Abstract
Export of proteins through type three secretion systems is critical for motility and virulence of many major bacterial pathogens. Three putative integral membrane proteins (FliP, FliQ, FliR) are suggested to form the core of an export gate in the inner membrane, but their structure, assembly and location within the final nanomachine remain unclear. We here present the structure of the Salmonella Typhimurium complex at 4.2 Å by cryo-electron microscopy. None of the subunits adopt canonical integral membrane protein topologies and common helix-turn-helix structural elements allow them to form a helical assembly with 5:4:1 stoichiometry. Fitting of the structure into reconstructions of intact secretion systems, combined with cross-linking, localize the export gate as a core component of the periplasmic portion of the machinery. This study thereby identifies the export gate as a key element of the secretion channel and implies that it primes the helical architecture of the components assembling downstream.
Type three secretion systems (T3SS) are nanomachines that span the bacterial cell envelope and provide a conduit for protein export from the bacterial cytoplasm 1. Two classes of T3SS exist. The first is dedicated to the export and assembly of bacterial flagella. The second, termed the injectisome, allows the delivery of effector proteins directly into the eukaryotic host cell cytoplasm 2. Both of these classes are associated with the pathogenicity of a wide range of clinically relevant bacteria 3. T3SS are assembled from a basal body consisting of a series of concentric oligomeric protein rings across the inner and outer membranes and periplasm, from which the helical hook and flagellum or needle structures project 2,4,5. Proteins associated with the cytoplasmic face of the basal body select proteins for export that are then transferred to a set of 5 membrane associated proteins located to center of the inner-membrane ring. These components (FliP, FliQ, FliR, FlhB, FlhA in the flagellar system and SctR, SctS, SctT, SctU, SctV in injectisomes, hereafter referred to as P, Q, R, B, A) are collectively termed the export apparatus (EA) and are absolutely required for the translocation of substrates across the bacterial envelope 6–8.
A combination of many structural techniques has led to atomic models for most of the soluble components, the circularly symmetric rings that compose the majority of the basal body and for both flagellar axial constituents and the injectisome needle 4. However, the EA remains poorly understood, with the topology and number of membrane helices of the three most hydrophobic proteins (P, Q and R) a subject of debate and with conflicting reports of stoichiometry between flagellar 7 and injectisome 9,10 T3SS (Fig. 1A). Given the high levels of structural homology amongst all T3SS structural components revealed to date, and the high levels of sequence conservation in the EA components in particular, it seemed likely that discrepancies between the systems reflected varying experimental approaches rather than true differences in the way in which this core component of the apparatus is assembled. P, Q & R are often encoded within a single operon and we decided to use a combination of biochemistry, native mass spectrometry and cryo-electron microscopy to investigate the structure and assembly of these core components in both flagellar and injectisome T3SS.
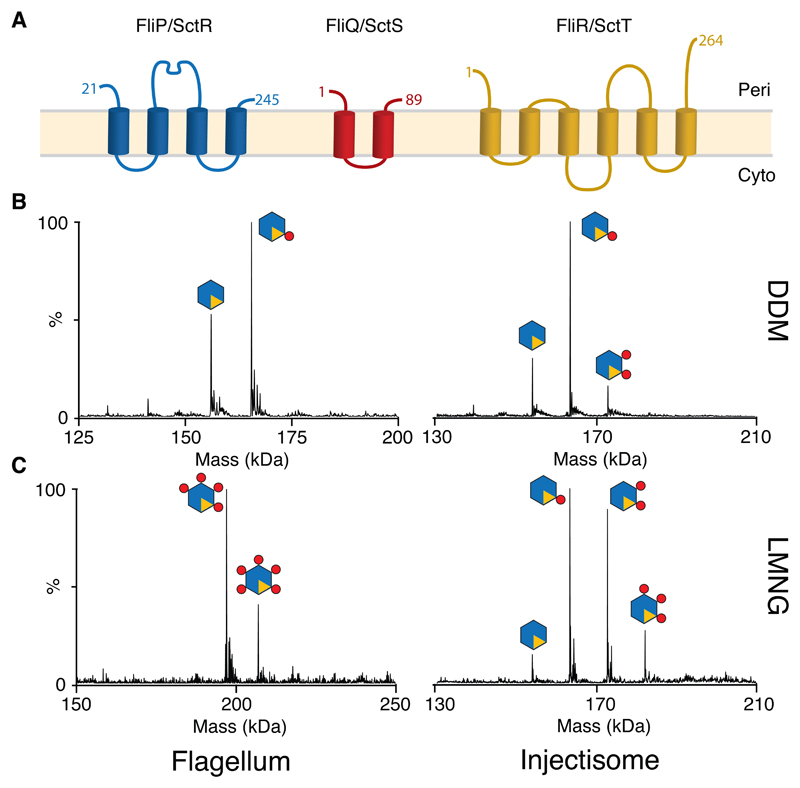
Fig. 1. Stoichiometry of the PQR complexes from both Flagellar and Injectisome T3SSs revealed by native mass spectromety (nMS).
(A) Consensus topology predictions for the flagellar (FliP, FliQ & FliR) and injectisome (SctR, SctS & SctT) export gate components P, Q & R, numbered according to the S. Typhimurium flagellar sequences. The orientation of the termini of Q and R with respect to the membrane has been previously debated and they are shown in here in the same orientation as P. (B) Deconvoluted native mass spectra of complexes extracted and purified in DDM reveals a P5R1 core complex with variable numbers of Q (C) Complexes purified in the less harsh detergent LMNG contain more Q subunits, with up to the five copies of Q seen in a flagellar species complex (Table S1). Raw data are available in Supplementary Data Set 1.
Results
Stoichiometry of PQR complexes
We previously used native mass spectrometry (nMS) to characterize a P5R1 complex from the Salmonella enterica serovar Typhimurium (S. Typhimurium) injectisome system 9, whilst others have used negative stain electron microscopy to define a flagellar P6 complex 7. To attempt to resolve whether flagellar and injectisome T3SS are differently assembled we expressed the P, Q and R components from a range of flagellar and injectisome systems, each from a single operon with a C-terminal dual strep-tag on R for purification. Using the detergent DDM for extraction, high resolution nMS demonstrated that the core of the export apparatus is formed by a P5R1 complex in all the systems studied (Fig. 1B, Table S1). Depending on the species expressed we observed a variable number of Q subunits associated with this core complex (Fig. 1B). Dissociating the intact core complex inside the MS using high levels of collisional activation resulted in formation of sub-complexes of all possible combinations of P and R (i.e. P4R1, P3R1, P2R1, P1R1, P5, P4, P3, P2, P1), suggesting that the R component is integrated into the complex rather than peripherally associated with a P5 ring (Fig. S1).
Since we observed variation in the copy number of the Q subunit we investigated different purification strategies and again examined the composition of the EA by nMS. Using the less harsh detergent LMNG to solubilize the complex revealed more associated Q subunits, with up to P5Q5R1 complexes being seen (Fig. 1C, Table S1). This suggests that multiple Q subunits are loosely associated with a P5R1 core.
Cryo-EM structure of a P5Q4R1 complex
Complexes purified in a range of detergents and amphipols were used to prepare cryo-EM grids with a focus on detergents revealed by nMS to be less destructive. A sample of S. Typhimurium FliPQR, purified in LMNG (Fig. S1), gave a range of different orientations (Fig. S2) and led to a reconstruction of the complex at 4.2Å (Fig. 2A, Fig. S3, Table 1). This allowed us to build models for all three components (Fig. 2B and Video S1). The complex is a P5Q4R1 assembly consistent with the dominant species in our earlier nMS data of the flagellar system (Fig. 1C, Fig. S1, Table S1): P and R are intimately associated in a pseudo-hexameric, closed structure, with the Q subunits peripherally associated around the outside of the core P5R1 complex. All three subunits form extended structures, built predominantly from pairs of kinked anti-parallel helices, with a common orientation of N and C termini, in conflict with earlier topology predictions. Further analysis reveals that R is a structural fusion of P and Q (Fig. 2C), further reinforcing the pseudo-hexameric nature of the complex. Each “monomer” of the pseudo-hexamer, with the exception of the fifth P subunit which is not associated with a Q subunit, can be described as three helical hairpins that are offset from one another by an approximately 25 Å rise along the long axis of the molecule. Hairpin 1 corresponds to P63-101 and R16-55, hairpin 2 corresponds to P178-245 and R76-157, while hairpin 3 corresponds to Q1-89 and R167-262. The major structural difference between the P–Q fusion and the R protein is seen to be an insertion in the P component that encompasses the second half of the second predicted trans-membrane helix and the previously identified “periplasmic domain” 7. This insertion forms an additional pair of shorter α-helical hairpins. An earlier crystal structure of this region from Thermotoga maritima overlays well with our structure (Fig. 2D) 7, despite lacking the first helix of the full insertion, presumably due to the mis-annotation of the trans-membrane helices. Five copies of this insertion domain form the top and outer-rim of the full PQR complex, and mapping of sequence conservation onto the surface of the complex reveals this to be the most variable region of the structure (Fig. 2E).
