Key Points
Question
Can next-generation sequencing reveal potentially actionable alterations in a significant proportion of patients with soft-tissue sarcoma?
Findings
In this analysis of next-generation sequencing data from 584 patients, actionable genomic alterations in up to 41% of cases were identified.
Meaning
Comprehensive genomic profiling can identify novel therapeutic strategies to address the limited options and poor prognoses of patients with soft-tissue sarcoma.
This study examines the use of next-generation sequencing for identification of potentially actionable alterations in patients with soft-tissue sarcoma.
Abstract
Importance
Patients with advanced soft-tissue sarcomas (STS) have a median overall survival of less than 18 months. Identification of molecular abnormalities for which targeted therapies are available or can be developed is critical for improving patient outcomes.
Objective
To characterize targetable genomic alterations (GAs) in patients with STS.
Design, Setting, and Participants
This cross-sectional study of next-generation sequencing results from 584 patients with STS included in the AACR GENIE Database.
Main Outcomes and Measures
Presence of targetable GAs in STS.
Results
Of 584 patients included in the analysis, 294 (50.3%) were men and 290 (49.7%) were women, with a median age of 56 years (range, 18-89 years). There were 331 (57%) patients with complex genomics sarcomas, 144 (25%) with translocation-related sarcomas, and 112 (18%) with other sarcomas (inactivating mutation, simple amplicon). A total of 2697 alterations were identified in 451 genes (1154 substitutions, 765 gene amplifications, 364 short indels and splicing variants, 346 gene homozygous deletions, and 68 gene rearrangements) with a median of 4 (1-53) per case. In order of frequency, the 20 genes most often altered were: TP53, MDM2, CDK4, RB1, ATRX, CDKN2A, PTEN, NF1, CDKN2B, KMT2D, GLI1, ATM, TERT, PI3KCA, NOTCH1, MAP2K4, ERBB4, ARID1A, TSC2, and TNFAIP3. At least 1 targetable GA was found in 239 cases (41%) with a statistically significant higher number in other and complex genomics sarcomas than in translocation-related sarcomas (respectively other: n=89, 82%, complex: n = 131, 40%, translocation: n = 19, 13%; χ2 test, P < .001).
Conclusions and Relevance
Up to 41% of STS harbored at least 1 clinically relevant GA with potential to influence and personalize therapy. Comprehensive genomic profiling can identify novel treatment paradigms to address the limited options and poor prognoses of patients with STS.
Introduction
Soft-tissue sarcomas (STS) are a heterogeneous group of rare tumors including more than 70 different subtypes and affecting approximately 200 000 individuals worldwide each year.1 Despite their biological and histologic differences, sarcomas are generally treated in the same manner, with a combination of surgery and radiation therapy. However, despite an adequate locoregional treatment, up to 40% of patients will develop metastatic disease. Chemotherapy is the cornerstone of treatment in the advanced setting with doxorubicin being the first-line standard of care for decades despite many attempts to identify better regimens. Median overall survival (OS) is poor, ranging between 12 and 18 months with only modest improvement within the past 20 years.2
From a molecular genetics perspective, STS are traditionally classified into 2 broad categories: sarcomas with specific genetic alterations and relatively simple karyotypes, most of them being translocation-related sarcomas (TRS); and sarcomas with complex and unbalanced karyotypes representing two-thirds of STS.1
Given the size and complexity of the human genome, having adequate numbers of specimens to study and access to annotated clinical databases will be critical to identify driver mutations as targets for new therapeutics; thus, next-generation sequencing (NGS) technologies are some of the most exciting tools for translational research today. This technology can identify changes in genes and their expression in tumors at the whole-genome level, and is currently driving genomic research in cancer. However, thus far, large-scale genomic characterization of cancers has focused on epithelial and hematologic malignant diseases.
In the present study, we report data from 584 STS analyzed through a comprehensive genomic profiling assay based on NGS and determine how the genome-derived drug targets identified could be used to guide personalized targeted treatment for this challenging disease.
