Abstract
This protocol is designed to quantify invadopodia formation and activity. Invadopodia are protrusive structures elaborated by cancer cells that mediate cell attachment and remodeling of the extracellular matrix. These structures contribute to the ability of cancer cells to invade and metastasize. In this protocol, both the presence of invadopodia and their activity is simultaneously assessed and quantified by a fluorescent microscopy-based assay.
Material and Reagents
Cells (i.e. SCC61 head and neck carcinoma cells)
Tissue culture media (i.e. DMEM) (Mediatech, catalog number: 10013CV)
Sterile PBS
Trypsin 0.05%/EDTA (Life Technologies, catalog number: 25300054)
Penicillin/Streptomycin solution (Omega Scientific, catalog number PS-20)
Ethanol (Decon Labs, catalog number: 2801)
Forceps Dumont #5 (Fine Science Tools, catalog number:11252-20)
25% Glutaraldehyde (Polysciences, catalog number: 00376-500)
Sucrose (Fisher Scientific, catalog number: S3500)
Sodium Borohydride (Fisher Scientific, catalog number: S67825)
16% Formaldehyde (Electron Microscopy Sciences, catalog number: 15710)
Gelatin from pig skin, Oregon green 488 conjugate (Life Technologies, catalog number: G13186)
Alexa Fluor 568 Phalloidin (Life Technologies, catalog number: A12380)
Mounting medium, Vectashield with DAPI (Vector Laboratories, catalog number: H-1200)
Bovine Serum Albumin fraction V (EMD Millipore, catalog number: 2980-1KG)
Triton X-100 (Promega corporation, catalog number: H5141)
Parafilm (Bemis Company, catalog number: PM996)
Equipment
Round 18 mm diameter glass coverslips (0.13-0.17 mm thickness) (Carolina Biological Supply Company, catalog number: 633033)
Microscope glass slides (75 × 25 mm) (Thermo Fisher Scientific, catalog number: 1255015)
12-well Microplates (Corning, catalog number: 3513)
Thermomixer or water bath set at 60 °C
Tissue culture incubator
Hemocytometer (LabSource, catalog number: 0267154) http://www.lifetechnologies.com/us/en/home/references/gibco-cell-culture-basics/cell-culture-protocols/counting-cells-in-a-hemacytometer.html
Fluorescence Microscope
Vacuum source
Procedure
A. Preparing fluorescent gelatin-coated coverslips
The objective of this step is to perform a gelatin coating as homogeneous as possible. To achieve that, it is better to coat no more than 4 coverslips at a time, repeating the process as many times as necessary to obtain the desired final number of coverslips. By doing that, the fluorescent gelatin stock solution is used recently warmed for each group of four coverslips. Also, it is important to work reasonably quickly (through steps A1 to A4 in Figure 1) to avoid that the gelatin starts drying before it has formed a thin uniform coating of the coverslip.
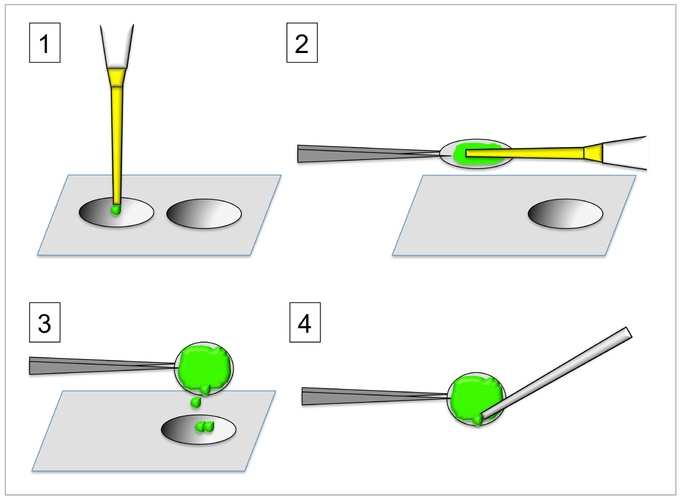
Figure 1. Steps for coating coverslips with fluorescent gelatin.
1-Add pre-warmed gelatin solution to the tip of sterilized coverslips; 2-Hold coverslip with forceps and spread gelatin with the help of a pipet tip. 3-Remove the excess gelatin solution over the next coverslip to prepare. 4-Carefully aspirate the excess gelatin from the lower side of the coverslip. Refer to text for further details.
