SUMMARY
Cellular reprogramming technology has created new opportunities in understanding human disease, drug discovery, and regenerative medicine. While a combinatorial code was initially found to reprogram somatic cells to pluripotency, a “second generation” of cellular reprogramming involves lineage-restricted transcription factors and microRNAs that directly reprogram one somatic cell to another. This technology was enabled by gene networks active during development, which induce global shifts in the epigenetic landscape driving cell fate decisions. A major utility of direct reprogramming is the potential of harnessing resident support cells within damaged organs to regenerate lost tissue by converting them into the desired cell type in situ. Here, we review the progress in direct cellular reprogramming with a focus on the paradigm of in vivo reprogramming for regenerative medicine, while pointing to hurdles that must be overcome to translate this technology into future therapeutics.
INTRODUCTION
The concept that differentiated cells are plastic and can be reprogrammed to alternate cell fates was first suggested by the cloning experiments of Gurdon (Gurdon et al., 1958) and later Wilmut (Campbell et al., 1996). In these studies, undefined factors in the oocyte cytoplasm were found to induce somatic cells to assume an embryonic state. Embryonic and fetal development ensued, culminating in live births and surprisingly normal postnatal development. This observation was the original form of “in vivo” cellular reprogramming.
Nearly 30 years later, a single myoblast cDNA encoding the transcription factor MyoD, expressed “where it is not normally”, was shown to convert fibroblasts directly to myoblasts (Davis et al., 1987). The cells did not revert to a pluripotent state before assuming their new fate—and the paradigm for what is now termed “direct reprogramming” was born, at least in vitro. These findings violated the prevailing view of somatic cell fate as inviolate and immutable, but were consistent with heterokaryon experiments that observed rapid nuclear reprogramming of fibroblasts upon fusion with myocytes (Blau et al., 1985). However, the observation that a single factor could completely convert cells into distantly-related cell fates turned out to be the exception, rather than the rule. As critical lineage-enriched transcription factors like MyoD were discovered for various cell types during development, each failed to exhibit a MyoD-like ability to convert fibroblasts into a new fate, although C/EBPα was notable for its sufficiency to convert lymphoid cells into closely-related myeloid cells of the hematopoietic system (Xie et al., 2004).
The notion that cell fate is in fact mutable and malleable finally took hold when Yamanaka showed that a cocktail of a few cell fate-changing transcription factors profoundly redirected somatic cells to a state of pluripotency (Takahashi and Yamanaka, 2006). This combinatorial approach paved the way to feverish activity in nuclear reprogramming. Much effort focused on refining methods to drive differentiated cells to a pluripotent state in various species and discovering the mechanisms. However, others began asking whether combinations of transcription factors could convert cell fates without first dedifferentiating the cells to pluripotency. In recent years, a combinatorial transcriptional “code” to directly reprogram cells toward specific lineages has emerged for many cell types. As a result, the Waddington model of cell differentiation as a determinant process has been revised to reflect an alternate view—that cell fate can readily be altered given appropriate conditions and cues (Fig. 1) (Ladewig et al., 2013).
Figure 1.
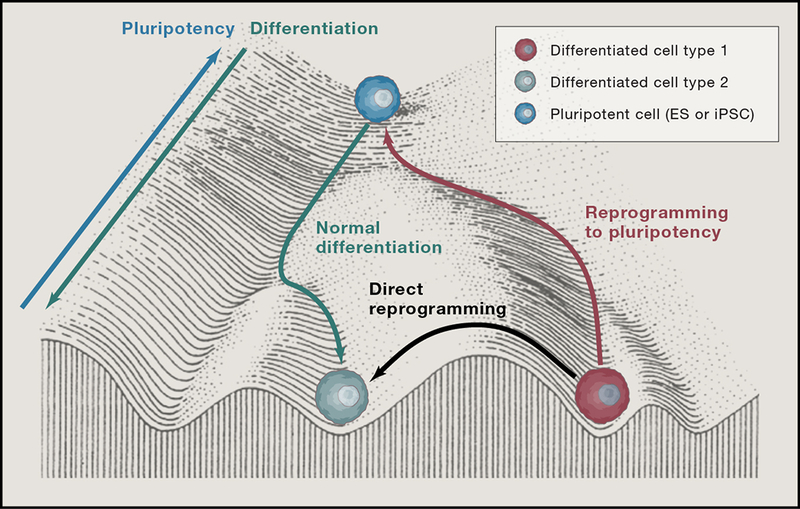
Conrad Waddington likened cell fate to a marble rolling downhill into one of several troughs representing fully differentiated cell types. Nuclear transfer and reprogramming showed that cells can be rolled back to the top of the hill by epigenetically altering the cell. Now, it is clear that cells can travel part way up the hill to roll back down a discrete number of troughs or even travel from one trough to another without going back up the hill at all, although the epigenetic barriers for such travel appear greater than traveling up hill.
In this review, we briefly summarize the path to such discoveries in vitro but largely focus on more recent advances in harnessing direct reprogramming strategies for in vivo regeneration, which is likely the most powerful use of this technology. Specifically, this strategy involves re-purposing cells in damaged tissue in situ to regenerate organs from within, providing an alternative to exogenous cell-based therapeutic approaches. A common theme in multiple tissue has emerged—the native environment often contains local unknown cues that enhance the quality and efficiency of direct reprogramming.
Direct Cellular Reprogramming In Vitro: Informing an In Vivo Strategy
Reprogramming to pluripotency.
In 2006, Takahashi and Yamanaka showed that a cocktail of four specific transcription factors could, ex vivo, convert differentiated fibroblasts to a pluripotent state resembling embryonic stem cells derived from the blastocyst inner cell mass (Takahashi and Yamanaka, 2006). Their pioneering studies of induced pluripotent stem (iPS) cells established “reprogramming” as a transformative technology for biomedicine. iPS cell technology is a robust and ethically acceptable way to convert differentiated cells to a pluripotent state; the iPS cells can then be directed, by factors important for development and differentiation, to form functional differentiated cells of a variety of lineages. These studies established the paradigm that differentiation is not a dead end. Rather, genetic and epigenetic cues can reverse cell fate to a more primitive state through large-scale alterations in gene expression and chromatin status that have been carefully mapped during reprogramming of somatic cells to iPS cells (reviewed in Zaret and Mango, 2016).
Combinatorial approaches for direct conversion.
Efforts to use a combinatorial approach for direct lineage conversion have been built on decades of developmental biology research. Numerous studies in flies, zebrafish, chicks, mice, and other model organisms have defined transcription factors that control cell fate during embryonic and fetal development, as well as experimentally tractable gene networks that regulate cell fate. However, apart from MyoD and C/EBPα, single factors have not been sufficient for cellular reprogramming for most tissues. Nevertheless, the field was poised to leverage the combinatorial screening approach first used for iPS cell reprogramming by Takahashi and Yamanaka. Combinatorial screening entails identifying a pool of candidate genes encoding, for instance, transcription factors or microRNAs (miRNAs) that regulate cell fate or differentiation, testing the ability of the pool to convert fibroblasts to a differentiated cell fate of interest, and then using a “minus-one” strategy to identify essential factors and pinpoint a minimal combination required for cell fate conversion. The first breakthroughs were reported for in vitro combinatorial reprogramming of fibroblasts to unrelated cell types, namely cardiomyocytes and neurons, and the advances in this area that set the stage for in vivo reprogramming are briefly summarized below.
