Abstract
Metabolites from type II fatty acid synthase (FAS) and polyketide synthase (PKS) pathways differ broadly in their identities and functional roles. The former are considered primary metabolites that are linear hydrocarbon acids, while the latter are complex aromatic or polyunsaturated secondary metabolites. Though the study of bacterial FAS has benefitted from decades of biochemical and structural investigations, type II PKSs have remained less understood. Here we review the recent approaches to understanding the protein-protein and protein-substrate interactions in these pathways, with an emphasis on recent chemical biology and structural applications. New approaches to the study of FAS have highlighted the critical role of the acyl carrier protein (ACP) with regard to how it stabilizes intermediates through sequestration and selectively delivers cargo to successive enzymes within these iterative pathways, utilizing the specificity conferred by protein-protein interactions to guide and organize enzymatic timing and specificity. Recent tools that have shown promise in FAS elucidation should find new approaches to studying type II PKS systems in the coming years.
1. Introduction
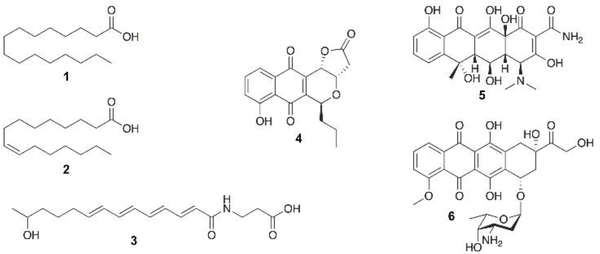
Type II fatty acid synthase (FAS) and polyketide synthase (PKS) share much in common in terms of their core enzymatic components, but their products couldn’t be more different. While fatty acids are metabolites primarily composed of one carboxyl group at the end of a linear, saturated or monounsaturated hydrocarbon, the secondary metabolites of type II PKSs, on the other hand, are highly functionalized polycyclic aromatic compounds, along with a few polyunsaturated natural products that have recently been identified. Among this class are first-line antibiotics, anticancer drugs, veterinary medicines, and agrochemicals (Fig. 1).1–3 An understanding of their biosynthesis offers the potential opportunity to redesign the pathways to produce novel aromatic and polyunsaturated compounds. However, while the type II FAS enzymatic pathway and protein structure have been extensively studied over the last century, many enzymes in type II PKS pathways remain mysterious, despite substantial interest in redesigning the pathways to produce custom designed molecules. Yet even the Escherichia coli FAS, the most well- studied acetate pathway, has remained recalcitrant to metabolic engineering and product control.4 It has become clear that a more fundamental understanding of both pathways will be the key to the long-term goals of controlling their biosynthesis.
Fig. 1.
Representative metabolites from type II fatty acid synthase (FAS) and polyketide synthase (PKS) pathways. Fatty acids: palmitic acid (1) and palmitoleic acid (2). Polyene (type II) polyketide: ishigamide (3). Aromatic polyketides: agricultural antibiotic frenolicin B (4), antibiotic oxytetracycline (5), and anticancer doxorubicin (6).
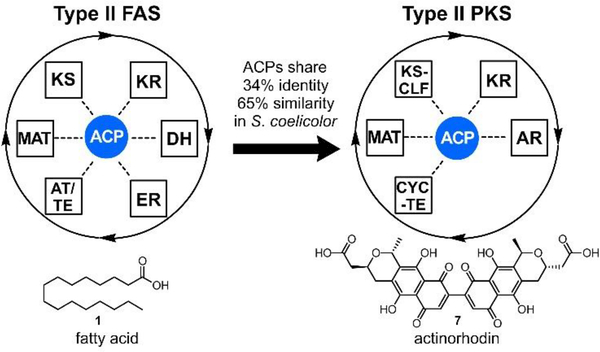
As evidenced by Fig. 2, Streptomyces coelicolor produces several natural products from ACP dependent pathways, including the de novo fatty acid synthesis and actinorhodin biosynthesis, a type II PKS pathway. The two ACPs from these pathways are highly homologous, sharing 34% amino acid identity (65% similarity) such that a casual BLAST searcher would deduce that they served the same function. Yet these pathways do not become scrambled; S. coelicolor does not produce hybrid or varied compounds. How do two highly homologous acetate pathways achieve orthogonality? It can only occur through precisely controlled protein-protein interactions.
Fig. 2.
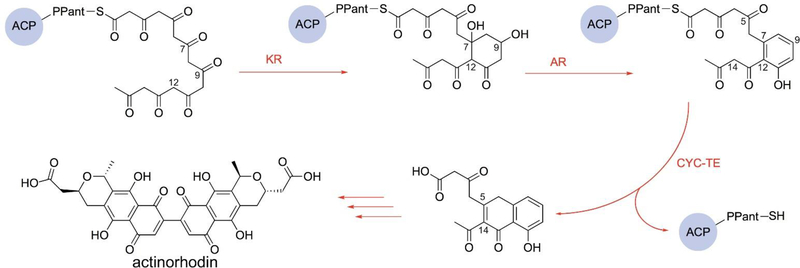
The central role of ACP in type II FAS and PKS pathways. Here, the actinorhodin type II PKS biosynthetic proteins in Streptomyces coelicolor are evolutionarily derived from type II FAS. Despite these similarities, the pathways remain isolated from each other – they do not scramble or exchange intermediates to provide chimeric products. This inherent fidelity, as well as known cross-reactivities, remains an important challenge to understanding and eventually reprogramming these pathways. ACP = acyl carrier protein; KS = β-ketoacyl-ACP synthase; MAT = malonylacyltransferase; KR = β-ketoacyl-ACP-reductase; DH = β-hydroxyacyl-ACP dehydratase; ER = enoyl reductase; AT/TE = acyltransferase/thioesterase; KS-CLF = ketosynthase-chain length factor; AR = aromatase; CYC-TE = cyclase-thioesterase.
The similarities and differences in the mechanism, structure, organization, and timing of these two pathways have offered challenges to their understanding. Given the evolutionary relationship between FAS and PKS,5 lessons learned in more well-known systems, including decades of research on bacterial FAS, can be brought to bear for less characterized pathways in order to more fully understand their mechanisms and processivities. Indeed, new tools that have been designed to interact with any carrier protein dependent pathway are commonly first applied to bacterial FAS enzymes, particularly those of E. coli. In this review, we discuss the basic enzymes and mechanisms of both pathways and introduce efforts to date that elucidate the enzyme structures and activities, with a focus on new tools and methods. Chief among these is a newfound appreciation and focus on protein-protein interactions. It has become apparent that protein-protein interactions are critical to the orchestration of all carrier protein dependent biosynthetic pathways, yet the details of these essential phenomena remain mostly uncharacterized. While we understand that the carrier proteins of each synthase must functionally interact with the enzymatic partners through specific recognition elements, a considerable amount of work remains to be done to complete this characterization, even in bacterial FAS. Although much of what is described here focuses on FAS, we address the type II systems that have been studied and draw parallels where possible.
2. Type II Fatty Acid Synthase and Polyketide Synthase Enzymology
2.1. Initiation in Type II FAS and PKS
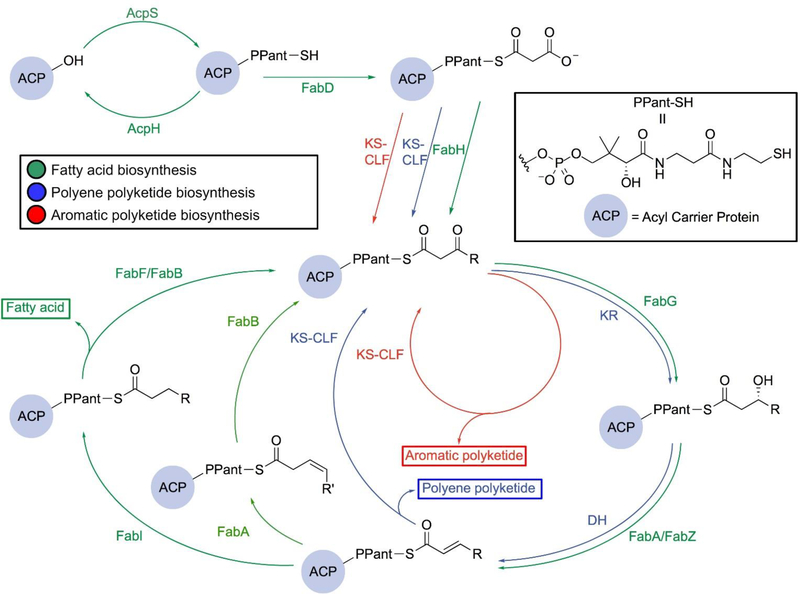
Type II fatty acid biosynthesis is an iterative cycle that relies on ACP to transport fatty acid precursors and intermediates through interactions with its partner enzymes, each of which takes on a different role in the pathway (Fig. 3, green path). We will focus on what is known about FAS from E. coli, given that the most biochemical, structural, and engineering data exist for this species. While several excellent reviews describe bacterial type II FAS in detail,6,7 here we provide an abridged description as a way to parallel FAS activity with type II PKS. To begin, ACP synthase (AcpS), a 4’ phosphopantetheinyl transferase (PPTase), post-translationally transfers the pantetheine prosthetic group from coenzymeA (CoA) to convert apo-ACP into its activated form, holo-ACP.8 This process has been found to be reversible with ACP hydrolase (AcpH), a Mn2+-dependent phosphodiesterase, which hydrolyzes the pantetheine group from holo-ACP to regenerate apo-ACP,9,10 presumably in the transition of the organism to stationary phase. Initially, malonyl-CoA is loaded onto the terminal sulfhydryl of holo-ACP via acyltransferase FabD, also known as the malonylacyltransferase (MAT) or malonyl-CoA:ACP transacylase, releasing CoA to generate malonyl-ACP. Next, FabH, the initiating β-ketoacyl-ACP synthase, a homodimer of the thiolase fold, drives the initiation of the elongation cycle by forming an acetyl-enzyme intermediate from acetyl-CoA and then condensing with malonyl-ACP via decarboxylative addition in the active site.9 This Claisen-like condensation generates acetoacetyl-ACP and serves as the first elongation step. FabH is solely involved in the initiation of fatty acid synthesis, mainly showing activity towards fatty acid substrates with four carbons or fewer. From here, ACP shuttles the intermediates to each of the enzymes in the fatty acid cycle via a thioester linkage to the terminal thiol of the pantetheine arm of ACP.