Sterilize coverslips by soaking them in 70% Ethanol solution, let them air dry. Gelatin-coated coverslips can be prepared in batches of 12 or 24 (one or two multiwell plates) and stored in the dark at 4 °C for up to 1-2 weeks.
Prepare fluorescent gelatin solution by diluting the gelatin stock (prepared according to manufacturer’s instructions at 1 mg/ml by adding 5 ml of distilled water to the commercial vial), in PBS containing 2% sucrose. Typically, gelatin is used at a final concentration of 0.2 mg/ml. Warm the fluorescent gelatin stock at 60 °C prior to dilution and keep the working solution protected from light and warmed during the coating process. Prepare around 500 μl of fluorescent gelatin working solution for 24-coverslip batch. Diluted fluorescent gelatin solution can be reused once upon frozen storage.
Cut a piece of parafilm of around 4 × 2 inches and tape it to the bench top. This will facilitate quick manipulation of the coverslips. Place a set of 4 coverslips in line on top of the parafilm. Have the gelatin working solution pre-warmed in-hand (i.e. by having a thermomixer or a water bath by your side on the bench top). Pipet 100 μl of warmed fluorescent gelatin solution on top of the first coverslip and return the stock to 60 °C (step A1 in Figure 1). Using forceps, lift the coverslip with the gelatin on top and, keeping it horizontal, use the other hand to spread the gelatin with the outside of the same pipet tip attached to a pipettor (step A2 in Figure 1). When spread, hold this coverslip over the next one to prepare and tilt the coverslip to a vertical position letting the excess of gelatin to fall on the next coverslip (step A3 in Figure 1). Still holding the first coverslip in a vertical position, use a soft vacuum source (only turned on lightly, just enough to hold a pipet tip at the end of the hose) to aspirate the extra-coating left on the bottom edge of the coverslip (step 4 in Figure 1). Using stronger vacuum (completely turned on) is not advised since it will leave a gelatin layer that is too thin. Place the coverslip inside the well of a 12-well plate protected from light. Immediately proceed to repeat the same procedure with the next coverslip.
When all coverslips are coated, let them dry (coating will look whitish when dry).
Add 1 ml of a pre-chilled solution of glutaraldehyde (diluted from the stock solution at 0.5% in PBS) to each well and incubate for 15 min on ice.
Remove the glutaraldehyde and dispose properly. Wash coverslips three times at room temperature with PBS.
Add 1 ml of a freshly prepared solution of Sodium Borohydride (5 mg/ml in PBS) and incubate for 3 minutes at room temperature. Stir the plate if necessary to avoid that the Hydrogen bubbles lift the coverslips to the surface of the liquid.
Remove the Sodium Borohydride and dispose properly. Wash coverslips three times at room temperature with PBS.
Transfer plates to a tissue culture hood. Using the forceps (an old pair of forceps with a bent tip may help in this process), transfer coverslips to a sterile 12-well plate and wash three times with sterile PBS. Coverslips may be used the same day or stored in PBS containing 200 units per ml of penicillin and 200 μg per ml of streptomycin (1:50 from the stock solution) at 4 °C in the dark for up to 2 weeks.
Notes:
An alternative method to cover the glass coverslips with the gelatin solution consists on pipetting the gelatin over the parafilm and invert the glass coverslip over the gelatin. This method may not achieve the same homogeneity but it may be easier for manipulation of coverslips.
Using up to 1 mg/ml of fluorescently labeled gelatin solution to coat glass coverslips might help to increase the number of invadopodia formed by certain cell lines. Some cells attach very strongly and “pull” the gelatin coating in addition to degrading it. The effect of pulling cannot be distinguished from the effect of degradation for quantification purposes on this assay. Using 1 mg/ml of fluorescently labeled gelatin solution to coat glass coverslips or decreasing FBS in the growing medium might decrease the ability of some cells to “pull” the gelatin coating.
B. Performing the fluorescent gelatin degradation assay
Working under sterile conditions, transfer the desired number of coated coverslips to a new sterile 12-well plate. If the fluorescent gelatin coated coverslips were already stored in antibiotic-containing PBS, wash them three times with sterile PBS and incubate in 1 ml of the same complete growth medium that you will use in the experiment. Place them inside the 37 °C tissue culture incubator.