Direct cardiac reprogramming.
After starting with nearly 20 transcription factors and a similar number of miRNAs, Ieda et al. reported that a combination of three cardiac developmental transcription factors—Gata4, Mef2c, and Tbx5 (GMT)—reprogrammed dermal or cardiac fibroblasts to induced cardiomyocyte-like cells (iCMs) (Fig. 2a) (Ieda et al., 2010). Ectopic expression of these factors was required for ~2 weeks, after which the reprogramming event was epigenetically stable. Interestingly, missense mutations in GATA4 and TBX5 cause similar congenital heart defects in humans. Moreover, the two factors they encode physically interact to regulate cardiac gene expression (Basson et al., 1997; Garg et al., 2003; Maitra et al., 2009), consistent with their combinatorial role in reprogramming.
Figure 2.
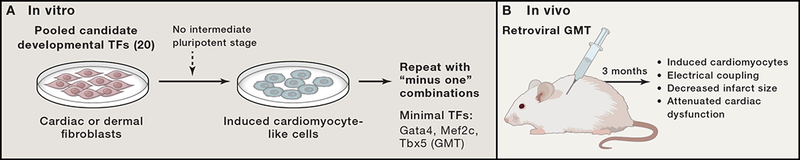
Schematic of approach to identify master regulatory factors capable of direct reprogramming in vitro and in vivo using cardiac reprogramming as example.
A) Method for in vitro screening of developmentally critical transcription factors (TFs) that directly converted fibroblasts to an induced cardiomyocyte-like state.
B) In vivo testing of reprogramming factors requires lineage tracing of cardiac fibroblasts as they transition into a new fate in the setting of injury. Introduction of cardiac reprogramming factors in vivo resulted in new conversion of resident fibroblasts into new cardiomyocyte-like that electrically integrated and contributed to improved cardiac function after injury.
Lineage tracing approaches demonstrated that during reprogramming with GMT, fibroblasts did not pass through a mesodermal or cardiac progenitor stage, suggesting a more direct conversion from one postnatal cell type to another. Consistent with this observation, the iCMs that were more fully reprogrammed had electrophysiological properties most similar to those of adult ventricular cardiomyocytes. The generation of iCMs with GMT addressed a nearly 25-year quest to achieve a MyoD-like event for cardiac muscle. However, the in vitro efficiency was limited, and most of the iCMs were only partially reprogrammed, suggesting that other factors may enhance reprogramming, at least in vitro.
As might be expected for a new technology, other combinations of factors in vitro were later found to convert fibroblasts to iCMs with greater efficiency (reviewed in Srivastava and Yu, 2015). Additional transcription factors such as Hand2 (Song et al., 2012; Srivastava et al., 1997) and miRNAs such as the muscle-specific miRNAs miR-1 and miR-133 (Chen et al., 2006; Heidersbach et al., 2013; Muraoka et al., 2014; Zhao et al., 2007; Zhao et al., 2005) increased the conversion rate in vitro. A combination of four miRNAs—miR-1, miR-133, miR-208, and miR-499—converted mouse fibroblasts to cardiac myocytes in the absence of any exogenous transcription factors; the efficiency of the conversion was improved by JAK inhibitor I (Jayawardena et al., 2012). Similarly, inhibiting TGF-β signaling (Ifkovits et al., 2014; Zhao et al., 2015) or the epigenetic regulator Bmi1 (Zhou et al., 2016) appeared to break down barriers to reprogramming and increase conversion efficiency. Conversely, activating Fgf and Vegf signaling with GMT greatly increased yield of beating cardiomyocytes by activating Akt (Yamakawa et al., 2015) while overexpression of Akt1 alone also resulted in efficient generation of beating cells, particularly in mouse embryonic fibroblasts (Zhou et al., 2015). In a different approach involving reprogramming of fibroblasts toward an early mesodermal progenitor, mouse embryonic fibroblasts were converted to differentiated cardiomyocytes by transient overexpression of the “Yamanaka factors” followed by expression of cardiogenic growth factors (Efe et al., 2011). However, the maturity of the cells was similar to that of cardiomyocytes derived from pluripotent stem cells.
Efforts to translate cardiac reprogramming technology from mice to humans proved difficult, as it became increasingly clear that human fibroblasts could not be converted by GMT or other combinations of factors capable of reprogramming mouse cells. Nonetheless, after screening for additional factors, several groups reported that overlapping cocktails of factors resulted in a degree of reprogramming comparable to that of mouse fibroblasts (Fu et al., 2013; Nam et al., 2013; Wada et al., 2013). A solely chemical approach using several small-molecule epigenetic regulators also efficiently converted human fibroblasts to beating cardiomyocytes—advancing the therapeutic potential of direct reprogramming strategies (Cao et al., 2016).
Direct neuronal reprogramming.
In parallel with advances in direct cardiac reprogramming, a similar combinatorial approach was being used to convert mouse embryonic and fetal fibroblasts to functional neurons ex vivo. In one study, the combination of transcription factors Ascl1, Brn2 (also called Pou3f2), and Myt1l converted fibroblasts to neurons that expressed neuron-specific proteins, generated action potentials, and formed functional synapses (Vierbuchen et al., 2010). In this combination Ascl1 functioned as a “pioneer” factor to initiate chromatin changes and recruit the other two factors (Wapinski et al., 2013). Soon thereafter, non-neurogenic astroglia from mouse cerebral cortex were converted by neuronal reprogramming to specific sub-types of neurons capable of forming synapses in culture (Heinrich et al., 2010). As with iCMs, the conversion occurred in the absence of cell division, and produced distinct neuronal subtypes, depending on which transcription factors were expressed. For example, expression of the dorsal telencephalic fate determinant neurogenin-2 directed cortical astroglia to generate synapse-forming glutamatergic neurons, whereas Dlx2, a ventral telencephalic fate determinant, induced a GABAergic identity. Under the appropriate culture conditions, a single factor, Sox2, converted fibroblasts to a neuronal fate, suggesting that optimizing culture conditions and signaling pathways within cells could simplify the reprogramming cocktail in certain settings, even with individual factors (Ring et al., 2012). Ultimately, several groups succeeded in converting human fibroblasts directly to dopaminergic neurons (Caiazzo et al., 2011; Pfisterer et al., 2011), spinal motor neurons (Son et al., 2011), and oligodendroglia (Yang et al., 2013).
Reprogramming to expandable progenitors.
The early direct conversion approaches induced a one-for-one exchange of cell types but did not provide a way to expand cell populations, as the converted cells rapidly exited the cell cycle. In 2012, several groups designed screens to generate expandable neural stem cells from fibroblasts by combinatorial direct conversion (Ring et al., 2012; Thier et al., 2012). This approach avoided reversion to pluripotency, which may carry risks for generating oncogenic cells in vivo. At the same time, it generated an expandable intermediate cell population of neural stem cells that could be then differentiated to form specific neuronal subtypes. Similarly, expandable cardiac progenitors were generated by forcing human dermal fibroblasts to express mammalian ETS2 and MESP, both homologues of genes essential for generating cardiac progenitors in the ascidian Ciona (Islas et al., 2012).