Methods
Patients
Clinical information such as sex and age were extracted from the GENIE database. The first GENIE data release includes genomic and clinical data from 8 cancer centers. At the time of data analysis, 18 486 patients were included in the GENIE registry. Of these, 635 samples were classified as STS. All patients had a single sample NGS sequenced and analyzed except 3 patients with 2 samples for which we used the primary tumor sample (GENIE-MSK-P-0002326-T01-IM3, GENIE-MSK-P-0001364-T01-IM3, GENIE-MSK-P-0000835-T01-IM3). Among the total cases, patients with diseases not considered STS1 (desmoid tumor/fibromatosis [n = 18], soft-tissue myoepithelioma [n = 8], histiocytic dendritic cell sarcoma [n = 7], follicular dendritic cell sarcoma [n = 5], inflammatory myofibroblastic tumor [n = 3], ossifying fibromyxoid tumor [n = 2], myxoma [n = 2], hemangioma [n = 1], tenosynovial giant cell tumor [n = 1], myofibroma [n = 1]) were excluded, leaving 584 patients for the analysis.
For the purpose of this analysis, histologic subtypes were classified in 3 main categories based on their known genetic characteristics1: (1) complex genomics sarcomas: sarcomas displaying multiple, complex karyotypic abnormalities with no specific pattern, including leiomyosarcoma, pleomorphic liposarcoma, and undifferentiated pleomorphic sarcomas; (2) translocation-related sarcomas: sarcomas with specific reciprocal translocations resulting in oncogenic fusion transcripts including synovialosarcoma and myxoid round cel liposarcomas; and (3) other sarcomas: sarcomas with specific oncogenic mutations such as epithelioid sarcomas or recurrent amplification, such as well-differentiated or dedifferentiated liposarcomas and intimal sarcomas.
The protocol was approved by the institutional review board of Institut Bergonié national ethics committee and was performed in accordance with the Good Clinical Practice guidelines of the International Conference on Harmonization. All the patients gave written informed consent.
Molecular Assays and Bioinformatic Analysis
Details related to the gene panels used, Pipeline for Annotating Mutations and Filtering Putative Germline SNPs are available on line: http://www.aacr.org/Documents/GENIEDataGuide.pdf.
Release 1.0.1 of GENIE data tables were downloaded from the Synapse data portal at https://www.synapse.org/#!Synapse:syn7844527.
Data tables consisted of the following files:
data_clinical.txt (patient annotation data)
data_CNA.txt (copy number)
data_fusions.txt (translocations)
data_mutations_extended.txt (snv/indel)
Single-point variants (missense, InDels, and Splice variants), copy number (hemi/homozygous deletions, gains, and amplifications), and fusions where aggregated in a single alteration table together with the annotations of the type of marker, its annotation (p.), the sample barcode, the sarcoma class, and the histologic subtype and used for bioinformatic analysis. Wrangling, mining, and reporting of GENIE data tables was performed using the R Statistical Data Analysis Environment (dplyr and tidyr packages; R foundation, Inc).
For the analysis of co-occurrence and mutual exclusivity of alterations, copy number gains and hemizygous deletions were removed from the alteration table and a contingency table of gene alterations per patient was built. Only gene alterations occurring in at least 5 patients and belonging to COSMIC CENSUS were kept (COSMIC CENSUS, version 81 2017; http://cancer.sanger.ac.uk/census/).
The test of proportion of alteration counts between sarcoma categories has been performed using the Pearson χ2 test implemented in the R prop.test module.
Scoring Targetable Alterations
Targetable alterations were defined in accordance with the OncoKB classification with 4 categories: (1) alterations associated with approved anticancer drugs in sarcomas; (2) standard care biomarker predictive of response to drugs not approved for treatment of sarcoma patients; (3) alterations associated with compelling clinical evidence of predictive value but neither biomarker nor drug are standard of care; and (4) alterations associated with compelling biologic evidence of predictive value but neither biomarker nor drug are standard of care.3
Results
Patient Characteristics
A total of 584 patients were included in the study (294 men and 290 women). Median age was 56 years (range, 18-89 years). Patient characteristics are described in the Table. Three hundred thirty-one (57%) cases were sarcoma with complex genomics, 144 (25%) sarcomas with translocation, and 112 (19%) other sarcomas. The 5 most frequent histologies were: leiomyosarcoma (112 [19%]), undifferentiated pleomorphic sarcoma/malignant fibrous histiocytoma/high-grade spindle cell sarcoma (74 [13%]), dedifferentiated liposarcoma (55 [9%]), angiosarcoma (43 [7%]), and synovial sarcoma (38 [7%]).
