Abstract
Drug-induced liver injury (DILI) is complex in mechanism. Different drugs could undergo different mechanisms but result in the same DILI type while the same drug could lead to different DILI types via different mechanisms. Therefore, predicting a drug’s potential for DILI should take its underlying mechanisms into consideration. To achieve that, we constructed a novel approach by incorporating drug’s Mode of Action (MOA) into Quantitative Structure-Activity Relationship (QSAR) modeling. This MOA-DILI approach was examined using a dataset of 333 drugs. The drugs were first grouped according to their MOA profiles (positive or negative in each MOA) based on the Tox21 qHTS assays. QSAR models for individual MOA assays were developed and subsequently combined to obtain the MOA-DILI model. A hold-out testing strategy (222 drugs for training and 111 drugs as a test set) was employed, which yielded the predictive accuracy of 0.711. For comparison, the MOA-DILI model was directly compared with the standard QSAR approach using the same hold-out strategy, and the QSAR model yielded an accuracy of 0.662. To minimize the random chance in splitting training/test sets, the hold-out testing process was repeated 1000 times, and the observed difference in prediction accuracy between MOA-DILI and QSARs was statistically significant (P-value<0.0001). Out of 17 MOAs used, 4 assays (i.e., antioxidant response elements, PPAR-gamma, estrogen receptor, and thyroid receptor assays) contributed most to the improved prediction of the MOA-DILI model over QSARs. In conclusion, the MOA-DILI approach has the potential to significantly improve predictive outcomes and to reveal complex relationships between MOAs and DILI, all of which would be helpful in developing DILI predictive models in drug screening and for risk assessment of industrial chemicals.
Graphical Abstract
Introduction
Drug-induced liver injury (DILI) is one of the major causes for drugs that failed in clinical trials and were withdrawn from the market1, 2. In order to identify drugs that are “bad actors” before they reach clinical trials, predictive models using in vitro data3, 4, in silico approaches5, 6 and toxicogenomics7 have been actively pursued. For example, Dawson et al. developed an in vitro DILI predictive model based on the inhibition of the bile salt export pump (BSEP) and demonstrated that BSEP inhibition was highly correlated with cholesteric DILI potentials1. A study by Usui et al. indicated that the combination of covalent binding levels of a drug to DNA and daily dose can be used to estimate the risk for DILI8. Howell et al. developed a mathematical predictive model called DILIsym based on the in vitro analysis of Methapyrilene9.
We are conducting the Liver Toxicity Knowledge Base (LTKB) project at the Food and Drug Administration (FDA) to utilize the data from preclinical studies for the prediction of DILI in humans10. The project has developed several predictive models11-14. For example, we developed a “rule-of-two” model based on the observations that oral medications of high daily dose and lipophilicity were associated with significant risk of DILI12; the model identified drugs that caused severe DILI but produced a number of false negatives. We also constructed a computational model that predicts human hepatotoxicity based on patterns of gene expression observed in rats following short-term in vivo exposures11. More recently, we developed a DILIScore model that is able to quantitatively assess a drug’s severity for DILI based on three sets of information (i.e., daily dose, lipophilicity, and the presence of reactive metabolites14.)
Quantitative Structure-Activity Relationship (QSAR) is another approach investigated in LTKB. For example, we developed the DILI Prediction System (DILIps)5, which took the consensus of 13 independent QSAR models corresponding to 13 hepatotoxicity-specific adverse events observed in clinical trials and/or post-marketing surveillances. We also developed a QSAR model using the in-house Decision Forest (DF) machine learning method15 in conjunction with the Mold2 descriptors16 with a large training set, which resulted in around 65% prediction accuracy on several large external validation sets7, 17. One of the key observations from these exercises was that a model for DILI prediction does not perform equally well across all drug classes13. We concluded that a “one-size-fits-all” approach for DILI prediction by treating all the drugs as a single group could fall short in predictivity due to the diverse mechanisms involved in DILI and incorporating the different mechanisms of DILI into a predictive model could improve prediction accuracy.
Grouping drugs by their mode of action (MOA) profiles for safety assessment as a general approach has been reported18 including the study of hepatotoxicity9, 19. For example, there are several methods that incorporated drug mechanisms into QSARs in predicting DILI5, 20-22, with two dominating approaches: (1) directly combining chemical descriptors and MOA features in modeling, or (2) using MOA as an endpoint for prediction, i.e., QSAR models are constructed to predict MOAs instead of DILI itself. In addition, some studies developed models for drugs with shared MOAs but different DILI outcomes. However, an approach involving sub-grouping of drugs by their MOAs in combination with QSAR modeling has not been investigated for DILI prediction.
The MOAs profile of a drug can be obtained using mechanism-specific in vitro assays, such as those implemented in Tox2123 and ToxCast24 programs. We hypothesized that an enhanced DILI predictive model could be achieved by considering a drug’s DILI mechanisms in the modeling process. For example, DILI can be initiated by reactive metabolites, which interact with large molecules (e.g. proteins, lipids, mitochondrion, and Nuclei Acids) and result in protein dysfunction, lipid peroxidation, mitochondria dysfunction, DNA damage, and oxidative stress25. Some Tox21 assays exhibit relevance to hepatotoxicity such as the antioxidant response element beta-lactamase reporter gene assay26.