Earlier this year, two groups independently developed a chemical approach to convert mouse fibroblasts to an early cardiac progenitor state that could be maintained as transient amplifying progenitors. The progenitors retained multipotency and developed into cardiomyocytes, endothelial cells, and smooth muscle cells (Lalit et al., 2016; Zhang et al., 2016). In another study, mouse fibroblasts were chemically converted to multipotent neural stem cells (Zhu et al., 2016). In these studies, the chemical cocktails appear to induce fibroblast conversion into an epigenetically unstable state closer to pluripotency followed by redirection into cardiac or neuronal fates. Not surprisingly, resulting cells were most similar in maturity to ones derived from pluripotent stem cells.
Overall, studies of direct conversion have identified many new combinations of factors that alter cell fate, including transcription factors described above, chemicals (Ladewig et al., 2012; Shi et al., 2008), microRNAs (Yoo et al., 2011), and combinations thereof (Wang et al., 2014), as well as single transcription factors with appropriate culture conditions (Ring et al., 2012). The various approaches share the common goal of making direct conversion more experimentally tractable, robust, and safe. Blood cells and other cell types have also been obtained in vitro by direct conversion (Szabo et al., 2010) (Xie et al., 2004) . The studies discussed above point to the utility of in vitro direct reprogramming, particularly as it pertains to cell-based therapies and disease modeling. Thus within less than a decade, in vitro reprogramming has become a rich and vigorous field, and covering it comprehensively is beyond the scope of this review but has been reviewed elsewhere (Xu et al., 2015).
In Vivo Reprogramming for Tissue Regeneration
In vivo reprogramming is an emerging field that is rightfully garnering attention for its therapeutic potential. The question of whether organs are amenable to direct conversion in vivo was first addressed in the pancreas, where some degree of plasticity exists between closely related cell types. Investigators in the cardiac and neural fields have advanced this concept further (Niu et al., 2013; Qian et al., 2012; Song et al., 2012; Torper et al., 2013). Distantly related cells in the adult heart and brain can be directly converted in vivo by appropriate combinations of developmentally relevant transcription factors. Numerous other cell types, such as liver, have been generated through direct in vivo reprogramming (Song et al., 2016). However, for the sake of brevity, we will focus on lessons learned from the more advanced studies of in vivo pancreatic, cardiac, and neuronal reprogramming and on the promising area of sensory neuronal regeneration. In addition, it is worth considering recent evidence that reprogramming of endogenous cells naturally occurs within organs as part of normal regeneration, as reported in mouse liver (Yanger et al., 2013),and zebrafish heart (Zhang et al., 2013). These processes could be leveraged for therapeutic approaches and have been reviewed elsewhere (Jessen et al., 2015).
Pancreatic Beta Cells
An early version of direct in vivo conversion involving highly related cell types was used to generate pancreatic beta cells from pancreatic exocrine cells and adult mouse pancreas (Zhou et al., 2008). Of approximately 20 transcription factors that were expressed in mature beta cells and their precursors, nine that resulted in a beta cell developmental phenotype when mutated were pooled and co-expressed with GFP as a marker, and injected into the pancreas of adult mice. Individual factors were then eliminated to identify three that increased the number of insulin-positive cells: specifically, adenoviral delivery of Ngn3, Pdx1, and Mafa (pAd-M3) reprogrammed pancreatic exocrine cells to a beta cell fate. The conversion was direct and not produced by dedifferentiation to a common progenitor, as determined by lineage tracing.
The newly reprogrammed beta cells were functional. They secreted insulin, synthesized vascular endothelial growth factor, and induced local angiogenic remodeling. In a mouse model of diabetes induced by streptozotocin injection, pAd-M3 produced a significant durable lowering of glucose levels and increased glucose tolerance and serum insulin levels. Although the three factors did not induce cellular conversion in vitro, the native in vivo environment apparently enhanced reprogramming, suggesting that endogenous signals play a role in coaxing beta cells to a functional state that is not possible in vitro.
This study sparked intense interest in gene therapy approaches to produce insulin-secreting beta cells by in vivo direct conversion from other cell types. Since beta cells are destroyed by immunological molecules in type I diabetes, restoring new beta cells without the need for allogeneic beta cell transplants offered a potentially powerful therapeutic strategy that would not rely on cadaveric donor tissue necessary for the Edmonton protocol, an early cell-replacement therapy. This protocol involved transplanting pancreatic islets from deceased donors into the livers of patients with type I diabetes whose insulin levels were difficult to control, followed by immunosuppressive therapy to prevent organ rejection (Shapiro et al., 2006). Multiple transplants are often required, and the immunosuppression has significant side effects.
Since the pancreas appears to possess inherent plasticity (Juhl et al., 2010), it was at first unclear whether in vivo direct conversion could be applied to other organs and tissues. For instance, after Melton’s 2008 study, Herrera and colleagues reported that pancreatic cells possess a previously unappreciated degree of plasticity (Thorel et al., 2010). Even without forced expression of transcription factors, adult mice survived after extreme beta cell loss induced by diphtheria toxin. Over time, their beta cells became more numerous, and a large proportion of the new beta cells were derived from alpha cells, as shown by lineage tracing.
The pancreas and liver arise from the same lineage during embryonic development, prompting others to test whether liver cells could be reprogrammed to insulin-secreting cells (Banga et al., 2012). Indeed, viral expression of the three pancreatic reprogramming factors in the livers of mice with streptozotocin-induced diabetes led to the growth of ectopic duct-like structures possessing markers and ultrastructural features of beta cells, and the diabetic phenotype was attenuated, even after reprogramming factors were no longer overexpressed. The liver cells that were converted to insulin-producing cells were Sox9+ cells, which are normally present in small bile ducts. This study suggested that disparate tissues that are related during embryonic development are amenable to in vivo direct conversion strategies. Indeed, a subsequent study showed that intestinal cells could also be converted into insulin-producing cells, although the optimal therapeutic approach remains unclear (Ariyachet et al., 2016).
A major challenge in direct conversion, in vitro or in vivo, is obtaining the correct cellular subtype. Based on the three-factor strategy to convert exocrine cells to beta cells (Zhou et al., 2008), Li et al. reported that by using the same adenoviral expression strategy to deliver different combinations of the three transcription factors to adult mice, pancreatic acinar cells could be converted to gamma-like and alpha-like cells, two other major islet endocrine subtypes (Li et al., 2014). Thus, combinatorial approaches can be further refined to establish highly specific populations of cell subtypes. Furthermore, it should be noted that knockdown of factors can also promote changes in cell fate, as observed with the switch from alpha to beta cells in the pancreas upon inhibition of the alpha cell-promoting transcription factor, Arx (Courtney et al., 2013).
