Abstract
How the epigenome of one cell type is remodeled during reprogramming into another unrelated type of cell remains unclear. Overexpression of transcription factors in somatic cells enables the induction of induced pluripotent stem cells (iPSCs). This process entails genome-wide remodeling of DNA methylation, chromatin, and transcription. Recent work suggests that the number of active X chromosomes present in a cell influences remodeling of DNA methylation during somatic cell reprogramming to mouse iPSCs. Female iPSCs with 2 active X chromosomes display global DNA hypomethylation, whereas male XY iPSCs show DNA methylation levels similar to the somatic cells they are derived from. Global DNA methylation erasure in female iPSCs takes place genome-wide and involves repression of DNA methyltransferases. However, on loss of one X chromosome, female iPSCs acquire a DNA methylation landscape resembling that of XY iPSCs. Therefore, it is the X chromosome dosage that dictates global DNA methylation levels in iPSCs. Here, we discuss the evidence that links X chromosome dosage with the regulation of DNA methylation in pluripotent stem cells. We focus on iPSCs reprogramming studies, where X chromosome status is a novel factor impacting our understanding of epigenetic remodeling.
Keywords: Induced pluripotent stem cells (iPSCs), X chromosome inactivation, DNA methylation, reprogramming, epigenetics
Comment on: Pasque V, Karnik R, Chronis C, et al. X chromosome dosage influences DNA methylation dynamics during reprogramming to mouse iPSCs. Stem Cell Reports. 2018;10:1537-1550. doi:10.1016/j.stemcr.2018.03.019. PMID: 29681539. https://www.ncbi.nlm.nih.gov/pubmed/29681539
Introduction
A fundamental discovery in biology is that differentiated cells can be reprogrammed into induced pluripotent stem cells (iPSCs) by transcription factor overexpression.1 Induced pluripotent stem cells resemble embryonic stem cells (ESCs) derived from the inner cell mass (ICM) of preimplantation embryos. They possess the capacity for indefinite self-renewal and differentiation into any cell type of the body. Therefore, reprogramming to iPSCs is a promising technology for regenerative medicine and a powerful tool for studying cellular identity. In a stepwise manner, reprogramming to iPSCs involves silencing of somatic genes, followed by sequential activation of pluripotency genes. Such rewiring of transcription involves remodeling of the epigenome.
The specialized identity of somatic cells is maintained by a variety of epigenetic mechanisms that ensure its stability. For reprogramming to succeed, these mechanisms must be reversed. This is an important point because failure to properly reset the epigenome has been associated with incomplete and aberrant reprogramming.2 DNA methylation is one example of modification associated with epigenetic mechanisms, which is remodeled during reprogramming.3 However, when and how DNA methylation is reset during transcription factor–induced pluripotency remains poorly understood.
Global and Sex-Specific Changes in DNA Methylation During Reprogramming to iPSCs
Efforts over the past decade have shed light onto the dynamics and the molecular mechanisms underlying DNA methylation reprogramming. Previous studies suggested that DNA methylation changes occur late during reprogramming.4–6 The iPSCs have also been shown to tolerate global DNA hypomethylation.3 Inhibition of the DNA methyltransferase 1, DNMT1, enhances the efficiency of iPSCs’ derivation,7 suggesting that DNA methylation acts as a barrier to reprogramming.
Although the sex of cells is often ignored in reprogramming studies, it was recently revealed to strongly influence the resetting of global DNA methylation in iPSCs.8,9 The iPSCs derived from male cells show levels of global DNA methylation comparable with somatic cells,8 such that the global level of DNA methylation does not change dramatically during reprogramming. However, when female cells are reprogrammed, the resulting iPSCs display global erasure of DNA methylation8 (Figure 1). The global hypomethylation takes place after the intermediate reprogramming stage marked by SSEA1. In support of sex-specific DNA hypomethylation in iPSCs, studies in ESCs have reported female-specific DNA hypomethylation.10 In summary, changes in global DNA methylation level are sex specific and take place late in reprogramming to iPSCs.
Figure 1.
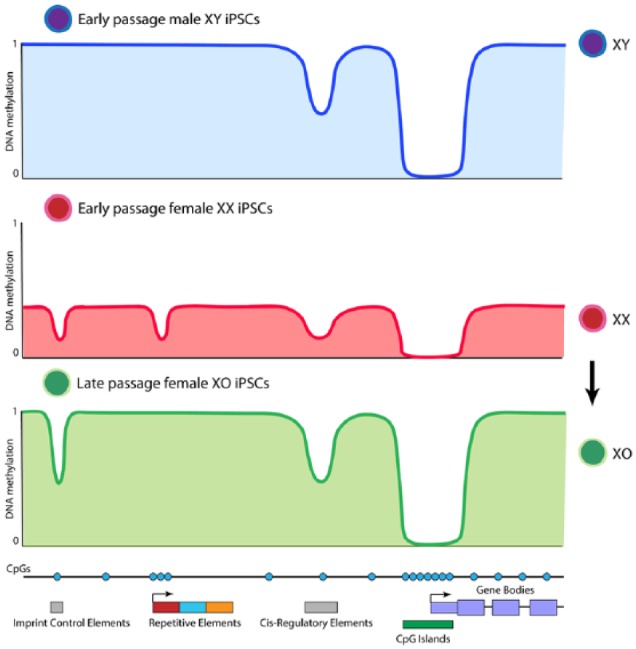
Representation of X dosage–dependent DNA methylation dynamics in male and female iPSCs. Global DNA methylation levels correlate with number of active X chromosomes. Imprint control elements are irreversibly hypomethylated in female iPSCs. Repetitive elements are demethylated in XX but not XY iPSCs. Cis-regulatory elements and CpG islands are demethylated irrespective of X dosage. iPSCs indicate induced pluripotent stem cells.