Drugs with different mechanisms may exhibit different DILI properties and thus should be predicted by mechanism-specific models. By incorporating MOA profiles, drugs could be more accurately categorized into subgroups specific for each DILI predictive model. Since a single drug might have several MOAs, the final prediction would be a consensus of several MOA-specific models. In this study, we applied MOAs profiled by the quantitative high-throughput (qHTS) assays in the Tox21 program27 followed by QSAR modeling. In order to construct a model that would effectively utilize both the bioassay results and the QSAR information, we integrated MOAs and QSARs into DILI prediction (called MOA-DILI). The approach is different from the conventional integrated approaches which often combined different types of features at the same level, for example, using a pooled feature-set consisting of features from different sources (See Discussion).
Specifically, the drugs were first grouped according to their MOA profiles (positive or negative in each MOA) based on the Tox21 qHTS assays. QSAR models for individual MOA groups were developed separately and all the QSAR models were then combined for a consensus prediction by voting. We found out that the MOA-DILI method has the potential to significantly improve predictive outcome with insight gained into the complex relationships between MOAs and DILI. Out of 17 MOAs used, four assays (i.e., antioxidant response elements, PPAR-gamma, estrogen receptor and thyroid receptor assays) contributed most to the improved prediction of the MOA-DILI model over QSARs. Thus, MOA-DILI could be helpful in drug screening for DILI detection in the early stage of drug development and for risk assessment of industrial chemicals
Material and methods
Dataset
The chemical structural information and DILI severity annotations for the drugs used to construct and test the MOA-DILI predictive model were obtained from the LTKB database10, 28. The dataset consists of 333 drugs with 155 drugs being “most-DILI-concern” and 178 drugs annotated as “no-DILI-concern”. “less-DILI-concern” drugs were not used in this study due to their ambiguous attributes. Of note, the objective of this study is to determine whether the proposed approach is going to improve the prediction by comparing to the standard QSARs. We reported a QSAR model for DILI prediction17, which used only “most-DILI concern” and “no-DILI concern” drugs for model evaluation. This QSAR approach was used side-by-side to compare the proposed methodology in this study. Therefore, only most- and no-DILI concern drugs were applied.
Bioassay activity data from toxicity relevant assays, such as ER (estrogen receptor), AR (androgen receptor), mitochondrial toxicity, p53, PPAR gamma, etc., was curated from the Tox21 program (https://tripod.nih.gov/tox21/) (Table 1). In particular, assays with less than 20 (arbitrary cut-off) active drugs were excluded in this study due to the fact that a small number of samples would result in less reliable QSAR models. As a result, 17 assays remained in this study. For each of the 17 assays used, a drug can be “active”, “inactive” or “unknown”. In general, an “active” drug means the drug showed as either active antagonist or active agonist of selected assay, and inactive drug means this drug didn’t show any active response of this assay (different assays might have different sensitivity and measurement, and we directly used this measurement from the Tox21 results (https://tripod.nih.gov/tox21/index)). “Unknown” is the ambiguous read-out from the assay that either contradicted each other from multiple experiments or not tested. If a drug is labeled as “unknown”, it would not be used in this study. A detailed list of drug information used in this study and their Tox21 bioassay activities is provided in Table S1.
Table 1.
Performance of QSARMOA model and assay alone predictions
| MOA ID | Assay Endpoint | Tox21 Assay Name | QSARMOA Model |
|---|---|---|---|
| MOA-1 | aryl hydrocarbon receptor | AHR | 0.663 (0.033) |
| MOA-2 | androgen receptor | AR-bla-agonist | 0.646 (0.032) |
| MOA-3 | androgen receptor | AR-bla-antagonist | 0.657 (0.034) |
| MOA-4 | antioxidant response element | ARE-bla | 0.649 (0.039) |
| MOA-5 | AR (MDA cell line) | AR-mda-kb2-luc-agonist | 0.653 (0.032) |
| MOA-6 | AR (MDA cell line) | AR-mda-kb2-luc-antagonist | 0.671 (0.032) |
| MOA-7 | aromatase inhibitors | aromatase | 0.651 (0.035) |
| MOA-8 | estrogen receptor alpha | ER-bla-antagonist | 0.656 (0.035) |
| MOA-9 | ER-alpha (BG1 cell line) | ER-luc-bg1-4e2-agonist | 0.633 (0.036) |
| MOA-10 | ER-alpha (BG1 cell line) | ER-luc-bg1-4e2-antagonist | 0.672 (0.032) |
| MOA-11 | Thyroid receptor | gh3-tre-antagonist | 0.671 (0.037) |
| MOA-12 | glucocorticoid receptor | GR-hela-bla-antagonist | 0.652 (0.034) |
| MOA-13 | heat shock response | HSE-bla | 0.647 (0.035) |
| MOA-14 | mitochondrial toxicity | Mitotox | 0.634 (0.036) |
| MOA-15 | p53 | p53 | 0.658 (0.033) |
| MOA-16 | peroxisome proliferator-activated receptor gamma | PPARG-bla-agonist | 0.677 (0.033) |
| MOA-17 | peroxisome proliferator-activated receptor gamma | PPARG-bla-antagonist | 0.636 (0.038) |
Quantitative Structure-Activity Relationships (QSARs)
One of the key strategies in this study was to conduct a direct comparison to the QSAR approach that was reported previously17. Thus, the same QSAR approach was applied that was constructed using Mold2 molecular descriptors16 and the Decision Forest algorithm15. Mold2 is an in-house chemical structure tool that rapidly calculates a large and diverse set of 2D molecular descriptors. A total of 777 descriptors were used in this study. Detailed descriptions of these 2D descriptors can be found in the Mold2 documentation, which is available at http://www.fda.gov/ScienceResearch/BioinformaticsTools/Mold2/.