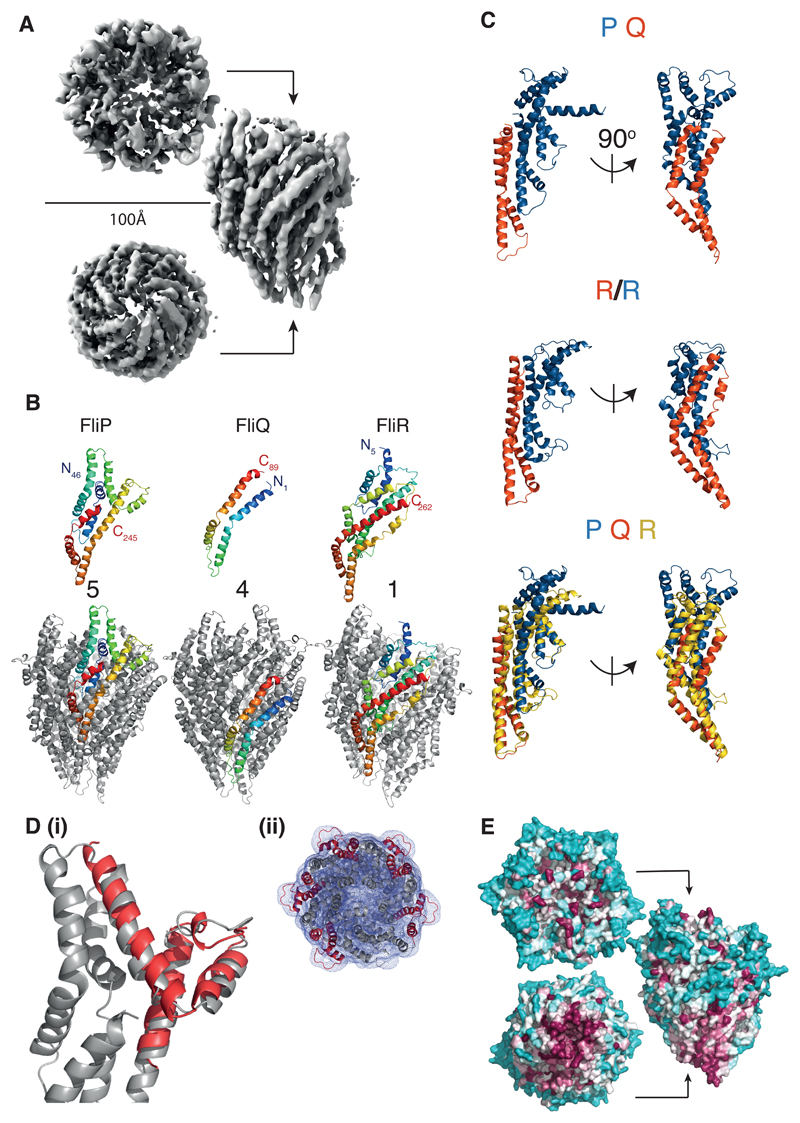
Fig. 2. Structure of the flagellar P5Q4R1 complex revealed at 4.2Å by cryo-electron microscopy.
(A) Cryo-EM map of the P5Q4R1 complex reconstructed from 98000 particles with C1 symmetry. The complex is ~120 Å in height and the top has a diameter of ~100 Å. (B) Structures of the monomeric chains and location of an example of each within the full assembly. Each monomer is colored from blue to red at the N and C termini respectively. (C) R is a fusion of the two shorter proteins. Upper panel shows a PQ complex (P-blue;Q-orange) in two orientations with an aligned copy of R below coloured to emphasise the structural homology. The lowest panel superimposes the P (blue):Q (orange) heterodimer onto R (yellow). (D) (i)Overlay of the crystal structure (5h72 7) of the “periplasmic domain” of Thermotoga maritima FliP (red) onto one copy of FliP (grey) in the FliPQR complex. (ii) Top down view of the FliP5Q4R1 model (grey cartoon) docked in the unsharpened 4.2 Å map (grey mesh), with the five copies of the “periplasmic domain” of FliP highlighted in red. (E) Analysis of conservation using the CONSURF server 29 reveals that the bottom of the complex is the major conserved external surface, while the region of P that adorns the outside is highly variable. The relative degree of conservation is colored dark purple for highly conserved to cyan for variable residues.
Table 1. Cryo-EM data collection, refinement and validation statistics.
| (EMDB-4173) (PDB 6f2e) |
|
|---|---|
| Data collection and processing | |
| Magnification | 165000 (K2), 96000 (Falcon3) |
| Voltage (kV) | 300 |
| Electron exposure (e–/Å2) | 47 (K2), 50 (Falcon3) |
| Defocus range (μm) | 0.5-4 |
| Pixel size (Å) | 0.86 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 474625 |
| Final particle images (no.) | 97718 |
| Map resolution (Å) | 4.2 |
| FSC threshold | 0.143 |
| Refinement | |
| Initial model used (PDB code) | None |
| Model resolution (Å) | 4.2 |
| FSC threshold | 0.143 |
| Map sharpening B factor (Å2) | -227 |
| Model composition | |
| Non-hydrogen atoms | 12541 |
| Protein residues | 1629 |
| Ligands | 0 |
| B factors (Å2) | |
| Protein | 227 |
| Ligand | 0 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.18 |
| Validation | |
| MolProbity score | 2.6 |
| Clashscore | 25.7 |
| Poor rotamers (%) | 0.1 |
| Ramachandran plot | |
| Favored (%) | 83.0 |
| Allowed (%) | 16.3 |
| Disallowed (%) | 0.7 |
The PQR complex forms a helical structure
Dissecting the complex further reveals that the subunits are arranged in a right-handed helix (Fig. 3A). The structural equivalence of a P–Q pair to R (Fig. 2C) means that the structure is effectively 6 copies of an R-like object forming a single helical turn, with the 5 copies of P further adorned by the inserted “periplasmic domain”. Analysis of the helical parameters that relate subunits reveals differences between the base and the tip of the structure (Table 2); the helical pitch is tighter at the top than at the bottom of the complex but the average values are consistent with those previously determined for both flagellar rod–hook–filament 11–13 and injectisome needle assemblies 14. As for these other axial helical assemblies, the PQR complex is constructed from pairs of helices arranged into a spiral, although the orientation and arrangement of the helical hairpins is distinct. Furthermore, analysis of the electrostatic surface of the PQR complex reveals that the inner surface, which would be predicted to form the export channel, is positively charged, a feature that is also shared by the known rod, hook, flagellum and needle structures.