Image reproduced with permission from Pasque et al.8
Global DNA Hypomethylation in iPSCs Correlates With the Presence of Two Active X Chromosomes
Reprogramming somatic cells to pluripotency induces reactivation of the silent X chromosome (XCR) in female cells.11 Therefore, early passage female iPSCs have 2 active X chromosomes, whereas male iPSCs have only 1 active X chromosome. This skewed ratio of X-to-autosome gene expression (X dosage) underlies DNA hypomethylation in female iPSCs.8 This is supported by experiments where on high passage, female iPSCs lose 1 of the 2 X chromosomes and as a result, global DNA methylation reverts to levels similar to XY iPSCs8 (Figure 1). Therefore, X dosage dictates global methylation levels in iPSCs.
Mechanistically, it is likely that, as a result of XCR in female iPSCs, the expression of X-linked gene Dusp9 increases and drives global hypomethylation by modulating the levels of DNA methyltransferases DNMT3A and DNMT3B, and most likely DNMT1 as well.12 In addition, a role for UHRF1, involved in recruiting DNMT1 for DNA methylation maintenance, and a noncatalytic role of AID were also reported to be involved in global DNA methylation erasure.9 Whether these act downstream of Dusp9 remains to be tested. In summary, differences in the genetic constitution of male and female cells, as well as in the epigenetic status of X chromosomes in somatic and pluripotent cells, lead to divergences in global DNA methylation level in iPSCs.
Influence of X Dosage on DNA Methylation of Key Regulatory Regions Associated With the Control of Cell Identity
Comprehensive genome-wide DNA methylation analyses have revealed that the dynamics of DNA methylation during reprogramming depend on the type of genetic elements considered and their location in the genome.
Imprinting is an epigenetic mechanism in mammals, required for proper development, in which differential expression of the maternal and paternal alleles of certain genes has been attributed to DNA methylation. The global erasure of DNA methylation in female iPSCs leads to the loss of this mark at imprint control regions8 (Figure 1). Moreover, these imprints are not reestablished after genome remethylation in XO female iPSCs.8 Thus, increased X dosage induces irreversible changes in DNA methylation associated with key mammalian epigenetic mechanisms.
Transcriptional programs are controlled by a set of key regulatory regions that include enhancers and promoters. Although the expression of somatic genes is downregulated early during reprogramming, somatic enhancers remethylation is initiated at intermediate reprogramming stages and completed only late during reprogramming in male iPSCs.8,9 In addition, the early wave of focal DNA demethylation at ESC enhancers and ESC super-enhancers is initiated early during reprogramming, independent of sex, before global DNA methylation erasure in female iPSCs5,8,9,13 (Figure 1). The importance of hypomethylation of regulatory elements for the function of enhancers in the context of reprogramming to iPSCs remains to be established. Focal DNA demethylation at ESCs enhancers early in reprogramming coincides with binding sites of OCT4 and SOX2.6,8 Taken together, these studies point to dynamic DNA methylation changes at key regulatory regions of the genome initiated at intermediate reprogramming stages and completed late during the induction of pluripotency.
Repetitive elements form a large portion of mammalian genomes. Processes such as DNA methylation and repressive histone modifications help to maintain repression of these elements, including potentially mobile transposable elements. Interestingly, several classes of repetitive elements such as LINEs, SINEs, and LTRs are demethylated in XX iPSCs, but not in XY iPSCs (Figure 1), indicating that also these elements are sensitive to X dosage in pluripotent stem cells.8
The overall picture that emerges is that distinct regulatory elements are dynamically methylated and unmethylated during reprogramming depending on their usage, with global erasure of DNA methylation as a result of increased X dosage in female iPSCs affecting most genomic regions tested.
Implications for Studying Reprogramming
These findings bring a set of important questions. Whether the pattern of DNA methylation in iPSCs reflects the state of pluripotent cells in the embryo and irrespective of that, what are the consequences of sex-specific DNA methylation for reprogramming studies and conclusions drawn from them?
X chromosome inactivation is a developmentally regulated process used in mammals to mediate gene dosage compensation between XX and XY cells. Early in development, pluripotent cells of the female ICM transiently acquire 2 active X chromosomes. However, it does not lead to clear differences in DNA methylation between female and male ICMs.12 Hence, we speculate that global DNA hypomethylation in iPSCs results from the long-term maintenance of what normally is a transient developmental state in the ICM.
