Abstract
The Ku70-Ku80 (Ku) heterodimer binds rapidly and tightly to ends of DNA double-strand breaks and recruits several factors of the Non-Homologous End Joining (NHEJ) pathway through molecular mechanisms that remain unclear. Here, we describe the crystal structures of the Ku-binding motifs (KBM) of the NHEJ proteins APLF (A-KBM) and XLF (X-KBM) bound to a Ku-DNA complex. The two KBMs motifs bind on remote sites of Ku80 α/β domain. The X-KBM occupies an internal pocket formed after an unprecedented large outward rotation of the Ku80 α/β domain. We reveal independent recruitment at laser-irradiated sites of the APLF-interacting protein XRCC4 and of XLF through the respective binding of A- and X-KBMs to Ku80. Finally, we show that mutations on the X-KBM and A KBM binding sites in Ku80 compromises efficiency and accuracy of end-joining and cellular radiosensitivity. A- and X-KBMs may represent two initial anchorage points necessary to build the NHEJ intricate interactions network.
Keywords: DSB repair, NHEJ, X-ray crystallography, induced fit, laser micro-irradiation, super resolution microscopy, switchSENSE, microcalorimetry
INTRODUCTION
In mammals the majority of DNA double-strand breaks (DSBs) is repaired by the non-homologous end-joining (NHEJ) pathway1,2. The Ku70-Ku80 heterodimer (Ku) rapidly and tightly interacts as a preformed ring with DSBs extremities in a non-sequence specific manner3. Ku serves as a hub for the recruitment of several NHEJ factors4,5. Among them, Ku recruits the XRCC4-LIG4-XLF ligation complex through interaction with XRCC4-LIG46,7 and with XLF, the latter relying on a Ku-binding motif (KBM) localized at the XLF extreme C-terminus (thereafter named X-KBM)8,9 (Figure 1a and Supplementary Figure 1). The ligation complex organizes into filaments both in vitro and in cells10–14.
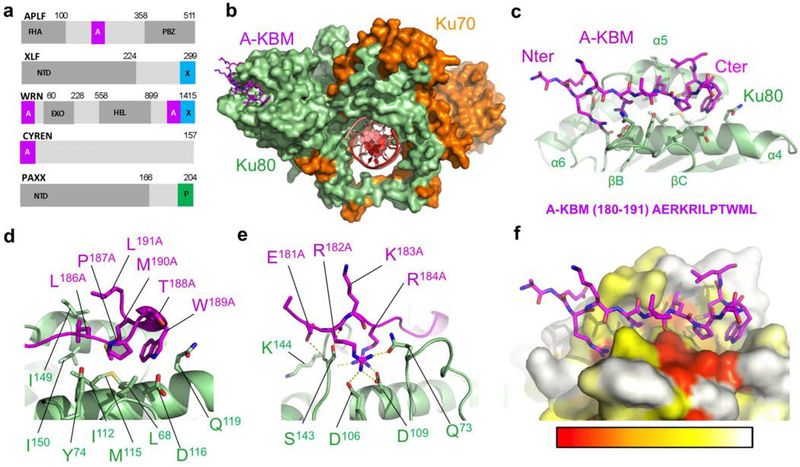
Figure 1. Crystal structure of the APLF KBM (A-KBM) bound to the Ku80 vWA domain.
(a) Positions of the A-KBM (magenta) and X-KBM (blue) motifs in APLF, XLF, WRN and CYREN. The C-terminal domain of PAXX contains a P-KBM that interacts with Ku70 subunit. NTD: N-terminal domain. (b) Overall view of the quaternary complex Ku70/Ku80/hDNA/(APLF peptide). The A-KBM (magenta) binds at the periphery of the Ku80 (light green) vWA domain. The Ku70 subunit and hDNA are represented respectively in orange and red. The hairpin part of the DNA has been removed for clarity. (c) The N-terminal part of the A-KBM motif has an extended conformation whereas the C-terminal residues form a turn. (d-e) Zoom of the interactions made by (d) the hydrophobic patch and (e) the basic patch of the A-KBM. (f) The A-KBM binding site is delineated by conserved residues of Ku80 vWA domain. The binding site is represented in surface mode with amino acids colored according to their conservation rate: red (highly conserved) to white (not conserved)). The conservation rate was measured using sequences of metazoan Ku80. The orientation is the same as in (c).
Interestingly, Ku interacts also with a number of accessory NHEJ factors. The APTX and PNKP-like factor (APLF) binds poly(ADP)-ribosylated proteins near DSBs sites15,16, and has been reported to have nuclease activity16,17. APLF tightly interacts with Ku through a KBM (thereafter named A-KBM) that is located in its central region9,18 (Figure 1a and Supplementary Figure 1). This interaction has been mapped to the periphery of the Ku80 von Willebrand A domain (vWA)5. Ku-APLF interaction was shown to facilitate recruitment of the APLF-partner XRCC4 at damaged sites9 and was proposed to stabilize the assembly of NHEJ factors around the DSB19. Notably, an A-KBM-like domain is present at the N-terminus of a recently identified inhibitor of the NHEJ pathway, CYREN(MRI), that also interacts with Ku8020 (Figure 1a). Ku also associates with the Werner syndrome protein (WRN) that is involved in many aspects of DNA metabolism including NHEJ21. Two motifs in the C-terminus of WRN cooperate for interaction with Ku, one being A-KBM like, and the other resembling the X-KBM present on XLF (Figure 1a). In addition, we and others showed recently that PAXX (Paralog of XRCC4 and XLF) interacts with the Ku70 subunit through a third type motif that is located in its C-terminus22,23 (Figure 1a). Despite identification of KBMs in several NHEJ factors, their respective contribution to the efficiency of DSB repair is not fully understood. For example, the puzzling observations that KBM deletion in XLF or APLF depletion in human cells lead to null or intermediate repair defect deserve further investigations9,24–27.
The interactome of Ku thus defines a large ensemble of motifs and proteins that could potentially compete or act synergistically. However, despite important structural and biophysical studies on NHEJ complexes28, the absence of high resolution structures of Ku-KBMs complexes limits our understanding of the roles and specificity of the different molecular interactions in the recruitment of NHEJ factors to DSBs. Mapping KBM-binding sites in structures is also needed to clarify potential competition of all the Ku interacting factors on limited positions (Supplementary figure 1).
Here, we establish the structural and functional basis of Ku unique modes of interaction with two factors within the NHEJ repair pathway. We describe the first crystal structures of Ku70-Ku80-DNA complex in interaction with the A-KBM (KBM of APLF) and with the X-KBM (KBM of XLF), revealing that the two KBMs occupy remote interaction sites on the Ku80 vWA domain. The functional context of our structural data was determined using cell based assays to visualize the recruitment of wild-type and mutant X-KBM motifs or XLF proteins as well as of the APLF-partner XRCC4 to DSBs sites induced by micro-irradiation in wild-type or mutant Ku backgrounds. Our data provide new mechanistic insights on the function of XLF and APLF in the NHEJ process.