The Decision Forest algorithm was used in this study to build all QSAR models, which is a classification algorithm that combines multiple decision tree models. In order to reduce over-fitting, each decision tree consists of a unique set of molecular descriptors15. A significant advantage of using the Decision Forest is the integration of feature selection in the modeling processes. In this study, the parameters used in the decision forest were set as follows: min # of tree=5, max # of tree=20, min reduction=0.01 and min drugs in one node=5. All constant features were filtered before entering the decision forest process. Since the number of samples in both positive (Most-DILI concern) and negative (No-DILI-concern) categories are quite balanced, the performance of QSAR model was mostly assessed by accuracy (Eq. 1), where TP is true positive, TN is true negative, FP is false positive, and FN is false negative. Additionally, Matthew’s Correlation Coefficient (MCC) and F1 score were also provided based on the measurement of Eq. 2 and 3, respectively.
| Eq. 1 |
| Eq. 2 |
| Eq. 3 |
Model Validation
Two validation procedures were used in this study, hold-out and cross-validation. Specifically, the 5-fold cross-validation (CV) procedure was used in QSARs, where drugs in the training set were randomly divided into 5 sets; 4 sets of drugs were used to develop the QSAR model which was then assessed by predicting the 5th set of drugs. This process was repeated 5 times where each set of drugs was left out once and only once. For the hold-out approach, the 333 drugs were randomly split to 2/3 (222 drugs) and 1/3 (111 drugs). The former was used to develop a model while the latter was used to validate the model. We repeated the hold-out process 1000 times to generate 1000 pairs of training/test sets. The average results from 1000 repetitions minimize the degree of chance correlation.
For comparison, label permutation test was applied in this study. In the label permutation test, the DILI severity annotations of all drugs were shuffled (keeping the same ratio of most-DILI-concern and no-DILI-concern) and then we applied the same modeling approach to the shuffled dataset.
Results
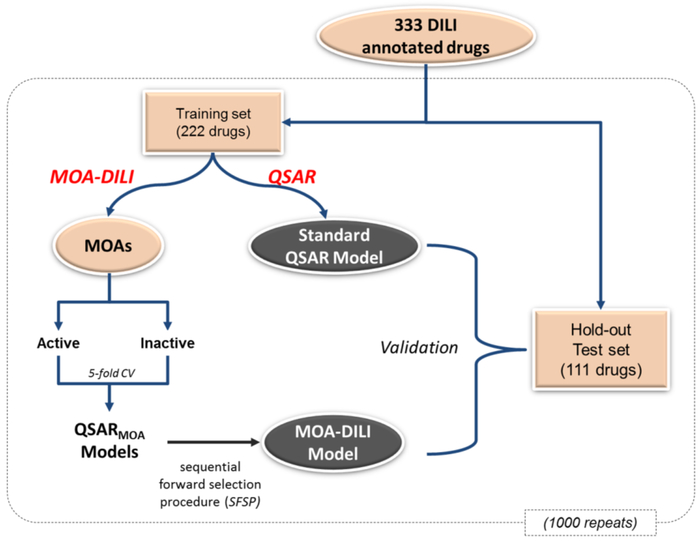
We have developed a modeling approach to predict potential DILI for a drug used 17 drug MOAs from the Tox21 qHTS assays. The models were based a set of 333 drugs with 155 drugs being “most-DILI-concern” and 178 drugs annotated as “no-DILI-concern”. The MOA-DILI modeling process is depicted in Figure 1. The 333 drugs were randomly divided into a training set (222 drugs) for model development and a test set (111 drugs) for hold-out testing. For model training, Tox21 assays were used to group training set compounds based on assay activity, and QSAR models were constructed for each assay respectively. For example, for the ARE-bla assay there were 53 number of active compounds with DILI calls (27 most-DILI-concern and 26 no-DILI-concern). A model was built that differentiated ARE-active compounds with and without DILI properties. In other words our models identify a subcomponent of ARE activation SAR features that are associated with DILI. For model testing, test compounds were first labeled by their activities in Tox21 assay and then predicted by relevant QSAR models. We also compared MOA-DILI with the standard QSAR procedure. All the results presented are the average of 1000 repetitions (i.e., 1000 pairs of training/test sets) to minimize the potential bias in splitting training/test sets.
Figure 1.
Overview of MOA-DILI modeling process. 333 drugs were randomly divided into a training set (222 drugs) for model development and a test set (111 drugs) for hold-out testing, and this process was repeated 1000 times to generate 1000 pairs of training/test sets. For each of 17 MOAs from Tox21 assays, the training set of drugs was split into two groups, one active in the assay and the other inactive in the assay. The 5-fold cross-validation was performed on each group and the results were combined to assess the prediction accuracy of each QSARMOA model. Sequential forward selection procedure was applied to assess the performance of the combinations of QSARMOA models and the best combination was namely the MOA-DILI model. The MOA-DILI was then evaluated using the hold-out test set and its performance was compared with the standard QSAR model using the same training set.