Fig. 3.
Comparison of the type II FAS and PKS pathways. Both the FAS and the PKS pathways begin with utilizing AcpS and FabD to generate its own malonyl-ACP. They start to diverge when entering the elongation cycle, in which the aromatic PKS pathway (red) only requires KS-CLF to produce polyketides while the polyene PKS pathway (blue) and FAS pathway (green) involve a NADPH-dependent ketoreductase (KR, or FabG in FAS) to reduce the β-carbonyl and a dehydratase (DH, or FabA and FabZ in FAS) to dehydrate the β–hydroxyacyl. The enoyl intermediate of the FAS pathway can subsequently be either reduced by the NADH-dependent FabI (dark green) or isomerized to its cis form by FabA (light green) before being further elongated by a ketoacylsynthase (FabF or FabB ). The latter pathway (light green) leads to the formation of unsaturated fatty acids.
To discuss type II PKS, we will focus on what is known about the S. coelicolor actinorhodin biosynthesis, since this is a canonical pathway for which most of the biochemical and structural information is known. The biosynthesis of type II PKSs follows a similar initiation strategy to FAS, with ACP loading performed by a stand-alone malonyltransferase before condensation. However, the starting unit can be derived from different acyl-CoAs. In the actinorhodin biosynthesis, the acetate starting unit can be derived from the decarboxylation of malonyl-ACP, and other aromatic PKS pathways can incorporate alternative acyl starting units.
Interestingly, it has been found that type II PKSs borrow some of the catalytic machineries from the de novo FAS pathway. For instance, actinorhodin actACP is post-translationally modified by the endogenous AcpS PPTase.11 Further, FabD has been shown to serve as an efficient malonyltransferase for holo-actACP.12
2.2. Elongation in Type II FAS
In both FAS and PKS, the acyl ACP continuously proceeds through the iterative pathway, elongating the fatty acid or polyketide chain by two carbons in each full cycle until it has reached its full length. In FAS, the enzyme that acts on acetoacetyl-ACP is FabG, a β-ketoacyl-ACP reductase (KR), that utilizes NADPH to selectively reduce the β-ketoacyl attached to the ACP to form (R)-β-hydroxyacyl-ACP with exclusively R stereochemistry.13 This β-hydroxyacyl-ACP is next dehydrated via elimination of water by a dehydratase (DH), either FabA or FabZ, the two key DHs in the de novo fatty acid synthesis cycle, to form a trans-enoyl-ACP. Their role is dependent on the carbon chain length in the pathway and will be discussed below. The trans-enoyl-ACP is reduced with NADH by enoyl reductase (ER), FabI. This yields an elongated, fully reduced acyl chain tethered to the ACP to complete the first turn of the FAS cycle.
For the second and subsequent turns, ACP delivers the nascent chain to an elongating ketosynthase (KS), FabB or FabF, thus entering the cycle again by condensing with a malonate moiety from malonyl-ACP to produce another β-ketoacyl elongated intermediate. This product is then followed by ketoreduction (FabG), dehydration (FabZ or FabA), and enoyl reduction (FabI) to complete another elongation cycle. Once the ketoacyl intermediate is reiterated through the cycle to reach its fully elongated length and undergoes enoyl reduction, the fatty acid is then cleaved and released by a thioesterase (TE) or transferred by an acyl transferase (AT) via thioesterification onto lipid precursors.7,14
Fatty acid chain elongation proceeds in two main routes to produce saturated or unsaturated fatty acids, with a couple of key differences between each path. Monounsaturated fatty acids are biosynthesized through the action of specialized DH (FabA) and KS (FabB) pairs in E. coli. The production of unsaturated fatty acids differs from their saturated counterparts through FabA’s additional ability to preferentially isomerize trans-2-decenoyl-ACP to form cis-3-decenoyl-ACP.15 In the case that a cis-enoyl-ACP is present in the chain, FabB acts after FabA to skip the ER step. Here, FabB catalyzes condensation of malonyl-ACP with cis-3-decenoyl-ACP to retain the cis-β,γ-unsaturation. FabF, however, catalyzes the final condensation of the unsaturated fatty acid cycle, since FabF is active towards C4-C16 intermediates and FabB is only active towards C4-C14 fatty acid substrates.16
2.3. Elongation in Type II PKS
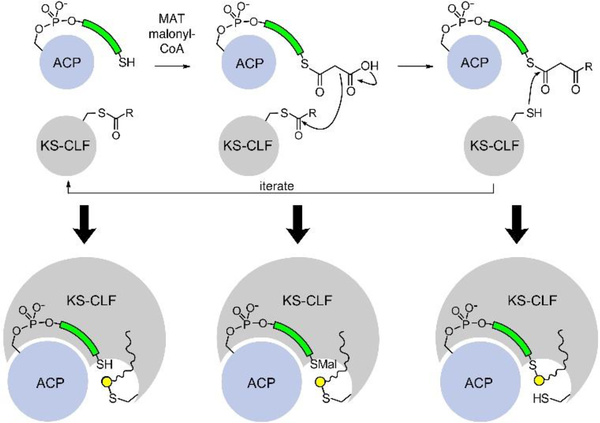
Following initiation, the first step of the elongation in type II PKS pathways mimics that of FAS, but the main difference is the replacement of a homodimeric KS with a heterodimeric complex known as the ketosynthase-chain length factor (KS-CLF). The KS-CLF shows homology to FabF; however, the CLF does not bear a catalytic cysteine residue like the KS. The KS-CLF elongates the acyl-ACP to a β-ketoacyl-ACP, and it is after this first elongation step that the major divergence from FAS is apparent. Rather than immediately reduce the nascent β-ketone after elongation, the KS-CLF instead catalyzes another elongation step (Fig. 3, red path), an action that is iterated for the in situ production of polyketones. In the actinorhodin biosynthesis, seven cycles of KS-CLF elongation occur one after the other, such that a full-length octaketide intermediate is formed. This fundamental change in activity drastically differs from the FAS model, in that the elongating intermediates no longer visit the same active sites in the same way (Fig. 4). Here, the elongating polyketone never needs to exit the active site of the KS-CLF. This highlights the subtle complexities of the KS-CLF mechanism in retaining and stabilizing inherently unstable polyketone intermediates, a topic that will be discussed below in the evaluation of structural biology.
Fig. 4.
The state of polyketones during aromatic PKS iterative elongation. The ACP and KS-CLF only adopt three states during aromatic polyketide biosynthesis: 1) holo-ACP and acylated KS-CLF; 2) Malonyl-ACP and acylated KS-CLF; 3) β-ketoacyl-ACP and holo-KS-CLF. Each of these states involves the elongated polyketone intermediate bound within the KS-CLF, indicating that the KS-CLF must stabilize these highly reactive elongated species via yet unknown mechanisms. The elongating polyketone above is depicted as R.
Following elongation of the actinorhodin octaketide precursor, the first cyclization step occurs between carbons C7 and C12 in an aldol fashion (Fig. 5). While the catalysis details are still unclear, this step likely occurs before activity by a ketoreductase (KR), which reduces the carbonyl at C9.17–19 Further aromatization of this first ring is catalyzed by an aromatase (AR).20 A second cyclization is next catalyzed by a bifunctional enzyme cyclase-thioesterase (CYC-TE), which concomitantly releases the substrate from ACP.21 The resulting bicyclic intermediate is then modified by post-PKS tailoring enzymes to eventually form actinorhodin (Fig. 3, red path).
Fig. 5.
The actinorhodin biosynthetic pathway post-elongation. The fully elongated polyketone undergoes a C7-C12 cyclization, and the carbonyl at the C9 position is reduced by the ketoreductase (KR). The aromatase (AR) catalyzes the formation of the first aromatic ring followed by the catalysis of the dual functional cyclase-thioesterase (CYC-TE) that cyclizes the intermediate and unloads it from actACP. The bicyclic substrate is then modified by post-PKS enzymes to form actinorhodin.
The elongation chemistries of FASs and PKSs differ significantly from each other due to the distinct traits of the intermediate species. While the acyl chains of elongated fatty acids are hydrophobic and chemically inert, the polyketide intermediates are amphiphilic and highly reactive. This implies a significant difference in the KS-CLF substrate binding pocket environment as well as the mechanisms of stabilization for the reactive polyketone intermediates to prevent unwanted reactions. While both systems have similar ACPs to shuttle the substrates and intermediates, organisms that possess one or more PKS pathways along with the ubiquitous FAS pathway have certain mechanisms to avert crosstalk between the systems. The protein-protein interactions between ACP and its partner proteins should play a crucial role here.
Type II PKSs had long been considered to only produce aromatic polyketides, but in recent years, pathways that produce polyenes have successively been discovered.22–24 Similar to the highly reducing type I PKS pathway, the elongation cycle of the polyene PKSs involves a KS-CLF to produce a 1,3-diketone, a KR to reduce the carbonyl at the β position, and a DH to form the α,β-unsaturated ketone (Fig. 3, blue path). These polyene PKSs are expected to have distinct substrate pockets from the typical type II PKSs. What is evident in the proposed biosynthesis of polyenes is that the mode of binding lies between that of type II FAS and classical aromatic type II PKS, in that the β-ketoacyl product of the KS-CLF must exit the pocket in order to be reduced by the KR and dehydrated by the DH at every turn of the cycle. How the channeling of the substrate differs from the other pathways remains to be explored.
2.4. Substrate sequestration
A previous understanding of type II fatty acid biosynthesis is the occurrence of substrate sequestration by the ACP,25 whereby each acyl intermediate that is C6 or longer is securely bound within the hydrophobic core of the four-alpha helical bundle that makes up the ACP.26 This phenomenon adds credence to the necessity for each enzyme to form protein-protein interactions with the ACP as a means to release the nascent fatty acid intermediate into the active site of each enzyme. Upon elucidation of this activity, several groups set about determining the existence of sequestration activity (or lack thereof) in different carrier protein-dependent pathways, and some examples of ACP sequestration in E. coli and S. coelicolor are illustrated in Fig. 6. As a result, it has been demonstrated that sequestration occurs primarily in type II pathways, including type II PKS and type II non-ribosomal peptide synthetases.28,30,31 However, sequestration does not appear to occur in type I FAS or type I PKS, which have large catalytic complexes with different domains in contrast to the discrete proteins in type II systems.32,33 This distinction makes intuitive sense, because type I synthases have the carrier proteins and catalytic domains within proximal reach, increasing the effective concentration through attachment within the module. Conversely, type II synthases require the ACP-bound substrate to encounter each appropriate enzyme within a pool of potential catalytic partners. Sequestration therefore serves the role of protecting elongating metabolites and delivering them to the appropriate enzyme at the proper time. How these pathways accomplish catalytic specificity remains unclear, and deciphering the role of protein-protein interactions will aid in our understanding of this phenomenon.