RESULTS
The KBM of APLF tightly interacts with a highly conserved site of Ku80 vWA domain.
The APLF factor contains a conserved Ku binding motif (A-KBM, aa 179–192) (Figure 1a). We co-crystallized a 18-mer A-KBM peptide (aa 174–191) with a Ku form deleted for the C-terminal regions (Kucc), and an hairpin DNA (hDNA)3 (Supplementary Figure 2a). In the crystal structure at 3.0 Å resolution (Table 1), the A-KBM peptide is well defined and is positioned at the periphery of the vWA domain of Ku80 (aa T6180-C15680) (thereafter, superscript 80 stands for Ku80) (Figure 1b, and Supplementary Figure 2b). It is located at more than 50 Å from the DNA binding site of Ku80. The A-KBM adopts an extended conformation in a pocket delineated by the helices α4 and α5 and the loop located between the β-strands B and C of Ku80 (Figure 1c). The hydrophobic part of the A-KBM, located in the C-terminal part of the motif, is composed by the amino acids I185LPTWML191 and is buried in a hydrophobic pocket formed by the Ku80 residues L6880, I11280, M11580, I14980 and I15080 (Figure 1d). The N-terminal part of the A-KBM contained a patch of three consecutive basic residues and an acid residue (E181RKR184 in human sequence) (Figure 1e). It forms salt bridges and charged hydrogen bonds with respectively the side chains of D10680, D10980, Q7380 and S14580, and the main chain of K14480 and S14380.
Table 1.
Data collection and refinement statistics
| Ku-pAPLF (PDB 6ERF) |
Ku-pXLF (PDB 6ERH) |
Ku-pXLFs (PDB 6ERG) |
|
|---|---|---|---|
| Data collection | |||
| Space group | Pi | P2i | P2i |
| Cell dimensions : | |||
| a, b, c (Å) | 98.9, 140.8, 150.3 | 111.8, 118.9, 128.2 | 111.7, 114.3, 127.2 |
| α, β, γ (°) | 68.6, 80.8, 81.2 | 90.0, 93.1, 90.0 | 90.0, 93.1, 90.0 |
| Resolution (Å) | 49.5–3.0 (3.28–3.0) | 49.01–2.8 (3.16–2.8) | 50–2.9 (3.09–2.9) |
| Anisotropy resolution limits (Å)§ | 2.9, 3.9, 3.3 | 2.7, 4.4, 3.4 | 2.8, 3.8, 3.0 |
| Resolution limit overall (Å)§ | 3.15 | 3.11 | 3.0 |
| Rmerge | 0.056 (0.79) | 0.164 (1.35) | 0.171 (2.48) |
| Rmeas | 0.072 (0.93) | 0.182 (1.47) | 0.182 (2.56) |
| Rpim | 0.051 (0.66) | 0.094 (0.76) | 0.068 (0.94) |
| I/σ (I) | 11.8 (1.3) | 9.1 (1.5) | 9.7 (1.0) |
| CC1/2 | 0.99 (0.60) | 0.997 (0.60) | 0.996 (0.51) |
| Completeness (spherical, %)§ | 66.5 (14.3) | 51.3 (8.5) | 72.9 (21.0) |
| Completeness (ellipsoidal, %)§ | 92.1 (70.1) | 92.4 (69.6) | 94.6 (68.5) |
| Redundancy | 3.4 (3.5) | 7.1 (6.9) | 14.0 (14.2) |
| Refinement | |||
| Resolution (Å) | 49.47–3.01 | 49.38–2.8 | 48.76–2.9 |
| No. reflections | 90993 | 41644 | 51644 |
| Rwork / Rfree | 0.209/0.227 | 0.225/0.252 | 0.218/0.244 |
| No. Atoms | 35410 | 18810 | 18989 |
| Protein | 33225 | 16649 | 16821 |
| DNA | 2178 | 2118 | 2133 |
| SO42- | 0 | 10 | 15 |
| Water | 7 | 33 | 20 |
| B factors (Å2) | |||
| Protein | 112 | 76 | 94 |
| DNA | 201 | 109 | 126 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | 0.007 | 0.008 |
| Bond angles (°) | 0.96 | 0.95 | 0.95 |
Values in parentheses are for highest-resolution shell.
Values from STARANISO, Global Phasing Ltd.
Isothermal Titration Calorimetry (ITC) showed that the interaction between A-KBM and Ku had a Kd of 33 ± 10 nM (Table 2, and Supplementary Figure 2e). We measured a nanomolar Kd for the interaction between Ku and a 18bp DNA as already reported29 and found a similar Kd for the interaction between the A-KBM and Ku alone or Ku bound to a 18bp DNA (Table 2). The interaction of the A-KBM with Kucc showed Kd and enthalpy values similar to full-length Ku (KuFL). Thus, the core heterodimeric region of Ku is likely sufficient for the interaction with the A-KBM. Notably, these affinities are stronger than the ones previously reported by fluorescence polarization with a labelled A-KBM peptide (Kd of 580 nM)9.
Table 2 :
Interactions measured by microcalorimetry between Ku70/Ku80, A-KBM, X-KBM and XLF
| Protein in the measurement cell | Ligand in the syringe | Kd (μM) | ΔH (kcal.M−1) | Remarks | |
|---|---|---|---|---|---|
| 1 | KuFL | pAPLF | 0.033 ± 0.01 | −13.3 ± 0.1 | A-KBM (APLF 174–191) |
| 2 | KuFL | DNA 18bp | 0.0041 ± 0.0007 | +5.3 ± 0.2 | DNA18bp versus Ku |
| 3 | KuFL+DNA-18bp | pAPLF | 0.023 ± 0.002 | −16.0 ± 0.4 | in presence of DNA18bp |
| 4 | KuCC | pAPLF | 0.020 ± 0.002 | −18.4 ± 0.7 | Ku without Cter domains |
| 5 | KuFL I112R | pAPLF | NI(a) | NI | Ku80 mutant on APLF site |
| 6 | KuFL | pXLF | 4.4 ± 0.2 | −2.8 ± 0.2 | X-KBM (XLF 281–299) |
| 7 | KuFL +DNA-18bp | pXLF | 2.4 ± 0.1 | −8.1 ± 0.6 | in presence of DNA18bp |
| 8 | KuCC | pXLF | 2.2 ± 0.9 | −3.1 ± 1.2 | Ku without Cter domains |
| 9 | KuFL | XLF | 1.0 ± 0.1 | −9.0(b) | XLF versus Ku |
| 10 | KuFL+DNA-18bp | XLF | 2.35 ± 0.1 | −8.1 ± 0.6 | XLF versus Ku/DNA18bp |
| 11 | KuFL | pXLF(LW) | 0.12 ± 0.03 | −12.1 ± 2.0 | X-KBM (L297W) |
| 12 | KuFL | pXLF(LE) | NI | NI | X-KBM (L297E) |
| 13 | KuFL+pAPLF | pXLF(LW) | NI | NI | L297W in presence of A-KBM |
NI means no interaction;
The enthalpy value was deduced from the first injection point in absence of lower plateau; The thermograms and isotherms of titration of the experiments corresponding to lines 1, 6 and 11 are presented in Supplementary Figure 2e–g. Interactions in lines 1, 4, 6, 8, 9, 11 were measured in triplicate, and the mean value with standard deviation is reported. Interactions in lines 2, 3, 4, 6, 10 were measured in duplicate, and the mean value with variation between min and max values is reported.