Table. Patient Characteristics.
| Characteristic | No. (%) |
|---|---|
| Genetic features | |
| Complex genomics | 331 (57) |
| Translocation-related | 144 (25) |
| Other | 109 (18) |
| Histotype | |
| Leiomyosarcoma | 112 (19) |
| Undifferentiated pleomorphic sarcoma | 74 (13) |
| Dedifferentiated liposarcoma | 55 (9) |
| Angiosarcoma | 43 (7) |
| Synovial sarcoma | 38 (7) |
| Solitary fibrous tumor | 34 (6) |
| Well-differentiated liposarcoma | 28 (5) |
| Sarcoma, not otherwise specified | 23 (4) |
| Desmoplastic small round cell tumor | 18 (3) |
| Myxofibrosarcoma | 16 (3) |
| Myxoid/round cell liposarcoma | 16 (3) |
| Embryonal rhabdomyosarcoma | 14 (2) |
| Rhabdomyosarcoma | 14 (2) |
| Epithelioid sarcoma | 11 (2) |
| Alveolar rhabdomyosarcoma | 10 (2) |
| Liposarcoma | 9 (2) |
| Pleomorphic liposarcoma | 9 (2) |
| Alveolar soft part sarcoma | 8 (1) |
| Clear cell sarcoma | 8 (1) |
| Intimal sarcoma | 8 (1) |
| Epithelioid hemangioendothelioma | 7 (1) |
| Fibrosarcoma | 6 (1) |
| Perivascular epithelioid cell tumor | 5 (1) |
| Radiation-associated sarcoma | 4 (1) |
| Pleomorphic rhabdomyosarcoma | 3 (1) |
| Sclerosing epithelioid fibrosarcoma | 3 (1) |
| Other | 8 (1) |
Genetic Landscape
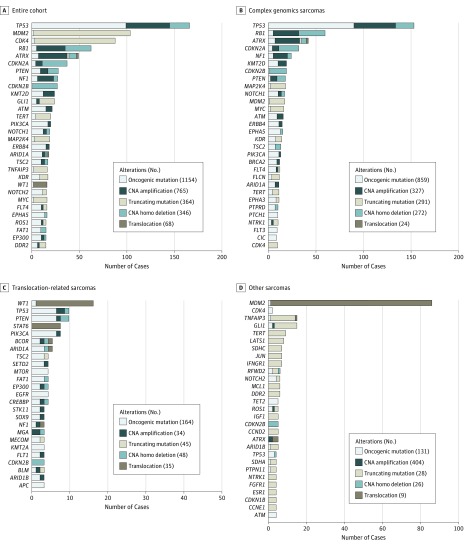
Among these 584 samples, 494 (84.6%) of the total STS cases harbored at least 1 alteration (eFigure 1 in the Supplement) and proportions of affected patients in each sarcoma group varied significantly among groups, with the other sarcomas group being the most altered (90.8%) and translocation-related sarcomas being the least mutated (77.8%; χ2 test, P = .10). The distribution of the number of alterations was different between complex genomics sarcomas, translocation-related sarcomas, and other sarcomas. For instance, up to 13% of complex genomic sarcomas and none of translocation-related sarcomas were associated with more than 10 alterations (eFigure 2 in the Supplement). A total of 2697 alterations were identified in 451 genes (1154 substitutions, 765 gene amplifications, 364 short indels plus splice variants, 346 gene homozygous deletions, and 68 gene rearrangements) (Figure 1A). The 20 genes most frequently altered were: TP53, MDM2, CDK4, RB1, ATRX, CDKN2A, PTEN, NF1, CDKN2B, KMT2D, GLI1, ATM, TERT, PI3KCA, NOTCH1, MAP2K4, ERBB4, ARID1A, TSC2, and TNFAIP3. Lollipop-style mutation diagrams for ATRX, PTEN, and PIK3CA genes are presented in eFigure 3 in the Supplement. Disparities in the distribution of genes altered in STS with complex genomics vs translocation-related sarcomas and other sarcomas were repeatedly observed (Figure 1B-D). The distribution of actionable alterations in the main 10 histologic subtypes (n=144) of STS are presented in Figure 2. The analysis of alteration co-occurrences and mutual exclusivity (eFigure 5 in the Supplement) shows that alterations in TP53, including both sequence alterations and gene deletions, were identified in 140 (49.5%) sarcomas with complex genomics but only in 8 (7.1%) translocation-related sarcomas and 4 (4%) in other sarcomas. A total of 101 (17%) patients with wild-type TP53 had dysregulation of the TP53 pathway as a result of MDM2 amplification (complex genomics sarcomas, 16 [3%]; translocation-related sarcomas, 1 [0.01%]; other sarcomas, 84 [14%]). A total of 96 STS with TP53 alteration also showed alteration of RB1 or CDKN2A of which 88 (58%) complex genomics sarcomas, 3 (2%) translocation-related sarcomas, and 5 (3%) other sarcomas (eFigure 5 in the Supplement). The RB1 and CDKN2A gene alterations were mutually exclusive.