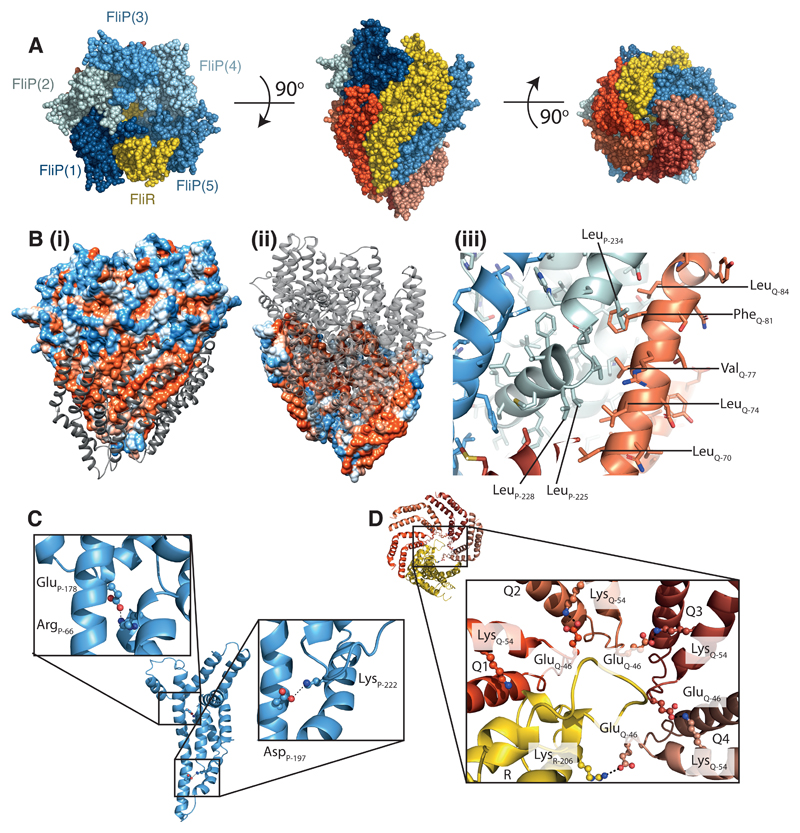
Fig. 3. The P5Q4R1 complex is a right-handed helical assembly with helical parameters consistent with flagellar and injectisome assemblies.
(A) Space filling representation of the P5Q4R1 complex with R (yellow), P (shades of blue) and Q (shades of red). (B) (i) the surface of the P5R1 complex is coloured according to hydrophobicity (orange-hydrophobic; blue-charged) whilst the four copies of Q packing against a hydrophobic surface on this complex are shown as grey ribbons in (ii) the scheme is reversed and viewed from the opposite side of the complex to reveal the hydrophobic surface on the back of Q against which the P5R1 complex packs (iii) zooms in on some of the specific hydrophobic residues buried on assembly of Q. (C) Intra-molecular salt bridges within P (D) Inter-molecular salt-bridges between copies of Q and R are shown.
Table 2. Helical parameters of the PQR export gate compared with flagellar and needle components.
| Subunits per turn | Axial rise (Å) | |
|---|---|---|
| FliP (1) to FliR | 5.1 | 5.6 |
| FliP (2) to FliP (1) | 5.6 | 4.9 |
| FliP (3) to FliP (2) | 5.8 | 3.2 |
| FliP (3) to FliP (4) | 6.0 | 2.3 |
| FliP (4) to FliP (5) | 6.0 | 3.8 |
| Average | 5.7 | 4.0 |
| Flagellar Filament | 5.5 | 4.7 |
| Flagellar Hook | 5.6 | 4.2 |
| Flagellar Rod | 5.6 | 4.1 |
| T3SS Needle | 5.6 | 4.3 |
Analysis of the interaction surfaces within the complex highlights the complexity of the object (Table S2), with each individual component making contacts with several other subunits. All of the interfaces involve residue pairs that have been shown to co-evolve 15, with homo-typic interaction surfaces explaining contacts that couldn’t be reconciled in a single protein model. The interaction surfaces between individual subunits are dominated by hydrophobic residues, and bury many of the amino acids that had previously been predicted to form trans-membrane helices. This means that, despite the robust prediction that all three subunits are integral membrane proteins, mapping hydrophobicity onto the surface of the model reveals a relatively small hydrophobic strip near the base of the structure. Most of the exposed hydrophobicity is contributed by residues of Q, with the majority of the P and R hydrophobic residues buried in the assembly (Fig. 3B). While the surface area buried in assembly of the complex is large for all subunits, presumably reflecting strong association (Table S2), we note that there are several hydrophobic cavities within the structure, between P and Q subunits, which we assume may be occupied by buried lipids or detergent that are not resolved at the resolution of our current maps.
It has previously been observed that P, Q and R contain conserved charged amino acids that lead to motility defects when mutated 16,17. Our structure demonstrates that these residues play key structural roles, forming intra- and inter-molecular salt bridge. The Asp197–Lys222 pair in P form an intra-molecular salt bridge between the hairpin 2 helices, while E178 of P is in salt-bridging distance of Arg66 (Fig. 3C). The pair of conserved charged residues in Q, Glu46 and Lys54, are seen to form an inter-subunit salt-bridge between neighboring Q subunits, i.e. Glu46 in the first 3 Q subunits (Q1-Q3) bridges to Lys54 in the neighboring Q (Q2-Q4). Intriguingly, the Glu46 of Q4, in the absence of a fifth Q, forms a salt bridge with the highly conserved Arg206 in R (Fig. 3D).
The PQR complex is a core structural component of the basal body
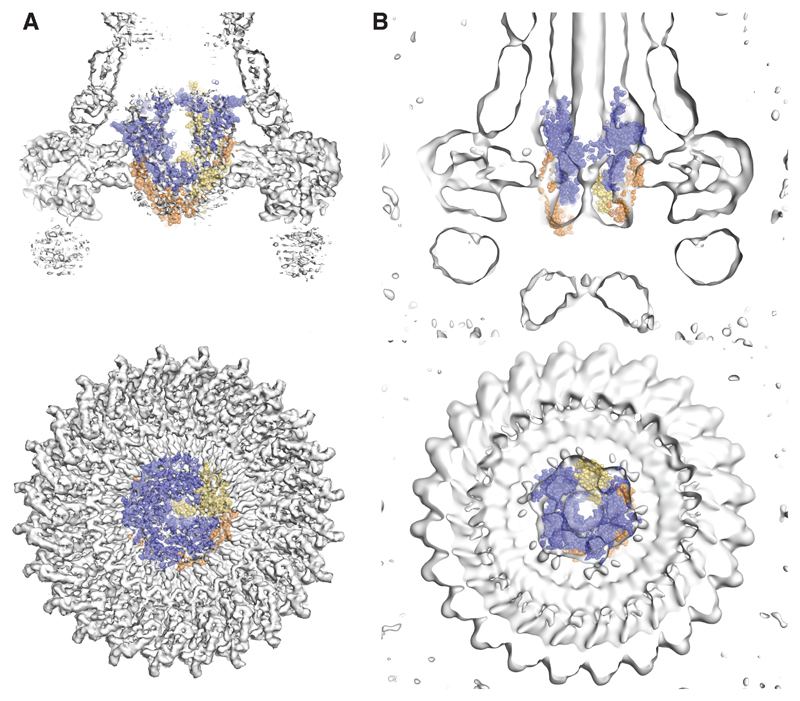
The overall dimensions, helicity and hydrophobicity patterning of the complex do not support a standard localization within the inner membrane. We therefore sought to determine the location of the PQR complex in the assembled T3SS. Inspection of earlier single particle reconstructions of isolated basal bodies 18 and in vivo tomograms from both flagellar 19 and injectisome 20 T3SSs demonstrated that the PQR complex forms the structure previously described as the “cup and socket” 8,21. This is particularly striking when fitting our model within the highest resolution structure of a basal body determined to date, where the height, diameter and shape of the density seen in the basal body are an excellent match (Fig. 4A) 18. This therefore suggests that, in this reconstruction of a rod-less and needle-less system, the PQR complex is in the closed conformation that we observe in isolation. Placing the PQR complex in the cup–socket location predicts close contacts between it and the circularly symmetric components (FliF and SctC–SctJ in flagellar and injectisome systems respectively). Using in vivo photo-crosslinking and chemical crosslinking-mass spectrometry in the S. Typhimurium injectisome system we were able to detect two residues in P on the outer surface of our complex that cross-linked to SctC (SpaP K132 and K135 to InvG K38) and a residue in SctJ (E138pBpa) that crosslinks to both P and R, providing further support for this location of the complex (Fig. 4A, B, Fig. S4-6).