To study DNA methylation in reprogramming, it is imperative to consider the sex of the cells used. For example, using only male cells would bypass female-specific global DNA demethylation in the pluripotent state. Hence, DNA methylation changes seen would mistakenly be attributed to reprogramming in general rather than X dosage effects. Conversely, pooling male and female cells in unknown ratios masks DNA methylation heterogeneity. Likewise, the sex of control cells should match the experimental ones, to avoid biased interpretations. Moreover, comparing male and female cells is not sufficient. One must also define the number of active X chromosomes, as, strictly speaking, it is X dosage that is important in female iPSCs for dictating global DNA methylation, and not the sex in its entirety. Therefore, comparing male and female iPSCs requires the knowledge of the number of active X chromosomes. Recent work by our group also shows that X dosage modulates the transcriptional and open chromatin landscape, as well as growth properties, of iPSCs.14 Altogether, these studies link sex chromosomes with DNA methylation and pluripotency and as a result bring novel, nonnegligible considerations to studying and understanding cell fate reprogramming.
Implications for Human Pluripotency
Are X dosage–dependent DNA methylation dynamics also present in human ESCs and iPSCs? Sex-specific differences in gene expression, but not in the global level of DNA methylation, have been reported in conventional primed human pluripotent stem cells.15 Currently, however, human naive pluripotent stem cells are thought to reside in an incomplete naive pluripotent state and have reduced X-linked gene expression compared with female cells of the early preimplantation embryo. We speculate that naive human pluripotent stem cells cultured in conditions that would properly induce X dosage increase may show additional sex-specific molecular and functional differences. With the emergence of improved culture conditions to induce and maintain human naive pluripotency, it will be interesting to assess potential X dosage effects.
Concluding Remarks
DNA methylation is a paradigmatic mark for the study of epigenetic processes. X dosage can have important effects on DNA methylation in iPSCs. Understanding the mechanisms by which DNA methylation is established and maintained is important to gain insights into health and diseases. Somatic cell reprogramming to iPSCs provides a useful system to study the reversal of epigenetic memory and the molecular mechanisms involved in cell state transitions.
Acknowledgments
The authors would like to thank our colleagues who made this research possible. They apologize to those authors for which we could not cite their work due to space constraint.
Footnotes
Author Contributions: AJ and VP wrote the manuscript with input from all authors.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: This work was supported by The Research Foundation – Flanders (FWO) (Odysseus return grant G0F7716N to V.P.), the KU Leuven Research Fund (BOFZAP starting grant StG/15/021BF to V.P., C1 grant C14/16/077 to V.P. and Project financing), and FWO PhD fellowships to A.J. (1158318N).
ORCID iD: Adrian Janiszewski  https://orcid.org/0000-0002-4156-5791
https://orcid.org/0000-0002-4156-5791
References
- 1. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. [DOI] [PubMed] [Google Scholar]
- 2. Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. [DOI] [PubMed] [Google Scholar]
- 4. Polo JM, Anderssen E, Walsh RM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee D-S, Shin JY, Tonge PD, et al. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat Commun. 2014;5:5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knaupp AS, Buckberry S, Pflueger J, et al. Transient and permanent reconfiguration of chromatin and transcription factor occupancy drive reprogramming. Cell Stem Cell. 2017;21:834–845.e6. [DOI] [PubMed] [Google Scholar]
- 7. Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pasque V, Karnik R, Chronis C, et al. X chromosome dosage influences DNA methylation dynamics during reprogramming to mouse iPSCs. Stem Cell Reports. 2018;10:1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Milagre I, Stubbs TM, King MR, et al. Gender differences in global but not targeted demethylation in iPSC reprogramming. Cell Rep. 2017;18:1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zvetkova I, Apedaile A, Ramsahoye B, et al. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet. 2005;37:1274–1279. [DOI] [PubMed] [Google Scholar]
- 11. Maherali N, Sridharan R, Xie W, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. [DOI] [PubMed] [Google Scholar]
- 12. Choi J, Clement K, Huebner AJ, et al. DUSP9 modulates DNA hypomethylation in female mouse pluripotent stem cells. Cell Stem Cell. 2017;20:706–719.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwarz BA, Cetinbas M, Clement K, et al. Prospective isolation of poised iPSC intermediates reveals principles of cellular reprogramming. Cell Stem Cell. 2018;23:289–305.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Song J, Janiszewski A, de Geest N, et al. Two active X-chromosomes modulate the growth, pluripotency exit and DNA methylation landscape of mouse naive pluripotent stem cells through different pathways. bioRxiv. 2018. doi: 10.1101/291450. [DOI] [Google Scholar]
- 15. Ronen D, Benvenisty N. Sex-dependent gene expression in human pluripotent stem cells. Cell Rep. 2014;8:923–932. [DOI] [PubMed] [Google Scholar]