Cardiomyocytes
Unlike pancreatic endocrine and exocrine cells, cardiac fibroblasts and cardiomyocytes arise from distinct progenitors, although they share a common mesodermal origin. Cardiac fibroblasts are abundant and can be activated and migrate to sites of injury, making them an attractive target for in vivo reprogramming to repair damaged hearts. In 2012, investigators who described the in vitro cocktail for cardiac reprogramming found that in vivo delivery of the GMT transcription factors directly into the heart by gene therapy converted endogenous mouse nonmyocytes, largely fibroblasts, into iCMs (Inagawa et al., 2012; Qian et al., 2012). The quality of reprogramming was much greater in vivo than in vitro: most cells were more fully reprogrammed into beating cells, and their transcriptomes were much more similar to those of endogenous cardiomyocytes than to those of cells generated in vitro (Qian et al., 2012). Furthermore, the reprogrammed myocytes were most similar to adult ventricular cardiomyocytes and electrically coupled both to other newly generated iCMs and to endogenous cardiomyocytes (Qian et al., 2012). After in vivo GMT delivery, the mice had decreased infarct size and attenuated cardiac dysfunction after coronary ligation (Fig. 2b) (Qian et al., 2012). As was the case for the stoichiometry of GMT in vitro, introduction of a polycistronic cassette of MGT, which produced the highest levels of Mef2c, resulted in more optimal reprogramming in vivo (Ueki et al., 2015), highlighting the importance of the dosage of each factor.
Other approaches for in vivo cardiac reprogramming have also been successful. Addition of the transcription factor Hand2 to GMT (GHMT) improved mouse cardiac reprogramming efficiency in vitro and improved efficiency of conversion in vivo along with improved cardiac function (Song et al., 2012). In vitro, GHMT appears to produce a spectrum of ventricular, atrial, and conduction cell types (Nam et al., 2014). The combination of miRNAs described earlier (miR-1, miR-133, miR-208, and miR-499) introduced with a lentivirus after infarct also appears to generate new myocytes and improve cardiac function (Jayawardena et al., 2015).
Generation of new iCMs in vivo was accompanied by greater capillary density. However, adjuvant therapy to promote angiogenesis appears to further enhance function after direct reprogramming. Co-administration of thymosin β4—a 43-amino-acid G-actin monomer-binding protein that promotes angiogenesis and cell survival, proliferation, and migration (Bock-Marquette et al., 2004; Smart et al., 2007)—enhanced GMT-mediated regeneration and further increased the fraction of blood ejected with each heart beat (Qian et al., 2012). Thymosin β4 also activates epicardial cells, which may have regenerative effects and promote regeneration through additional mechanisms. Supporting the notion that enhanced angiogenesis with reprogramming may improve function, delivery of vascular endothelial growth factor with GMT had similarly positive effects (Mathison et al., 2012).
Besides generating new myocytes, in vivo reprogramming in each study was associated with a significant reduction in fibrosis. The newly emerged iCMs may secrete factors that inhibit collagen expression and matrix metalloproteinase activity, thereby reducing fibrosis. Furthermore, fibroblasts that were infected by reprogramming factors but failed to reprogram may be intrinsically altered and therefore may have impaired ability to promote fibrosis. It is likely that a combination of these effects is responsible for improving heart function and decreasing scar formation after injury.
Beyond the use of reprogramming to create beating cardiomyocytes, there is interest in generating cells of the specialized cardiac conduction system through direct reprogramming in vivo. For example, sino-atrial node cells are specialized non-contracting myocytes that serve as the pacemaker cells. Expression of the transcription factor Tbx18 apparently induced a cell fate switch of cardiomyocytes into cells with pacemaker-like activity (Kapoor et al., 2013). Adenoviral delivery of Tbx18 in vivo in a guinea pig model of bradycardia helped restore a more normal heart rate, suggesting a potential alternative to mechanical pacemakers. Although much refinement is needed, the notion of regenerating the small number of pacemaker or other specialized conduction cells to correct rhythm disturbances is an attractive area for research (reviewed in MacRae, 2016). More precise knowledge of the transcriptome of such cells, as reported for pacemaker cells (Vedantham et al., 2015), will be required and should emerge from new single-cell RNA-sequencing approaches in the near future.
Neurons
In parallel with the cardiac researchers, neurobiologists were establishing an analogous paradigm for the adult brain. Their efforts focused mainly on ectopic expression of single transcription factors rather than a combinatorial approach. The adult heart has few active progenitors and little regenerative capacity, as illustrated by its inability to replace tissue damaged by ischemic events. In contrast, specific zones of the adult brain have a marked degree of plasticity and migratory capacity (reviewed in Kriegstein and Alvarez-Buylla, 2009; Zhao et al., 2008). For example, in rodent models of brain injury, neuroblasts derived from adult neural stem cells migrate to damaged areas and differentiate into specific neural lineages (Arvidsson et al., 2002; Lugert et al., 2012; Zhao et al., 2008). Much of the discussion here focuses on efforts to convert glial cells to functional synaptic neurons. Glial cells have features of progenitors and are the most abundant cells in adult brain and could be a therapeutic avenue for repairing diseased or injured brains.
An initial study to test whether ectopic transcription factor expression can convert neurons from one subtype to another was done in mouse embryos and early neonates. Delivery of Fezf2—a transcription factor specific to layer 5B output neurons—reprogrammed postmitotic neocortical neurons to LB5 neurons, as judged by morphological and electrophysiological criteria (De la Rossa et al., 2013). Similarly, delivery of Fezf2 by in utero electroporation converted postmitotic layer II/III callosal projection neurons to layer-V/VI corticofugal projection neurons, a different neuronal subtype (Rouaux and Arlotta, 2013). Clearly, postmitotic neurons can undergo lineage conversion, at least early in mouse development.
In a subsequent study, delivery of the neural transcription factor Sox2 to adult mouse striatum reprogrammed endogenous astrocytes to proliferating neuroblasts (Niu et al., 2013). Further, the growth factors brain-derived neurotrophic factor and noggin, or the histone deacetylase inhibitor valproic acid, coaxed the induced neuroblasts to form electrophysiologically functional neurons that integrated into neural networks. This approach effectively converted spinal cord astrocytes to proliferating neural stem cells, which matured into synapse-forming interneurons in spinal cords of mouse models with or without severe injury (Su et al., 2014). Later, the mechanism for Sox2-mediated conversion of resident astrocytes to neural progenitors was shown to progress through Ascl1+ and Dcx+ adult neuroblasts as intermediate progenitors (Niu et al., 2015). In fact, Ascl1 alone was sufficient in vivo to reprogram astrocytes into induced neurons (Liu et al., 2015). Sox2 was also able to reprogram pericytes in the brain into induced neurons, suggesting its effects were not unique to astrocytes (Karow et al., 2012).Moreover, cortical glial cells rendered reactive by stab wound injury or Alzheimer’s disease pathology were reprogrammed by NeuroD1 to form glutamatergic neurons and GABAergic neurons (Guo et al., 2014). Thus, specific factors can induce unique neuronal fates.
While progress has been made in reprogramming to discrete neuronal subtypes in vivo, demonstration of functional consequences of the reprogrammed cells has been elusive. To this end, Parmar and colleagues have developed refined tools to study the conversion of Ng2 glia to GABAergic and glutamatergic neurons and their integration into the host brain (Torper et al., 2015). Using Cre-recombinase-dependent AAV vectors, they delivered Ascl1, Lmx1a, and Nurr1 specifically to NG2 glia in the striatum of adult mice and monitored neuronal conversion and circuit integration through a novel neuron-specific reporter. Their vectors improved the efficiency of neural conversion in vivo, and permitted long-term phenotypic and functional analysis, including integration of neurons into local circuitry. Further testing of neuronal physiology and behavior will be necessary to advance this area of in vivo reprogramming.