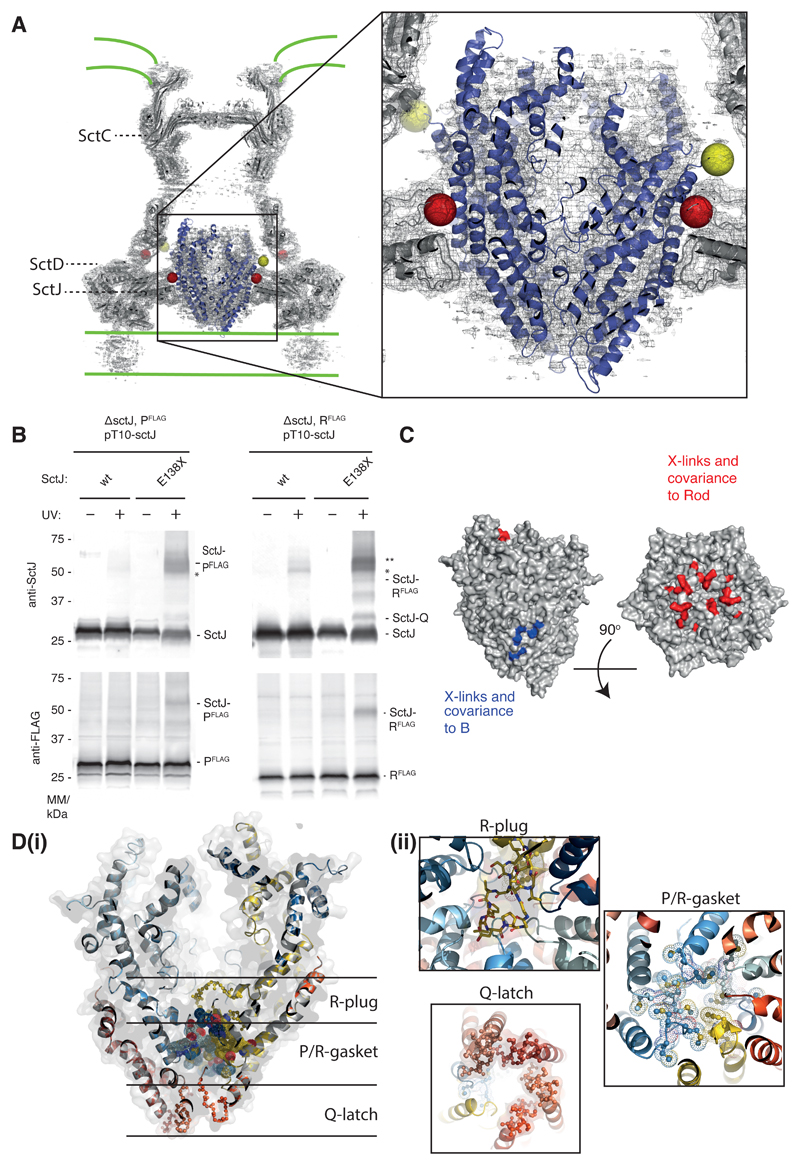
Fig. 4. The P5Q4R1 complex is a core component of the basal body and forms a platform for assembly of the Rod.
(A) Positioning our structure within an earlier high resolution reconstruction of the injectisome basal body 18 (grey surface and cartoon) reveals that the P5Q4R1 complex (blue cartoon) fits the un-occupied density in the center of the basal body. This region of the basal body has previously been called the “cup and socket” and sits above the proposed inner membrane location (shown as green lines). Residues on P that can be cross-linked to the basal body are highlighted by yellow spheres at the Cα position, while residues on SctC and SctJ of the basal body that cross-link to the PQR complex are shown in red. (B) In vivo photo-crosslinking studies reveal cross-links between P5Q4R1 and the inner membrane ring component SctJ in the S. Typhimurium injectisome. The residues involved are highlighted in (A). * SctJ-SctJ pBpa-independent cross-links. ** pBpa-dependent SctJ-ScJ crosslinks result in a crosslink-ladder, of which the homodimeric SctJ-SctJ crosslink is indicated by **. Representative Western blots (upper panel probed with anti-SctJ, lower panel with anti-FLAG tag), n=3. (C) Mapping of earlier cross-linking and our co-variation data (Table S3) onto P5Q4R1 reveal probable binding sites for B (FlhB or SctU) and the (inner) rod components. (D) The export gate is constricted at multiple points (i) a slab section of the entire assembly shows the levels at which the different constriction points operate (ii) views from either above or below each constriction point highlight the structural elements involved.
Previous in vivo photo cross-linking studies in our laboratory 9, combined with co-variance (Table S3) 15 analyses, have also implied specific and direct interactions between PQR and another putative integral membrane protein termed B (FlhB or SctU). Mapping the residues implicated in the B interaction onto our structure suggests it assembles at the interface between R and the Q-less P at the base of our helical assembly (Fig. 4C).
The socket location of the complex also suggests that the rod, which has previously been suggested to assemble directly onto the annular FliF ring 19,22 in the flagellar system, will actually assemble onto or into the already helical PQR complex. In order to investigate this further, we mapped the location of residues on PQR that have been shown to cross-link to the rod proteins 9. We also identified residues that strongly co-evolve between PQR and rod subunits (Table S3) 15. All of these amino acids map to the N-terminal helices on P and R and to the extreme C-termini of the rod proteins, including the Val99 residue of FliE that drastically reduces flagellation when mutated 22. This places the rod binding site at the top of the EA complex, inside the walls formed from the “periplasmic domains” of the P subunits (Fig. 4C).
A further important observation is that the PQR complex is seen to be constricted at multiple points within the helical assembly (Fig. 4D). The top of the structure is closed to an <10 Å aperture by the N-terminal helices of P and R before opening out into a small lumen with a diameter of 25 Å. At the base of this lumen a 15 residue insertion in R (residues 106-121) forms a plug that completely occludes the channel. Immediately below this, five copies of the highly conserved Met-Met-Met loop in P (Fig. S7) form a molecular gasket that again constricts the channel to <10 Å. Finally, the base of the structure is held closed by a salt bridge network between Q-subunits and the Q-like portion of R, involving the highly conserved hydrophilic loop of Q (Fig. S7).
Discussion
We have presented here the first structure of the core of a T3SS Export Apparatus. In addition we have provided corroborating nMS data that provides strong evidence for the assembly being common to all T3SS, whether flagellar or injectisome in nature, giving a common structural framework on which to interpret earlier data and derive future mechanistic models. Our structure has revealed a number of interesting features that not only increase our understanding of T3SS, but also have more general implications for predictions of membrane protein structure-function relationships.
One of the more striking observations is that the PQR complex assembles into a pseudo-hexameric helical structure. The P and Q components together structurally mimic the R subunit, and the P5Q4R1 stoichiometry we observe means that this portion of the EA is formed of 6 copies of an R-like object, decorated on the outside by an insertion in the P subunits. In our structure one of the P subunits lacks an associated Q, and it will remain to be seen whether this has functional relevance, especially given the proximity of the potential fifth Q binding site to the predicted site of binding of FlhB(SctU) (Fig. 4C). Positioning the complex in “cup and socket” density of a previously published injectisome cryo-EM map reveals that the PQR complex sits at the heart of the basal body, a fact that we confirm using cross-linking studies. This localization, combined with the helical nature of the structure, therefore answers the long posited question of how the flagellum-needle helix nucleates: the asymmetric 5+1 structure of the EA drives the formation of the helical pitch, via the asymmetric nature of the different P binding sites on each side of the unique R subunit.