Figure 1. Prevalent Genomic Alterations .
The most prevalent genomic alterations in 584 samples of soft-tissue sarcomas in the entire cohort (A); with complex genomics sarcomas (B); with translocation-related sarcomas (C); and with other sarcomas (D).
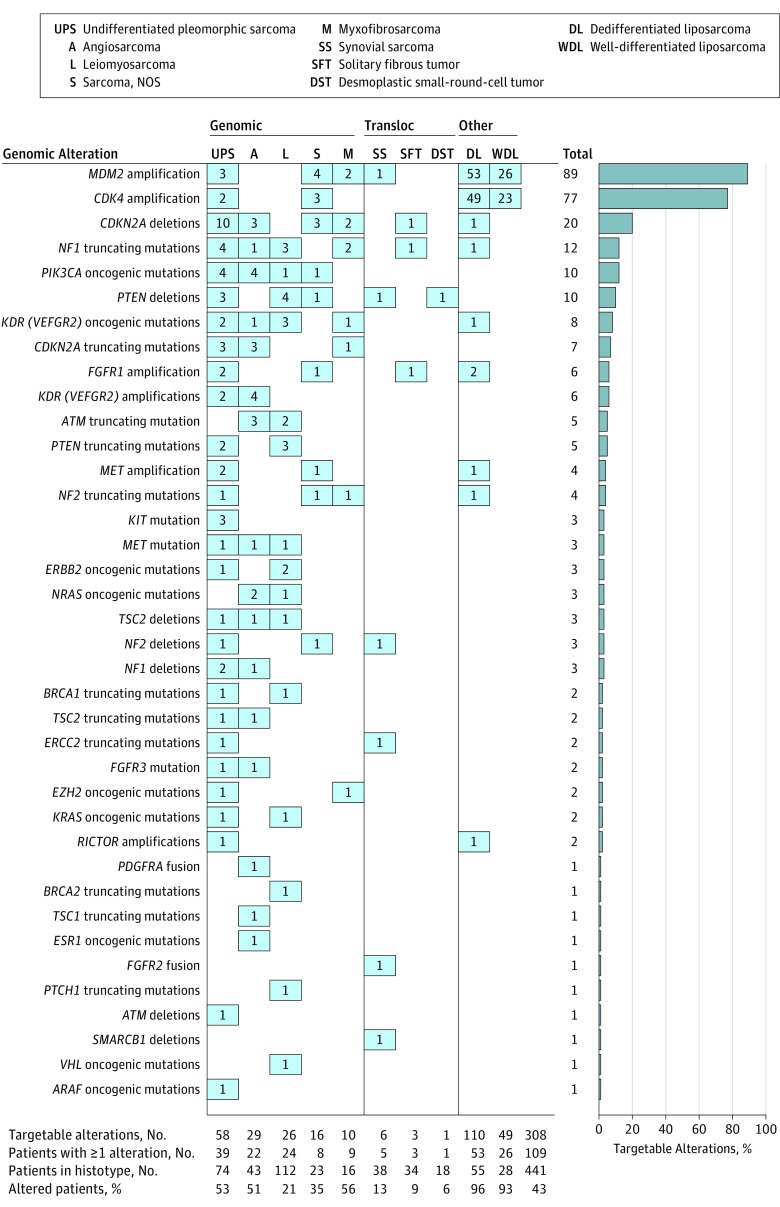
Figure 2. Distribution of Targetable Alterations in the 10 Most Prevalent Sarcoma Histotypes.
