously defined.13 The present RPA confirmed the importance of gene expression signature as an independent prognostic factor for PFS among patients with normal baseline LDH, but not for OS. Furthermore, the immune signature was significantly enriched in favorable clinical prognostic subgroups in the vemurafenib and cobimetinib plus vemurafenib cohorts, but not in the dacarbazine cohort. These observations are consistent with recent reports demonstrating immunosuppressive effects of oncogenic BRAF signaling on the tumor-host interaction and beneficial effects of MEK inhibition on response to immune checkpoint inhibitors,18,19,20,21 supporting further study of the interaction of BRAF/MEK inhibition with the tumor microenvironment and immune response.
Limitations
This retrospective analysis used pooled data from multiple studies conducted across an approximately 8-year span. Variations in regulatory approval and access to new treatments over time and across various geographical regions could have influenced survival outcomes in a nonuniform manner. In addition, the pooled data set included patients enrolled in nonrandomized studies with potential introduction of selection bias for particular treatments.
Conclusions
In summary, findings from this analysis are robust and generally consistent with those of other analyses, indicating that baseline LDH, performance status, extent of disease, and gene expression signature are key determinants of survival outcomes in patients with BRAF V600–mutated metastatic melanoma treated with BRAF and/or MEK inhibitors. The results of this exploratory pooled analysis are consistent with previously observed PFS and OS benefits of cobimetinib plus vemurafenib over vemurafenib monotherapy in the pivotal phase 3 study (coBRIM; NCT01689519). The PFS and OS were prolonged in patients within good prognostic subgroups, which may explain the observed tail in survival curves in recent long-term analyses.6,7,8,9 The results of this analysis provide a framework that may be used to compare outcomes across trials and regimens in patients with metastatic melanoma. Understanding treatment effects of different regimens across prognostic groups, as described by us and others, may aid clinicians in clinical decision making for individual patients.
eTable 1. Genes Included in the Cell Cycle and Immune Gene Expression Signature
eTable 2. Baseline Characteristics
eTable 3. Baseline Characteristics in Patients With Available Gene Expression Signature Data
eTable 4. Distribution of Gene Expression Signatures by Prognostic Subgroups Identified for PFS in All Pooled Patients With Gene Expression Data (n = 608)
eTable 5. Distribution of Gene Expression Signatures by Prognostic Subgroups Identified for OS in All Pooled Patients With Gene Expression Data (n = 608)
eTable 6. Objective Response Rate by Prognostic Subgroups Identified for PFS and OS in All Pooled Patients Across Treatment Cohorts
eTable 7. Duration of Response by Prognostic Subgroups Identified for PFS and OS in All Pooled Patients Across Treatment Cohorts
eFigure 1. PFS by Prognostic Subgroup Identified by the Recursive Partitioning Model in All Pooled Patients
eFigure 2. Recursive Partitioning Decision Tree and OS Outcomes by Identified Prognostic Subgroups in All Pooled Patients.
eFigure 3. OS by Prognostic Subgroup Identified in the Recursive Partitioning Model in All Pooled Patients.
eFigure 4. Recursive Partitioning Decision Tree and PFS Outcomes by Identified Prognostic Subgroups in All Patients With Gene Expression Data.
eFigure 5. Recursive Partitioning Decision Tree and Outcomes by Identified Prognostic Subgroups for OS in All Patients With Gene Expression Data.
eFigure 6. Prevalence of Immune or Cell Cycle Gene Expression Signatures by Prognostic Subgroups For PFS (Panel A) and OS (Panel B) in all Pooled Patients With Gene Expression Data (n = 608).
eFigure 7. Objective Response by Prognostic Subgroups.
References
- 1.Chapman PB, Hauschild A, Robert C, et al. ; BRIM-3 Study Group . Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507-2516. doi: 10.1056/NEJMoa1103782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McArthur GA, Chapman PB, Robert C, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol. 2014;15(3):323-332. doi: 10.1016/S1470-2045(14)70012-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ribas A, Gonzalez R, Pavlick A, et al. Combination of vemurafenib and cobimetinib in patients with advanced BRAF(V600)-mutated melanoma: a phase 1b study. Lancet Oncol. 2014;15(9):954-965. doi: 10.1016/S1470-2045(14)70301-8 [DOI] [PubMed] [Google Scholar]
- 4.Larkin J, Ascierto PA, Dréno B, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371(20):1867-1876. doi: 10.1056/NEJMoa1408868 [DOI] [PubMed] [Google Scholar]
- 5.Ascierto PA, McArthur GA, Dréno B, et al. Cobimetinib combined with vemurafenib in advanced BRAF(V600)-mutant melanoma (coBRIM): updated efficacy results from a randomised, double-blind, phase 3 trial. Lancet Oncol. 2016;17(9):1248-1260. doi: 10.1016/S1470-2045(16)30122-X [DOI] [PubMed] [Google Scholar]
- 6.Chapman PB, Robert C, Larkin J, et al. Vemurafenib in patients with BRAFV600 mutation-positive metastatic melanoma: final overall survival results of the randomized BRIM-3 study. Ann Oncol. 2017;28(10):2581-2587. doi: 10.1093/annonc/mdx339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McArthur G, Dréno B, Atkinson V, et al. Efficacy of long-term cobimetinib (C) plus vemurafenib (V) in advanced BRAFV600-mutated melanoma: 3-year follow-up of the phase 3 coBRIM study and 4 year follow-up of the phase 1b BRIM7 study. Presented at: Thirteenth International Congress of The Society for Melanoma Research; November 6-9, 2016; Boston, MA. [Google Scholar]