For each of the 17 MOAs from the Tox21 assays, we first separated drugs into two groups, one is active in the assay and the other is inactive in the assay. Next, the 5-fold cross validation (5-fold CV) procedure for each group was independently conducted using the standard QSAR procedure with Decision Forest and Mold2 descriptors (Materials and Methods). The CV results from both groups were then combined to obtain the prediction performance of the QSAR model for the assay, called QSARMOA model. The averaged prediction performance of QSARMOA models is summarized in Table 1. The averaged prediction accuracy of QSARMOA models ranges from 0.633 to 0.677, where pparg-bla-agonist (MOA-16) showed the best performance over all MOAs. All QSARMOA models showed significantly better performance than the sample prevalence (0.535, p<0.0001).
Next, we investigated whether the combination of QSARMOA models from 17 MOAs would be able to achieve a better performance. In this investigation, a sequential forward selection procedure was applied to generate various combinations of the QSARMOA models, where QSARMOA were sequentially added one at a time by their performance, and the final prediction of the combined models was obtained based on voting. We found that the best combination, called MOA-DILI model, outperformed all the QSARMOA models (Table 2). The MOA-DILI model also outperformed the standard QSAR model; the averaged accuracy of MOA-DILI models by 5-fold CV (0.757) was significantly (P-value <0.0001) higher than the averaged accuracy of QSAR models which was 0.658. For the hold-out assessment using the test set, the averaged predicting accuracy of MOA-DILI models was 0.695. Although with a considerable drop, this predictive performance was still significantly better than the corresponding QSAR models, which was only 0.662 in average (P-value <0.0001). Similarly, the label permutation test yielded the prediction accuracy of 0.582 which was significantly lower than MOA-DILI models (P-value<0.0001), indicating that DILI prediction were enhanced by using the MOA-DILI model. Similar performance was obtained by using MCC and F1 score, where MOA-DILI model showed significantly better performance than these from QSAR models.
Table 2.
Overall Performance of MOA-DILI model in training and test set.
| ACC | MCC | F1score | ||
|---|---|---|---|---|
| MOA-DILI model | Training Set | 0.757 (0.020) | 0.505 (0.045) | 0.717 (0.032) |
| Test Set | 0.695 (0.043) | 0.385 (0.087) | 0.640 (0.058) | |
| MOA-DILI model with Top 4 MOAs | Training Set | 0.703 (0.027) | 0.397 (0.056) | 0.649 (0.042) |
| Test Set | 0.711 (0.040) | 0.416 (0.080) | 0.659 (0.052) | |
| Standard QSAR model | Training Set | 0.658 (0.031) | 0.310 (0.062) | 0.626 (0.040) |
| Test Set | 0.662 (0.041) | 0.322 (0.082) | 0.627 (0.049) | |
| Label Permutated model | Training Set | 0.591 (0.039) | 0.200 (0.080) | 0.609 (0.046) |
| Test Set | 0.582 (0.042) | 0.182 (0.084) | 0.604 (0.046) |
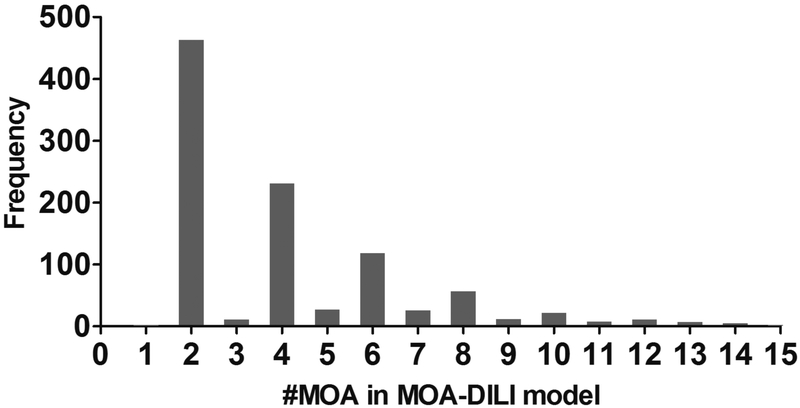
The 1000 repetitions resulted in 1000 different MOA-DILI models, which allow assessment of the frequency of each MOA selected by MOA-DILI models. As shown in Figure 2, most MOA-DILI models only used 2 or 4 MOAs; the average number of MOAs was around 3.56. Four MOAs (MOA-4, 10, 11, and 16), labeled as ARE-bla (antioxidant response element), ER-luc-bg1-4e2-antagonist (ER-alpha, BG1 cell line), gh3-tre-antagonist (thyroid receptor) and PPARG-bla-agonist (peroxisome proliferator-activated receptor gamma), were used by more than 30% of MOA-DILI models, much more often than other MOAs (Supplementary Table S2). Consequently, we constructed the MOA-DILI model using only these 4 MOAs with the same process outlined in Figure 1. As summarized in Table 2, this final model yielded prediction accuracies of 0.697 in 5-fold CV and 0.711 in hold-out testing, which was comparable with other MOA-DILI models and much better than the QSAR model (P-value <0.0001). The detailed prediction results of the top 4 assay specific models are provided in Supplementary Table S3.
Figure 2.
Distribution of the number of QSARMOA models used in 1000 MOA-DILI models. Most MOA-DILI models used an even number of MOAs.