On the top row of the figure, histotypes are grouped into 3 classes: complex genomics, translocation-related, and other sarcomas. The column total on the right-end side represents the number of alterations per gene. The total number of targetable alterations per histotype, the number of patients per histotype with at least 1 alteration, the total number of patients per histotype and the ration of the number of patients with 1 or more alterations over the total number of patients for each histologic subtype are represented at the bottom of the Figure.
Ninety (15%) of 584 sarcoma samples did not show detectable alterations in our study. To check the hypothesis that this could be owing to differences in the gene composition between small-panel and large-panel NGS assays, we analyzed the distribution of cases by NGS assay, split into 2 groups: (1) without alterations (n = 90) and (2) with alterations (n = 494) (eFigure 6 in the Supplement). A total of 45 (50%) cases without alterations were analyzed by using small-panel NGS assays (46-74 genes) and the ratio of tumors with no GA was significantly higher in smaller panels than in large panels, suggesting an impact of the gene panel size in the probability to identify GA in STS.
Because mutations of the SWI/SNF complex, which is a multisubunit chromatin remodeling complex that performs fundamental roles in gene regulation and cell lineage specification, have been identified in up to 20% of epithelial and hematologic malignant abnormalities, we wanted also to investigate their incidence in STS. We analyzed therefore the status of ARID1A, ARID2, PBRM1, SMARCA4, and SMARCB1. Twelve (2.0%) of 584 patients had inactivating mutations or genomic deletions in SWI/SNF subunits, of which 7 (1.1%) complex genomics sarcomas, 3 (0.5%) translocation-related sarcomas, 2 (0.3%) other sarcomas. The most common mutated subunits were ARID1A (4 [0.7%], of which 3 [0.5%] were complex genomics sarcomas), followed by ARID2 (2 [0.3%], of which 2 [0.3%] were complex genomics sarcomas), PBRM1 (3 [0.5%], of which 2 [0.3%] were complex genomics sarcomas), and SMARCB1 (3 [0.5%], of which 2 [0.3%] were other sarcomas).
At least 1 clinically relevant GA that could potentially guide decisions for targeted treatment was identified in 239 (41%) of 584 cases: 132 (40%) of 331 cases in the complex genomics sarcoma group, 19 (13%) of 144 in the translocation-related sarcomas group, and 89 (82%) of 109 in the other sarcoma groups (χ2 test, P < .001). The most common clinically relevant alterations potentially affecting treatment decisions are described in eFigure 4 and eTable 1 in the Supplement. Strikingly, actionable alterations in the receptor tyrosine kinase (RTK)/Ras signaling pathway were identified in 53 (16%) of the complex genomics sarcoma cases, compared with 8 (5.6%) in the translocation-related sarcoma cases, and 10 (9.2%) in the other sarcoma cases (χ2 test, P = .003) and affected the following genes: ALK, ARAF, ERBB2, FGFR1, FGFR2, FGFR3, FGF3, FGF4, KRAS, MET, NF1, NF2, and NRAS. In almost all cases, alterations in the RTK/Ras pathway were mutually exclusive.
Clinical Impact of Alterations Targeting
The American Association for Cancer Research (AACR) GENIE database does not contain treatment and outcome data. To illustrate the potential impact of targeting genomic alterations in sarcoma patients we analyzed the outcome of sarcoma patients included in the BIP study, which is an institution-wide screening program to identify patients referred to Institut Bergonié (Bordeaux, France) with somatic alterations that can be matched to targeted therapies in early phase clinical trials (https://clinicaltrials.gov/ct2/show/NCT02534649). A total of 58 patients with advanced STS were included in the study. Among them 17 (29.5%) had an actionable alteration. Seven (12%) were treated in a drug-matched clinical trial. Among them, 5 had clinical benefit including 2 partial response and 3 stable disease at 24 weeks or longer (eTable 2 and eFigure 7 in the Supplement).