Sensory Receptor Cells
Sensory receptor cells, which reside in the retina, olfactory epithelium, and inner ear, are another promising target cell type for therapeutic in situ reprogramming (Fig. 3). In mammals, cells of the retina and inner ear are not regenerative; thus, inducing cell fate changes could provide a unique mechanism for restoring functional cell types in the setting of visual or hearing loss. The developmental mechanisms and transcription factors controlling differentiation of sensory epithelia from the three sensory tissues share common elements (reviewed by Bermingham-McDonogh and Reh, 2011). For instance, sensory receptor cells arise from Sox2-expressing epithelial progenitor cells that express proneural basic helix-loop-helix transcription factors. These cells are important for the differentiation of receptor cells and associated neurons. The progenitor cells give rise to both sensory and supporting cells. In the inner ear and retina, Notch signaling controls the differentiation of supporting cells. Manipulating gene expression in the progenitors or their derivatives by direct reprogramming could provide a strategy to convert supporting cells to a sensory or neuronal fate, similar to converting astrocytes to neurons discussed above. Alternatively, direct conversion to drive dedifferentiation of resident cells to progenitors in diseased or injured tissues could provide a source of regenerative cells to restore sensory tissues.
Figure 3.
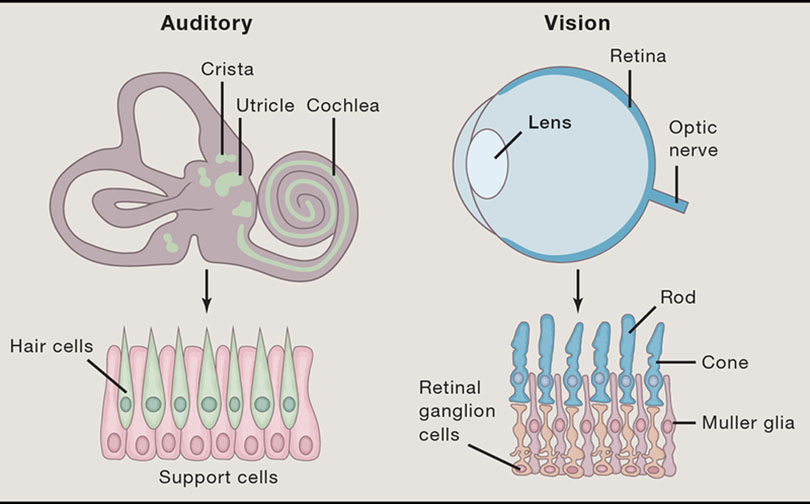
Schematic of sensory organ cells that could be harnessed for regenerative potential.
To examine this possibility, Reh and colleagues exploited findings in non-mammalian vertebrates capable of regenerating retinal tissue after injury. In zebrafish, quiescent Müller glia respond to chemical- or light-induced damage by dedifferentiating to form multipotent progenitors that give rise to all retinal neural subtypes (Pollak et al., 2013). In contrast, mammalian retina responds to injury by undergoing reactive gliosis. Ascl1a, which is required for retinal regeneration in fish and is rapidly upregulated after injury, is not upregulated in mammalian retina after chemical-induced damage (Karl et al., 2008). Thus, the regenerative capacity of mammalian retina might be limited by its inability to activate Ascl1a expression in response to injury.
Reh and colleagues tested this hypothesis by virally delivering Ascl1 to Müller glia in dissociated cultures (Pollak et al., 2013). The Ascl1-reprogrammed cells expressed retinal progenitor–specific genes and lost their glial identity, as judged by morphology and gene expression. They also acquired a neuronal appearance, displayed neuron-like responses to neurotransmitters, and developed robust expression of pan-neuronal markers and specific markers of retinal neurons.
Ascl1 expression in retinal Müller glia in vivo also appeared to reprogram them to a neuronal fate (Ueki et al., 2015). Adult glial cells were reprogrammed to a neurogenic state based on gene expression and morphological criteria, but only in mice with retinal damage induced by chemicals or excessive light. The Müller glia–derived cells expressed markers of bipolar cells, amacrine cells, and photoreceptors. Interestingly, young mice responded more efficiently to Ascl1 overexpression than older mice. It appeared that chromatin changes associated with aging rendered progenitor gene loci less accessible in Müller glia as mice aged. This may be a general concern in direct reprogramming of cells for therapeutic purposes, but this possibility remains to be tested in other cell types.
Sensory hair cells of the inner ear are another promising avenue for in vivo direct reprogramming. Loss of hair cells from genetic mutations, aging, or exposure to noise or certain drugs causes permanent hearing loss (Kuo et al., 2015). In humans, hair cells do not regenerate. However, like retinal cells, hair cells in birds, fish, and frogs do regenerate, suggesting that the molecular mechanisms could be exploited to regenerate human hair cells by in vivo reprogramming.
Studies of mouse knockout mutants have identified several transcription factors that function during inner ear development, specifically during hair cell morphogenesis, survival, cell fate, patterning, and proliferation (Schimmang, 2013). Factors responsible for cell fate control are of keen interest for their potential for reprogramming and transdifferentiation. The best known is Atoh1 (Math1), a basic helix-loop-helix transcription factor that when misexpressed in the rat inner ear drives ectopic generation of hair cells (Woods et al., 2004; Zheng and Gao, 2000). These early studies suggested an intriguing concept—that hair cells can indeed be regenerated in mammals by gene modification approaches. However, only small numbers of hair cells were generated, and their survival was poor.
Combined expression of Atoh1 and a constitutively active form of β-catenin achieved robust generation of hair cells in the mouse inner ear cochlea; cell survival was much higher than when Atoh1 was expressed alone (Kuo et al., 2015). In cells expressing Lrg5, a marker of stem cells in the intestine and hair follicle, co-expression of β-catenin and Atoh1 with a Cre-based transgenic approach markedly increased proliferation. Moreover, the newly generated cells differentiated into hair cells containing stereocilia bundles, which are a marker of hair cells; however, they were not innervated. This approach generated 10-fold more new hair cells than delivery of Atoh1 alone (Woods et al., 2004; Zheng and Gao, 2000). Despite the immaturity of the newly generated cells, this study demonstrated the potential effectiveness of delivering key transcription factors and signaling pathway components to regenerate hair cells in vivo.
In a similar approach, ectopic expression of Atoh1 expression and the transcription factors Gfi1 and Pou4f3 was used to drive hair cell development from somatic cells in chick embryonic otic epithelium (Costa et al., 2015). Gfi1 and Pouf4f3—zinc-finger and POU-domain transcription factors, respectively—are transcriptional regulators that are essential for the differentiation and survival of all vestibular and auditory hair cells. They appear to function with Atoh1 to determine hair cell fate in the inner ear. In chick embryos, co-expression of the three genes by a tetracycline-induced transposon system to control spatial and temporal expression resulted in robust development of hair cells from various otic progenitors. Commitment to the hair cell fate was independent of developmental stage and identity of the transfected cells and resulted in polarized cells containing rudimentary stereociliary bundles at their luminal surface. The reprogrammed hair cells expressed genes relevant to the development and function of inner ear hair cells, as shown by transcriptional profiling. The cells also appeared to express functional mechanoreceptor channels but lacked certain morphological characteristics of mature hair cells, such as highly organized stereociliary bundles. Thus, complete functional maturation of the reprogrammed hair cells likely requires additional intrinsic or extrinsic factors.