Discussion
In this study, we introduced the MOA-DILI modeling algorithm to improve DILI prediction, by integrating the MOA information into QSARs. Using a drug’s chemical structure to predict DILI has been reported with limited success due to the complex mechanisms involved in DILI. Therefore, considering MOA profiles of a drug in addition to its chemical structural information could improve DILI prediction. There are several similar approaches reported along this line of thinking. We conducted a PubMed search, using “QSAR” and “Liver toxicity”, which returns 173 hits (by January, 2017). The general approaches of integrating drug mechanisms and MOAs into toxicity prediction with QSARs falls into two categories. One is feature combination, which feeds different types of features (e.g., chemical descriptors and in vitro assay data) into the modeling process as independent variables in an equal setting21, 29. Another approach is consensus modeling, which takes the majority votes of several models constructed with different features respectively5, 20, 22. Different from these approaches, the MOA-DILI algorithm uses the MOA features to divide drugs into different groups and, for each group, QSAR models were constructed and subsequently combined.
A model’s performance is largely dependent on three factors, the dataset size, the active/inactive distribution, and how the actives/inactives are determined which relates to the noise levels in the assays that measure the drug activities. Thus, the absolute accuracy for a predictive model has to be evaluated in the context of these three factors. Currently, QSARs have been well adopted in drug screening with no wet-lab experiments involved. Our approach does involve wet-lab data (i.e., Tox21 data). Thus, to demonstrate the value of our proposed methodology, we conducted a direct comparison between our approach and standard QSARs. The MOA-DILI model exhibited the prediction accuracy of 0.757 in 5-fold cross validation and 0.695 in hold-out testing. Compared to our previously established QSAR models using the same approach, the MOA-DILI modeling showed improved performance in DILI prediction.
It is worthwhile to mention that the criteria applied to label drugs that induce liver injury are a significant consideration when developing the prediction model. In fact, to determine whether a drug truly causes liver injury or not is difficult since the occurrence of DILI is usually rare or idiosyncratic. There are three factors that need to be considered for DILI classification28, 30: 1) Causality – is the observed liver injury from this drug? 2) incidence – how many DILI cases resulted from this drug are sufficient to incriminate it as DILI? and 3) severity – how severe is severe enough to call a drug as DILI (e.g., the clinical impact for the patient ranging from transient liver enzyme elevations to liver failure requiring transplantation or even death)? These three factors have to be considered together in order to identify the potential DILI risk for a drug. The drug labeling documents provide a balanced assessment by considering all these three factors in assessing the likelihood of a drug causing DILI. This DILI classification label was used in this study.
In this study, we applied the Tox21 assays as drug MOAs. Tox21 assays are not specifically designed for DILI prediction. Other assays such as Oxidative Stress, Glutathione Depletion, Formation of Reactive Metabolites, and Mitochondrial Dysfunction are more important and relevant to DILI. We decided to use the Tox21 data in this proof-of-concept study because it does provide the large dataset which is required to demonstrate the utility of the proposed method. At the time when this study was conducted, Tox21 provides 17 assays mainly related to nuclear receptors and stress response pathway assays, which were tested in this investigation. We found that four assays, labeled as ARE-bla (antioxidant response element, MOA-4 assay in Table 1), ER-luc-bg1-4e2-antagonist (ER-alpha, BG1 cell line, MOA-9 assay in Table 1), gh3-tre-antagonist (thyroid receptor, MOA-11 assay in Table 1), and PPARG-bla-agonist (peroxisome proliferator-activated receptor gamma, MOA-16 assay in Table 1), contributed the most to the DILI prediction. There are a number of reports indicating the relevance of these assays to liver disease and DILI22, 31-34.
For example, the PPAR family is linked with liver disease32, 35, 36. Troglitazone, a typical PPAR-gamma ligand drug for type II diabetes, is withdrawn due to its severe hepatotoxicity37, 38. We noticed that, out of 21 PPAR-gamma antagonists, 18 of them are most DILI-concern drugs, indicating PPAR-gamma ligands would have a high correlation with DILI. The ER-alpha antagonist assay (MOA-9) measures estrogenic activity of a compound. Tamoxifen, a well-known ER-alpha antagonist for the treatment of breast cancer has been associated with liver injury including fatty liver39, steatohepatitis40-43, etc. In our dataset, 24 of 38 ER-alpha antagonists are labeled as most-DILI-concern drugs. MOA-4 assay (antioxidant response element) indicates activation of the antioxidant response element (ARE) by measuring indirect glutathione depletion that is an important mechanism for DILI. ARE signaling pathway plays an important role in the amelioration of oxidative stress, of which Nrf2 (NF-E2-related factor 2, a major transcription factor) plays critical roles in protecting the cell from oxidative damage occurring because of injury and inflammation44 and acts as a primary regulator of the glutathione detoxification system. Targeting Nrf2 is a promising strategy for the prevention of toxin-induced liver damage45, and Nrf2-deficient mice are reported to be highly susceptible to APAP-induced liver injury46. It is considered that if a drug activates Nrf2 that does not cause glutathione depletion, it is mostly protective against liver damage47. Many DILI compounds such as diclofenac and carbamazepine have strong Nrf2 activation48. In our dataset, over half of (27 of 53) ARE agonists used are “most-DILI-concern” drugs, including many withdrawn drugs due to risk of hepatotoxicity. For example, Celecoxib and Cilostazol are both active in the antioxidant response assay. Celecoxib may cause cholestatic liver failure and may also be associated with severe hepatotoxicity49-51, whereas Cilostazol may attenuate cholestatic liver injury52.