Discussion
To our knowledge, the work reported here is the most comprehensive genomic analysis of STS performed to date. Our results indicate that the p53 pathway abrogation appears as a pivotal event in complex genomics and other sarcomas in agreement with previous sanger sequencing-based studies.4 Importantly, a high proportion of STS with TP53 alteration, also showed alteration of RB1 or CDKN2A suggesting that simultaneous disruption of the RB1/CDKN2A and the TP53 pathways contribute to their tumorigenesis. Further studies are needed to assess the impact of TP53 alterations in terms of prognosis and resistance to therapy. Indeed, several studies demonstrated the prognostic relevance of TP53 for many epithelial tumor types but none of them focused on STS.5 Moreover, overexpression of mutated TP53 has been associated with resistance to several cytotoxic therapies including drugs used for patients with STS, such as anthracyclines (doxorubicin) or antimetabolites (gemcitabine).5
Within the past 5 years, the oncology community has observed an explosion in technological abilities to rapidly and efficiently sequence tumor tissue to identify actionable mutations and provided personalized therapies to individual patients. This study aimed to assess the usefulness of such tests in STS.
Previous studies based on a smaller series of cases suggested that high-depth sequencing may reveal actionable alterations6,7 or may help to refine the prognosis of patients with STS.8 The present study uncovers an unexpectedly high frequency of clinically relevant GAs in STS.
Limitations
Despite its potential impact for the clinical outcome of patients with advanced STS, the present study has 2 definite limitations related to the absence of outcome data in the AACR GENIE database: the exact number of patients whose therapy was changed on the basis of the genomic profiling results is not known nor is the number of cases in which it is known that the genomic analysis led to a major clinical response to targeted therapy. In this regard, the data reported by Gounder et al9 in abstract form during the 2017 Annual Meeting of the American Society of Clinical Oncology are quite informative. The authors reported a series of 118 sarcoma patients who underwent genomic testing with the purpose of identifying an actionable genomic alteration. Among the 118, 60 (50%) patients had treatment-linked alteration. A total of 31 patients were enrolled in a genotype-matched clinical trial, whereas 29 were not included in a clinical trial for 3 main reasons: no clinical trial available, poor performance status, or results of genomic testing too late in the course of their treatment. Although the authors did not report detailed data related to the outcome of patients included in genotype-matched clinical trials, they show, as in our study, several instances of clinical benefit including: 2 partial responses with an MDM2 inhibitor in patients with MDM2-amplified sarcoma (1 dedifferentiated liposarcoma, 1 intimal sarcoma), 1 chordoma with CDKN2A deletion had a durable response with a CDK4/6 inhibitor, 1 patient with undifferentiated pleomorphic sarcoma bearing INI1 deletion had durable response with an EZH2 inhibitor, 1 patient with a BRAF-mutated undifferentiated pleomorphic sarcoma had a durable response with a combination of trametenib and dabrafenib, and 1 patient with metastatic angiosarcoma with a mutation of KDR (T771R) had a durable partial response with a VEGR inhibitor; a patient with rapidly progressive metastatic osteosarcoma and an ATM truncating mutation had 1-year disease stabilization with a regimen combining a PARP inhibitor and an ATR inhibitor.
Another limitation of this study was that no data were available regarding the incidence of germline mutation. Indeed, an international team of researchers has uncovered multiple new germline mutations that may influence the development of sarcomas.10 Notably, they found that variants in several DNA damage-sensing and repair genes contribute greatly to sarcoma risk, including BRCA2, ATM, ATR, and ERCC2. Given the successful development of PARP inhibitors in BRAC1 or 2 mutated ovarian and breast cancer, further efforts are needed to evaluate the clinical usefulness of germline testing in patients with sarcoma.
Finally, besides its therapeutic impact, implementation of NGS may also have an important value to refine diagnosis of sarcoma. Indeed, sarcomas are a heterogeneous ensemble of rare tumor subtypes with specific histologic features. Both rarity and heterogeneity make their diagnosis especially difficult even for expert pathologists. The consequence is a high rate of misdiagnosis. Our group has shown that the use of molecular cytogenetics to identify fusion transcripts and gene rearrangement may refine the diagnosis sarcoma in a considerable proportion of cases with a clear impact from a clinical point of view.11 However, these methods are limited by low resolution or narrow target range. By providing a base-by-base view of the genome, NGS can identify single-nucleotide variants (SNV), small structural changes, and balanced translocations, increasing information while decreasing costs with a genome-wide view of variation. Recently, a study showed that implementation of NGS in the diagnostic workflow of sarcoma is able to refine the diagnosis in 8% of cases with a potential impact on therapeutic strategies.