Mapping the conservation rate of the residues at the surface of Ku80 shows that this pocket is the main conserved pocket together with the DNA binding pocket (Figure 1f). The residues L6880, Y7480 and I11280 make tight interactions with the hydrophobic part of the A-KBM motif. Mutations at these positions were reported to greatly reduce or disrupt the interactions with APLF in yeast two-hybrid experiments or in EMSA9. We produced the Ku I112R mutant that, as expected had no residual interaction with the A-KBM motif by ITC (Table 2).
X-KBM creates an outward rotation of the vWA domain and a large groove in Ku80.
We then determined the crystal structures of the Ku70-Ku80-hDNA complex bound with peptides derived from the XLF X-KBM ((L281X to S299X) and (S287X-S299X) peptides) (Supplementary Figure 2c). The crystal structures at 2.8 and 2.9 Å resolution (Table 1) show an unprecedented large outward rotation of the Ku80 vWA (Figure 2a, b). This movement forms a large groove between the Ku80 vWA and the rest of the Ku heterodimer. We therefore termed this conformation the open state of Ku, in contrast to the closed state observed in the three other crystal structures reported (Ku alone (1JEQ), Ku-hDNA (1JEY) and Ku-hDNA-A-KBM (this study)). The conformational change of Ku80 vWA does not affect Ku interaction with the duplex DNA (Supplementary Figure 2d)
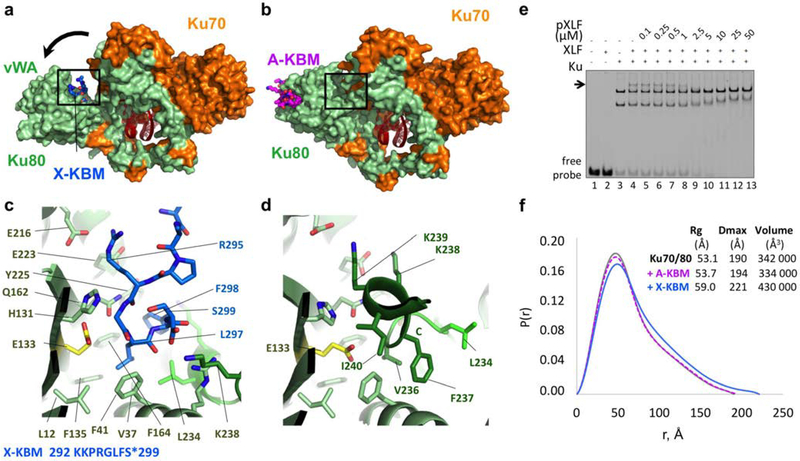
Figure 2. Crystal structure of the XPLF KBM (X-KBM) bound to the Ku80.
(a) Crystal structure of the quaternary complex Ku70-Ku80-DNA-(X-KBM peptide). The X-KBM (blue) binds in an internal site of the Ku80 subunit created upon an outward rotation of the vWA domain. The Ku80 vWA opening creates a large groove between the Ku80 vWA and the rest of the heterodimer. (b) The crystal structure of Ku70/Ku80/DNA in presence of the A-KBM is shown with the same orientation. (c-d) Comparison of the X-KBM binding site in presence of X-KBM (c) or A-KBM (d) peptides. The X-KBM interacts with Ku80 residues involved in Ku intramolecular contacts in the closed state of Ku observed with the A-KBM or with no peptide. The last GLFS residues of the X-KBM interact with the bottom of the groove formed in the open state. The glutamic acid presents an atypical hydrophobic environment and could be at the origin of the vWA instability. The X-KBM residues occupy the position of the helix 236–241 of Ku80 in the closed conformation and some X-KBM side chains (R295X, L297X and F298X) mimic the intramolecular interactions made by Ku80 residues with the vWA domain. (e) Gel shift assay with XLF and Ku in presence of a 50bp DNA with a FAM in 5’and competition with pXLF containing the X-KBM motif. The arrow indicates the XLF Ku-DNA complex. Uncropped gel image is shown in Supplementary Data Set 1. (f) The pair distributions P(r) obtained in solution by SAXS analysis indicates an opening of the Ku70/Ku80/DNA complex with higher Dmax and Rg in presence of the X-KBM (blue line) compared to the Ku/DNA complex without peptide (grey line) and to the A-KBM complex (magenta line). Values deduced from SAXS analysis are reported beside the curves.
The X-KBM is located on the Ku80 vWA face of the newly created groove in a pocket delineated by 4 strands (βA, βD, βE and βE’) and 3 helices (α2, α7’, α7”). The motif is positioned closer to the DNA (at 12Å) than the A-KBM, though not in direct contact. We can model eight residues of the X-KBM (292KKPRGLFS299) in the crystal obtained with the 19mer or 14mer X-KBM. These residues are the last eight residues of the XLF sequence. The residues 296GLFS299 of the X-KBM occupy a hydrophobic pocket delineated by Ku80 residues L1280, V3780, F4180, F13580, F16480, Y22580, and L23480 (Figure 2c). In the closed state, these Ku residues are buried and mediate intramolecular contacts (Figure 2d). The X-KBM may thus stabilize a transient open conformation of Ku80 in equilibrium with the closed state under basal conditions. In addition, we observed that the Ku80 vWA opening comes along with important secondary structure changes in the linker region (R23280-E24180) that separates the vWA and the rest of the Ku heterodimer (Figure 2c, d).
ITC measurements showed a moderate affinity of X-KBM for Ku, with a Kd of 4.4 ± 0.2 μM (about 200-fold weaker than the affinity of the A-KBM) (Table 2, and Supplementary Figure 2f). We observed similar affinities and thermodynamic parameters for the X-KBM with a Ku-DNA complex or with Kucc (Table 2). Notably, this micromolar interaction was not detected in a previous study using fluorescence polarization with labelled peptides9. We also measured similar affinities for the interactions between XLF homodimer and Ku alone or Ku bound to a 18bp DNA (Table 2). These data show that in absence of DNA or with a short DNA protected by Ku ring, XLF and its X-KBM interact similarly with Ku.
EMSA analyses confirmed an interaction between XLF and Ku complexed with a 50bp DNA in the μM range (Supplementary Figure 3a–b). Competitions experiments showed that the X-KBM peptide (pXLF) competes in the μM range with the XLF protein and that the A-KBM peptide (pAPLF) does not displace XLF, supporting remote sites of interactions (Figure 2e and Supplementary Figure 3c, d). Also, the C-terminus of PAXX (pPAXX) does not compete with XLF binding, in agreement with previous studies that report an interaction between PAXX C-terminus and Ku70 subunit22,23 (Supplementary Figure 3e).