Fig. 6.
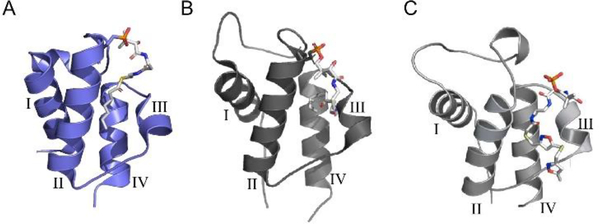
ACP sequestration of elongating intermediates. ACPs from both type II FAS and PKS pathways have been shown to sequester intermediates within the hydrophobic cavity at the center of the four-helix bundle. (A) E. coli heptanoyl-AcpP (PDB: 2FAD).27 (B) S. coelicolor octanoyl-actACP (PDB: 2KGC).28 (C) S. coelicolor actACP with tethered probe 11, based on NMR (Fig. 9B) and in silico docking.29
3. Current approaches to study protein-protein and protein-substrate interactions
Many chemical tools have been developed to probe protein-protein and protein-substrate interactions of FAS enzymes, ACP, and the fatty acid intermediates, many of which have been previously reviewed.34 The same tools can also be applied when studying type II PKS to better understand the structure-function relationships of the enzymes involved in the pathway. Structural elucidation to visualize these interactions has also been enabled by using new chemical biology tools and techniques. In conjunction with these tools, manipulation of the enzymes involved in type II FAS and PKS can shed light onto previously undescribed protein-protein and protein-substrate interactions, which are summarized in this section.
3.1. Tools to visualize proteins
Structural biology tools such as X-ray crystallography, NMR, and computational methods are leading techniques to visualize protein structure and are regularly employed in studies to probe protein-protein interactions. While these tools are frequently used to complement biochemical studies, this section will focus on their methodology rather than on their application.
3.1.1. Crystallography.
X-ray crystallography can be used to visualize proteins at the atomic level and thus serves as a primary tool to elucidate protein structure in structural biology. A concentrated protein sample of high purity is required to form fine crystals, and unpredictable crystallization conditions can cause barriers for protein crystallography. Many FAS and PKS protein structures have been solved in this manner, and additional co-crystallized structures have been solved with a bound substrate to highlight protein-substrate interactions. The information gained from crystallography lacks details about protein dynamics, but combined with other structural tools, such as NMR spectroscopy, valuable knowledge regarding protein structure and function can be obtained to better understand protein interactions.
3.1.2. NMR techniques.
NMR serves as a powerful tool to determine protein structure, study the dynamics of small proteins, and investigate protein interactions without prior knowledge of structural information. While X-ray crystallography relies on the formation of crystals to elucidate protein structure, NMR spectroscopy can yield structural information of a protein in solution or in its solid state, a major advantage to this technique. However, protein preparation and full elucidation of a structure with solution phase NMR can be laborious and requires the use of NMR active isotopes. The use of 2D spectra, specifically heteronuclear single quantum coherence (HSQC) NMR spectroscopy, offers a convenient way to solely look at the N-H bond of a protein backbone and observe changes due to protein interactions with substrates or other protein partners.35 3D experiments can be performed to correlate the N-H signals with adjacent carbons of the protein backbone to assign the peaks within the HSQC spectra.36 Often thought of as a protein “fingerprint”, HSQC has been used as an ideal method to analyze the effect of titrating increasing concentrations of enzyme partners on the chemical environment of carrier proteins.37 Here, calculating the chemical shift perturbation (CSP) tracks the HSQC changes for each residue upon interaction with an interacting partner. This allows for the identification of key residues that play a role in these interactions, as well as giving insight into the conformational changes that occur. Salt bridge formation between the interacting proteins can also be studied by shifts in the titration curves, also aiding in pinpointing the protein binding sites, which can be advantageous in engineering the pathways.38 NMR spectroscopy can complement X-ray crystallography, because the protein sample conditions can be varied to observe the changes in chemical shifts and their effects on protein activity. This could reduce the barriers associated with crystallization by providing information needed to optimize the conditions for crystal formation.
3.1.3. Computational methods.
Computational studies of protein activity are an increasingly important field for studying the FAS and PKS biosynthetic systems, as they can provide invaluable structural details about protein dynamics and predict critical interactions through techniques such as docking experiments and molecular dynamic (MD) simulations.34 A docking approach is beneficial when simulations are needed to predict how a protein would interact with a substrate or inhibitor prior to its application and determines the favorable binding conformation of the substrate within an enzyme’s binding site. Therefore, docking can provide structural clues that shed light onto protein-substrate interactions. MD is a powerful tool that computationally simulates the movements and interactions of atoms as a function of time.39 By propagating Newton’s laws of motion, the trajectories of atoms affected by protein-protein interactions can be evaluated to predict the binding and conformational energies of proteins. Coupled with structural data such as NMR or X-ray crystallography, MD simulations provide a unique glimpse into protein activity.
3.2. Manipulation of proteins to probe protein-protein and protein-substrate interactions
The complexity of the proteins in these type II FAS and PKS systems often makes it difficult to study the protein-protein and protein-substrate interactions, but through their manipulation, these interactions can be better understood.
3.2.1. Inhibition/mutagenesis studies.
Though inhibition and mutagenesis studies are not new methods for researching proteins, they are continually leveraged to understand protein interactions. Targeting the enzymes of the fatty acid cycle with inhibitors can aid in understanding how the enzymes interact with pathway substrates. While several inhibitors have been reported for type II FAS systems,40 the best characterized are inhibitors of E. coli FAS, a model for the highly conserved type II FAS.41 Further details of these inhibitors can be found in a previous review on the structure-function relationships in FAS.34 Additionally, mutagenesis studies of specific residues of FAS and PKS enzymes can help to elucidate their role.
While most inhibitors mimic natural substrates or ligands in their interactions within enzyme active sites, covalent inhibitors have the unique ability to trap protein-substrate interactions, often through active site residues, which can then be observed by crystallography or NMR. Cerulenin is a well-studied covalent inhibitor of both PKS and FAS ketosynthases, where the attack by the KS active site cysteine upon the epoxide of cerulenin mimics the condensation transition state, forming an irreversible covalent complex with the enzyme. This interaction was used to illuminate the hydrophobic pocket at the dimer interface of FabF where the acyl chain of the inhibitor and natural substrate lie.42 Another example is the diazaborine inhibitor of the FAS ER, which covalently bonds to the 2’-hydroxyl of the cofactor’s ribose, mimicking the binding of the substrate through hydrogen bonding and hydrophobic interactions.43 Competitive inhibitors can also be used to illuminate active site chemistry. Triclosan, a synthetic competitive inhibitor of the ER, also interacts with the 2’-hydroxyl of the ribose. However, this inhibitor forms a tight, non-covalent ternary complex with the bound FabI-NAD+ through hydrogen bonding in the substrate site, demonstrating protein-substrate interactions.44,45
Previous studies of mutants of the ketosynthases, reductases, and dehydratases in the FAS cycle have provided insight into the residues that play important roles in the enzyme mechanisms and their interactions with substrates.34 Mutagenesis studies of the PKS KS-CLF have also provided understanding of their important residues and confirmed the role of the CLF subunit as the determinant of the polyketide chain length.46 In order to gain a more complete understanding of these systems, a combination of data from several different tools, including mutagenesis, inhibitors, and structural biology techniques, is required to highlight different protein-protein interactions.
3.2.2. Crosslinking methods to probe ACP-partner protein interactions.
Bifunctional probes can be used to crosslink proteins with a binding protein as a means to capture protein-protein interactions. Such crosslinking probes are designed to tether enzymes together using many techniques: they can leverage mechanism-based inactivators, known inhibitors, or non-selective agents. Given that the ACP interacts and binds with its partner enzymes in each step of catalysis, crosslinking has been used to help evaluate protein-protein interactions in FAS, and the same methods can be applied when studying PKS. Used effectively, crosslinking acts as one of the main ways to capture these transient interactions.
Mechanism-based crosslinkers have led to advancements in understanding interactions of FAS proteins in complex with ACP. Analogues of the natural pantetheine arm appended to the ACP are prepared synthetically and then chemoenzymatically attached to the ACP using a “one pot” modification strategy.47 Briefly, a pantetheine probe can be synthesized and converted by enzymatic transformation to a full coenzyme A analog, which is then coupled to a conserved serine of ACP by a phosphopantetheinyl transferase.47,48 Commonly designed crosslinking probes contain a terminal warhead which, once tethered to ACP, can react with a partner enzyme to covalently bond and form a crosslinked complex. Crosslinkers often mimic the natural substrate so that both protein-protein and protein-substrate interactions can be observed.
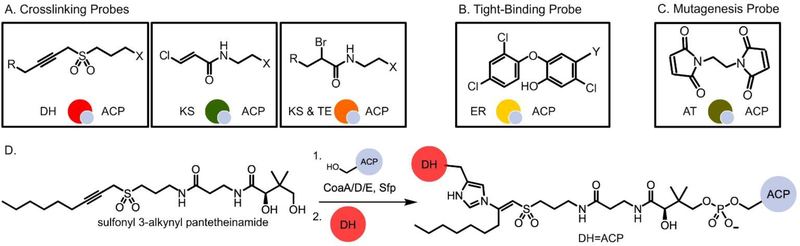
Different mechanism-based crosslinkers have been studied to probe ACP interactions with a few partner enzymes in the FAS system, but many FAS and PKS enzymes still lack elucidated crosslinked structures. Examples of these pantetheine analogues include epoxides based on the cerulenin inhibitor and simple acrylamide analogues, such as chloroacrylate (Fig. 7A), that could undergo an addition-elimination reaction by a nucleophilic active site residue, such as the cysteine in KS.49 Another example is the sulfonyl 3-alkynyl pantetheinamides (Fig. 7A) that were used to crosslink ACP with the FAS DH, FabA. The active site residue, His70, of FabA is proposed to deprotonate the α-hydrogen of sulfone to form an allene intermediate, which then interacts with the same residue to form the crosslinked complex (Fig. 7D).50,51 This crosslinker was first shown to best mimic the native substrate (R)-β-hydroxydecanoyl-ACP using fluorescent labeling experiments.51 The interactions observed from this crosslinking will be discussed in detail in the “Protein-protein interactions” section.
