Abstract
How organisms retain a memory of ancestral environmental exposure is a phenomenon that is still poorly understood. Recently published work by our group and others, regarding environmentally induced transgenerational effects, have identified an array of mechanisms, with a particular focus on histone modifications, that shed some light on the underlying epigenetic processes driving long-term generational effects.
Keywords: C. elegans, transgenerational inheritance, reproductive function, Bisphenol A, histone demethylase
Comment on: Camacho J, Truong L, Kurt Z, Chen YW, Morselli M, Gutierrez G, Pellegrini M, Yang X, Allard P. The memory of environmental chemical exposure in C. elegans is dependent on the Jumonji demethylases jmjd-2 and jmjd-3/utx-1. Cell Rep. 2018;23:2392-2404. doi:10.1016/j.celrep.2018.04.078. PubMed PMID: 29791850; PubMed Central PMCID: PMC6003705. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6003705/
The elicitation and inheritance of phenotypes from environmental cues has been researched and debated for decades.1 There is now ample evidence that the epigenome is a key mediator of biologic response and adaptation of organisms to a wide array of natural environmental changes such as starvation,2 diet,3–5 temperature,6,7 and hyperosmotic stress.8 However, whether these changes could be passed down to future generations and whether non-natural, i.e., man-made, cues could also elicit a transgenerational response largely remain to be clarified, especially from a mechanistic standpoint.
Since the identification of a transgenerational impact of exposure to the pesticide Vinclozolin,9,10 followed by similar findings with Bisphenol A (BPA) and phthalates,11 DNA methylation has been proposed to be a central mediator of effect transmission since the various phenotypes observed in later generations beyond where a direct exposure could have occurred were correlated with an alteration of DNA methylation patterns. These findings also indicated a prominent role for germ cells since these marks could only be transferred to later generations via the germline. However, the extensive epigenetic reprogramming of primordial germ cells during early embryogenesis, where most CpG methylation is erased,12 has been seen as a challenge in explaining how DNA methylation alone could be the mechanism of inheritance. Thus, these studies left a gap in our mechanistic understanding of environmental epigenetic inheritance and did not explore the involvement of other epigenetic marks beside DNA methylation.
Our work13 sought to clarify the mechanisms of transgenerational effects of man-made environmental chemicals, while focusing on epigenetic marks other than DNA methylation. This was made possible utilizing the Caenorhabditis elegans model, which lacks definitive 5 mC DNA methylation, but shares a remarkable degree of conservation of histone modifications and of the machinery that regulates them. In C. elegans, core histones share 80% identical amino acid sequence when compared with human histones.14 Additionally, some of the post-translational modifications of the C. elegans H3 (CeHIS3) and C. elegans H4 (CeHIS4) proteins have been shown to be identical to those in human H3 and H4 proteins.15–17 Because of sequence conservation, it is hypothesized that C. elegans have direct orthologues of all the mammalian modification enzymes.18 C. elegans can therefore provide further information to fill the existent gap in knowledge regarding environmental exposures and inherited effects. In our work, we decided to focus on exposure to BPA, a well-known plastic manufacturing chemical and endocrine disruptor. Previous studies showed that its exposure is linked to epigenetic alterations such as decreased DNA methylation in mice,19 decreased DNA methylation in preadolescent girls,20 as well as reduced concentration of H3K9me3 in mouse germinal vesicle oocytes.21 These studies indicated BPA’s ability to affect the epigenome in a variety of ways, and our research therefore aimed to clearly identify BPA’s short- and long-term generational effects and the mechanisms behind them.