Conclusions
This study demonstrates that implementation of next-generation sequencing provides a framework for comprehensively interrogating actionable mutations in STS. Given the poor prognosis of STS treated by nontargeted conventional therapies, comprehensive genomic profiling shows promise to identify targeted therapeutic approaches to improve outcomes for this devastating disease. These results support the MULTISARC trial that will be launched in early 2018. This prospective randomized study is based on the hypothesis that implementation of NGS for the treatment of patients with advanced STS may improve their outcome. Patients with advanced STS will be randomized between 2 groups: in the experimental group, exome and RNA sequencing will be performed and their results discussed in a molecular tumor board to tailor the treatment of patients; in the control group, no molecular profiling will be done and patients will be treated in a conventional way (eFigure 7 in the Supplement). Thanks to a public-private partnership, 16 targeted therapies will be included in the program. One of the objectives of the MULTISARC study will be to demonstrate that use of NGS data can improve OS of advanced STS by allowing in at least a proportion of them the identification of 1 or more additional appropriate lines of treatment.
eMethods.
eTable 1. Actionable alterations in a series of 584 STS
eTable 2. Clinical outcome of STS patients treated with targeted therapies
eFigure 1. Proportion of genetic alterations according to genetic class of STS
eFigure 2. Distribution of the number of alterations
eFigure 3. Lollipop-style mutation diagrams for frequently altered genes
eFigure 4. Distribution of actionable alterations in the 10 most frequent histological subtypes of STS
eFigure 5. Co-occurrence and mutual exclusivity of genetic alterations in STS
eFigure 6. Objective responses to targeted therapies in STS
eFigure 7. Design of the MULTISARC study
References
- 1.Fletcher CD, Bridge JA, Hogendoorn PCW, Mertens F. WHO classification of Tumours of Soft Tissue and Bone IARC. International Agency for Research on Cancer; Lyon, France; 2013. [Google Scholar]
- 2.Italiano A, Mathoulin-Pelissier S, Cesne AL, et al. Trends in survival for patients with metastatic soft-tissue sarcoma. Cancer. 2011;117(5):1049-1054. [DOI] [PubMed] [Google Scholar]
- 3.Chakravarty D, Gao J, Phillips S, et al. OncoKB: a precision oncology knowledge base [published online May 16, 2017]. JCO Precision Oncology. doi: 10.1200/PO.17.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pérot G, Chibon F, Montero A, et al. Constant p53 pathway inactivation in a large series of soft tissue sarcomas with complex genetics. Am J Pathol. 2010;177(4):2080-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol. 2010;2(3):a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barretina J, Taylor BS, Banerji S, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42(8):715-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network. Electronic address: elizabeth.demicco@sinaihealthsystem.ca; Cancer Genome Atlas Research Network . Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171(4):950-965.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chibon F, Lagarde P, Salas S, et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med. 2010;16(7):781-787. [DOI] [PubMed] [Google Scholar]
- 9.Gounder MM, Ali SM, Robinson V, et al. Impact of next-generation sequencing (NGS) on diagnostic and therapeutic options in soft-tissue and bone sarcoma. J Clin Oncol. 2017;35(suppl; abstr 11001) [Google Scholar]
- 10.Ballinger ML, Goode DL, Ray-Coquard I, et al. ; International Sarcoma Kindred Study . Monogenic and polygenic determinants of sarcoma risk: an international genetic study. Lancet Oncol. 2016;17(9):1261-1271. [DOI] [PubMed] [Google Scholar]
- 11.Italiano A, Di Mauro I, Rapp J, et al. Clinical effect of molecular methods in sarcoma diagnosis (GENSARC): a prospective, multicentre, observational study. Lancet Oncol. 2016;17(4):532-538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods.
eTable 1. Actionable alterations in a series of 584 STS
eTable 2. Clinical outcome of STS patients treated with targeted therapies
eFigure 1. Proportion of genetic alterations according to genetic class of STS
eFigure 2. Distribution of the number of alterations
eFigure 3. Lollipop-style mutation diagrams for frequently altered genes
eFigure 4. Distribution of actionable alterations in the 10 most frequent histological subtypes of STS
eFigure 5. Co-occurrence and mutual exclusivity of genetic alterations in STS
eFigure 6. Objective responses to targeted therapies in STS
eFigure 7. Design of the MULTISARC study