Fig. 7.
Probes for the investigation of PPIs. (A) Mechanism-based crosslinking probes. X = pantetheine analog; R = carbon chain. (B) The tight-binding probe for ER. Y = linker. (C) The BMOE probe. (D) Sulfonyl-3-alkynyl pantetheinamide undergoes chemoenzymatic one pot reaction and crosslink reaction to form crosslinked complex with DH.
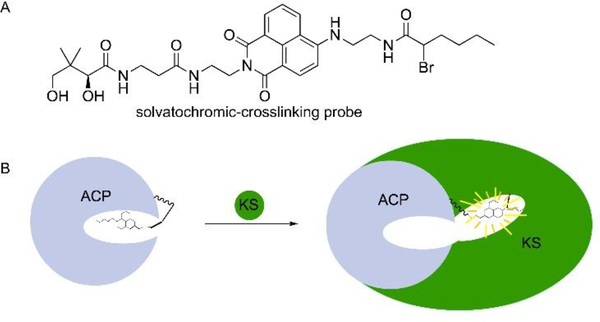
Another recent study shows how fluorescent, solvatochromic fluorophores can be used to visualize ACP sequestration and released into partner enzyme active sites through dual fluorescent-crosslinking probes (Fig. 8A).52 While they do not mimic the native substrates, the small dyes fit in the hydrophobic pocket of ACP where cargo is generally sequestered. Once the probes are loaded onto ACP via the one pot chemoenzymatic reaction and reacted with the active site cysteine residue of FabF through a reaction with the α-bromo moiety of the probe, a crosslinked complex was produced. An increase in fluorescence upon crosslinking of the crypto-ACP, which has a loaded pantetheine-probe, with FabF was believed to confirm the chain flipping mechanism in which the probe leaves the ACP pocket and extends into the active site of FabF (Fig. 8B). These solvatochromic pantetheine probes could be used for sequestration studies of the PKS ACP, as well as study the protein-protein interactions with other enzymes in the FAS and PKS cycles.
Fig. 8.
Solvatochromic-crosslinking probe. (A) The probe. (B) Flipping of the probe from the pocket of ACP to the pocket of KS occurred when the two proteins are bound, causing an increase in fluorescence. This is known as the chain flipping mechanism.
Tight-binding probes have also been developed for interaction studies. Triclosan (Fig. 7B) forms a tight complex in the FAS ER active site, and when appended to the pantetheine on ACP, it illuminates the protein-protein interactions. Details of these interactions will be further described in “The FabI-AcpP co-crystal structure” section.
Non-selective agents are another crosslinking tool that have been tested in type I PKS but can be applied to type II systems. Vicenistatin is a type I PKS product in which an in-trans AT is responsible for substrate loading. Crosslinking was employed using a bifunctional maleimide reagent, 1,2-bismaleimidoethane (BMOE), to trap the VinK (AT)-VinL (ACP) complex (Fig. 7C).53 BMOE is a non-selective crosslinker, reacting with any free thiol. To trap VinK, Ser266 of VinK, a non-catalytic residue located at the base of the substrate-binding tunnel, was mutated to cysteine to enable the crosslinking reaction. The sulfhydryl of the mutated VinK cysteine reacts with one of the maleimide groups of BMOE, while the terminal sulfhydryl on the phosphopantetheine arm of VinL reacts with the other maleimide group to form a crosslinked complex. The crystal structure confirmed that residues Arg153, Met206, and Arg299 of VinK are essential for the salt bridges and hydrophobic interactions with VinL, similar to the interactions seen in FabA and ACP in the FAS system.54 This reagent could be useful when crosslinking different proteins to obtain protein-protein interactions due to its non-selective nature; however, it is important that a sulfhydryl group is present for crosslinking to occur. Cysteines present at other locations may be prone to reacting with this reagent and may need to be mutated to prevent this.
While few crosslinked complexes exist for the enzymes of FAS and PKS, we expect to see a lot of progress in the coming months and years to be made in this field to open a window into the protein-protein and protein-substrate interactions of these systems.
3.2.3. Atom replacement strategy for type II PKS.
The inherent instability of polyketone intermediates in type II PKSs poses challenges for examining its mechanism. One way to overcome this is by using atom replacement strategies, in which polyketone surrogates that mimic the natural intermediates in length, polarity, and hydrophobicity are used to provide insight into the biosynthesis of the PKS products.29,55–57 Such strategies can be accomplished by substituting the carbonyl group with carbonyl bioisosteres that possess similar size or characteristics.58
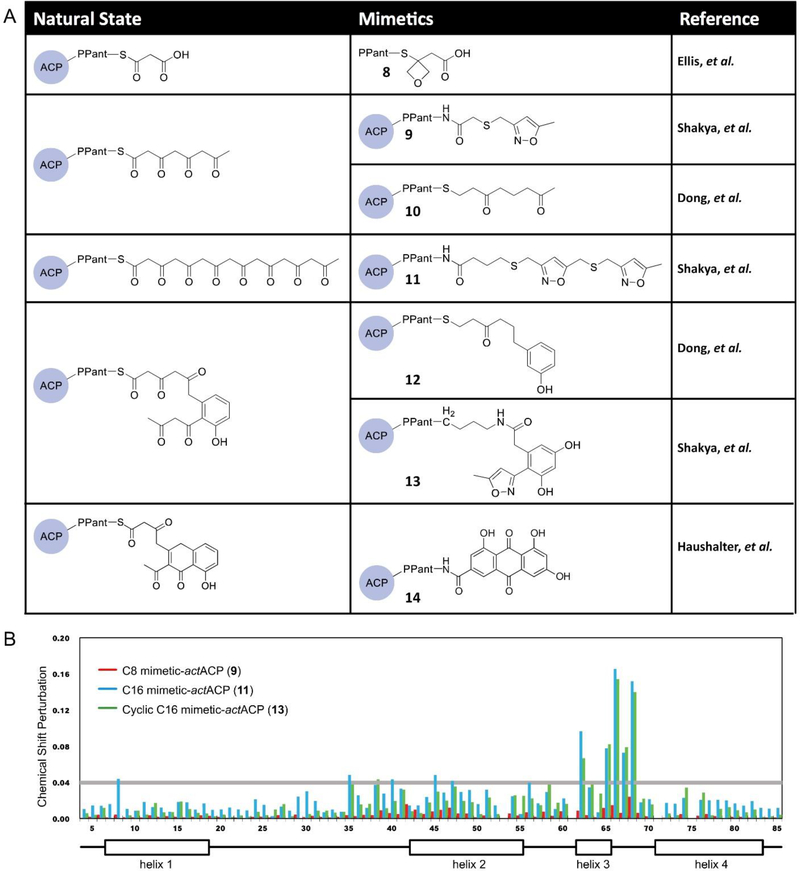
A recent atom replacement approach uses an oxetane-based polyketide mimetic (8) that was demonstrated to probe the substrate binding and mechanism of the priming KS, DpsC, of the daunorubicin biosynthesis (Fig. 9A).56 In this study, oxetanes, a well-known carbonyl bioisostere, replaced the carbonyl of the thioester to mimic malonyl-phosphopantetheine.56 Although oxetane is a slightly larger group compared to carbonyls, the lone pairs of the oxygen in both groups are positioned similarly, providing evidence for the use of the oxetane mimetic. Co-crystallization was performed with DpsC and the oxetane probe to characterize the substrate interaction with the priming KS. MD simulations on the oxetane replacement probe and the natural substrate, malonyl-phosphopantetheine, show comparable binding affinities and movements of binding site residues, confirming that the mimetic is similar enough to provide mechanistic insights of DpsC. 56
Fig. 9.
Atom replacement strategy. (A) Different carbonyl isosteres are utilized to generate stable mimetics in order to probe the protein-substrate interactions of PKSs. (B) Chemical shift perturbation plot of crypto-actACP using the chemical shift of holo-actACP as the base. The tetraketide mimetic shows no significant perturbation on actACP while the octaketide and cyclic polyketone mimetics show strong perturbation in helices II and III, suggesting that only the latter two substrates are sequestered by actACP.41
Another atom replacement strategy involves replacing the carbonyl and diketide units that are not involved in the cyclization steps with thioethers and isoxazoles, respectively. Fig. 9A shows the linear octaketide and cyclic atom replacement probes that have been synthesized with these moieties to mimic the native substrates and loaded onto actACP via chemoenzymatic methods.47 While no probe can exactly mimic the natural substrate, this atom replacement strategy acts as a useful tool to allow the study of protein-substrate interactions of ACP. Solution-phase protein NMR was applied to study how these isoxazole atom replacement probes that are loaded onto 15N-labeled actACP (9, 11, 13) compare to holo-actACP. As shown in the CSP plot collected (Fig. 9B), significant chemical perturbations of actACP are present with the octaketide and cyclic polyketone mimetics between helices II and III. This suggests sequestration of the substrates by actACP in a similar fashion to FAS ACP.29,30,55 Docking experiments were also used to simulate the same atom replacement probes to confirm that sequestration only occurs with the fully elongated mimetics.29
In addition to the isoxazole atom replacement probes, other atom replacement strategies have been developed to probe the actPKS pathway, including strategically removing carbonyl units to produce mimetics of the linear octaketide and cyclic PKS substrates,55 as well as using the tricyclic emodic acid to study the PKS pathway.30 These mimetics are loaded onto ACP (Fig. 9A) and all three replacement strategies confirmed via HSQC NMR experiments that the linear polyketide and the bicyclic intermediate of actPKS are sequestered between helices II and III of actACP, demonstrating the efficacy of using this strategy.
4. Important findings from structural biology and mutational analysis
In the past two decades, extensive progress has been made on the structural study of type II FASs. The structures of almost all E. coli FAS enzymes have been studied by X-ray crystallography, which provides the base for mechanistic study.13,42,59–64 Co-crystal structures of proteins with native substrates provide additional information of substrate sequestration and give more solid evidence of reaction mechanism. Excellent reviews by White et al. and Finzel et al. summarize these accomplishments.6,34 In contrast to the FASs, few crystal structures of type II PKSs exist.17,65–67 This is due in part to the instability of PKS proteins and the high reactivity of the PKS intermediates. This section will focus on what is known about the structural biology of type II PKSs as well as the findings from mutational studies.