Primary auditory neurons are another target for direct reprogramming with the goal of reversing hearing loss due to disease and exposure to noise. Bipolar auditory neurons innervate cochlear hair cells and convey signals from the hair cells to the brain. Similar to its effect in retinal neurons, Ascl1 expressed in postnatal mice directly converted non-neuronal cochlear cells into a neuronal phenotype expressing the synaptic markers SNAP25 and synapsin I (Nishimura et al., 2014). Co-introduction of a second basic helix-loop-helix transcription factor, NeuroD1, promoted neuronal differentiation and survival but did not improve the distribution and electrophysiological properties of the cells. Thus, additional factors are likely necessary to achieve full maturation of the transdifferentiated auditory neurons. Nevertheless, these advances suggest it may be possible to generate auditory neurons by direct reprogramming of non-neural cells in the cochlea.
Challenges and Future Directions for in Vivo Direct Reprogramming
Direct in vivo reprogramming for local in situ conversion of cells is emerging as an alternative approach to regenerative medicine that would not require cell transplantation. Promising proof-of-concept studies have recently been reported in small animals, however numerous challenges must be overcome for this technology to impact human health. New technologies are rapidly paving the way for the needed breakthroughs.
Although discrete combinations of lineage-restricted regulatory factors can reprogram cells in many tissues, improved knowledge of the gene networks that drive cell fate will be necessary to intelligently improve reprogramming efficiency. Modern “omics” approaches that delineate the epigenetic events necessary for cells to acquire distinct fates will provide insight into the cues that will improve cell conversion. The discovery that removal of epigenetic barriers enhances cardiac reprogramming points to methods that may increase efficiency and explain paths of reprogramming (Zhou et al., 2016). Alternatively, unbiased screens using chemical libraries or CRISPR interference should reveal barriers to cell fate conversion that must be overcome to promote cellular plasticity. These areas of research are being facilitated by single-cell approaches to monitor transcriptome changes and epigenomic shifts. Recent single cell RNA-Seq analyses of induced neuron production suggests that most fibroblasts are competent to reprogram and alter gene expression rapidly, but silencing of reprogramming factors, death from an epigenetically unstable state, and reprogramming toward alternative fates limits the number of cells that successfully reprogram (Treutlein et al., 2016). These analyses revealed that expression of Ascl1, which appears to function as a “pioneer” factor in initiating neuronal reprogramming (Wapinski et al., 2013), initiates exit from the cell cycle and neuronal gene expression. However, many cells that initiate a cell fate switch undergo apoptosis due to oxidative stress given the dramatic change in redox state, and recent evidence demonstrates that use of antioxidants dramatically improved neuronal reprogramming (Gascon et al., 2016). These are several examples of how mechanistic understanding of the reprogramming process can lead to improvements in efficiency and quality of reprogramming, with some approaches potentially being applicable to multiple cell types.
Another major obstacle to address is delivery. As methods of reprogramming are optimized, safe and efficient delivery of the proper cues to the desired cell types may be the rate-limiting step. Gene therapy approaches are promising, and the advent of next-generation vectors with improved safety profiles has led to a resurgence in clinical trials of gene delivery for many diseases. Local delivery and cell-type-specific promoters may be useful for targeting distinct cells within organs. Unintended ectopic reprogramming is a theoretical risk. However, cellular reprogramming requires high levels of ectopic gene expression and has not been reported in ectopic tissues after local delivery, possibly because of low levels of the reprogramming factors at distant sites. The potential for chemical reprogramming is enticing, but achieving it will require engineering strategies to efficiently deliver compounds locally for extended periods of time. Advances in nanotechnology may allow such an approach. Finally, progress in the use of modified mRNA may allow vector-free gene delivery; however, this will require increasing the half-life of mRNA and accelerating cellular reprogramming events.
Ultimately, safety and efficacy trials in large animals will be necessary, particularly for organs such as the heart, where exponentially more cells will be needed for regeneration than in small animals. Safety issues will involve not only those related to delivery, but also the potentially detrimental consequences of partially reprogrammed cells. For conditions where there are no currently efficacious approaches, even small improvements will be successful outcomes.
Finally, the regulatory landscape will need to be addressed as this technology advances. Optimized reprogramming cocktails for many tissue will likely contain multiple genes, secreted proteins, or chemicals, and new delivery devices may be required. Considering each as a separate entity would slow regulatory approval. Acknowledgement that the combination is the product and that individual factors alone may be relatively inert should lead to a discussion that may expedite the design and evaluation of in vivo reprogramming, particularly for desperate populations with no current medical options.
In summary, a promising new approach for regenerative medicine has emerged from advances in developmental biology and the combinatorial approach to cellular reprogramming. The challenges ahead to improve this technology and translate it to clinical applications are not insignificant, but they appear to be tractable, given the rapidly changing landscape in biology. We look forward to the day when simple cues can be administered to harness the regenerative potential of cells within our organs, giving them, literally in some cases, a change of heart or mind.
ACKNOWLEDGMENTS
We thank Stephen Ordway for his editorial support and members of the Srivastava lab for their contributions and insight. D.S. was supported by grants from NHLBI/NIH, L.K. Whittier Foundation, William Younger Family Foundation, Eugene Roddenberry Foundation, and the California Institute for Regenerative Medicine (CIRM).