Our isolated PQR complex adopts a closed conformation with multiple constriction points, including one formed by the highly conserved hydrophobic Met-Met-Met loop in the P subunit. This loop has previously been proposed to act as a molecular gasket, based on the observation that mutation of the Met residues leads to a measurable increase in ion conductance across the cell membrane 17. This P-gasket is bookended by an R-plug and a Q-latch. The R-plug is a large hydrophobic insertion that completely closes the channel and mutation of this insertion can overcome secretion defects caused by mutations in other components of the EA 23. The Q-latch is more open and hydrophilic than either the P-gasket or the R-plug, but it wraps around the P-subunits and holds them closed via highly conserved inter-chain salt-bridges. The tight closure of the PQR complex is likely to be important for maintaining bacterial viability, by ensuring the complex does not form breaches in the cell membrane prior to full assembly of the basal body and control of gating by the entire T3SS. As noted above, the shape of our closed complex is consistent with the shape of the socket–cup seen in a rod-less basal body and this, coupled with the relatively tight fit of the complex against the FliF(SctJ) ring, suggests that the initially assembled basal body is impermeable to substrate (Fig. 5A). However, we noted that the shape of the socket–cup density in reconstructions of more intact basal bodies containing needles 21,24 was more open at the base of the density now ascribed to PQR. We therefore sought to model this open state and observed that the simplest mechanism to open the constriction points at the base of structure is to straighten the kinked helices in the individual subunits. We accomplished this with the minimum manipulation by allowing the conformation of the P and R subunits to relax back to the much straighter arrangement of helices found in the models derived in silico from co-evolution data. This had the effect of opening the constrictions at the base of the complex via a concerted iris mechanism, producing a model without significant clashes at the secondary structure level (Fig. 5B and Video S2). Such a conformation would therefore allow substrate access to a secretion channel through the centre of the PQR complex and permit assembly of the rod on the top of the export gate, consistent with cross-linking and co-evolution data (Fig. 4C).
Fig. 5. Modelling opening of the export gate.
Earlier basal body reconstructions show that the P5Q4R1 complex is closed in the absence of rod components ((A) 18) and open in their presence ((B) 21). The upper panels show slabs of the side views of these earlier maps and the lower panels shows slabs from just above the complex. In (A) the structure of the P5Q4R1 complex is fit as in Figure 4(A). An open state of the P5Q4R1 complex can be modelled (B) by superposition of models of P and R produced from co-evolution data onto our structure of the closed complex.
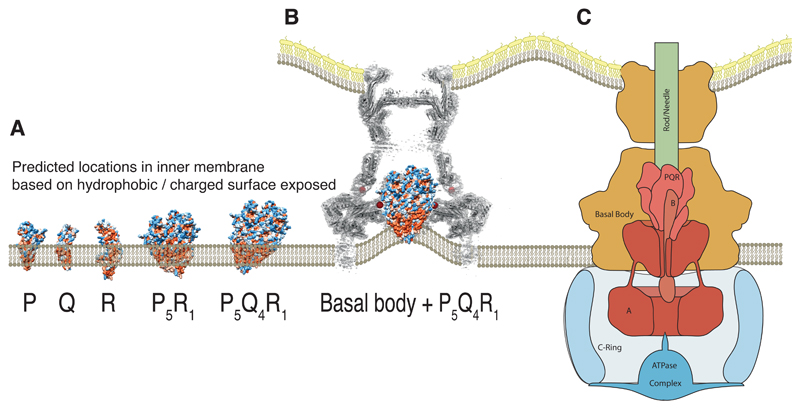
Of course this observation of a closed PQR complex leads on to the question of the mechanism of the opening of the channel and the potential role of other T3SS components. One consequence of the cup–socket localization of the PQR portion of the EA is that it places it above the predicted inner membrane position (Fig. 4A) in the native assembly. Only the very tip of the Q-ring would be in contact with a predicted bilayer, despite the clear hydrophobic band that extends around the outer surface of the Q-subunits (Fig. 6A). This would suggest that the bilayer inside the basal body is significantly distorted, or that these hydrophobic patches are covered by further protein interactions with the other two putative integral membrane proteins, A (FlhA or SctV) & B (FlhB or SctU). The A protein has been demonstrated to form a circular, nonameric complex, with a large domain of known structure localized in the cytoplasm immediately below the basal body 25,26. The integral membrane domain of this protein would therefore be predicted to lie in the inner membrane around the base of our PQR complex. We note that in situ tomograms of the S. Typhimurium injectisome show a distinct density in this location, but that cells deficient in A not only lack this density but also demonstrate a distorted membrane leaflet that reaches up to location that we now propose is occupied by the base of the Q subunits 20 (Fig. S8, Fig 6B-C). This localization of A relative to the PQR complex, combined with its proposed role in coupling protonmotive force to protein export 16,27, would be consistent with the other EA components playing an active role in opening the PQR channel.
Fig. 6. Placing the complex within the context of the full type three secretion system.
(A) Locating the isolated monomers of P, Q and R within a lipid bilayer is not trivial due to the extended nature of the hydrophobic surface and the large number of charged patches within this surface. However, the assembled object is likely to project into the periplasmic space. (B) In the absence of the other export apparatus components (A and B), earlier tomograms show the inner membrane, within the basal body, is deformed towards the region we now assign to the hydrophobic surface of Q 20. (C) Proposed relative locations of the five export apparatus components within the type three secretion system places the transmembrane portions of the nonameric A at the base of the P5Q4R1 complex. B is likely to form part of the helical export gate complex with the previously assumed “transmembrane” helices driving assembly and with the cytoplasmic domain hanging below a helical P5Q4R1B1 complex.
Our structure also poses interesting questions when it comes to prediction of complex helical trans-membrane protein structures. All three of the P, Q and R components were long predicted to be standard trans-membrane proteins, with the Q and R subunits frequently predicted to have inverted topology to the P subunit, i.e. with N- and C-termini in the cytoplasm. Our structure not only re-assigns the topology, showing that all 3 subunits have their termini on the periplasmic side of the structure, but the overall packing of the final object also demonstrates the difficulties of predicting trans-membrane regions in complex, multimeric membrane proteins. When each monomer is extracted from the complex it is difficult to identify an intra-membrane location that could bury the exposed hydrophobic surfaces without also burying significant hydrophilic patches (Fig 6A), as the previously predicted transmembrane helices are offset from one another along the long axis of the molecule. This suggests that efficient complex assembly may be directly coupled to membrane insertion. These structures also highlight the power of using co-evolutionary contacts in structure prediction, as models produced using such methods predict the offset helices in P 28. However, it was noted by the authors of this study that it was difficult to reconcile this large displacement with positioning in the bacterial membrane; a conundrum our structure now solves.
It is also possible that the assembly process may help drive removal of the structure from the lipid bilayer, with existing hydrophobic surfaces becoming covered by newly inserted components, creating new hydrophobic surfaces. One hypothesis would be that this process is initiated by the unique R component to ensure assembly of the correct object, since R becomes the “top” component of the helix. However, we note that the P component is able to form homo-hexamers when over-expressed in the absence of R 7, which may suggest that self-association of P subunits initiates assembly, with R inserting into the site created between the fifth and first copies of P. Either way, the Q-like portion of R then becomes the lowest hydrophobic patch and completes the first Q-subunit binding site, providing key lateral contacts in addition to the existing P surface. This Q-subunit now becomes the lowest hydrophobic component and provides the next lateral Q binding surface and so on. When expressed without the T3SS annular ring forming proteins, the complex clearly remains associated with the inner membrane 8, presumably with the hydrophobic band across the bottom of the complex embedded in the bilayer (Fig 6A). It is tempting to speculate that, during native assembly, one of the forces acting to extract the complex from the bilayer is the “wedging” of the inserted domain in P (previously termed the “periplasmic” domain) above the assembling FliF or SctD–SctJ inner ring, since the dimensions of the object mean that an assembled P5Q4R1 object could not pass through this ring once it is assembled. This again suggests that tight coupling is required between assembly of all the Sec transported components of T3SS systems in order to generate a secretion competent T3SS, consistent with earlier data 8.
In summary, our study has demonstrated the following regarding the PQR export gate complex: 1) that it has a core stoichiometry that is conserved across flagellar and injectisome T3SS; 2) that it forms a structure with an unpredicted helical symmetry; 3) that it sits above the likely location of the bacterial inner membrane as a core component of the basal body. The data suggest that the core export gate complex is contiguous with the helical axial components that culminate in the flagellum or needle, and therefore that the export pathway for secreted substrates will be through the centre of this complex, directly into the channel within the rod and filament or needle. Clearly, therefore, the central position and core role of this complex in the secretion pathway of many virulent bacteria make it an attractive target for future drug development, especially in light of increased antibiotic resistance.