4.1. The priming ketosynthase
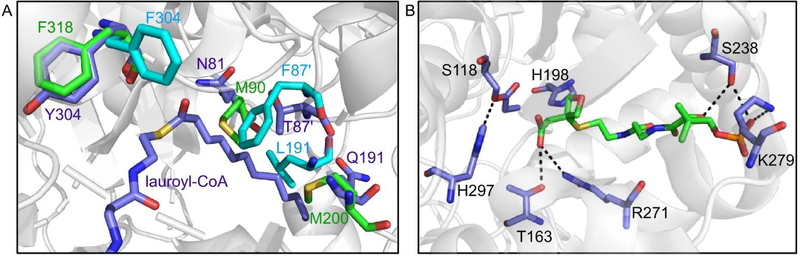
In FAS, the elongation cycle starts with the formation of acetoacetyl-ACP from malonyl-ACP by a priming KS, FabH. However, there are three possible types of priming mechanisms in type II PKS, and only one of them involves a priming KS.68 The two priming KSs that have been structurally characterized are ZhuH and DpsC, from the antibiotic R1128 and daunorubicin biosynthesis respectively.56,65 ZhuH and FabH are structurally similar to each other but have very distinct substrate specificities. Both ZhuH and ecFabH (from Escherichia coli) uptake substrates with C2-C4 chain lengths, but only ZhuH can sequester branched chain substrates. More uniquely, the type II mtFabH (from Mycobacterium tuberculosis) uptakes long chain fatty acids generated by a type I FAS and can tolerate substrates with C8-C20 chain lengths.69 Crystal structure overlays of ecFabH and ZhuH, as well as mtFabH sequestering its natural substrate lauroyl-CoA, reveals that the gatekeeper residue is Phe87’ from another subunit in ecFabH and Met90 in ZhuH (Fig. 10A).65 Compared to the phenylalanine residue, the more flexible side chain of methionine may contribute to the ability of ZhuH to sequester branched acyl-CoA. In addition, the rotamer conformation of a highly conserved phenylalanine residue (Phe304 in ecFabH and its equivalent in other priming KSs) located at the base of the CoA binding cleft determines the shape of the pocket, thus contributing to the substrate specificity.70
Fig. 10.
The binding pocket of priming KS. (A) Overlay of the crystal structures of ZhuH (green), ecFabH (light blue), and mtFabH with its substrate lauroyl-CoA (purple) reveals the gatekeeper residues M90 (ZhuH) and F87’(ecFabH). The phenylalanine rotamers that influence the pocket shape are also shown. (B) Malonyl-CoA mimetic (green, 8) in the DpsC binding pocket. The oxetane is not involved in any hydrogen bond network. It is proposed that the thioester carbonyl will turn and form a hydrogen bond with H198 after decarboxylation.
The priming KS DpsC catalyzes the transfer of a propionyl group from propionyl-CoA to the active site serine. It subsequently interacts with malonyl-ACP to initiate the cycle. This process involves an intermediate state in which a malonate is in the proximity of the propionated serine but was challenging to characterize by X-ray crystallography or other structural biology methods. Previous attempts of visualizing the same intermediate state of ecFabH by co-crystallizing with malonyl-CoA failed due to the instability of the malonyl group.71 To overcome this barrier, a recent work replaced the thioester carbonyl with an oxetane ring (Fig. 9A and Fig. 10B).72 The resulting mimetic was successfully co-crystallized with DpsC, yielding the structure of the intermediate state. Given that DpsC has a Ser-His-Asp catalytic triad instead of the Cys-His-Asn triad in canonical priming KS, and that the structure of DpsC is quite unique, it is not a surprise to see that the substrate is oriented differently in the binding pocket compared to the hypothesized FabH orientation. A key difference is the lack of an oxyanion hole that stabilizes the thioester carbonyl during decarboxylation in FabH. It is hypothesized that the substrate reorients itself after decarboxylation to form a hydrogen bond with His198. The success of this carbonyl surrogate points out a new road to investigating the catalytic mechanism of type II PKSs.
4.2. Chain length control by the extending ketosynthase
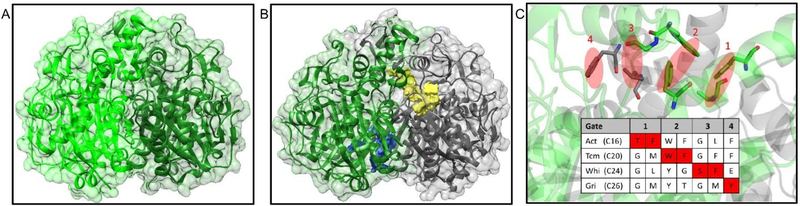
It is hypothesized that the reactive polyketone intermediates in the elongation cycle of the aromatic PKS pathway remain in the pocket of KS-CLF during all steps of elongation (Fig. 4). The only crystal structure of KS-CLF to date is the actKS-CLF (Fig. 11B).17 This heterodimer is structurally similar to the homodimeric extending KS of FAS, such as FabF (Fig. 11A). The substrate pocket can be identified in the structure, which extends from the catalytic Cys169 of the KS subunit to the Phe116 of the CLF subunit. Surface view of the tunnel shows a narrow and amphipathic substrate binding pocket, suggesting that the pocket has a high specificity for the substrate and that a strong sequestration interaction is expected. Despite the fact that the CLF can act as a decarboxylase,73 it is primarily known to control the chain length of the substrate through its gatekeeper residues.46 Mutations on these residues, suggested by the sequence alignment of several CLFs, have successfully engineered some KS-CLFs to produce substrates of different chain lengths. The F109A/F116A double mutant of actCLF produces decaketides (C24) that make up 96% of total polyketides in vitro, confirming that these phenylalanine residues (in gate 1 and 2 of Fig. 11C) prevent the natural substrate, octaketides (C16), from extending further. In addition, when Gly116 (Fig. 11C, gate 1) of tcmCLF is mutated into threonine, a larger residue, the mutant synthesizes more octaketides than nonaketides (C20). Combining these results with the crystal structure of actKS-CLF reveals the gates for chain length control (Fig. 11C).
Fig. 11.
The structural information of extending ketosynthase. (A) Crystal structure of FabF. (PDB: 2GFW) (B) Crystal structure of actKS-CLF with KS colored in gray and CLF colored in green. (PDB: 1TQY) The substrate pocket (yellow) extends from the KS subunit into the CLF subunit. Another pocket with unknown purpose is found in the CLF subunit (blue). (C) Four gates in KS-CLF that determine the chain length, using actKS-CLF crystal structure as an illustration (KS in gray and CLF in green). The bulky gatekeeper residues are colored red in the protein sequence of different KS-CLFs. Act = actinorhodin, Tcm = tetracenomycin, Whi = WhiE spore pigment, and Gri = griseorhodin. The parenthesis after each KS-CLF indicates the chain length of the respective polyketide product. The substrate extends from the right to the left of the figure, thus, the earlier the gate is blocked, the shorter the product is.
The FAS ACP is known to sequester the acyl intermediates when shuttling them between the partner proteins. This sequestration serves as one of the driving forces of the elongation cycle. In the aromatic PKS system, on the other hand, ACP only interacts with one enzyme, KS-CLF, during elongation. actACP sequesters its substrate between helix II and III, and it has been shown that the sequestration happens only after the polyketone intermediate is fully elongated.29,55 This result, in combination with the tight binding pocket of KS-CLF, supports the hypothesis that KS-CLF keeps the substrate inside its pocket during elongation.
4.3. The role of ketoreductase in the first ring cyclization
The KR of type II PKS shows regiospecificity by reducing only one carbonyl group of the elongating polyketone. In the case of actKR, such specificity is at the C9 carbonyl (Fig. 5). Comparison of the actKR structure with FabG, the FAS KR that reduces each β-carbonyl group of a growing fatty acid chain, reveals a flexible region around helices VI and VII in FabG that contributes to the tolerance of substrates with different chain lengths.18
An important inquiry of the actinorhodin biosynthesis is the timing of the first ring cyclization. Early studies that led to the “design rule” of engineering type II PKSs show that reduction at the C9 position by actKR drives the first cyclization to occur between C7 and C12, implying that the cyclization happens after the reduction.74 However, docking simulation of the full length linear substrate in the actKR binding pocket shows a loss of regiospecific reduction at C9, indicating that the cyclization is more likely to happen before the reduction.18 Further docking experiments of the cyclic substrate also suggest that constraints from the ring are necessary to position the substrate for C9 reduction.75 These results show a direct causal relationship between C7-C12 cyclization and C9 reduction, but with the former activity as the cause.
However, the first ring cyclization in aromatic PKSs can be mediated by either the KS-CLF or the KR. Given that the minimal actPKS (actACP and actKS-CLF)76 only produces C7-C12 and C10-C15 cyclic compounds (SEK4 and SEK4b, respectively)77 instead of the many other possible structures, it is possible that the KS-CLF can catalyze the ring formation. The crystal structure of the actKS-CLF shows a water molecule in close proximity to the C7 carbonyl when a substrate is bound. It is possible that this water molecule donates a proton to the substrate and catalyzes the C7-C12 ring formation. The pocket also has room to accommodate the cyclized substrate.17 However, it has been shown that the addition of actKR to the minimal tcmPKS affects the regiospecificity of cyclization, indicating that KR could play a role in the ring forming catalysis.78 Given that the actACP sequesters the linear polyketone,29 it is possible that the unstable intermediate can be protected and transferred to the actKR to be cyclized. Mutational studies of actKR reveal the highly conserved residue T145 that is necessary for mediating cyclization. It has been shown that T145 can form a hydrogen bond with the C11 carbonyl and subsequently facilitates the nucleophilic attack of C12.78 The T145A mutant fails to direct cyclization, leading to the loss of ability to form mutactin, a molecule produced by co-expressing minimal PKS and actKR. As a result, the actKR catalyzes the first ring cyclization, whereas the actKS-CLF may contribute to it in some degree.