REFERENCES
- Ariyachet C, Tovaglieri A, Xiang G, Lu J, Shah MS, Richmond CA, Verbeke C, Melton DA, Stanger BZ, Mooney D, et al. (2016). Reprogrammed stomach tissue as a renewable source of functional beta cells for blood glucose regulation. Cell Stem Cell 18, 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson A, Collin T, Kirik D, Kokaia Z, and Lindvall O (2002). Neuronal replacement from endogenous precursors in the adult brain after stroke. Nat Med 8, 963–970. [DOI] [PubMed] [Google Scholar]
- Banga A, Akinci E, Greder LV, Dutton JR, and Slack JM (2012). In vivo reprogramming of Sox9+ cells in the liver to insulin-secreting ducts. Proc Natl Acad Sci U S A 109, 15336–15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al. (1997). Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 15, 30–35. [DOI] [PubMed] [Google Scholar]
- Bermingham-McDonogh O, and Reh TA (2011). Regulated reprogramming in the regeneration of sensory receptor cells. Neuron 71, 389–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau HM, Pavlath GK, Hardeman EC, Chiu CP, Silberstein L, Webster SG, Miller SC, and Webster C (1985). Plasticity of the differentiated state. Science 230, 758–766. [DOI] [PubMed] [Google Scholar]
- Bock-Marquette I, Saxena A, White MD, Dimaio JM, and Srivastava D (2004). Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature (article) 432, 466–472. [DOI] [PubMed] [Google Scholar]
- Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, et al. (2011). Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227. [DOI] [PubMed] [Google Scholar]
- Campbell KH, McWhir J, Ritchie WA, and Wilmut I (1996). Sheep cloned by nuclear transfer from a cultured cell line. Nature 380, 64–66. [DOI] [PubMed] [Google Scholar]
- Cao N, Huang Y, Zheng J, Spencer CI, Zhang Y, Fu J, Nie B, Wang H, Ma T, Xu T, et al. (2016). Pharmacological conversion of human fibroblasts into functional cardiomyocytes. Science, In press. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, and Wang DZ (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38, 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Sanchez-Guardado L, Juniat S, Gale JE, Daudet N, and Henrique D (2015). Generation of sensory hair cells by genetic programming with a combination of transcription factors. Development 142, 1948–1959. [DOI] [PubMed] [Google Scholar]
- Courtney M, Gjernes E, Druelle N, Ravaud C, Vieira A, Ben-Othman N, Pfeifer A, Avolio F, Leuckx G, Lacas-Gervais S, et al. (2013). The inactivation of Arx in pancreatic alpha-cells triggers their neogenesis and conversion into functional beta-like cells. PLoS Genet 9, e1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, and Lassar AB (1987). Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000. [DOI] [PubMed] [Google Scholar]
- De la Rossa A, Bellone C, Golding B, Vitali I, Moss J, Toni N, Luscher C, and Jabaudon D (2013). In vivo reprogramming of circuit connectivity in postmitotic neocortical neurons. Nat Neurosci 16, 193–200. [DOI] [PubMed] [Google Scholar]
- Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, and Ding S (2011). Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol 13, 215–222. [DOI] [PubMed] [Google Scholar]
- Fu J, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau BG, and Srivastava D (2013). Direct reprogramming of human fibroblasts toward the cardiomyocyte lineage. Stem Cell Reports 1, 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al. (2003). GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424, 443–447. [DOI] [PubMed] [Google Scholar]
- Gascon S, Murenu E, Masserdotti G, Ortega F, Russo GL, Petrik D, Deshpande A, Heinrich C, Karow M, Robertson SP, et al. (2016). Identification and successful negotiation of a metabolic checkpoint in direct neuronal reprogramming. Cell Stem Cell 18, 396–409. [DOI] [PubMed] [Google Scholar]
- Guo Z, Zhang L, Wu Z, Chen Y, Wang F, and Chen G (2014). In vivo direct reprogramming of reactive glial cells into functional neurons after brain injury and in an Alzheimer’s disease model. Cell Stem Cell 14, 188–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, and Fischberg M (1958). Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 182, 64–65. [DOI] [PubMed] [Google Scholar]
- Heidersbach A, Saxby C, Carver-Moore K, Huang Y, Ang Y, de Jong PJ, Ivey KN, and Srivastava D (2013). microRNA-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. eLife 2, e01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C, Blum R, Gascon S, Masserdotti G, Tripathi P, Sanchez R, Tiedt S, Schroeder T, Gotz M, and Berninger B (2010). Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol 8, e1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu J, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, and Srivastava D (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifkovits JL, Addis RC, Epstein JA, and Gearhart JD (2014). Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PloS one 9, e89678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagawa K, Miyamoto K, Yamakawa H, Muraoka N, Sadahiro T, Umei T, Wada R, Katsumata Y, Kaneda R, Nakade K, et al. (2012). Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circulation research 111, 1147–1156. [DOI] [PubMed] [Google Scholar]
- Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D, et al. (2012). Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci U S A 109, 13016–13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, and Dzau VJ (2012). MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res 110, 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena TM, Finch EA, Zhang L, Zhang H, Hodgkinson CP, Pratt RE, Rosenberg PB, Mirotsou M, and Dzau VJ (2015). MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circulation research 116, 418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R, and Arthur-Farraj P (2015). The role of cell plasticity in tissue repair: Adaptive cellular reprogramming. Dev Cell 34, 613–620. [DOI] [PubMed] [Google Scholar]
- Juhl K, Bonner-Weir S, and Sharma A (2010). Regenerating pancreatic beta-cells: plasticity of adult pancreatic cells and the feasibility of in-vivo neogenesis. Curr Opin Organ Transplant 15, 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N, Liang W, Marban E, and Cho HC (2013). Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol 31, 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl MO, Hayes S, Nelson BR, Tan K, Buckingham B, and Reh TA (2008). Stimulation of neural regeneration in the mouse retina. Proc Natl Acad Sci U S A 105, 19508–19513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow M, Sanchez R, Schichor C, Masserdotti G, Ortega F, Heinrich C, Gascon S, Khan MA, Lie DC, Dellavalle A, et al. (2012). Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell 11, 471–476. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, and Alvarez-Buylla A (2009). The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci 32, 149–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo BR, Baldwin EM, Layman WS, Taketo MM, and Zuo J (2015). In vivo cochlear hair cell generation and survival by coactivation of beta-catenin and Atoh1. J Neurosci 35, 10786–10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig J, Koch P, and Brustle O (2013). Leveling Waddington: The emergence of direct programming and the loss of cell fate hierarchies. Nat Rev Mol Cell Biol 14, 225–236. [DOI] [PubMed] [Google Scholar]
- Ladewig J, Mertens J, Kesavan J, Doerr J, Poppe D, Glaue F, Herms S, Wernet P, Kogler G, Muller FJ, et al. (2012). Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods 9, 575–578. [DOI] [PubMed] [Google Scholar]
- Lalit PA, Salick MR, Nelson DO, Squirrell JM, Shafer CM, Patel NG, Saeed I, Schmuck EG, Markandeya YS, Wong R, et al. (2016). Lineage reprogramming of fibroblasts into proliferative induced cardiac progenitor cells by defined factors. Cell Stem Cell 18, 354–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Nakanishi M, Zumsteg A, Shear M, Wright C, Melton DA, and Zhou Q (2014). In vivo reprogramming of pancreatic acinar cells to three islet endocrine subtypes. Elife 3, e01846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Miao Q, Yuan J, Han S, Zhang P, Li S, Rao Z, Zhao W, Ye Q, Geng J, et al. (2015). Ascl1 converts dorsal midbrain astrocytes into functional neurons in vivo. J Neurosci 35, 9336–9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugert S, Vogt M, Tchorz JS, Muller M, Giachino C, and Taylor V (2012). Homeostatic neurogenesis in the adult hippocampus does not involve amplification of Ascl1(high) intermediate progenitors. Nat Commun 3, 670. [DOI] [PubMed] [Google Scholar]
- MacRae CA (2016). In vitro and in vivo reprogramming for the conduction system. Trends Cardiovasc Med 26, 21–22. [DOI] [PubMed] [Google Scholar]