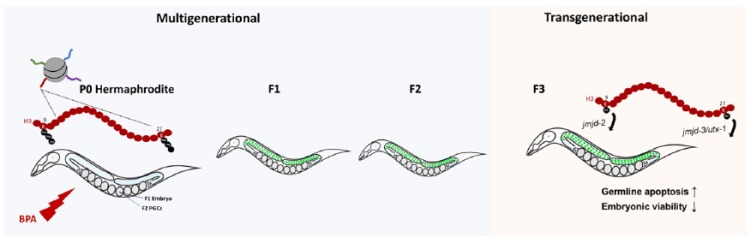
In sum, we uncovered a transgenerational effect on reproduction stemming from exposure to BPA that was mediated in part by a deregulation of repressive histone modifications.13 To come to this conclusion, we first used a strain carrying a highly repetitive GFP transgene that is epigenetically silenced in the germline in a fashion that is reminiscent of the silencing of endogenous heterochromatin in the germline. Our assessment focused on the ability of BPA to disrupt the silenced state of the transgene in the germline that we could monitor and measure over several generations (see Figure 1). Results indicated a significantly higher repetitive array de-silencing effect in the germline after BPA exposure that endured for five generations. To further investigate the impact of ancestral BPA exposure on the germline, we performed RNA-seq analysis on germline tissue, which identified 264 transcripts that were significantly upregulated or downregulated in F3 germlines ancestrally exposed to BPA compared with dimethyl sulfoxide (DMSO). Gene ontology analyses of functional categories represented highlighted reproduction as a representative functional category, indicating a profound transgenerational impact of ancestral BPA exposure on the germline transcriptome, and ultimately germline function.
Figure 1.
BPA exposures in C. elegans reduces the levels of the repressive histone marks H3K9me3 and H3K27me3, regulated by the demethylases jmjd-2 and jmjd-3/utx-1, respectively. This disruption causes a de-silencing effect and reproductive dysfunction observed from the P0 generation until the F4. The F3 generation represents the first generation where there was no direct contact with the environmental toxicant (BPA).
In addition to RNA-seq, we also performed ChIP-seq analysis in whole worms, which showed that the repressive marks H3K9me3 and H3K27me3 predominantly occupy the gene body of silenced genes and that H3K27me3 is significantly reduced in the gene body of BPA ancestrally exposed worms compared with the control groups. Comparing distribution of repressive marks along the chromosome axes showed reduction of both marks from the distal chromosomal regions, largely heterochromatic, and a slight enrichment in the chromosome centers when comparing BPA to DMSO. Noting these differences in the epigenome occur transgenerationally, our focus shifted back to the germline where the changes must occur in order to be inherited. Pachytene germline nuclei imaged by immunofluorescence showed a significant ~25% reduction in global H3K9me3 and H3K27me3 between DMSO and BPA. This transgenerational impact on repressive marks in the germline was not solely confined to the repetitive array but was also detectable on the autosomes and the X-chromosome. Since F3 germlines showed a strong alteration of their chromatin and transcriptome, we investigated whether these were associated with transgenerational reproductive defects. We indeed measured an increase in embryonic lethality in worms ancestrally exposed to BPA when compared with DMSO control. Additionally, we measured germline health by monitoring induction of germline apoptosis and observed a significant increase in apoptotic germline nuclei in F3 worms ancestrally exposed to BPA. This indicated that ancestral BPA exposure elicits a clear transgenerational reproductive dysfunction effect.
Finally, getting to mechanisms, we tested the causal relationship between the reduction in H3K9me3 and H3K27me3 germline levels and BPA-induced transgenerational outcomes. We believed that the dependence of these marks might involve the activity of enzymes that regulate them. This was supported by our own aforementioned RNA-seq data in which seven differentially expressed chromatin factors were identified. Since BPA appeared to reduce repressive marks, we focused on histone demethylases targeting H3K9me3 and H3K27me3 to attempt to rescue BPA’s transgenerational effects. Using a feeding RNAi strategy targeting jmjd-2 (H3K9me3/H3K36me3 KDM)22,23 or jmjd-3/utx-1 (H3K27me3 KDM),24 we were able to modulate and rescue BPA’s transgenerational effects. The downregulation of jmjd-2 or jmjd-3/utx-1 through RNAi at the F1 to F2 transition increased the levels of H3K9me3 and H3K27me3 in the F3 germlines. The RNAi treatment also led to a rescue of BPA-induced reproductive dysfunction in the F3. To validate these results, we also performed drug rescue experiments using KDM4/JMJD-2 inhibitor IOX-1, which has been shown to elevate H3K9me3 levels in vitro and in cell culture settings,25–27 and the potent selective Jumonji JMJD-3/UTX-1 H3K27 demethylase inhibitor GSK-J4.28 Using the chemicals individually or in combination to inhibit both demethylases significantly decreased the germline array desilencing and embryonic lethality effects. These two distinct methods of rescuing BPA’s transgenerational effects indicate that the activity of either JMJD2 or JMJD3/UTX1 is required for inheritance of BPA-induced reproductive effects. Our approach is what distinguishes our study from others. We did not use mutants where the initial response to the environmental cue would be abrogated. Instead, we used RNAi or drug exposure in such a way that the worms could respond appropriately to the cue first but then were prevented from transferring that information to the following generations. Therefore, our study is unique in its ability to discriminate between altered response and altered inheritance of effect.