Methods
Materials
Chemicals were from Sigma-Aldrich unless otherwise specified. Detergents n-dodecyl-maltoside (DDM), Lauryl Maltose Neopentyl Glycol (LMNG) and pentaethylene glycol monooctyl ether (C8E5) were from Anatrace. para-benzoylphenylalanine was from Bachem. SERVAGel™ TG PRiME™ 4-20% and 8-16 % precast gels were from Serva and 4–20% Mini-PROTEAN® TGX™ were from Bio-Rad. Primers are listed in Table S4 and were synthetized by Eurofins, Invitrogen or Sigma-Aldrich. Polyclonal rabbit anti-PrgK antibody was a kind gift of Thomas Marlovits, Vienna. Monoclonal M2 anti-FLAG antibody was from Sigma-Aldrich. Monoclonal anti-SipB and anti-InvJ antibodies were raised by Abmart. Secondary antibodies goat anti-mouse IgG DyLight 800 conjugate and goat anti-rabbit IgG DyLight 680 conjugate were from Thermo Fisher Scientific.
Bacterial strains and plasmids
Bacterial strains and plasmids used in this study are listed in Table S5. Primers for construction of strains and plasmids are listed in Table S4. Plasmids were generated by Gibson assembly of fragments using the NEBuilder HiFi Master Mix (NEB). Fragments were created by PCR with the relevant primers using Q5 polymerase (NEB). Gibson assembly and PCR were carried out following the manufacturer’s recommendations. All Salmonella strains were derived from S. Typhimurium strain SL1344 30. Bacterial cultures were supplemented as required with streptomycin (50 μg/mL), tetracycline (12.5 μg/mL), ampicillin (100 μg/mL), kanamycin (25 μg/mL or 60 μg/mL for large scale expression in TB media), or chloramphenicol (10 μg/mL).
Export gate purification
FliOPQR or SctRST were expressed in E. coli MT56 (DE3) 31 as a single operon with a C-terminal Twin-Strep tag on the R component from a pT12 vector (Table S5). Cells were spun down at 4,000 g and resuspended in buffer A (100 mM Tris, 150 mM NaCl, 1 mM EDTA, pH 8). After lysis in an EmulsiFlex homogeniser (Avestin), cell debris was removed by spinning at 24,000 g for 20 minutes. The cell membranes were isolated from the cleared lysate by ultracentrifugation at 200,000 g for one hour. Membranes were resuspended in buffer A in a hand-held homogeniser and detergent (DDM or LMNG) was added to 1 % (w/v). After stirring for one hour, unsolubilized material was spun down at 50,000 g for 30 minutes and the supernatant was applied to a StrepTrap column (GE Healthcare). The resin was washed overnight by recirculating in buffer A with 0.1 % (w/v) detergent. Purified protein was eluted with 10 mM desthiobiotin in buffer A containing 0.01% LMNG or 0.02 % (w/v) DDM and further polished by size-exclusion chromatography (S200 10/300 increase, GE Healthcare). When expressing FliOPQR, pure FliPQR was separated from residual FliO chaperone in the size-exclusion step.
FliPQR for electron microscopy was purified as above from E. coli MT56 (DE3) expressing FliOPQR from a pT12 vector and FlhBA from a pBAD vector using LMNG while omitting the overnight wash step. The leading edge of the resulting peak in size-exclusion chromatography was concentrated to 1 mg/ml and flash frozen in aliquots. SEC-MALS analysis was carried out by injecting 100 μL of sample onto an S200 increase 10/300 column (GE Healthcare) equilibrated in gel filtration buffer plus the appropriate detergent. Light scattering and refractive index changes were measured using a Dawn Heleos-II light scattering detector and an Optilab-TrEX refractive index monitor. Analysis was carried out using ASTRA 6.1.1.17 software assuming a protein dn/dc value of 0.186 ml/g and a detergent dn/dc value of 0.132 ml/g.
Native mass spectrometry
Prior to mass spectrometry aliquots of purified export gate were exchanged into 200 mM ammonium acetate supplemented with 0.5 % (v/v) C8E5 detergent using P6 Biospin columns (Bio-Rad). Extensive detergent screening was performed to find the optimal detergent for native MS. Spectra were acquired using a Q Exactive mass spectrometer (Thermo Fisher Scientific) modified for transmission and detection of high m/z ions and optimized for native mass spectrometry of membrane proteins 32. Samples were introduced into the mass spectrometer using gold coated borosilicate capillaries prepared in house. For measurement of the intact PQR complex, instrumental conditions were optimized to achieve micelle removal with minimal complex dissociation. Capillary voltage was +1.2-1.6 kV, capillary temperature 40-80 °C, AGC target 1×106, higher-energy collisional dissociation (HCD) energy 150-200 V, collision gas was argon and UHV pressure 1-2×10-9 mbar. Detection was performed with transient times of 64 ms. The noise parameter was set to 3 rather than the default of 4.64. Spectra were acquired for 10-100 scans with 1-10 microscans and averaged in Xcalibur 2.1 before being processed and deconvoluted using Unidec 33.
EM sample preparation
3 μl of purified export gate complex (0.7 to 1 mg/ml) were applied onto glow-discharged holey carbon coated grids (Quantifoil 400 mesh, Au R1.2/1.3), adsorbed for 10 s, blotted for 3 s at 100% humidity at 22 °C and frozen in liquid ethane using a Vitrobot Mark IV (FEI).
EM data acquisition and processing
An initial 7.8 Å map was calculated from data collected on a Talos Arctica (FEI) operating at 200 kV and equipped with a Falcon 3 detector (FEI). A total of 11,650 movies were collected at a sampling of 0.78 Å/pixel with a total dose of ~50e- in linear mode. Motion correction was carried out using Simple-unblur 34 and contrast transfer functions (CTFs) of the summed micrographs were calculated using CTFFIND4 35. Particles were picked using Gautomatch 36 and Simple 34, extracted in a 240 x 240 Å box, and the subsequent dataset was subjected to multiple rounds of reference-free 2D classification in Relion 2.0 37. An initial model was generated ab initio using Simple-Prime3D 34 and used as a reference for 3D classification in Relion 37. Particles from the highest resolution class were then subjected to auto-refinement.
Subsequently, further data were collected on a Titan Krios (FEI) operating at 300 kV (Table S6). 401 movies were collected on a K2 Summit detector (Gatan) in counting mode at a sampling of 0.86 Å/pixel, 3.9 e-/Å2/s, 12 s exposure, total dose 47e-, 40 fractions written. 1,687 movies were collected on a Falcon 3 detector (FEI) in counting mode with a sampling of 0.86 Å/pixel, 1 e-/Å2/s, 50 s exposure, total dose 50e-, 40 fractions written. Motion correction and dose weighting were carried out using Simple-unblur 34, a variation of the approach used in Unblur that applies automatic weighting of the frames using a correlation-based M-estimator and continuous optimisation of the shift parameters. Input is a textfile with absolute paths to movie files in addition to a few input parameters, some of which deserve a comment. Individual frames were low-pass filtered according to the dose-weighting strategy, as determined by the dose rate and exposure time for each dataset. CTFs were calculated using CTFFIND4 35. Particles were picked using Gautomatch, extracted using a 256 x 256 Å box, and subjected to multiple rounds of reference-free 2D classification in Relion 2.0 37. The earlier 7.8 Å map was used as a reference for a first round of 3D classification. Particles from the highest resolution class were subjected to auto-refinement using an oblate spheroid as a reference. The resulting model was then used as a reference in a masked auto-refinement job, followed by a further 3D classification run. The final auto-refinement was carried out in Relion 2.0 using 97,718 particles and a soft mask. Post-processing was carried out in Relion using a soft mask and a B-factor of -227 Å2 was applied. Gold standard Fourier shell correlations using the 0.143 criterion led to a resolution estimate of 4.15 Å. Local resolution estimations were calculated using ResMap 38.