In conclusion, MOA-DILI modeling could significantly improve model performance for DILI prediction by combining MOA and chemical structure information. Importantly, the MOA-DILI identifies the targets that might be involved in the mechanisms of DILI. It is an excitement to point out that the Tox21 project has generated data from many assays for a large number of compounds, the majority of which are industrial chemicals. This list will continue to grow in the future, which will inevitably become a tremendous resource for toxicity study including DILI. Specifically, Tox21 covered more than 10,000 compounds tested against 47 different types of assays. Most drugs in its inventory are annotated with DILI by LTKB26, 29. Thus, the model developed with Tox21 data has the utility for not only drug screening but also for predicting the DILI potential of industrial chemicals.
Supplementary Material
Acknowledgements
The authors thank Dr. Jun Zhang and Dr. BinSheng Gong for their helpful comments on the manuscript.
Abbreviations
- DILI
Drug-induced liver injury
- MOA
Mode of Action
- QSAR
Quantitative Structure-Activity Relationship
- DF
Decision Forest
- MCC
Matthew’s Correlation Coefficient
- CV
Cross Validation
Footnotes
Supporting information
Provide detailed Tox21 assay activity information of 333 DILI annotated drugs; basic statistics of 17 tox21 assay used in this study; and the detailed predicting performance of the top 4 assay specific models
Disclaimer: The views presented in this article do not necessarily reflect current or future opinion or policy of the US Food and Drug Administration. Any mention of commercial products is for clarification and not intended as an endorsement.
References
- 1.Dawson S; Stahl S; Paul N; Barber J; Kenna JG In Vitro Inhibition of the Bile Salt Export Pump Correlates with Risk of Cholestatic Drug-Induced Liver Injury in Humans. Drug Metab. Dispos 2012, 40, 130–138. [DOI] [PubMed] [Google Scholar]
- 2.Watkins PB Drug Safety Sciences and the Bottleneck in Drug Development. Clin. Pharmacol. Ther 2011, 89, 788–790. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J; Doshi U; Suzuki A; Chang CW; Borlak J; Li AP; Tong W Evaluation of Multiple Mechanism-Based Toxicity Endpoints in Primary Cultured Human Hepatocytes for the Identification of Drugs with Clinical Hepatotoxicity: Results from 152 marketed drugs with known liver injury profiles. Chem Biol Interact 2016, 255, 3–11. [DOI] [PubMed] [Google Scholar]
- 4.Kim MT; Huang R; Sedykh A; Wang W; Xia M; Zhu H Mechanism Profiling of Hepatotoxicity Caused by Oxidative Stress Using Antioxidant Response Element Reporter Gene Assay Models and Big Data. Environ. Health Perspect 2016, 124, 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Z; Shi Q; Ding D; Kelly R; Fang H; Tong W Translating Clinical Findings into Knowledge in Drug Safety Evaluation-Drug Induced Liver Injury Prediction System (DILIps). PLoS Comput Biol 2011, 7, e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekins S; Williams AJ; Xu JJ A Predictive Ligand-Based Bayesian Model for Human Drug-Induced Liver Injury. Drug Metab. Dispos 2010, 38, 2302–2308. [DOI] [PubMed] [Google Scholar]
- 7.Zhang M; Chen M; Tong W Is Toxicogenomics A More Reliable and Sensitive Biomarker Than Conventional Indicators from Rats to Predict Drug-Induced Liver Injury in Humans? Chem. Res. Toxicol 2011, 25, 122–129. [DOI] [PubMed] [Google Scholar]
- 8.Usui T; Mise M; Hashizume T; Yabuki M; Komuro S Evaluation of the Potential for Drug-Induced Liver Injury Based on In Vitro Covalent Binding to Human Liver Proteins. Drug Metab. Dispos 2009, 37, 2383–2392. [DOI] [PubMed] [Google Scholar]
- 9.Howell BA; Yang Y; Kumar R; Woodhead JL; Harrill AH; Clewell III HJ; Andersen ME; Siler SQ; Watkins PB In Vitro to In Vivo Extrapolation and Species Response Comparisons for Drug-Induced Liver Injury (DILI) Using Dilisym™: A Mechanistic, Mathematical Model of DILI. J. Pharmacokinet. Pharmacodyn 2012, 39, 527–541. [DOI] [PubMed] [Google Scholar]
- 10.Chen M; Zhang J; Wang Y; Liu Z; Kelly R; Zhou G; Fang H; Borlak J; Tong W The Liver Toxicity Knowledge Base: A Systems Approach to A Complex End Point. Clin. Pharmacol. Ther 2013, 93, 409–412. [DOI] [PubMed] [Google Scholar]
- 11.Chen M; Zhang M; Borlak J; Tong W A Decade of Toxicogenomic Research and Its Contribution to Toxicological Science. Toxicol Sci 2012, 130, 217–228. [DOI] [PubMed] [Google Scholar]
- 12.Chen M; Borlak J; Tong W High Lipophilicity and High Daily Dose of Oral Medications are Associated with Significant Risk for Drug-Induced Liver Injury. Hepatology 2013, 58, 388–396. [DOI] [PubMed] [Google Scholar]