The outward rotation of the vWA domain of Ku80 is mediated by Glu133.
To evaluate if the outward rotation of Ku80 observed in the crystal structure with the X-KBM peptide was present in solution, we performed SAXS analyses. The SAXS data with the A-KBM peptide were comparable with those of the Ku-hDNA sample without any peptide (respective Rg of 53.1Å and 53.7) (Figure 2f). We measured an increase of the Rg to 59.0Å in presence of the X-KBM, corresponding to the opening of Ku80 observed in the crystal (Figure 2f). Comparison of the Dmax values provided further corroboration for the opening of the Ku molecule with an increase of 30 Å.
The acid residue E13380 buried in Ku80 is well positioned to act as a spring facilitating the Ku80 opening (Figure 2c, d). The glutamate E13380 is buried in the closed conformation of the Ku80 vWA and its pKa value is estimated by the PDB2PQR-2.0 server30 at a value of 9.1, far from the normal pKa of 4.5 for a glutamate in solvent. The outward rotation of Ku80 vWA should be energetically facilitated by the solvation of this Glu following the displacement of Ku80 residues V23680, F23780 and I24080 away from E13380 carboxylate function (Figure 2d). Multiple sequence alignments show that E13380 position and the residues surrounding are well conserved in mammalian and saurian and that Ku70 has no equivalent buried acidic residue at this position (Supplementary Figure 4a–b).
Molecular bases of the specificities of A-KBM and X-BKM motifs binding to Ku80.
The A-KBM and X-KBM present sequence similarities with a basic patch in their N-terminus followed by a hydrophobic patch9 (Supplementary Figure 1). Comparison of the crystal structures of Ku bound to these two motifs suggests that the high affinity of the A-KBM relies on the tryptophan W189A in place of the Leu297X in X-KBM. Thus, we used ITC to measure the interaction of the X-KBM motifs with the mutation L297W (LW) or with a non-conservative L297E mutation (LE). The (LW) peptide has a Kd of 0.12 ± 0.03 μM, an interaction 40-fold tighter than wild-type X-KBM (Table 2 and Supplementary figure 2g). The LE mutant presents no detectable interaction with Ku (Table 2). Competition experiments with Ku saturated with the A-KBM showed that the X-KBM (L297W) no longer interacts with Ku, suggesting that the sole L297W mutation is able to redirect the X-KBM towards the A-KBM binding site on Ku80 (Table 2).
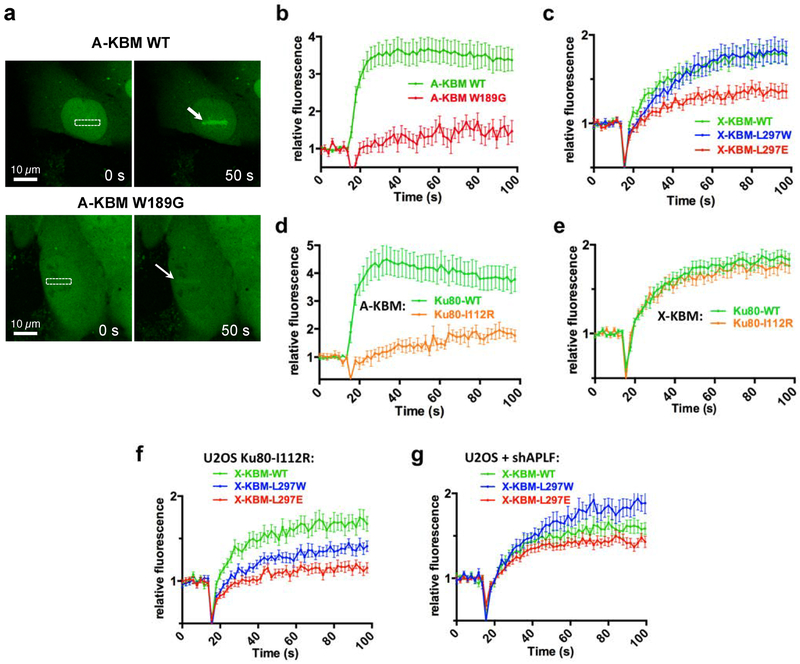
Then live cell imaging was used to monitor the recruitment of CFP-fused A-KBM and X-KBM fragments to DSBs sites induced with laser micro-irradiation. Under conditions of similar damage yield (Supplementary Figure 5e), the A-KBM motif transfected in U2OS cells was strongly recruited but the W189G mutation impaired both its nuclear localization and recruitment to laser sites (Figure 3a–b), as reported9,18. X-KBM live recruitment was impaired by L297E mutation but not L297W mutation (Figure 3c), corroborating ITC data. We then used U2OS cells expressing an inducible shRNA against Ku8031 (Supplementary Figure 5a) that were complemented with wild-type or I112R mutant Ku80 (Supplementary Figure 5c–d). The I112R mutation impaired A-KBM recruitment, as expected, but not that of X-KBM (Figure 3d, e). Notably, I112R Ku80 mutant specifically lowered the recruitment of L297W X-KBM (compare Figures 3c and 3f). Conversely, APLF knock-down boosted the recruitment of the LW mutant peptide above that of wild-type X-KBM (Figure 3g and Supplementary Figure 5b for control of shAPLF efficiency). Together, these data in cells support that the LW mutation redirects the X-KBM fragment to the APLF-binding site in Ku80 and point out the W189 residue as a key determinant for APLF specific interaction with Ku80.
Figure 3. Life cell imaging of A-KBM and X-KBM recruitment after nuclear micro-irradiation.
(a) Wild-type (WT) and mutant CFP-(A-KBM) behaviour at 0 s and 50 s after laser nuclear micro-irradiation. The white rectangle and arrows mark irradiated areas. Magnification: X40. (b) Dynamics of wild-type and mutant CFP-(A-KBM) at laser-induced damage sites in U2OS cells. Mean values of relative fluorescence with s.e.m. were calculated from data obtained in several individual cells: n=23 and 19 cells for WT and mutant A-KBM, respectively). p values at last time point were calculated using unpaired two-tailed t-test: WT vs W189G p<0.0001. (c) Dynamics of wild-type and mutant CFP-(X-KBM) at laser-damaged sites as in b). n=27, 21, and 24 cells for WT, L297E and L297W X-KBM, respectively. p values at last time point : WT vs L297W p=0.8574; WT vs L297E p=0.0021. (d-e) Dynamics of CFP-(A KBM) (d) and (X-KBM) (e) at laser damaged sites in cells expressing wild-type or I122R mutant Ku80 as in b). n=20, 13 cells for A-KBM in WT or I122R Ku80, and n=48 and 39 cells for X-KBM in WT or I122R Ku80, respectively. p values at last time point : (d) WT vs I112R p=0.0002; (e) WT vs I112R p=0.5692 (f-g) Dynamics of wild-type and mutant CFP-(X-KBM) at laser-damaged sites in cells expressing I112R mutant Ku80 (f) or treated with a shAPLF (g) as in b). n=26, 28, and 21 cells for WT, L297E and L297W X-KBM in (f), and n=15 cells for each of WT, L297E and L297W X-KBM in (g). p values at last time point : (f) WT vs L297W p=0.023; WT vs L297E p=<0.0001; (g) WT vs L297W p=0.0144; WT vs L297E p=0.2654.