Model building
Atomic models of FliP, FliQ and FliR were built using Coot 39 (Table S6). Initially, the crystal structure of the “periplasmic domain” of Thermotoga maritima FliP (5h72, 40) was fit into the 5 peripheral loop structures of the map (Fig. 2D). This was used to guide docking of the equivalent region of a FliP model that was built and refined using Rosetta guided by co-evolutionary restraints (downloaded from the Gremlin database - https://gremlin2.bakerlab.org/meta.php) 41. Visual inspection of the FliP model demonstrated that the relative orientations of pairs of helices were consistent with the connectivity of the map but significant hinging of the helices was required (Video S1). Rigid-body movements of helical segments were carried out in Coot followed by rebuilding of local segments guided by bulky sidechain density. Five copies of the rebuilt FliP were then placed. A FliR model from the Gremlin database was then fit in the 6th unique position of the map using the same procedure. Finally, a Gremlin FliQ model was docked into density, using co-evolutionary contacts between the N-terminal quarter of FliQ and FliR to guide orientation, followed by rigid body hinging of the 2 helices. Four copies of the rebuilt FliQ were then docked into density. The overall P5Q4R1 model was then subjected to real-space refinement in Phenix using stereochemical and secondary structure restraints. The final model is consistent with 88% (60 out of 68) of co-evolutionary contacts between different chains, excluding those used to place Q. It is also of note that, despite the moderate resolution of the map precluding certainty of rotamer choice, all the conserved, charged residues in P, Q and R are well placed to form salt-bridges with one another suggesting a structural rationale for earlier studies demonstrating that mutation of residues within this group uniformly results in loss of function 42 43.
Sequence analysis
Coevolutionary contacts between FliPQR and FliE or FlhB were determined by submitting the relevant sequences to the Gremlin server 44. The following parameters were used: Δgene was set to infinity and a multiple sequence alignment was generated using the Jackhmmer algorithm with an E-value threshold of 10-20. For contacts between different chains of FliP or FliQ, precalculated alignments from the Gremlin database were used 41,44. Contacts with a Gremlin score greater than 0.9 and a scaled sore greater than 0.5 were regarded as significant.
Secretion assay
Analysis of type III-dependent secretion of proteins into the culture medium (Fig. S10) was carried out as described previously 45.
In vivo photocrosslinking
In vivo photocrosslinking was performed as described previously 46 except that S. Typhimurium strains MIB3147 (SpaPFLAG, ΔprgK) and MIB3148 (SpaRFLAG, ΔprgK) were used, complemented with expression of the prgK wild type or E138pBpa mutant, respectively, from the low copy number pTACO10 plasmid 47. Preparation of crude bacterial membranes and subsequent analysis of the crosslinking by SDS PAGE and Western blotting were performed as described previously 46. Antibodies were those previously described and validated in this system 46.
Chemical crosslinking of purified needle complexes
Purification of needle complexes from MIB3118 (InvCK165E) was performed as described previously 46. This InvC mutant disrupts the ATPase activity in order to make the system secretion incompetent without disrupting other components and it mimics the approach taken to produce the 6.3 Å S. Typhimurium basal body cryo-EM map 48. After collection from the CsCl-gradient, needle complexes were subjected to size exclusion chromatography (Superdex 200 10/300 GL, GE Healthcare) and collected from the void volume. Needle complexes were concentrated to 1 mg/ml using Amicon Ultra 100 k cutoff spin concentrators (Merck Millipore) and subsequently subjected to chemical cross-linking. Disuccinimidyl suberate (DSS; 50 mM; Thermo Fisher Scientific) in DMSO was added to a final concentration of 1 mM to the complex solution of 20 µg/ml. The reaction mixture was incubated with agitation at 4 °C for 3 h. The reaction was quenched by addition of 1 M Tris-HCl, pH 7.5, to a final concentration of 50 mM for 15 min at room temperature. The crosslinked needle complexes were analyzed by SDS PAGE and Coomassie staining. For mass spectrometry, the crosslinked needle complex was run 1 cm into a 4-20% SDS PAGE gel, stained by Coomassie, and all stained material in the lane was cut out for further analysis.
Crosslink sample preparation and LC-MS/MS data acquisition
Samples were in-gel digested using trypsin and chymotrypsin. The resultant peptides were analysed on an EASY-nLC 1200 (Thermo Fisher Scientific) coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific). Peptides were loaded onto a 75 μm (ID), 15 cm column packed in-house with reversed-phase ReproSil-Pur 120 C18-AQ 1.9 μm resin (Dr. Maisch GmbH). Peptides were then eluted using a 43 min linear gradient of solvent B (80% ACN in 0.1% formic acid) from 10 % - 33 %. Full-scans were recorded between 300-1650 Thompson at a resolution of 60,000 with an AGC target of 1e6. The 7 most intense ions from each full scan were selected for fragmentation (MS/MS) by HCD using an NCE of 27 and an AGC target of 1e5 in 110 ms at a resolution of 60,000.
XL-MS data analysis
Crosslinks were identified using pLink (http://pfind.ict.ac.cn/software/pLink) 49. Mass error tolerance for precursors was set to 4 ppm for 5 isotopes including the monoisotopic mass. Minimum peptide length was set to 6 amino acids with oxidation (Met) and carbamidomethylation (Cys) set as variable and fixed modifications respectively. Both trypsin and chymotrypsin cleavage sites were defined for the digestion enzyme and 4 missed cleavages were allowed. Based on the measured precursor masses, heavy and light crosslink pairs were identified after searching for and identifying both heavy and light DSS crosslinked peptides (mass shift: 12.075 Da). The interaction maps between complex proteins were generated via xiNET-Crosslink Viewer (http://crosslinkviewer.org). Digestion efficiency and raw mass accuracy were determined in a dedicated processing and detection of unmodified peptides by MaxQuant software 50 using default settings.
Data Availability Statement
Protein Coordinates and EM Volume are available from the Protein Data Bank (PDB 6f2e) and EM Data Bank (EMDB-4173) respectively. Source data for Figure 1, 3d, 4b and 4c are available with the paper online. All other data available on request.
Supplementary Material
Acknowledgments
We thank E. Johnson & A. Costin of the Central Oxford Structural Microscopy and Imaging Centre and all staff of the ARC Centre of Advanced Imaging, Monash University, for assistance with data collection. H. Elmlund (Monash) is thanked for assistance with data collection, processing strategies and access to SIMPLE code ahead of release. For collection of preliminary data we acknowledge Diamond for access and support of the Cryo-EM facilities at the UK national electron bio-imaging centre (eBIC), proposal EM15354, funded by the Wellcome Trust, MRC and BBSRC. We thank T. Marlovits, Centre for Structural Systems Biology, Hamburg, for providing PrgK (SctJ) antisera. The Central Oxford Structural Microscopy and Imaging Centre is supported by the Wellcome Trust (201536), The EPA Cephalosporin Turst, the Wolfson Foundation and a Royal Society/Wolfson Foundation Laboratory Refurbishment Grant (WL160052). Work performed in the lab of S. M. Lea was supported by a Wellcome Trust Investigator Award (100298) and an MRC programme grant (M011984). Work performed in the lab of C. V. Robinson is funded by a Wellcome Trust Investigator Award (104633), an ERC Advanced Grant ENABLE (641317) and an MRC programme grant (N020413). J. Gault is a Junior Research Fellow of The Queen’s College, Oxford. Work performed in the laboratory of S. Wagner was supported by the Alexander von Humboldt Foundation in the framework of the Sofja Kovalevskaja Award endowed by the Federal Ministry of Education and Research (BMBF) and by the Deutsche Forschungsgemeinschaft (DFG) as part of the Collaborative Research Center (SFB) 766 Bacterial cell envelope, project B14.
Footnotes
Author Contributions: All authors contributed to editing of manuscript and figures.
L.K.: Performed experiments. Strain and plasmid construction, complex purification, native mass spectrometry, cryoEM grid optimisation, cryoEM data analysis and model building and analysis.
P.A.: Performed experiments. Strain and plasmid construction, complex purification initial cryoEM grid optimisation and cryoEM data analysis.
S.J.: Performed experiments. Designed and supervised experiments. Characteristion of protein complexes. CryoEM data analysis, structure determination and analysis. Wrote first draft of manuscript and figures with SML.
J.G.: Performed and supervised native mass spectrometry experiments.
J.D.: Performed experiments. CryoEM grid optimisation and data collection
J.C.: Performed experiments. Optimised computational analysis of cryo-EM data.