- 13.Chen M; Tung C-W; Shi Q; Guo L; Shi L; Fang H; Borlak J; Tong W A Testing Strategy to Predict Risk for Drug-Induced Liver Injury in Humans Using High-Content Screen Assays and the ‘Rule-Of-Two’ Model. Arch. Toxicol 2014, 88, 1439–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M; Borlak J; Tong W A Model to Predict Severity of Drug-Induced Liver Injury in Humans. Hepatology 2016, 64, 931–940. [DOI] [PubMed] [Google Scholar]
- 15.Tong W; Hong H; Fang H; Xie Q; Perkins R Decision Forest: Combining the Predictions of Multiple Independent Decision Tree Models. J. Chem. Inf. Model 2003, 43, 525–531. [DOI] [PubMed] [Google Scholar]
- 16.Hong H; Xie Q; Ge W; Qian F; Fang H; Shi L; Su Z; Perkins R; Tong W Mold2, Molecular Descriptors from 2D Structures for Chemoinformatics and Toxicoinformatics. J. Chem. Inf. Model 2008, 48, 1337–1344. [DOI] [PubMed] [Google Scholar]
- 17.Chen M; Hong H; Fang H; Kelly R; Zhou G; Borlak J; Tong W Quantitative Structure-Activity Relationship Models for Predicting Drug-Induced Liver Injury based on FDA-Approved Drug Labeling Annotation and Using A Large Collection of Drugs. Toxicol Sci 2013, 136, 242–249. [DOI] [PubMed] [Google Scholar]
- 18.Abernethy D; Woodcock J; Lesko L Pharmacological Mechanism-Based Drug Safety Assessment and Prediction. Clin. Pharmacol. Ther 2011, 89, 793–797. [DOI] [PubMed] [Google Scholar]
- 19.O'Connell T; Watkins PB The Application of Metabonomics to Predict Drug-Induced Liver Injury. Clin. Pharmacol. Ther 2010, 88, 394–399. [DOI] [PubMed] [Google Scholar]
- 20.Mulliner D; Schmidt F; Stolte M; Spirkl H-P; Czich A; Amberg A Computational Models for Human and Animal Hepatotoxicity with A Global Application Scope. Chem. Res. Toxicol 2016, 29, 757–767. [DOI] [PubMed] [Google Scholar]
- 21.Zhu X-W; Xin Y-J; Chen Q-H Chemical and In Vitro Biological Information to Predict Mouse Liver Toxicity Using Recursive Random Forests. SAR QSAR Environ. Res 2016, 27, 559–572. [DOI] [PubMed] [Google Scholar]
- 22.Mellor CL; Steinmetz FP; Cronin MT Using Molecular Initiating Events to Develop A Structural Alert Based Screening Workflow for Nuclear Receptor Ligands Associated with Hepatic Steatosis. Chem. Res. Toxicol 2016, 29, 203–212. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt CW TOX 21: New Dimensions of Toxicity Testing. Environ. Health Perspect 2009, 117, A348–A353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dix DJ; Houck KA; Martin MT; Richard AM; Setzer RW; Kavlock RJ The Toxcast Program for Prioritizing Toxicity Testing of Environmental Chemicals. Toxicol. Sci 2007, 95, 5–12. [DOI] [PubMed] [Google Scholar]
- 25.Lee WM Drug-Induced Hepatotoxicity. N. Engl. J. Med 1995, 333, 1118–1127. [DOI] [PubMed] [Google Scholar]
- 26.Kim MT; Huang R; Sedykh A; Wang W; Xia M; Zhu H Mechanism Profiling of Hepatotoxicity Caused by Oxidative Stress Using Antioxidant Response Element Reporter Gene Assay Models and Big Data. Environ. Health Perspect 2016, 124, 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tice RR; Austin CP; Kavlock RJ; Bucher JR Improving The Human Hazard Characterization of Chemicals: A Tox21 Update. Environ. Health Perspect 2013, 121, 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen M; Suzuki A; Thakkar S; Yu K; Hu C; Tong W Dilirank: the Largest Reference Drug List Ranked by the Risk for Developing Drug-Induced Liver Injury in Humans. Drug discovery today 2016, 21, 648–653. [DOI] [PubMed] [Google Scholar]
- 29.Liu J; Mansouri K; Judson RS; Martin MT; Hong H; Chen M; Xu X; Thomas RS; Shah I Predicting Hepatotoxicity Using Toxcast in Vitro Bioactivity and Chemical Structure. Chem. Res. Toxicol 2015, 28, 738–751. [DOI] [PubMed] [Google Scholar]
- 30.Chen M; Vijay V; Shi Q; Liu Z; Fang H; Tong W FDA-Approved Drug Labeling for The Study of Drug-Induced Liver Injury. Drug discovery today 2011, 16, 697–703. [DOI] [PubMed] [Google Scholar]
- 31.Wagner M; Zollner G; Trauner M Nuclear Receptors in Liver Disease. Hepatology 2011, 53, 1023–1034. [DOI] [PubMed] [Google Scholar]
- 32.Mahmoud AM; Germoush MO; Alotaibi MF; Hussein OE Possible Involvement of Nrf2 and Pparγ Up-Regulation in the Protective Effect of Umbelliferone Against Cyclophosphamide-Induced Hepatotoxicity. Biomed. Pharmacother 2017, 86, 297–306. [DOI] [PubMed] [Google Scholar]
- 33.Yu L; Liu X; Li X; Yuan Z; Yang H; Zhang L; Jiang Z Protective Effects of SRT1720 via the HNF1α/FXR Signalling Pathway and Anti-Inflammatory Mechanisms in Mice with Estrogen-Induced Cholestatic Liver Injury. Toxicol. Lett 2016, 264, 1–11. [DOI] [PubMed] [Google Scholar]