- 8.Menzies AM, Wilmott JS, Drummond M, et al. Clinicopathologic features associated with efficacy and long-term survival in metastatic melanoma patients treated with BRAF or combined BRAF and MEK inhibitors. Cancer. 2015;121(21):3826-3835. doi: 10.1002/cncr.29586 [DOI] [PubMed] [Google Scholar]
- 9.Long GV, Weber JS, Infante JR, et al. Overall survival and durable responses in patients with BRAF V600-mutant metastatic melanoma receiving dabrafenib combined with trametinib. J Clin Oncol. 2016;34(8):871-878. doi: 10.1200/JCO.2015.62.9345 [DOI] [PubMed] [Google Scholar]
- 10.Dickson PV, Gershenwald JE. Staging and prognosis of cutaneous melanoma. Surg Oncol Clin N Am. 2011;20(1):1-17. doi: 10.1016/j.soc.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balch CM, Gershenwald JE, Soong SJ, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27(36):6199-6206. doi: 10.1200/JCO.2009.23.4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long GV, Grob J-J, Nathan P, et al. Factors predictive of response, disease progression, and overall survival after dabrafenib and trametinib combination treatment: a pooled analysis of individual patient data from randomised trials. Lancet Oncol. 2016;17(12):1743-1754. doi: 10.1016/S1470-2045(16)30578-2 [DOI] [PubMed] [Google Scholar]
- 13.Wongchenko MJ, McArthur GA, Dréno B, et al. Gene expression profiling in BRAF-mutated melanoma reveals patient subgroups with poor outcomes to vemurafenib that may be overcome by cobimetinib plus vemurafenib. Clin Cancer Res. 2017;23(17):5238-5245. doi: 10.1158/1078-0432.CCR-17-0172 [DOI] [PubMed] [Google Scholar]
- 14.Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366(8):707-714. doi: 10.1056/NEJMoa1112302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: a conditional inference framework. J Comput Graph Stat. 2006;15(3):651-674. doi: 10.1198/106186006X133933 [DOI] [Google Scholar]
- 16.Schadendorf D, Long GV, Stroiakovski D, et al. Three-year pooled analysis of factors associated with clinical outcomes across dabrafenib and trametinib combination therapy phase 3 randomised trials. Eur J Cancer. 2017;82:45-55. doi: 10.1016/j.ejca.2017.05.033 [DOI] [PubMed] [Google Scholar]
- 17.Robert C, Larkin J, Ascierto PA, et al. Characterization of complete responses (CRs) in patients with advanced melanoma (MEL) who received the combination of nivolumab (NIVO) and ipilimumab (IPI), NIVO or IPI alone [ESMO abstract 1213O]. Ann Oncol. 2017;28(suppl 5):v428-v448. doi: 10.1093/annonc/mdx377 [DOI] [Google Scholar]
- 18.Mandalà M, De Logu F, Merelli B, Nassini R, Massi D. Immunomodulating property of MAPK inhibitors: from translational knowledge to clinical implementation. Lab Invest. 2017;97(2):166-175. doi: 10.1038/labinvest.2016.132 [DOI] [PubMed] [Google Scholar]
- 19.Ebert PJR, Cheung J, Yang Y, et al. MAP kinase inhibition promotes T cell and anti-tumor activity in combination with PD-L1 checkpoint blockade. Immunity. 2016;44(3):609-621. doi: 10.1016/j.immuni.2016.01.024 [DOI] [PubMed] [Google Scholar]
- 20.Miller WH, Kim TM, Lee CB, et al. Atezolizumab (A) + cobimetinib (C) in metastatic melanoma (mel): updated safety and clinical activity [ASCO abstract 3057]. J Clin Oncol. 2017;25(suppl):3057. [Google Scholar]
- 21.Sullivan RJ, Gonzalez R, Lewis KD, et al. Atezolizumab (A) + cobimetinib (C) + vemurafenib (V) in BRAFV600-mutant metastatic melanoma (mel): updated safety and clinical activity [ASCO abstract 3063]. J Clin Oncol. 2017;25(suppl):3063. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Genes Included in the Cell Cycle and Immune Gene Expression Signature
eTable 2. Baseline Characteristics
eTable 3. Baseline Characteristics in Patients With Available Gene Expression Signature Data
eTable 4. Distribution of Gene Expression Signatures by Prognostic Subgroups Identified for PFS in All Pooled Patients With Gene Expression Data (n = 608)
eTable 5. Distribution of Gene Expression Signatures by Prognostic Subgroups Identified for OS in All Pooled Patients With Gene Expression Data (n = 608)
eTable 6. Objective Response Rate by Prognostic Subgroups Identified for PFS and OS in All Pooled Patients Across Treatment Cohorts
eTable 7. Duration of Response by Prognostic Subgroups Identified for PFS and OS in All Pooled Patients Across Treatment Cohorts
eFigure 1. PFS by Prognostic Subgroup Identified by the Recursive Partitioning Model in All Pooled Patients
eFigure 2. Recursive Partitioning Decision Tree and OS Outcomes by Identified Prognostic Subgroups in All Pooled Patients.
eFigure 3. OS by Prognostic Subgroup Identified in the Recursive Partitioning Model in All Pooled Patients.
eFigure 4. Recursive Partitioning Decision Tree and PFS Outcomes by Identified Prognostic Subgroups in All Patients With Gene Expression Data.
eFigure 5. Recursive Partitioning Decision Tree and Outcomes by Identified Prognostic Subgroups for OS in All Patients With Gene Expression Data.
eFigure 6. Prevalence of Immune or Cell Cycle Gene Expression Signatures by Prognostic Subgroups For PFS (Panel A) and OS (Panel B) in all Pooled Patients With Gene Expression Data (n = 608).
eFigure 7. Objective Response by Prognostic Subgroups.