T.D.: Performed experiments. Strain and plasmid construction, in vivo photocrosslinking
M.T.M.: Performed experiments. NC purification, chemical crosslinking
T.G.: Performed experiments. MS of DSS crosslinked samples. Analysed data.
B.M.: Analysed DSS MS data. Wrote paper: DSS MS methods section.
S.W.: Analysed data: in vivo photocrosslinking, chemical crosslinking, DSS MS. Wrote first draft of photo- and chemical-crosslinking sections.
C.V.R.: Supervised design and implementation of native mass spectrometry experiments
S.M.L.: Performed and supervised experiments. CryoEM grid optimisation and data collection, data and structure analysis. Wrote first draft of paper with SJ.
References
- 1.Abrusci P, McDowell MA, Lea SM, Johnson S. Building a secreting nanomachine: a structural overview of the T3SS. Curr Opin Struct Biol. 2014;25:111–117. doi: 10.1016/j.sbi.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erhardt M, Namba K, Hughes KT. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol. 2010;2:a000299. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng W, et al. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. 2017;15:323–337. doi: 10.1038/nrmicro.2017.20. [DOI] [PubMed] [Google Scholar]
- 5.Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 6.Fabiani FD, et al. A flagellum-specific chaperone facilitates assembly of the core type III export apparatus of the bacterial flagellum. PLoS Biol. 2017;15:e2002267. doi: 10.1371/journal.pbio.2002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukumura T, et al. Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex. PLoS Biol. 2017;15:e2002281. doi: 10.1371/journal.pbio.2002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner S, et al. Organization and coordinated assembly of the type III secretion export apparatus. Proc Natl Acad Sci U S A. 2010;107:17745–17750. doi: 10.1073/pnas.1008053107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietsche T, et al. Structural and Functional Characterization of the Bacterial Type III Secretion Export Apparatus. PLoS pathogens. 2016;12:e1006071. doi: 10.1371/journal.ppat.1006071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zilkenat S, et al. Determination of the Stoichiometry of the Complete Bacterial Type III Secretion Needle Complex Using a Combined Quantitative Proteomic Approach. Mol Cell Proteomics. 2016;15:1598–1609. doi: 10.1074/mcp.M115.056598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 12.Fujii T, et al. Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook. Nat Commun. 2017;8 doi: 10.1038/ncomms14276. 14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsunami H, Barker CS, Yoon YH, Wolf M, Samatey FA. Complete structure of the bacterial flagellar hook reveals extensive set of stabilizing interactions. Nat Commun. 2016;7 doi: 10.1038/ncomms13425. 13425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cordes FS, et al. Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem. 2003;278:17103–17107. doi: 10.1074/jbc.M300091200. [DOI] [PubMed] [Google Scholar]
- 15.Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife. 2014;3:e02030. doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erhardt M, et al. Mechanism of type-III protein secretion: Regulation of FlhA conformation by a functionally critical charged-residue cluster. Molecular microbiology. 2017;104:234–249. doi: 10.1111/mmi.13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward E, et al. Type-III Secretion Pore Formed by Flagellar Protein FliP. Molecular microbiology. 2017 doi: 10.1111/mmi.13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Worrall LJ, et al. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature. 2016 doi: 10.1038/nature20576. [DOI] [PubMed] [Google Scholar]
- 19.Zhao X, et al. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2013;110:14390–14395. doi: 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu B, Lara-Tejero M, Kong Q, Galan JE, Liu J. In Situ Molecular Architecture of the Salmonella Type III Secretion Machine. Cell. 2017;168:1065–1074 e1010. doi: 10.1016/j.cell.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schraidt O, Marlovits TC. Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science. 2011;331:1192–1195. doi: 10.1126/science.1199358. [DOI] [PubMed] [Google Scholar]
- 22.Minamino T, Yamaguchi S, Macnab RM. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J Bacteriol. 2000;182:3029–3036. doi: 10.1128/jb.182.11.3029-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara N, Namba K, Minamino T. Genetic characterization of conserved charged residues in the bacterial flagellar type III export protein FlhA. PloS one. 2011;6:e22417. doi: 10.1371/journal.pone.0022417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radics J, Konigsmaier L, Marlovits TC. Structure of a pathogenic type 3 secretion system in action. Nature structural & molecular biology. 2014;21:82–87. doi: 10.1038/nsmb.2722. [DOI] [PubMed] [Google Scholar]
- 25.Abrusci P, et al. Architecture of the major component of the type III secretion system export apparatus. Nature structural & molecular biology. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, et al. Structural diversity of bacterial flagellar motors. EMBO J. 2011;30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minamino T, Morimoto YV, Hara N, Aldridge PD, Namba K. The Bacterial Flagellar Type III Export Gate Complex Is a Dual Fuel Engine That Can Use Both H+ and Na+ for Flagellar Protein Export. PLoS pathogens. 2016;12:e1005495. doi: 10.1371/journal.ppat.1005495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor WR, Matthews-Palmer TR, Beeby M. Molecular Models for the Core Components of the Flagellar Type-III Secretion Complex. PloS one. 2016;11:e0164047. doi: 10.1371/journal.pone.0164047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashkenazy H, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 31.Baumgarten T, et al. Isolation and characterization of the E. coli membrane protein production strain Mutant56(DE3) Sci Rep. 2017;7 doi: 10.1038/srep45089. 45089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gault J, et al. High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat Methods. 2016;13:333–336. doi: 10.1038/nmeth.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marty MT, Hoi KK, Gault J, Robinson CV. Probing the Lipid Annular Belt by Gas-Phase Dissociation of Membrane Proteins in Nanodiscs. Angew Chem Int Ed Engl. 2016;55:550–554. doi: 10.1002/anie.201508289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reboul CF, Eager M, Elmlund D, Elmlund H. Single-particle cryo-EM-Improved ab initio 3D reconstruction with SIMPLE/PRIME. Protein Sci. 2017 doi: 10.1002/pro.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urnavicius L, et al. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–1446. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5 doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swint-Kruse L, Brown CS. Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics. 2005;21:3327–3328. doi: 10.1093/bioinformatics/bti511. [DOI] [PubMed] [Google Scholar]
- 39.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukumura T, et al. Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex. PLoS Biol. 2017;15:e2002281. doi: 10.1371/journal.pbio.2002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ovchinnikov KV, et al. The Leaderless Bacteriocin Enterocin K1 Is Highly Potent against Enterococcus faecium: A Study on Structure, Target Spectrum and Receptor. Front Microbiol. 2017;8:774. doi: 10.3389/fmicb.2017.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabiani FD, et al. A flagellum-specific chaperone facilitates assembly of the core type III export apparatus of the bacterial flagellum. PLoS Biol. 2017;15:e2002267. doi: 10.1371/journal.pbio.2002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward E, et al. Type-III Secretion Pore Formed by Flagellar Protein FliP. Molecular microbiology. 2017 doi: 10.1111/mmi.13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife. 2014;3:e02030. doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monjaras Feria JV, Lefebre MD, Stierhof YD, Galan JE, Wagner S. Role of autocleavage in the function of a type III secretion specificity switch protein in Salmonella enterica serovar Typhimurium. MBio. 2015;6:e01459–01415. doi: 10.1128/mBio.01459-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dietsche T, et al. Structural and Functional Characterization of the Bacterial Type III Secretion Export Apparatus. PLoS pathogens. 2016;12:e1006071. doi: 10.1371/journal.ppat.1006071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner S, et al. Organization and coordinated assembly of the type III secretion export apparatus. Proc Natl Acad Sci U S A. 2010;107:17745–17750. doi: 10.1073/pnas.1008053107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Worrall LJ, et al. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature. 2016 doi: 10.1038/nature20576. [DOI] [PubMed] [Google Scholar]
- 49.Yang B, et al. Identification of cross-linked peptides from complex samples. Nat Methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 50.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protein Coordinates and EM Volume are available from the Protein Data Bank (PDB 6f2e) and EM Data Bank (EMDB-4173) respectively. Source data for Figure 1, 3d, 4b and 4c are available with the paper online. All other data available on request.