- 34.Mellor CL; Steinmetz FP; Cronin MT The Identification of Nuclear Receptors Associated with Hepatic Steatosis to Develop and Extend Adverse Outcome Pathways. Crit. Rev. Toxicol 2016, 46, 138–152. [DOI] [PubMed] [Google Scholar]
- 35.Kuboki S; Shin T; Huber N; Eismann T; Galloway E; Schuster R; Blanchard J; Zingarelli B; Lentsch AB Peroxisome Proliferator-Activated Receptor-Gamma Protects against Hepatic Ischemia/Reperfusion Injury in Mice. Hepatology 2008, 47, 215–224. [DOI] [PubMed] [Google Scholar]
- 36.Kersten S; Desvergne B; Wahli W Roles of PPARs in Health and Disease. Nature 2000, 405, 421–424. [DOI] [PubMed] [Google Scholar]
- 37.Gale EA Lessons from The Glitazones: A Story of Drug Development. Lancet 2001, 357, 1870–1875. [DOI] [PubMed] [Google Scholar]
- 38.Kohlroser J; Mathai J; Reichheld J; Banner BF; Bonkovsky HL Hepatotoxicity due to Troglitazone: Report of Two Cases and Review of Adverse Events Reported to the United States Food And Drug Administration. Am. J. Gastroenterol 2000, 95, 272–276. [DOI] [PubMed] [Google Scholar]
- 39.Ogawa Y; Murata Y; Nishioka A; Inomata T; Yoshida S Tamoxifen-Induced Fatty Liver in Patients with Breast Cancer. Lancet 1998, 351, 725. [DOI] [PubMed] [Google Scholar]
- 40.Pinto HC; Baptista A; Camilo ME; de Costa EB; Valente A; de Moura MC Tamoxifen-associated Steatohepatitis-Report of Three Cases. J. Hepatol 1995, 23, 95–97. [DOI] [PubMed] [Google Scholar]
- 41.Nemoto Y; Saibara T; Ogawa Y; Zhang T; Xu N; Masafumi O; Akisawa N; Iwasaki S; Maeda T; Onishi S Tamoxifen-induced Nonalcoholic Steatohepatitis in Breast Cancer Patients Treated with Adjuvant tamoxifen. Intern Med. 2002, 41, 345–350. [DOI] [PubMed] [Google Scholar]
- 42.Yorikomurata YO; Saibara T; Nishioka A; Yoshimasafujiwara MF; Inomata T; Enzan H; Onishi S; Yoshida S Unrecognized Hepatic Steatosis and Non-Alcoholic Steatohepatitis in Adjuvant Tamoxifen for Breast Cancer Patients. Oncol Rep. 2000, 7, 1299–1304. [DOI] [PubMed] [Google Scholar]
- 43.Bruno S; Maisonneuve P; Castellana P; Rotmensz N; Rossi S; Maggioni M; Persico M; Colombo A; Monasterolo F; Casadei-Giunchi D Incidence and Risk Factors for Non-Alcoholic Steatohepatitis: Prospective Study of 5408 Women Enrolled in Italian Tamoxifen Chemoprevention Trial. bmj 2005, 330, 932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan K; Han X-D; Kan YW An Important Function of Nrf2 in Combating Oxidative Stress: Detoxification of Acetaminophen. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 4611–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu W; Hellerbrand C; Köhler UA; Bugnon P; Kan Y-W; Werner S; Beyer TA The Nrf2 Transcription Factor Protects from Toxin-Induced Liver Injury and Fibrosis. Lab Invest. 2008, 88, 1068–1078. [DOI] [PubMed] [Google Scholar]
- 46.Enomoto A; Itoh K; Nagayoshi E; Haruta J; Kimura T; O'Connor T; Harada T; Yamamoto M High Sensitivity of Nrf2 Knockout Mice to Acetaminophen Hepatotoxicity Associated with Decreased Expression of ARE-Regulated Drug Metabolizing Enzymes and Antioxidant Genes. Toxicol. Sci 2001, 59, 169–177. [DOI] [PubMed] [Google Scholar]
- 47.Okawa H; Motohashi H; Kobayashi A; Aburatani H; Kensler TW; Yamamoto M Hepatocyte-Specific Deletion of the Keap1 Gene Activates Nrf2 and Confers Potent Resistance Against Acute Drug Toxicity. Biochem. Biophys. Res. Commun 2006, 339, 79–88. [DOI] [PubMed] [Google Scholar]
- 48.Herpers B; Wink S; Fredriksson L; Di Z; Hendriks G; Vrieling H; de Bont H; van de Water B Activation of The Nrf2 Response By Intrinsic Hepatotoxic Drugs Correlates with Suppression of NF-Kb Activation and Sensitizes Toward Tnfα-Induced Cytotoxicity. Arch. Toxicol 2016, 90, 1163–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nachimuthu S; Volfinzon L; Gopal L Acute Hepatocellular and Cholestatic Injury in A Patient Taking Celecoxib. Postgrad. Med. J 2001, 77, 548–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'beirne J; Cairns S Cholestatic Hepatitis in Association with Celecoxib. Bmj 2002, 325, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El H II; Malik SM; Alwakeel HR; Shaikh OS; Sasatomi E; Kandil HM Celecoxib-Induced Cholestatic Liver Failure Requiring Orthotopic Liver Transplantation. World J Gastroenterol 2009, 15, 3937–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawy HSA Cilostazol Attenuates Cholestatic Liver Injury and its Complications in Common Bile Duct Ligated Rats. Eur J Pharmacol. 2015, 752, 8–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.