Collect the cells to use in the assay using standard techniques (i.e. trypsinization), and count them using the hemocytometer or other suitable method.
Plate 1 ml of a cell suspension, typically containing between 20-40,000 cells on top of each well containing 1 ml of medium.
Return cells to the incubator for 8-16 h. The length of the assay should be determined empirically for each cell line and conditions.
C. Processing cells for invadopodia detection and fluorescent microscopy
At endpoint, wash cells once with PBS and quickly fix in a formaldehyde solution (4% in PBS) for 10-15 minutes at room temperature and protected from light.
Remove formaldehyde and dispose properly. Wash three times with PBS and incubate in a BSA solution (3% in PBS containing 0.1% Triton X-100) for 15-30 min at room temperature and protected from light.
Remove BSA solution and stain F-actin with Alexa Fluor 568 Phalloidin (diluted around 1:500 to 1:100 in PBS containing 0.3% BSA and 0.1% Triton X-100). Incubate for 0.5 to 1 h at room temperature protected from light. The optimal phalloidin dilution and incubation time depends on the cell line used and should be determined empirically.
Remove the phalloidin solution and wash three times with PBS containing 0.1% Triton-X100 and three times with PBS alone.
Mount the coverslips by inverting them over a glass slide containing a drop of mounting medium containing DAPI. Slides can be stored in the dark at 4 °C for several weeks.
D. Imaging and quantifying invadopodia formation and activity
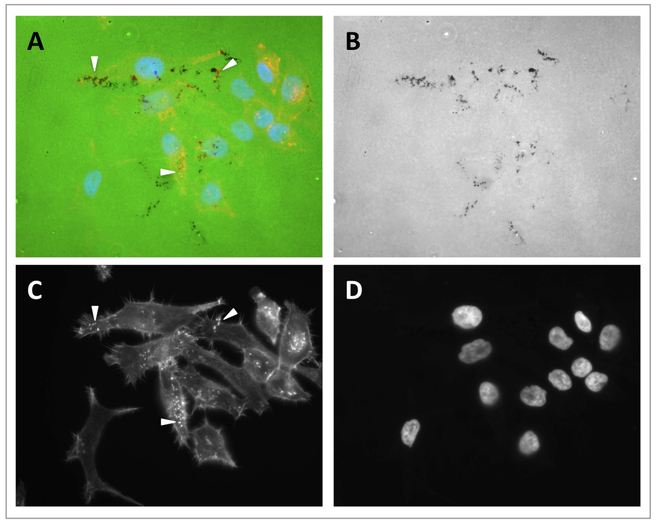
Slides can be imaged with a fluorescent microscope equipped for detection of Alexa 488, Alexa 568 and DAPI. Invadopodia are detected in the “red” channel as F-actin rich puncta in the ventral surface of the cell in contact with the gelatin (Figure 2 C). Gelatin degradation is detected in the “green” channel as dark areas over the green background (Figure 2 B). Typically, a subset of F-actin rich structures will co-localize with discrete gelatin degradation spots, indicating that these F-actin rich structures are likely invadopodia (Figure 2 A).
Quantification of invadopodia formation and activity may be performed on the same set of digital images. To perform a representative image collection, it is advisable to image areas that are representative to the whole coverslip in order to account for differences in the thickness of the gelatin coating. A possibility is to divide the coverlip into three imaginary columns and image five rows per column covering the whole height of each column. At least 15 fields per coverslip should be imaged, typically under 40x magnification taking all three channels (“red”, “green” and “blue”). Moving from one field to the next when in the channel for DAPI will help you identify areas containing similar number of cells while you are “blinded” for any other experimental outcome in a particular field to be imaged.
To quantify invadopodia formation, count the number of cells forming invadopodia on each digital image and normalize to the number of total cells in that same image. After counting all your images, collected data may be represented as “percent of cells forming invadopodia”. Perform the appropriate statistical analysis to determine significant differences among treatments.
- To quantify invadopodia activity, black and white images of gelatin degradation are analyzed using ImageJ (NIH). The objective is to measure “area fraction” (the percent of area that corresponds to degradation) on a given image. The “area fraction” value will then be normalized to the number of nuclei in each image as measured from the DAPI channel in the same field (Figure 3). The steps for obtaining the “area fraction” value are:
- Set Image J to measure “area fraction”: Analyze>Set Measurements>select “Area Fraction”.