- Maitra M, Schluterman MK, Nichols HA, Richardson JA, Lo CW, Srivastava D, and Garg V (2009). Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol 326, 368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathison M, Gersch RP, Nasser A, Lilo S, Korman M, Fourman M, Hackett N, Shroyer K, Yang J, Ma Y, et al. (2012). In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. Journal of the American Heart Association 1, e005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, Nakashima H, Akiyama M, Wada R, Inagawa K, et al. (2014). MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. The EMBO journal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam YJ, Lubczyk C, Bhakta M, Zang T, Fernandez-Perez A, McAnally J, Bassel-Duby R, Olson EN, and Munshi NV (2014). Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development 141, 4267–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, et al. (2013). Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci U S A 110, 5588–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Weichert RM, Liu W, Davis RL, and Dabdoub A (2014). Generation of induced neurons by direct reprogramming in the mammalian cochlea. Neuroscience 275, 125–135. [DOI] [PubMed] [Google Scholar]
- Niu W, Zang T, Smith DK, Vue TY, Zou Y, Bachoo R, Johnson JE, and Zhang CL (2015). SOX2 reprograms resident astrocytes into neural progenitors in the adult brain. Stem Cell Reports 4, 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zang T, Zou Y, Fang S, Smith DK, Bachoo R, and Zhang CL (2013). In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat Cell Biol 15, 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, and Parmar M (2011). Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A 108, 10343–10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak J, Wilken MS, Ueki Y, Cox KE, Sullivan JM, Taylor RJ, Levine EM, and Reh TA (2013). ASCL1 reprograms mouse Muller glia into neurogenic retinal progenitors. Development 140, 2619–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, and Srivastava D (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, and Huang Y (2012). Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 11, 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaux C, and Arlotta P (2013). Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat Cell Biol 15, 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmang T (2013). Transcription factors that control inner ear development and their potential for transdifferentiation and reprogramming. Hear Res 297, 84–90. [DOI] [PubMed] [Google Scholar]
- Shapiro AM, Ricordi C, Hering BJ, Auchincloss H, Lindblad R, Robertson RP, Secchi A, Brendel MD, Berney T, Brennan DC, et al. (2006). International trial of the Edmonton protocol for islet transplantation. N Engl J Med 355, 1318–1330. [DOI] [PubMed] [Google Scholar]
- Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, and Ding S (2008). A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2, 525–528. [DOI] [PubMed] [Google Scholar]
- Smart N, Risebro CA, Melville AA, Moses K, Schwartz RJ, Chien KR, and Riley PR (2007). Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature 445, 177–182. [DOI] [PubMed] [Google Scholar]
- Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, and Eggan K (2011). Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Pacher M, Balakrishnan A, Yuan Q, Tsay HC, Yang D, Reetz J, Brandes S, Dai Z, Putzer BM, et al. (2016). Direct reprogramming of hepatic myofibroblasts into hepatocytes in vivo attenuates liver fibrosis. Cell Stem Cell. [DOI] [PubMed] [Google Scholar]
- Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, and Olson EN (1997). Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet 16, 154–160. [DOI] [PubMed] [Google Scholar]
- Srivastava D, and Yu P (2015). Recent advances in direct cardiac reprogramming. Curr Opin Genet Dev 34, 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Niu W, Liu ML, Zou Y, and Zhang CL (2014). In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat Commun 5, 3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, and Bhatia M (2010). Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468, 521–526. [DOI] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Thier M, Worsdorfer P, Lakes YB, Gorris R, Herms S, Opitz T, Seiferling D, Quandel T, Hoffmann P, Nothen MM, et al. (2012). Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 10, 473–479. [DOI] [PubMed] [Google Scholar]
- Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, and Herrera PL (2010). Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature 464, 1149–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torper O, Ottosson DR, Pereira M, Lau S, Cardoso T, Grealish S, and Parmar M (2015). In vivo reprogramming of striatal NG2 glia into functional neurons that integrate into local host circuitry. Cell Rep 12, 474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torper O, Pfisterer U, Wolf DA, Pereira M, Lau S, Jakobsson J, Bjorklund A, Grealish S, and Parmar M (2013). Generation of induced neurons via direct conversion in vivo. Proc Natl Acad Sci U S A 110, 7038–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Lee QY, Camp JG, Mall M, Koh W, Shariati SA, Sim S, Neff NF, Skotheim JM, Wernig M, et al. (2016). Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534, 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki Y, Wilken MS, Cox KE, Chipman L, Jorstad N, Sternhagen K, Simic M, Ullom K, Nakafuku M, and Reh TA (2015). Transgenic expression of the proneural transcription factor Ascl1 in Muller glia stimulates retinal regeneration in young mice. Proc Natl Acad Sci U S A 112, 13717–13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedantham V, Galang G, Evangelista M, Deo RC, and Srivastava D (2015). RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for Islet-1 in cardiac pacemaker cells. Circ Res 116, 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, and Wernig M (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada R, Muraoka N, Inagawa K, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Kaneda R, Suzuki T, Kamiya K, et al. (2013). Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci U S A 110, 12667–12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cao N, Spencer CI, Nie B, Ma T, Xu T, Zhang Y, Wang X, Srivastava D, and Ding S (2014). Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep 6, 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, et al. (2013). Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods C, Montcouquiol M, and Kelley MW (2004). Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci 7, 1310–1318. [DOI] [PubMed] [Google Scholar]
- Xie H, Ye M, Feng R, and Graf T (2004). Stepwise reprogramming of B cells into macrophages. Cell 117, 663–676. [DOI] [PubMed] [Google Scholar]
- Xu J, Du Y, and Deng H (2015). Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell 16, 119–134. [DOI] [PubMed] [Google Scholar]
- Yamakawa H, Muraoka N, Miyamoto K, Sadahiro T, Isomi M, Haginiwa S, Kojima H, Umei T, Akiyama M, Kuishi Y, et al. (2015). Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Reports 5, 1128–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Zuchero JB, Ahlenius H, Marro S, Ng YH, Vierbuchen T, Hawkins JS, Geissler R, Barres BA, and Wernig M (2013). Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol 31, 434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, and Stanger BZ (2013). Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev 27, 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, and Crabtree GR (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, and Mango SE (2016). Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev 37, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Han P, Yang H, Ouyang K, Lee D, Lin YF, Ocorr K, Kang G, Chen J, Stainier DY, et al. (2013). In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 498, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cao N, Huang Y, Spencer CI, Fu JD, Yu C, Liu K, Nie B, Xu T, Li K, et al. (2016). Expandable cardiovascular progenitor cells reprogrammed from fibroblasts. Cell Stem Cell 18, 368–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, and Gage FH (2008). Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Londono P, Cao Y, Sharpe EJ, Proenza C, O’Rourke R, Jones KL, Jeong MY, Walker LA, Buttrick PM, et al. (2015). High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun 6, 8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, and Srivastava D (2007). Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell 129, 303–317. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Samal E, and Srivastava D (2005). Serum response factor regulates a muscle-specific mircroRNA that targets Hand2 during cardiogenesis. Nature 436, 214–220. [DOI] [PubMed] [Google Scholar]
- Zheng JL, and Gao WQ (2000). Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci 3, 580–586. [DOI] [PubMed] [Google Scholar]
- Zhou H, Dickson ME, Kim MS, Bassel-Duby R, and Olson EN (2015). Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci U S A 112, 11864–11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, and Melton DA (2008). In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455, 627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang L, Vaseghi HR, Liu Z, Lu R, Alimohamadi S, Yin C, Fu JD, Wang GG, Liu J, et al. (2016). Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell 18, 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Russ HA, Wang X, Zhang M, Ma T, Xu T, Tang S, Hebrok M, and Ding S (2016). Human pancreatic beta-like cells converted from fibroblasts. Nat Commun 7, 10080. [DOI] [PMC free article] [PubMed] [Google Scholar]