5. Protein-protein interactions
In almost all type II systems, the ACP is a 4-helical protein of about 10 kDa that shuttles substrate intermediates between each enzyme in the pathway. The remarkable ability for such a small protein to be recognized by and interact with 12 partner enzymes in the E. coli de novo fatty acid biosynthesis and at least 9 others from peripheral pathways belies a subtle communication strategy that we are only beginning to understand. The prevailing hypothesis is based upon specific salt bridges and hydrophobic interactions between the ACP and each enzyme, which serves to coordinate communication and control. Sequestration of substrates within the ACP results in small changes in the surface residues of the ACP, thus varying the binding specificity with pathway enzymes. It is believed that these protein-protein interactions accelerate the overall kinetics of the metabolic pathway by avoiding stochastic binding and sampling of each metabolite by every enzyme. At the same time, this mechanism prevents crosstalk from non-cognate enzymes and other ACPs.
To visualize such interactions via crystallography, it is necessary to trap the dynamic ACP as it interacts with partner enzymes. This can be accomplished by co-crystallization of unmodified ACP or by a crosslinking method. To date, only four crystal structures of ACP-bound FAS have been reported, and no such structures for type II PKSs exist. The following section will focus on the existing crosslinked/co-crystal structures of FASs and the information drawn from these studies about protein-protein and protein-substrate interactions.
5.1. The FabA=AcpP crosslinked complex
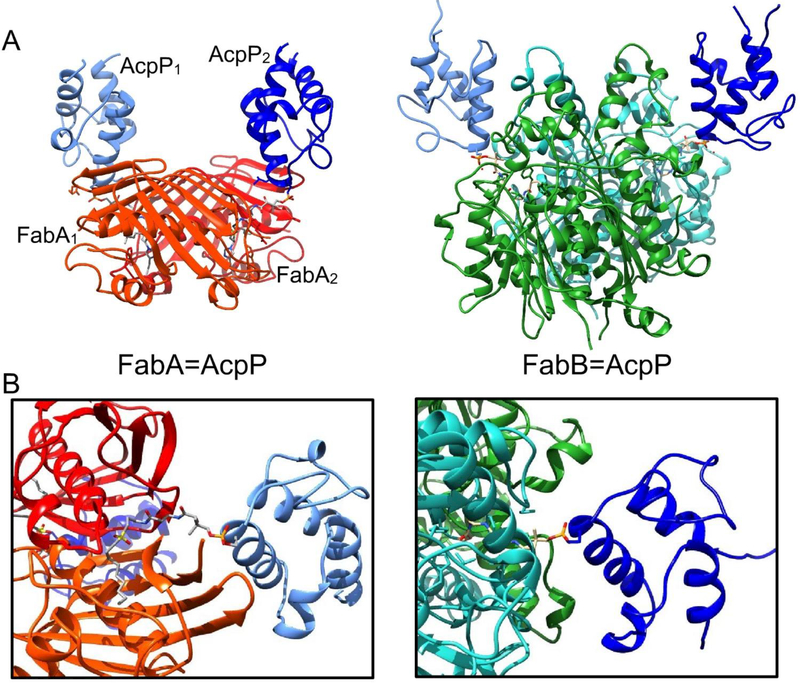
FabA, a DH of FAS, is known to control fatty acid chain length and saturation levels.79 It catalyzes the dehydration of β-hydroxy-decanoyl-ACP as well as the isomerization of the subsequent trans-2-decenoyl-ACP into cis-3-decenoyl-ACP. In E. coli, this is the only pathway for the biosynthesis of unsaturated fatty acids. The FabA catalysis involves a His-Asp dyad in which the His70 serves as a base to first remove the α-proton from C2. The enolate is delocalized via conjugation with the adjacent carbonyl, followed by elimination of a protonated β-hydroxyl group. This mechanism led to the discovery of the first reported mechanism-based inhibitor and subsequent design of the sulfonyl alkyne crosslinker.80 Depicted in Fig. 7D, this moiety targets the His70 residue of FabA, which catalyzes deprotonation and forms a covalent bond with the β-carbon. The crosslinked complex in Fig. 12A, left, shows the FabA homodimer bound with two ACPs, one covalently bound to each active site His70.54 Interestingly, the two FabA=AcpP protomers are not identical, possibly representing different stages in catalysis (Fig. 13). NMR titration of increasing the ratio of FabA to octanoyl-AcpP shows a migration of cross peaks toward those peaks identified by the HSQC of the crosslinked complex, indicating that the bound conformation of ACPs represent the most accurate depiction of the natural, transient binding event.
Fig. 12.
Crystal structures of crosslinked complexes. (A) Comparison of the FabA=AcpP (PDB: 4KEH) and FabB=AcpP (PDB:5KOH) complexes. Both homodimers have two ACPs attached at the entrance to each active site, located at the interface of the monomers. However, the FabA=AcpP complex shows two different binding events, and are distinguished by numbering. (B) Comparison of the protein-ACP interfaces.
Fig. 13.
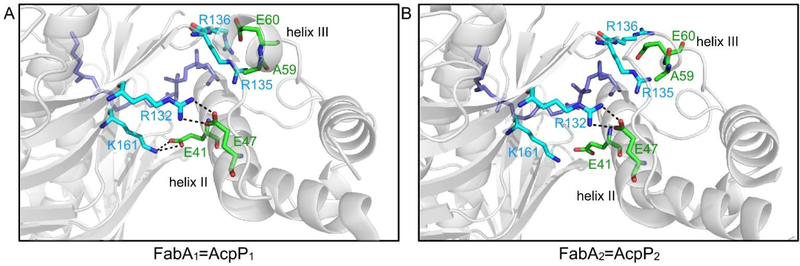
Important residues on the protein interface of the FabA=AcpP crosslinked complex. FabA residues (light blue) interact with the residues (green) in helices II and III of AcpP in order to transfer the substrate (purple). (A) The FabA1=AcpP1 protomer and (B) the FabA2=AcpP2 protomer show different interface interactions. It is proposed that they represent different stages in catalysis
The NMR titration study also identified important residues in helices II and III of AcpP that interact with FabA, which is in accurate agreement with those found in the crosslinked complex. Along with the interacting residues of FabA revealed by the crystal structure, a series of binding events can be proposed to mediate the transition of substrates (Fig. 13). First, the arginine-rich positive residues (Fig. 13, light blue) on the FabA surface interact with the anionic phosphopantetheine group attached to ACP. Subsequent mutagenesis studies of these surface residues show significantly lower efficiencies in crosslinking, and thus incomplete binding.81 Next, Arg132 and Lys161 of FabA form salt bridges with Glu41 and Glu47 of ACP, respectively, to stabilize the complex. Thirdly, helix III of ACP is pried open by the interaction of FabA surface arginines with the Ala59 and Glu60 residues of ACP. Finally, hydrophobic interactions between FabA and ACP helix II further anchor the complex. This process demonstrates the course of the chain-flipping mechanism, in which the protein-protein interactions flip the substrate out from sequestration within the ACP to enter the enzyme partner’s active site for subsequent catalysis.
Further comparison of the crosslinked and the apo-FabA structures reveals two gatekeeper residues, Phe165 and Phe171, located at the entrance of the active site.81 Mutations of these residues to alanines eliminate the ability of FabA to act selectively with substrates of different acyl chain lengths, confirming their gatekeeper role. Rotation of Phe171 can partially open the entrance to accept small molecule substrates, such as the inhibitor 3-decynoyl-N-acetyl cysteamine.59 With the crypto-ACP bound, however, Phe165 also must turn away to fully open the gate for sequestration.
5.2. The FabB=AcpP crosslinked complex
Recently, the crystal structure for a crosslinked E. coli FabB=AcpP has been deposited (Fig. 12A, right).82 The crosslinking structure has been accomplished through the use of a chloroacrylate crosslinking probe (Fig. 7A), with both monomers of the FabB homodimer capturing an ACP via covalent linkage to the active site Cys163. While a detailed study of this structure remains to be published, a simple comparison with the FabA=AcpP crosslinked structure can be made by fixing the positions of ACP in each (Fig. 12B). While both structures represent homodimers that are crosslinked to two ACPs, the binding motifs are surprisingly different. For instance, in FabA=AcpP, the FabA monomer that pries away helix III to release the substrate from sequestration is not the monomer that is covalently crosslinked to ACP. Conversely, in FabB=AcpP, the crosslinked monomer is the same, which engages helix III. Such surprising comparisons will likely be revealed as more crosslinked structures emerge.
5.3. The FabZ-ACP co-crystal structure from Helicobacter pylori
FabZ is the second DH from the bacterial FAS pathway shared with E. coli and lacks the ability to catalyze isomerization, but it can accept substrates with a larger variety of chain lengths. Unfortunately, efforts to overexpress FabZ from E. coli have failed to date to produce soluble protein. The recent co-crystal structure of a H. pylori FabZ homodimer with a single holo-ACP reveals that the binding of ACP induces the movement of the β-sheet layer, resulting in a Y-shaped tunnel that reveals an additional sequestration site.83 Further docking experiments using fatty acids with different chain lengths show that shorter substrates (C4-C12) prefer the original pocket I, whereas longer substrates (C14 and C16) prefer the temporal pocket II. This induced Y-shaped tunnel partially explains the difference in substrate specificity of FabZ and FabA, with FabA still possessing the narrow L-shaped tunnel even with ACP bound, thus sequestering substrates with chain lengths only up to C12. Docking of an unsaturated fatty acid shows that the Y-shaped tunnel sterically hinders a cis-substrate, explaining the lack of isomerization ability for FabZ.
The solution of the FabZ-ACP co-crystal structure with only one bound ACP offers a curious phenomenon in the company of several 2:2 structures. Further examination of the structure reveals that binding of an ACP induces the unbound monomer to form a closed conformation that prevents the second ACP from binding. This leads to the proposal of the seesaw-like mechanism in which two subunits alternate to bind ACP, although crystal packing could also explain this unusual stoichiometry. In addition, Tyr100 is identified as a gatekeeper residue working similarly to those recognized in FabA.