Together, these results demonstrate the key role of repressive histone modifications, namely, H3K9me3 and H3K27me3, in the inheritance of reproductive dysfunctions induced by a well-defined environmental exposure. These findings shine a light on how artificial environmental exposures can be biologically integrated and transgenerationally inherited. Our work highlights the importance of comprehensively examining our chemical environmental for its potential effects on our germline epigenome, which can in turn allow us to find interventional means to prevent transmission of effects to future generations.
Although our work begins to answer questions in the field of environmental exposure effects on future generations, currently there is not a single epigenetic mark that can be considered responsible for the transfer of environmental exposure effects from one generation to the next, and H3K4me3, H3K9me3, and H3K27me3 have all been implicated in that process.29 This could be due to potential redundancy or crosstalk between specific marks although we cannot exclude the possibility that specific exposures may use distinct epigenetic mechanisms for their inheritance. Recent studies have highlighted the challenge of identifying a unifying mechanism of inheritance, if it indeed exists. For example, other C. elegans studies showed that starvation can cause transgenerational effects mediated through the generation of small RNAs that target genes important for nutrition.30 Interestingly, histone modifications were also functionally connected to transgenerational effects and small RNA transfer. Indeed, in met-2 C. elegans mutants, which are defective in H3K9 methyltransferase, there is a progressive reduction in fertility that unfolds over 10 to 30 generations.31 The argonaute factor hdre-1, associated with small RNAs, is required for the progressive sterility phenotype in the met-2 mutant. Thus, the authors proposed a model where MET-2 functions to suppress the transgenerational transfer of small RNAs via the regulation of H3K9me3, as a model of inheritance. Together, these recent studies show the importance of epigenetic mechanisms other than DNA methylation as vehicles for transgenerational effects and highlight the need to comprehensively examine distinct exposures and epigenetic mechanisms to improve our understanding of environmental memory.
Finally, our work as well as that of others point to another important question. For phenotypes that are present in the soma of later generations, how is the information, likely epigenetically encoded as demonstrated by our efforts, transferred from germ cells across developmental and differentiation stages to affect adult cell types, altering their cellular programs and function. Such a complex question is also best addressed in C. elegans where the location and timing of each cellular differentiation event are well described. Thus, while there is still much work to do, our current studies are helping to create guidelines based on model organisms and standardized approaches that will in turn allow us to understand the underlying intricate mechanisms of environmental exposure effects unraveling across generations.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: JC received support from NIH/NIEHS T32 ES015457 Training in Molecular Toxicology, the North American Graduate Fellowship, the NSF AGEP Competitive Edge, the NSF Graduate Research Fellowship, and the Eugene-Cota Robles Fellowship. PA is supported by NIH/NIEHS R01 ES02748701 and the Burroughs Wellcome Foundation.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: JC and PA wrote the manuscript.