- Open a black and white image of gelatin degradation (green channel) in Image J and then go to Image>Adjust>Threshold. If the black areas over white background are representative of the degradation in the original image, no further adjustment is necessary. If the program detects black areas that are broader than the actual degradation (i.e. because of irregular gelatin coating), the threshold has to be manually adjusted. On the new window move the bottom on the lower bar towards the left until the dark areas are representative of the real degradation observed in the original image.
- When the final image is representative of the gelatin degradation on the original image (as in Figure 3 C) click “set”. Measure “area fraction” by: Analyze>Measure. The values obtained in Image J may be exported to Microsoft Excel.
Figure 2. Representative images of invadopodia and gelatin degradation assay.
Digital images from a gelatin degradation assay performed on SCC61 cells. Images were obtained using a 40x objective in a fluorescence microscope. A. Merged channels showing fluorescent gelatin (green), F-actin staining (red) and nuclei (blue). B. Green channel (gelatin). C. Red channel (F-actin). D. Blue channel (nuclei). Arrowheads point to invadopodia in A and C.
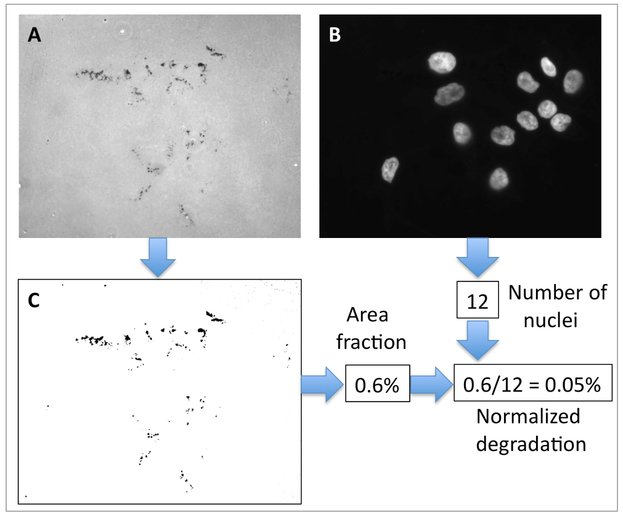
Figure 3. Quantification of invadopodia activity.
Images from the green channel (gelatin) and blue channel (nuclei) from the same microscopy field are used. The original image from the green channel (A) is processed using Image J to obtain the image in (B), and the area fraction value. The original image from the blue channel (C) is used to calculate cell number. These two values are used to obtain the “normalized degradation” value of each microscopy field from a sample.
The measurements corresponding “to area fraction” are normalized to the number of nuclei in the DAPI channel image from the same field. The final value corresponds to “normalized degradation”. The same analysis is repeated for each image of each sample. Values can be represented as mean and standard deviations. Perform the appropriate statistical analysis to determine significant differences among treatments. An example is showed in Figure 3.
Note: When assessing the formation of invadopodia in your cell line of interest for the first time, it is advisable to perform additional immunofluorescent experiments to assess the presence of invadopodia components such as Tks5, cortactin and Arp2/3 at the F-actin rich structures.
Acknowledgments
This protocol was initially adapted from: Mueller et al. (1992). The first adaptation was published in: Berdeaux et al. (2004). It was later implemented in: Berdeaux et al. (2013). Funding during first adaptation: National Institutes of Health grant CA17542 to G.S. Martin. Funding during implementation: National Institutes of Health CA129686 to S. A. Courtneidge. Other acknowledgements: The author would like to acknowledge Dr. G.S. Martin (UC Berkeley, California) and S.A. Courtneidge (UHSU, Portland, Oregon) for their support over the time this protocol was adapted and implemented.
References
- 1.Berdeaux RL, Diaz B, Kim L and Martin GS (2004). Active Rho is localized to podosomes induced by oncogenic Src and is required for their assembly and function. J Cell Biol 166(3): 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diaz B, Yuen A, Iizuka S, Higashiyama S and Courtneidge SA (2013). Notch increases the shedding of HB-EGF by ADAM12 to potentiate invadopodia formation in hypoxia. J Cell Biol 201(2): 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller SC, Yeh Y and Chen WT (1992). Tyrosine phosphorylation of membrane proteins mediates cellular invasion by transformed cells. J Cell Biol 119(5): 1309–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]