5.4. The FabI-AcpP co-crystal structure
The ER (FabI) of FAS is responsible for reducing the enoyl-ACP to its acyl-ACP form, depending on its binding with the NADH cofactor. While no crosslinked complex yet exists for FabI, many tools, such as co-crystallization, inhibition, and MD, have been employed to study the protein interactions with ACP. An earlier co-crystal structure of FabI with dodecanoyl-ACP showed its structure to be a tetramer with each monomer having a central β-sheet that contains eight helices surrounding seven β-strands.84 The crystallography data showed that once FabI and ACP interacts, FabI’s structure does not change drastically, but the substrate binding loop does change, becoming more ordered. Additionally, the basic residues of FabI’s helix VIII form hydrogen bonds with the acidic residues in helix II of ACP, forming a stable structure. Mutation of these basic residues significantly decrease the enzyme’s ability to reduce the dodecanoyl-ACP, confirming the importance of the electrostatic interactions of these basic residues with ACP’s acidic residues. Additional stabilization is provided by hydrogen bonding of the ACP pantetheine with FabI.84 The transient nature of this interaction caused the crystal structure to not be fully resolved, and so the same group turned to MD simulations to observe the interactions of the proteins and substrate. While the catalytic triad of FabI is made up of Tyr146-Tyr156-Lys173, the MD simulations, along with mutagenesis studies, suggest that the active site residue Tyr156 does not directly play a role in stabilizing the substrate’s carbonyl group through hydrogen bonding. Rather, Tyr146, due to its closer proximity to the substrate, helps to catalyze the reduction of the substrate by hydrogen bonding with its carbonyl group.84 From these studies, it is inferred that the active site serine of ACP carries the substrate to the active site of FabI, consisting of helix VIII, the substrate binding loop, and a mobile loop of five residues. The latter loops move and become ordered to allow the phosphopantetheine arm of ACP to transfer the substrate to FabI’s active site.84,85
A recent inhibition study to develop tight-binding probes to examine the FabI and ACP interactions was developed by chemoenzymatically modifying an apo-ACP to tether to triclosan, generating crypto-ACP. This selective inhibitor of bacterial FAS ERs was linked to the 4’-phosphopantetheine arm of ACP (Fig. 7B).85 Because of the known competitive inhibition of FabI by triclosan, the inhibitor was used as the ER specific motif to provide tight binding of the inhibitor within the enzyme active site. Data indicated that the binding of ACP to FabI only occurs when triclosan was present on the pantetheine arm and presented greater inhibition as a crypto-ACP probe than as a probe without ACP. This shows how important the binding of ACP to FabI is.85 In the same study, the probe was loaded onto the PKS actACP to test E. coli FabI’s specificity toward AcpP. The crypto-actACP with the triclosan probe shows no inhibition of FabI by itself, confirming the necessity of crypto-AcpP to observe the protein-protein interactions. This design can prove useful when studying other enzymes in the type II FAS and PKS systems to understand the key interactions of ACP with its partner proteins.
6. Future directions
The more we understand about protein-protein and protein-substrate interactions in type II FAS and PKS biosynthesis, the sooner we will be capable of designing these pathways for new applications. Understanding the fundamental principles underlying ACP interactions with substrates and enzyme partners will inform a set of rules guiding their processivity in these pathways. Such knowledge will direct efforts to modify and engineer these pathways for the production of novel metabolites tailored to specific applications.
FAS engineering provides an immediate example. To date, groups have struggled with metabolic engineering of type II FAS due to the lack of knowledge about protein interactions. Indeed, most published articles provided anecdotal rationales for success or failure upon expression of exogenous FAS enzymes in model organisms.86 For instance, in an effort to engineer increased production of medium-chain fatty acids for improved biodiesel feedstocks, researchers expressed exogenous medium chain fatty acid TEs from plants into E. coli, resulting only in increased production of short chain fatty acids.87 Despite multiple attempts, medium chain fatty acid production remained elusive until the recent rational mutagenesis of these TEs to better interact with the E. coli ACP.88 Indeed, similar challenges have been faced in other organisms with type II FAS, such as microalgae,89 pointing to immediate opportunities to engineer the protein-protein interface of ACP with exogenous enzymes as a durable solution to the synthetic biology challenges in these pathways. We can expect to see significant contributions in this area very soon. Crosslinked complexes of the E. coli ACP and most, if not all, of its enzymatic partners will likely be elucidated within coming months and years, and this information will help inform next-generation FAS engineering efforts. Likewise, type II PKS pathways are sure to experience a parallel increase in activity. Enabled by crosslinking and structural biology tools, the rules governing protein-protein interactions in aromatic PKSs will become clearer, and these pathways will become more available for modification and engineering.27
7. Conclusion
The details of type II FAS and PKS biosynthetic pathways continue to be uncovered. New developments in structure, activity, and protein-protein interactions continue apace, as does the development of new tools to probe these pathways. We are hopeful that the coming decade will offer significant advances in our understanding of these pathways, where protein-protein and protein-substrate interactions will no doubt play a central role. With the underlying rules and mechanisms in place, manipulation of these pathways will rapidly follow.
8. Acknowledgements
This work was supported by NIH RO1 GM095970 and NSF IOS1516156. We thank T. Bartholow, K. Charov, and Dr. L. Misson for assistance in editing the manuscript. We also thank Dr. D. John Lee for the CSP plot data.
9 References
- 1.Griffin MO, Fricovsky E, Ceballos G and Villarreal F, Am. J. Physiol.- Cell Physiol, 2010, 299, C539–C548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minotti G, Menna P, Salvatorelli E, Cairo G and Gianni L, Pharmacol. Rev, 2004, 56, 185–229. [DOI] [PubMed] [Google Scholar]
- 3.Das A and Khosla C, Acc. Chem. Res, 2009, 42, 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handke P, Lynch SA and Gill RT, Metab. Eng, 2011, 13, 28–37. [DOI] [PubMed] [Google Scholar]
- 5.Jenke-Kodama H, Sandmann A, Müller R and Dittmann E, Mol. Biol. Evol, 2005, 22, 2027–2039. [DOI] [PubMed] [Google Scholar]
- 6.White SW, Zheng J, Zhang Y-M and Rock CO, Annu. Rev. Biochem, 2005, 74, 791–831. [DOI] [PubMed] [Google Scholar]
- 7.Beld J, John Lee D and Burkart MD, Mol. Biosyst, 2015, 11, 38–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beld J, Sonnenschein EC, Vickery CR, Noel JP and Burkart MD, Nat Prod Rep, 2014, 31, 61–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas J and Cronan JE, J. Biol. Chem, 2005, 280, 34675–34683. [DOI] [PubMed] [Google Scholar]
- 10.Kosa NM, Pham KM and Burkart MD, Chem Sci, 2014, 5, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox RJ, Crosby J, Daltrop O, Glod F, Jarzabek ME, Nicholson TP, Reed M, Simpson TJ, Smith LH, Soulas F, Szafranska AE and Westcott J, J. Chem. Soc. [Perkin 1], 2002, 1644–1649. [Google Scholar]
- 12.Revill WP, Bibb MJ and Hopwood DA, J. Bacteriol, 1995, 177, 3946–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price AC, Zhang Y-M, Rock CO and White SW, Biochemistry, 2001, 40, 12772–12781. [DOI] [PubMed] [Google Scholar]
- 14.Spencer AK, Greenspan AD and Cronan JE, J. Biol. Chem, 1978, 253, 5922–5926. [PubMed] [Google Scholar]
- 15.Kass LR and Bloch K, Proc. Natl. Acad. Sci, 1967, 58, 1168–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garwin JL, Klages AL and Cronan JE, J. Biol. Chem, 1980, 255, 3263–3265. [PubMed] [Google Scholar]
- 17.Keatinge-Clay AT, Maltby DA, Medzihradszky KF, Khosla C and Stroud RM, Nat. Struct. Mol. Biol, 2004, 11, 888–893. [DOI] [PubMed] [Google Scholar]
- 18.Korman TP, Hill JA, Vu TN and Tsai S-C, Biochemistry, 2004, 43, 14529–14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Javidpour P, Bruegger J, Srithahan S, Korman TP, Crump MP, Crosby J, Burkart MD and Tsai S-C, Chem. Biol, 2013, 20, 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDaniel R, Ebert-Khosla S, Hopwood DA and Khosla C, J. Am. Chem. Soc, 1994, 116, 10855–10859. [Google Scholar]
- 21.Taguchi T, Awakawa T, Nishihara Y, Kawamura M, Ohnishi Y and Ichinose K, ChemBioChem, 2017, 18, 316–323. [DOI] [PubMed] [Google Scholar]
- 22.Du D, Katsuyama Y, Shin-ya K and Ohnishi Y, Angew. Chem. Int. Ed, 2018, 57, 1954–1957. [DOI] [PubMed] [Google Scholar]
- 23.Pohle S, Appelt C, Roux M, Fiedler H-P and Süssmuth RD, J. Am. Chem. Soc, 2011, 133, 6194–6205. [DOI] [PubMed] [Google Scholar]