X-KBM mutations impair XLF recruitment and XRCC4-XLF filament stability.
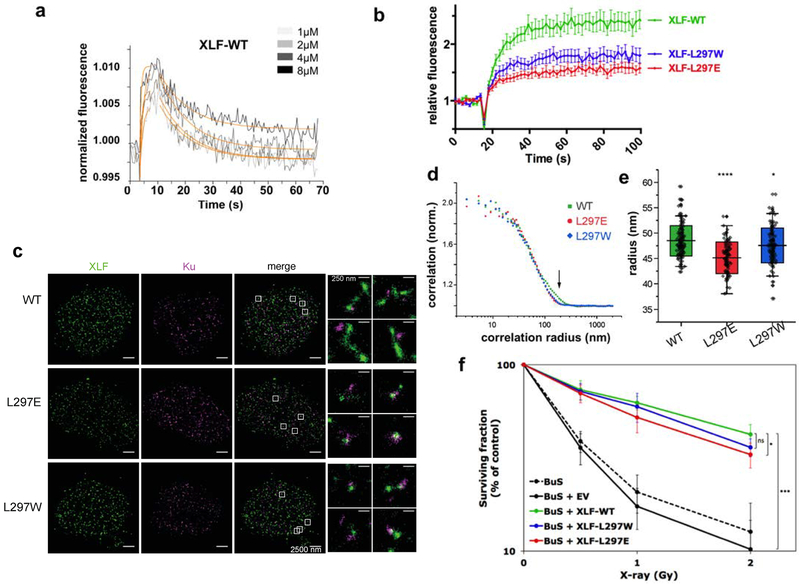
We then investigated the properties of the interaction between full-length XLF protein and Ku. We first used the SwitchSENSE approach32 in which oligonucleotide nanolevers labelled with a fluorescence probe are bound to a gold surface (Supplementary Figure 3f). Ku bound onto 48bp DNA nanolevers with a Kd in the nM range as already reported29 and a long dissociation time (Supplementary Figure 3g). Wild-type XLF onto the Ku-DNA complex showed a rapid kon (4.7 ± 1.7 105 M−1s−1) followed by a rapid dissociation (koff = 0,09 ± 0,004s−1) and a corresponding Kd of 0.19 ± 0.07 μM (Figure 4a). This affinity is about 10 fold stronger than the one measured by ITC with a smaller DNA and may reflect additional interactions of XLF with DNA emerging from Ku ring as observed with PAXX22. LW and LE mutants showed a 2.3- and 5.1-fold weaker affinity than WT protein, respectively (Kd of 0.45 ± 0.26 μM for LW and Kd of 0.98 ± 0.15 μM for LE) Supplementary Figure 3h). As compared with ITC and recruitment data with X-KBM LW mutant peptide, this suggests that the LW mutation cannot redirect the XLF protein to the APLF binding site in Ku80.
Figure 4. Biophysical and cellular analyses of XLF mutants in X-KBM.
(a) SwitchSENSE kinetic analysis of the WT XLF interaction with Ku-DNA complexes. Solid grey lines represent raw data (from 1 to 8 μM; light grey to dark grey; averages of triplicates). Global fitting was performed, following a single-exponential function (solid orange lines) yielding kinetic rate constants; kON=4.7 ± 1.7 105 M−1s−1 and kOFF = 9.1 ± 0.4 10−2s−1 for XLF(wt). (b) Dynamics of wild-type and mutant CFP-XLF at laser-damaged sites in BuS cells as in Figure 3b. n=20 cells for WT, L297E and L297W XLF. p values at last time point : WT vs L297W p=0.0093; WT vs L297E p<0.0001. (c) Representative super-resolution images of WT, L297E mutant, and L297W mutant BuS nucleus, with XLF and Ku displayed in green and magenta, respectively (scale = 2500 nm). Right: zoomed-in areas (scale = 250 nm). (d) Representative pair correlation function calculated from the 8×8 μm2 center square of one XLF nucleus image of WT (green), L297E (red), and L297W (blue) mutants. WT XLF shows bigger correlation radius (arrow). (e) Statistics of XLF foci size. Each plot represents the average XLF foci size (indicated as radius translated from the correlation radius) in one nucleus. Box’s height displays the s.d. with the mean value labelled in the middle. n=116, 95, 104 nuclei for WT, L297E, and L297W. The two-sample unpaired t-test between WT and L297E is p=10−13 while that between WT and L297W is p=0.03. (f) Cell survival of BuS cells complemented with vector (EV) or WT or mutated XLF. y axis is log scale. Error bars represent s.d., n=5 to 6 independent experiments. p values were calculated using unpaired two-tailed t-test: WT vs EV p=1.788e-06; WT vs LW p=0.068; WT vs LE p=0.021. (*p< 0.05, **p < 0.01, ***p < 0.001).
We then measured the recruitment of the CFP-tagged full-length XLF protein (CFP-XLF) expressed in human XLF-defective BuS cells (SV40T-transformed, telomerase immortalized fibroblasts derived from the XLF-deficient P2 patient - homozygous C622T nonsense mutant (R178X))33 (Figure 4b). Wild-type XLF showed a rapid mobilization to irradiated nuclear sites, as reported34. Both LE and LW mutations impaired to various extents XLF protein recruitment (Figure 4b), again indicating that LW mutation cannot redirect the XLF protein to the APLF binding site in Ku80. In addition, we observed an important reduction of the recruitment of G296W, S299E or F298G XLF mutants in the extreme C-terminus (Supplementary Figure 5f–h). We also questioned the contribution of APLF-XRCC4 complex to XLF recruitment by expressing L115D XLF mutant unable to interact with XRCC435,36. We measured an efficient recruitment of L115D XLF to laser-induced DSBs that was insensitive to APLF knock-down but impaired with the XLF L115D/L297E XLF double mutant (Supplementary Figure 5i). Together, these data support a major role for Ku80 interaction with L297 and extreme C-terminal residues for XLF recruitment at DSBs in cells.
Multi-color super-resolution localization microscopy (STORM) allows characterizing formation of XRCC4-XLF filaments close to Ku foci and DSBs ends14. On DSBs induction with the radiomimetic drug neocarzinostatin, extended XLF filaments close to Ku80 foci were observed in XLF complemented BuS cells, whereas cells harboring (LE) and (LW) mutants showed slightly smaller and more punctuated XLF structures (Figure 4c and Figure 4d–e for quantification). In contrast to these data, evaluation of the effect of mutations in the X-KBM on cell radiosensitivity showed that (LW) or (LE) XLF mutants were associated with respectively no or minor radiosensitization, compared to high radiosensitivity of BuS cells and full restoration of radioresistance on expression of wild-type XLF (Figure 4f).