References
- 1. Burbank L. The training of the human plant. The Century Magazine. May 1907, 1907:127–137. [Google Scholar]
- 2. Ravelli GP, Stein ZA, Susser MW. Obesity in young men after famine exposure in utero and early infancy. N Engl J Med. 1976;295:349–353. [DOI] [PubMed] [Google Scholar]
- 3. Lambrot R, Xu C, Saint-Phar S, et al. Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat Commun. 2013;4:2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ng S-F, Lin RCY, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. [DOI] [PubMed] [Google Scholar]
- 5. Carone BR, Fauquier L, Habib N, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13:627–639. [DOI] [PubMed] [Google Scholar]
- 7. Song J, Angel A, Howard M, Dean C. Vernalization: a cold-induced epigenetic switch. J Cell Sci. 2012;125:3723– 3731. [DOI] [PubMed] [Google Scholar]
- 8. Seong KH, Li D, Shimizu H, Nakamura R, Ishii S. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011;145:1049–1061. [DOI] [PubMed] [Google Scholar]
- 9. Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006;147:5515–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS ONE. 2013;8:e55387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang WWC, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nat Rev Genet. 2016;17:585–600. [DOI] [PubMed] [Google Scholar]
- 13. Camacho J, Truong L, Kurt Z, et al. The memory of environmental chemical exposure in C. elegans is dependent on the Jumonji demethylases jmjd-2 and jmjd-3/utx-1. Cell Rep. 2018;23:2392–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vanfleteren JR, Bun SMV, Delcambe LL, Beeumen JJV. Multiple forms of histone H2B from the nematode Caenorhabditis elegans. Biochem J. 1986;235:769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanfleteren JR, Bun SMV, Beeumen JJV. The primary structure of histone H2A from the nematode Caenorhabditis elegans. Biochem J. 1987;243:297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vanfleteren JR, Van Bun SM, Van Beeumen JJ. The primary structure of histone H3 from the nematode Caenorhabditis elegans. FEBS Lett. 1987;211:59–63. [DOI] [PubMed] [Google Scholar]
- 17. Vanfleteren JR, Van Bun SM, Van Beeumen JJ. The primary structure of histone H4 from the nematode Caenorhabditis elegans. Comp Biochem Physiol. 1987;87:847–849. [DOI] [PubMed] [Google Scholar]
- 18. Cui M, Han M. Roles of chromatin factors in C. elegans development. In: The C. elegans Research Community, ed. WormBook; May 2007:1-16. Available at: https://www.ncbi.nlm.nih.gov/pubmed/?term=Roles+of+chromatin+factors+in+C.+elegans+development. [DOI] [PMC free article] [PubMed]
- 19. Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104:13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim JH, Rozek LS, Soliman AS, et al. Bisphenol A-associated epigenomic changes in prepubescent girls: a cross-sectional study in Gharbiah, Egypt. Environ Health. 2013;12:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trapphoff T, Heiligentag M, Hajj NE, Haaf T, Eichenlaub-Ritter U. Chronic exposure to a low concentration of bisphenol A during follicle culture affects the epigenetic status of germinal vesicles and metaphase II oocytes. Fertil Steril. 2013;100:1758.e1–1767.e1. [DOI] [PubMed] [Google Scholar]
- 22. Greer EL, Beese -Sims SE, Brookes E, et al. A histone methylation network regulates transgenerational epigenetic memory in C. Elegans. Cell Rep. 2014;7:113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whetstine JR, Nottke A, Lan F, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. [DOI] [PubMed] [Google Scholar]
- 24. Agger K, Cloos PAC, Christensen J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. [DOI] [PubMed] [Google Scholar]
- 25. King ONF, Li XS, Sakurai M, et al. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors. PLoS ONE. 2010;5:e15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu Q, Chen J, Zhang J, Xu C, Yang S, Jiang H. IOX1, a JMJD2A inhibitor, suppresses the proliferation and migration of vascular smooth muscle cells induced by angiotensin II by regulating the expression of cell cycle-related proteins. Int J Mol Med. 2016;37:189–196. [DOI] [PubMed] [Google Scholar]
- 27. Schiller R, Scozzafava G, Tumber A, et al. A cell-permeable ester derivative of the JmjC histone demethylase inhibitor IOX1. ChemMedChem. 2014;9:566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kruidenier L, Chung C, Cheng Z, et al. A selective Jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weinhouse C, Truong L, Meyer JN, Allard P. Caenorhabditis elegans as an emerging model system in environmental epigenetics. Environ Mol Mutagen. 2018;59:7560–7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rechavi O, Houri-Ze’evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ, Hobert O. Starvation-induced transgenerational inheritance of small RNAs in C. Elegans. Cell. 2014;158:2277–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lev I, Seroussi U, Gingold H, Bril R, Anava S, Rechavi O. MET-2-dependent H3K9 methylation suppresses transgenerational small RNA inheritance. Curr Biol. 2017;27:81138–81147. [DOI] [PubMed] [Google Scholar]