- 24.Bilyk O, Brötz E, Tokovenko B, Bechthold A, Paululat T and Luzhetskyy A, ACS Chem. Biol, 2016, 11, 241–250. [DOI] [PubMed] [Google Scholar]
- 25.Zornetzer GA, Fox BG and Markley JL, Biochemistry, 2006, 45, 5217–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kosa NM, Haushalter RW, Smith AR and Burkart MD, Nat. Methods, 2012, 9, 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roujeinikova A, Simon WJ, Gilroy J, Rice DW, Rafferty JB and Slabas AR, J. Mol. Biol, 2007, 365, 135–145. [DOI] [PubMed] [Google Scholar]
- 28.Evans SE, Williams C, Arthur CJ, Płoskoń E, Wattana-amorn P, Cox RJ, Crosby J, Willis CL, Simpson TJ and Crump MP, J. Mol. Biol, 2009, 389, 511–528. [DOI] [PubMed] [Google Scholar]
- 29.Shakya G, Rivera H, Lee DJ, Jaremko MJ, La Clair JJ, Fox DT, Haushalter RW, Schaub AJ, Bruegger J, Barajas JF, White AR, Kaur P, Gwozdziowski ER, Wong F, Tsai S-C and Burkart MD, J. Am. Chem. Soc, 2014, 136, 16792–16799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haushalter RW, Filipp FV, Ko K, Yu R, Opella SJ and Burkart MD, ACS Chem. Biol, 2011, 6, 413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaremko MJ, Lee DJ, Opella SJ and Burkart MD, J. Am. Chem. Soc, 2015, 137, 11546–11549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ploskoń E, Arthur CJ, Evans SE, Williams C, Crosby J, Simpson TJ and Crump MP, J. Biol. Chem, 2008, 283, 518–528. [DOI] [PubMed] [Google Scholar]
- 33.Tran L, Broadhurst RW, Tosin M, Cavalli A and Weissman KJ, Chem. Biol, 2010, 17, 705–716. [DOI] [PubMed] [Google Scholar]
- 34.Finzel K, Lee DJ and Burkart MD, ChemBioChem, 2015, 16, 528–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodenhausen G and Ruben DJ, Chem. Phys. Lett, 1980, 69, 185–189. [Google Scholar]
- 36.Bax A, Curr. Opin. Struct. Biol, 1994, 4, 738–744. [Google Scholar]
- 37.Macomber RS, J. Chem. Educ, 1992, 69, 375. [Google Scholar]
- 38.Williamson MP, Prog. Nucl. Magn. Reson. Spectrosc, 2013, 73, 1–16. [DOI] [PubMed] [Google Scholar]
- 39.McCammon JA, Gelin BR and Karplus M, Nature, 1977, 267, 585–590. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y and Ma S, ChemMedChem, 2013, 8, 1589–1608. [DOI] [PubMed] [Google Scholar]
- 41.Heath RJ, White SW and Rock CO, Appl. Microbiol. Biotechnol, 2002, 58, 695–703. [DOI] [PubMed] [Google Scholar]
- 42.Moche M, Schneider G, Edwards P, Dehesh K and Lindqvist Y, J. Biol. Chem, 1999, 27, 1574–1574. [DOI] [PubMed] [Google Scholar]
- 43.Roujeinikova A, Sedelnikova S, de Boer G-J, Stuitje AR, Slabas AR, Rafferty JB and Rice DW, J. Biol. Chem, 1999, 274, 30811–30817. [DOI] [PubMed] [Google Scholar]
- 44.Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME and Rock CO, J. Biol. Chem, 1999, 274, 11110–11114. [DOI] [PubMed] [Google Scholar]
- 45.Levy CW, Roujeinikova A, Sedelnikova S, Baker PJ, Stuitje AR, Slabas AR, Rice DW and Rafferty JB, Nature, 1999, 398, 383–384. [DOI] [PubMed] [Google Scholar]
- 46.Tang Y, Tsai S-C and Khosla C, J. Am. Chem. Soc, 2003, 125, 12708–12709. [DOI] [PubMed] [Google Scholar]
- 47.Worthington AS and Burkart MD, Org Biomol Chem, 2006, 4, 44–46. [DOI] [PubMed] [Google Scholar]
- 48.Quadri LEN, Weinreb PH, Lei M, Nakano MM, Zuber P and Walsh CT, Biochemistry, 1998, 37, 1585–1595. [DOI] [PubMed] [Google Scholar]
- 49.Worthington AS, Rivera H, Torpey JW, Alexander MD and Burkart MD, ACS Chem. Biol, 2006, 1, 687–691. [DOI] [PubMed] [Google Scholar]
- 50.Ishikawa F, Haushalter RW, Lee DJ, Finzel K and Burkart MD, J. Am. Chem. Soc, 2013, 135, 8846–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishikawa F, Haushalter RW and Burkart MD, J. Am. Chem. Soc, 2012, 134, 769–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beld J, Cang H and Burkart MD, Angew. Chem. Int. Ed Engl, 2014, 53, 14456–14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyanaga A, Iwasawa S, Shinohara Y, Kudo F and Eguchi T, Proc. Natl. Acad. Sci, 2016, 113, 1802–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen C, Haushalter RW, Lee DJ, Markwick PRL, Bruegger J, Caldara-Festin G, Finzel K, Jackson DR, Ishikawa F, O’Dowd B, McCammon JA, Opella SJ, Tsai S-C and Burkart MD, Nature, 2014, 505, 427–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong X, Bailey CD, Williams C, Crosby J, Simpson TJ, Willis CL and Crump MP, Chem. Sci, 2016, 7, 1779–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ellis BD, Milligan JC, White AR, Duong V, Altman PX, Mohammed LY, Crump MP, Crosby J, Luo R, Vanderwal CD and Tsai S-C, J. Am. Chem. Soc, 2018, 140, 4961–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barajas JF, Shakya G, Moreno G, Rivera H, Jackson DR, Topper CL, Vagstad AL, La Clair JJ, Townsend CA and Burkart MD, Proc. Natl. Acad. Sci, 2017, 201609001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meanwell NA, J. Med. Chem, 2011, 54, 2529–2591. [DOI] [PubMed] [Google Scholar]
- 59.Leesong M, Henderson BS, Gillig JR, Schwab JM and Smith JL, Structure, 1996, 4, 253–264. [DOI] [PubMed] [Google Scholar]
- 60.Roujeinikova A, Baldock C, Simon WJ, Gilroy J, Baker PJ, Stuitje AR, Rice DW, Slabas AR and Rafferty JB, Structure, 2002, 10, 825–835. [DOI] [PubMed] [Google Scholar]
- 61.Arabolaza A, Shillito ME, Lin T-W, Diacovich L, Melgar M, Pham H, Amick D, Gramajo H and Tsai S-C, Biochemistry, 2010, 49, 7367–7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qiu X, Janson CA, Konstantinidis AK, Nwagwu S, Silverman C, Smith WW, Khandekar S, Lonsdale J and Abdel-Meguid SS, J. Biol. Chem, 1999, 274, 36465–36471. [DOI] [PubMed] [Google Scholar]
- 63.Serre L, Verbree EC, Dauter Z, Stuitje AR and Derewenda ZS, J. Biol. Chem, 1995, 270, 12961–12964. [DOI] [PubMed] [Google Scholar]
- 64.Kimber MS, Martin F, Lu Y, Houston S, Vedadi M, Dharamsi A, Fiebig KM, Schmid M and Rock CO, J. Biol. Chem, 2004, 279, 52593–52602. [DOI] [PubMed] [Google Scholar]
- 65.Pan H, Tsai S, Meadows ES, Miercke LJW, Keatinge-Clay AT, O’Connell J, Khosla C and Stroud RM, Structure, 2002, 10, 1559–1568. [DOI] [PubMed] [Google Scholar]
- 66.Hadfield AT, Limpkin C, Teartasin W, Simpson TJ, Crosby J and Crump MP, Structure, 2004, 12, 1865–1875. [DOI] [PubMed] [Google Scholar]
- 67.Mori T, Shimomura K, Saito Y, Yang D, Awakawa T, Morita H and Abe I, BE Publ, DOI: 10.2210/pdb5gk1/pdb. [DOI] [PubMed] [Google Scholar]
- 68.Hertweck C, Luzhetskyy A, Rebets Y and Bechthold A, Nat. Prod. Rep, 2007, 24, 162–190. [DOI] [PubMed] [Google Scholar]
- 69.Choi K-H, Kremer L, Besra GS and Rock CO, J. Biol. Chem, 2000, 275, 28201–28207. [DOI] [PubMed] [Google Scholar]
- 70.Gajiwala Ketan S, Margosiak Stephen, Lu Jia, Cortez Joseph, Su Ying, Nie Zhe and Appelt Krzysztof, FEBS Lett, 2009, 583, 2939–2946. [DOI] [PubMed] [Google Scholar]
- 71.Qiu X, Janson CA, Smith WW, Head M, Lonsdale J and Konstantinidis AK, J. Mol. Biol, 2001, 307, 341–356. [DOI] [PubMed] [Google Scholar]
- 72.Ellis BD, Milligan JC, White AR, Duong V, Altman PX, Mohammed LY, Crump MP, Crosby J, Luo R, Vanderwal CD and Tsai S-C, J. Am. Chem. Soc, 2018, 140, 4961–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bisang C, Long PF, Corte´s J, Westcott J, Crosby J, Matharu A-L, Cox RJ, Simpson TJ, Staunton J and Leadlay PF, Nature, 1999, 401, 502–505. [DOI] [PubMed] [Google Scholar]
- 74.McDaniel R, Ebert-Khosla S, Hopwood DA and Khosla C, Nature, 1995, 375, 549–554. [DOI] [PubMed] [Google Scholar]
- 75.Korman TP, Tan Y, Wong J, Luo R and Tsai S-C, Biochemistry, 2008, 47, 1837–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carreras CW and Khosla C, Biochemistry, 1998, 37, 2084–2088. [DOI] [PubMed] [Google Scholar]
- 77.Fu H, Hopwood DA and Khosla C, Chem. Biol, 1994, 1, 205–210. [DOI] [PubMed] [Google Scholar]
- 78.McDaniel R, Ebert-Khosla S, Fu H, Hopwood DA and Khosla C, Proc. Natl. Acad. Sci. U. S. A, 1994, 91, 11542–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heath RJ and Rock CO, J. Biol. Chem, 1996, 271, 27795–27801. [DOI] [PubMed] [Google Scholar]
- 80.Helmkamp GM and Bloch K, J. Biol. Chem, 1969, 244, 6014–6022. [PubMed] [Google Scholar]
- 81.Finzel K, Nguyen C, Jackson DR, Gupta A, Tsai S-C and Burkart MD, Chem. Biol, 2015, 22, 1453–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Milligan JC, Jackson DR, Barajas JF and Tsai SC, BE Publ, DOI: 10.2210/pdb5kof/pdb. [DOI] [Google Scholar]
- 83.Zhang L, Xiao J, Xu J, Fu T, Cao Z, Zhu L, Chen H-Z, Shen X, Jiang H and Zhang L, Cell Res, 2016, 26, 1330–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rafi S, Novichenok P, Kolappan S, Zhang X, Stratton CF, Rawat R, Kisker C, Simmerling C and Tonge PJ, J. Biol. Chem, 2006, 281, 39285–39293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tallorin L, Finzel K, Nguyen QG, Beld J, La Clair JJ and Burkart MD, J. Am. Chem. Soc, 2016, 138, 3962–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fujita Y, Matsuoka H and Hirooka K, Mol. Microbiol, 2007, 66, 829–839. [DOI] [PubMed] [Google Scholar]
- 87.Yan H, Wang Z, Wang F, Tan T and Liu L, Eng. Life Sci, 2016, 16, 53–59. [Google Scholar]
- 88.Feng Y, Zhang Y, Wang Y, Liu J, Liu Y, Cao X and Xue S, Appl. Microbiol. Biotechnol, 2018, 102, 3173–3182. [DOI] [PubMed] [Google Scholar]
- 89.Blatti JL, Beld J, Behnke CA, Mendez M, Mayfield SP and Burkart MD, PLoS ONE, 2012, 7, e42949. [DOI] [PMC free article] [PubMed] [Google Scholar]