APLF and XLF binding to Ku80 promote DSB repair and cell survival to IR.
Finally, we questioned the discrepancy between subnormal cell survival and defective XLF recruitment and filaments formation associated with mutations in X-KBM. The outward rotation in Ku80 upon X-KBM binding more likely relies on E13380, the equivalent position of which in Ku70 is a methionine (M167) (Supplementary Figure 4b). Therefore, we designed E133M mutant in Ku80 and Q162E mutation that may alter the charge environment of E133 (Supplementary Figure 5c). E133M or Q162E mutation in Ku80 negatively impacted X-KBM recruitment but not that of control A-KBM (Supplementary Figures 6a and 6b). This result further supports the independent binding sites on Ku80 of the A-KBM and X-KBM motifs and validates E133 and Q162 positions in Ku80 as key residues for X-KBM binding.
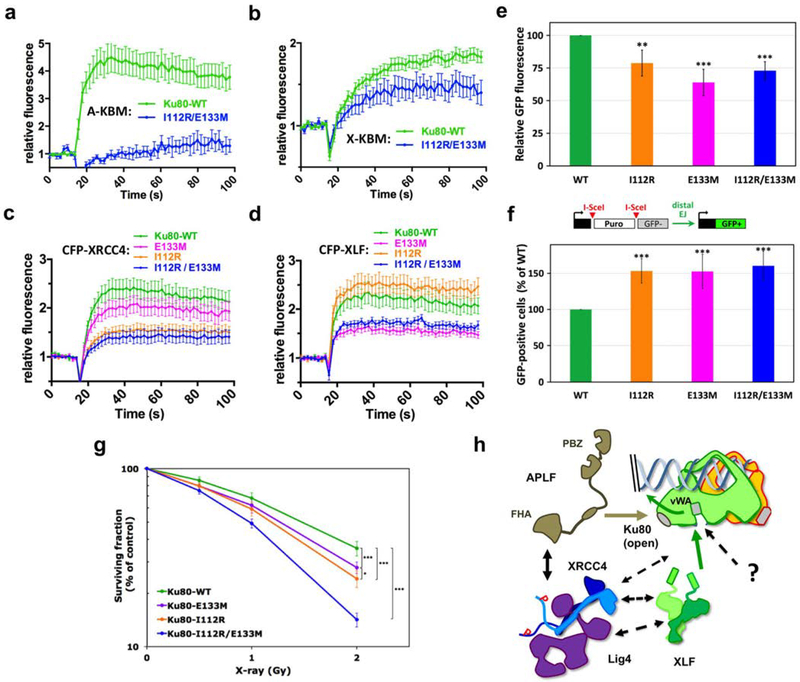
Then, mutations in the Ku80 binding sites for X- and A-KBM were combined in the I112R/E133M Ku80 double mutant (Supplementary Figure 5d). This combination of mutations clearly impaired the recruitment of both CFP-(X- and A-KBM) peptides (Figures 5a and 5b) while the recruitment of the mutant CFP-Ku fusions was preserved (Supplementary Figure 6c–d). We also measured the recruitment of the APLF partner XRCC4 and of XLF, expressed as CFP-tagged full-length proteins (Figure 5c–d). Strikingly, only the double E133M/I112R mutation strongly but not completely impaired the recruitment of both proteins (Figure 5c–d). This result supports the independent binding of APLF-XRCC4 and XLF proteins on Ku80.
Figure 5. Effects of Ku80 mutations in APLF and XLF binding sites.
(a-b) Dynamics of CFP-(A-KBM) (a) and (X-KBM) (b) at laser damaged sites, in U2OS cells expressing wild-type or I112R/E133M mutant Ku80. n=20 and 9 cells in (a) and n=48 and 11 cells in (b) for WT and mutant Ku80. p values at last time point: (a) WT vs I112R/E133M p=0.001; (b) WT vs I112R/E133M p=0.0111. (c-d) Dynamics of CFP-XRCC4 (c) and XLF (d) in cells expressing wild-type, I112R, E133M or I112R/E133M mutant Ku80. n=38, 27, 28, and 24 cells for WT, E133M, I112R and I112R/E133M Ku80 conditions in (c) and n=24, 26, 20 and 23 cells for I112R, WT, I112R/E133M and E133M Ku80 conditions in (d). p values at last time point: (c) WT vs E133M p=0.532; WT vs I112R p=0.0133; WT vs I112R/E133M p=0.0048; (d) WT vs I112R p=0.246; WT vs E133M p=0.0048; WT vs I112R/E133M p=0.0248. (e) End-joining activity in U2OS cells expressing mutated or WT Ku80. Error bars represent s.d., n=4 independent experiments. p values (unpaired two-tailed t- test): WT vs E133M p=0.0004; WT vs I112R p=0.0052; WT vs I112R/E133M p=0.0002. (f) Distal end-joining in U2OS cells containing mutated or WT Ku80. Error bars represent s.d., n=7 independent experiments. p values (unpaired two-tailed t-test): WT vs E133M p=7.49 e-05; WT vs I112R p=2.21 e-06; WT vs I112R/E133M p=4.05 e-06. (g) Survival of U2OS cells expressing WT or mutated Ku80. y axis is log scale. Error bars represent s.d., n=7 to 10 independent experiments. p (unpaired two-tailed t-test): WT vs I112R p=1.47 e-06; WT vs E133M p=6.32 e-05; WT vs I112R/E133M p=2.52 e-13; I112R vs E133M p=0.011. Significant p-values are indicated as follows: *p< 0.05, **p < 0.01, ***p < 0.001. (h) Model for APLF and XLF KBMs function during NHEJ.
Ku80 mutants expressed in U2OS cells had no detectable impact on XLF filaments (Supplementary Figure 6e–f) but lowered end-joining efficiency on a linear plasmid transfected in U2OS cells (Figure 5e). To assess repair accuracy, we used an assay in which GFP expression from a cassette integrated in cells relied on the loss of a DNA fragment between two cut sites (modified from37) (Figure 5f), reported to be favoured upon NHEJ inhibition38,39. Strikingly, we observed an increase in fragment loss with the three Ku80 mutants (Figure 5f), indicating that loose APLF-XRCC4 or XLF interactions with Ku80 promote genomic instability. Finally, the combination of E113M and I112R Ku80 mutations had a strong impact on cell radiosensitivity with an additive effect of both mutations (Figure 5g). These results indicate that APLF-XRCC4 and XLF interactions with Ku80 cooperate to promote cell survival to IR.
DISCUSSION
Here, we show that each of the A- and X-KBM binds Ku independently of other APLF or XLF protein domains. This is in contrast with the absence of binding reported for the isolated X-KBM in recent studies with fluorescent polarization9,40 that may be due to lower sensitivity, steric hindrance by the fluorescence probe or differences in the Ku constructs used. Therefore, the present results rule out the conclusion that Ku-XLF interaction necessarily needs XLF binding to DNA (even though DNA could stabilise the interaction, as deduced from our switchSENSE data) or to DNA-PKcs40.
Despite the A-KBM and X-KBM motifs have important sequence similarities (Supplementary figure 1), they target different regions of Ku80 with different modes of actions, a more rigid one for the A-KBM and an induced fit one for the X-KBM. In contrast to the L297W X-KBM peptide, the (LW) full-length XLF mutant protein cannot be redirected to the APLF-binding site on Ku80. This may be explained by steric constraints either intrinsic to the protein structure or due to interactions with other partners in the NHEJ complex. It was reported that a X-KBM in WRN protein functions cooperatively with an A-KBM located upstream (1403–1412) in binding Ku complexes40 (Figure 1a). Our structural data allow proposing the first model of Ku interacting with the tandem sequence of the WRN A-KBM and X-KBM, including the central linker (Supplementary Figure 4c). This model now awaits validation by the crystal structure of the tandem motif of WRN on Ku.
XRCC4 and XLF organize into filaments both in vitro and in cells10–14 and can also assemble as sliding sleeves-like structures on broken DNA in vitro even without Ku41. In this study, cells containing single or double mutations on the A-KBM and/or X-KBM binding sites of Ku80 show intact filaments while mutations on the X-KBM of XLF induce a slight reduction in the filaments size. These data suggest that interactions additional to Ku80-XLF may participate in filament formation, like the XLF C-terminal DNA-binding domain that spans the X-KBM42.
How is NHEJ ligation complex assembled at broken DNA? APLF FHA domain interacts with XRCC417,26 and XLF establishes head to head contact with XRCC410–13. Here, we show that the two remote APLF- and XLF-KBM binding sites in Ku80 promote the independent recruitment of XRCC4 and XLF at broken DNA and that disruption of XRCC4-XLF interaction (through L115D mutation) does not compromise XLF recruitment. Destabilisation of APLF or XLF interactions with Ku80 impairs repair efficiency and cell survival to DSBs and also favours genome instability associated with distal end-joining. These effects are most likely explained by loose assembly of the NHEJ apparatus at break ends. Therefore, we propose a model in which APLF and XLF KBMs represent two initial anchorage points for the rapid and independent recruitment of APLF-XRCC4 and XLF on Ku (Figure 5h). After initial recruitment, interactions additional to Ku80-XLF contacts may stabilize XLF at DBS sites (Figure 5h): our recruitment data show a stable interaction of XLF at DSB sites in cells that differs from the rapid dissociation from Ku-DNA observed in vitro by switchSENSE; also, although XRCC4 is dispensable for XLF initial recruitment, it has been shown to stabilize XLF at damaged sites34. Thus, additional contacts stabilizing XLF may include XLF-DNA34,42,43 and/or XLF-XRCC4 interactions, including filaments formation10–14 and/or interaction with the LIG4 BRCT1 domain44. In addition, after XRCC4 recruitment through APLF binding to Ku, XRCC4 may also stabilize in turn APLF at damaged sites since FHA mutants of APLF that do not interact with XRCC4 show a reduced retention after laser micro-irradiation18. Moreover, links with DNA-PK may further properly stabilize and/or position the NHEJ ligation complex at DSBs (Figure 5h): Ku directly interacts with the XRCC4-LIG4 complex7 through either XRCC445 or LIG46 and DNA-PKcs also directly contacts XRCC446–48. Intimate links between the DNA-PK and ligation complexes are illustrated by the requirement of an intact XLF-XRCC4-LIG4 complex to ensure optimal DNA ends synapsis49,50.
The intricate network linking end-recognition and ligation NHEJ complexes may allow compensation of partially defective individual components. Indeed, we found that separate or even combined Ku80 mutations do not completely abolish XRCC4 and XLF recruitment and do not radiosensitize cells as much as XLF complete defect that abolishes all XLF functions in NHEJ51. In that view, only a mild, if any, repair defect in human cells has been associated with X-KBM deletion27 or APLF depletion9,24–26: in case of individual absence of XLF C-terminal tail or of APLF, the other intact partner would still bind to Ku80 and be able, although with a slower kinetics, to recruit the other components to achieve ligation.
Finally, our present study adds a new aspect to the DNA-PK-ligation complex interaction network that is the swing of the Ku80 vWA domain upon XLF binding (Figure 5h). From our structural and mutagenesis approaches, we propose that the outward swing of the vWA domain of Ku80 is mainly dependent on the conserved acidic residue E13380. Although this swing does not impact the A-KBM binding site nor the DNA binding domain, it exposes a large groove between the vWA and the ring domain of Ku80 that may non-exclusively reinforce XRCC4-LIG4 and/or XLF interaction with Ku or attract yet unknown components. Interestingly, XLF deficiency impacts on the activity of the Ku partner terminal deoxynucleotidyl transferase during V(D)J recombination52. Mutagenesis studies on the conserved positions that delineated the unmasked surface of this groove will help to define precisely the role of this swing in the NHEJ reaction.
In conclusion, the present data further substantiate the emerging model that several NHEJ factors bearing a limited repertoire of KBMs recognize a limited number of KBM-binding sites on Ku16. The complex regulation of Ku sites occupancy by NHEJ factors during the repair process deserves further investigations.
ONLINE METHODS
DNA preparation
The 500 bp linear dsDNA molecules were amplified from the 3516–4016 region of pBR322 plasmid with biotinylated primers (5’-bGGATCTCAACAGCGGTAA-3’ and 5’ bCTTTATCCGCCTCCATCC-3’). DNA fragments were purified on a MiniQ anion exchange column with a chromatography SMART system (GE Healthcare), ethanol precipitated and resuspended in a 10 mM Tris-HCl, pH 7.5, 1 mM EDTA buffer.
Oligonucleotides for ITC and EMSA
CN1 34bp : CGCGCCCAGCTTTCCCAGCTAATAAACTAAAAAC
CN2 21bp : GTTTTTAGTTTATTGGGCGCG
CN3 18bp up : GTTATCCGAGCGTGAGAC
CN4 18bp down : GTCTCACGCTCGGATAAC
NLB48 : TAG TCG TAA GCT GAT ATG GCT GAT TAG TCG GAA GCA TCG AAC GCT GAT
MM 50bp up : FAM-TAAATGCCAATGCTGCTGATACGTACTCGGACTGATTCGGAACTGTAACG
MM 50bp do : CGTTACAGTTCCGAATCAGTCCGAGTACGTATCAGCAGCATTGGCATTTA
Peptides and Proteins:
The synthetic peptides containing the KBM and XLM motifs were purchased from Genecust at 95% purity, and the concentrations of the stock peptide solutions were determined by amino acid composition. The oligonucleotides used for ITC and crystallization were synthesized by Sigma-Aldrich and Eurogentec.
The full length Ku70(1–609)/Ku80(1–732) heterodimer and a truncated version of the heterodimer deleted Ku70(1–544)/Ku80(1–551) were cloned in the Multibac vectors with a 10-His tag and a TEV site on the Ku80 N-terminus53. Each plasmid was integrated in a Yellow Green Protein (YFP) containing bacmid by transformation in EMBACY E.coli stain (kind gift from Imre Berger, Bristol University). The resulting recombinant bacmids were used to transfect Sf21 insect cells giving the V0 virus generation. After amplification, stocks of viruses were titrated by the dilution limit method using YFP as marker for infected cells and Mac Grady table. Production was initiated in Sf21 cells culture by infection with baculovirus at MOI of 5×10−3. Insect cells were collected 5–6 days after the infection (3–4 days after the proliferation arrest). Cells were sonicated and the supernatant was incubated with Benzonase (300 units for 30 min at 4°C). The Ku heterodimer was purified on a NiNTA-Agarose affinity column (Protino, Macherey Nagel) with a 1M NaCl wash step to remove DNA excess. The eluted Ku was then bound onto an anion exchange column (Resource Q, GE Healthcare) equilibrate with buffer Q (20 mM Tris pH 8.0, 50 mM NaCl, 50 mM KCl, 10 mM β-mercaptoethanol). Final yield of the Ku heterodimer was typically 35 mg of purified heterodimer by liter of culture. The full length 10-His tagged XLF protein was produced in insect cells with a similar protocols with a yield of 50mg of purified protein by L of culture. A truncated version of XLF(1–224) was also produced in E. coli
Crystallization of the Ku70/Ku80/hDNA complexes with A-KBM and X-KBM peptides
The DNA used in this study is the hairpin DNA previously used by Walker et al3. It was obtained using HPLC-purified oligonucleotide of 34 and 21 nucleotides (see oligonucleotides above). The oligonucleotides were annealed and added in 1.1-fold molar excess to Ku heterodimer. The peptides containing the A-KBM motif (18mer, 174–192), the long X-KBM motif (19mer, 281–299) or the short X-KBM motif (13mer 287–299) were added respectively with a 1.1, 2 and 2 fold excess. Crystallization screenings on the KuCC-hDNA-peptides were performed on the HTX platform (EMBL, Grenoble) with an automatic visualization at 4°C. The crystals of Kucc-hDNA-pAPLF were reproduced and optimized in the laboratory at 20 °C using the sitting drop method by mixing 1.5μL of the 20 mg/mL Ku-DNA-peptides complexes with 1.5 μL of the solution containing 13% polyethylene glycol (PEG) 3350, 150 mM NaNO3, and 100 mM Bis-Tris-Propan (pH 6.5). The crystals (100×150×1000μm) grew in 5–6 hours and were frozen in a solution of the mother liquor with 20% glycerol. The Kucc-hDNA-pXLF and Kucc-DNA-pXLFs complexes were crystallized at 20 °C by mixing 1.5μL of the 7 mg/mL complex solution with 1.5 μL of a solution containing 18% polyethylene glycol (PEG) 3350, 150 mM Na2SO4, and 100 mM Bis-Tris-Propane (pH 8,5). The crystals (100×200×50μm) grew in 5–6 days and were frozen with 20% glycerol.
Determination of the crystal structures
Diffraction data were collected at the Proxima 1 and Proxima 2 beamlines at the synchrotron SOLEIL. The datasets were indexed and integrated using the XDS package54, the XDSME package (XDS Made Easier, https://github.com/legrandp/xdsme) and the CCP4 suite55. The crystals present a highly anisotropic diffraction (between 2.85Å and 4.25Å resolution according to the axes). The anisotropy of Kucc-hDNA-pAPLF crystals was treated with the STARANISO program (http://staraniso.globalphasing.org/). The software performs an anisotropic cut-off of merged intensity data, a Bayesian estimation of the structure amplitudes, and applies an anisotropic correction to the data. The structure of the Ku70/80-hDNA-pAPLF was determined by molecular replacement with the program MOLREP using the structure of Ku70/80 (pdb 1JEY) without the DNA coordinates3. Four molecules of Ku were consecutively positioned. Electron density for the hDNA was clearly visible in the position previously reported by Walker. Refinement was performed using BUSTER56 and PHENIX57. The models were built with Coot58. After DNA building, an electron density was visible on the Ku80 vWA near the Ku80 amino acids identified by Grundy et al by mutagenesis. The final statistics are presented below. In the final model, the following regions of Ku70/Ku80 are not visible: Ku70 1–33, 535–544 and Ku80 1–5, 170–181, 190–191, 543–551. The quality of the model was assessed using Molprobity59.
The crystal structures of the Ku70/80-hDNA complexed with the long and short pXLF were solved by molecular replacement. Firstly, the coordinates of Ku70/80 and the hairpin DNA present in pdb 1JEY were used as model. We positioned two molecules in the asymmetric unit with clear electron density except on the vWA region of Ku80 (region aa 6 to 242). We then performed the molecular replacement with the same coordinates deleted of the Ku80 vWA region. The electron density for the secondary structure elements of the Ku80 vWA was clearly visible though at a different position than in 1JEY. A second molecular replacement step was performed to position the vWA domain of the two Ku80 molecules in the asymmetric unit. The structure of the Ku70/80-DNA-short pXLF was solved in a similar manner. An electron density was visible in an internal position of Ku80 vWA that could be attributed to pXLF. The final statistics are presented in Table1. In the final model with the long pXLF, the following regions of Ku70/Ku80 are not visible (Ku70 1–34, 535–554 and Ku80 543–544). In the final model with the short pXLF, the following regions of Ku70/Ku80 are not visible (Ku70 1–34; 535–554 and Ku80 171–194, 300–301, 542–544).
An additional electron density was observed in the long and short pXLF complex structures located close to the extreme N-terminus of Ku80 and to the hairpin DNA. This electron density was successfully modeled with the first missing residues of Ku80, the TEV site sequence preceding (ENLYFQG) and seven histidines from the 10-His tag. To evaluate the influence of the tag present on the N-terminus of Ku80 on the Ku-XLF interaction, we prepared a digested form of Ku with the TEV protease (KuTEV) and measured by ITC its interaction with the X-KBM. The ITC shows that thermodynamic parameters of the KuTEV are similar to the Ku and that the tag does not significatively influence the binding of the XLF motif.
Small-angle Xray scattering:
Several data of Kucc-hDNA complex with a protein-DNA ratio (1:1.2) were collected alone or in presence of pXLF or pAPLF peptides. Data were collected on the SWING beamline (SOLEIL synchrotron) at a 1.8m sample-detector distance. The complexes were prepared at 1.0, 3.0 and 5mg/mL and spun for 10 minutes at 13000 rpm prior to SAXS analysis to eliminate aggregates. Volumes of 40μL of each sample and buffer (20 mM Tris pH 8.0, 150mM NaCl, 5mM β-mercaptoethanol) were injected into the SAXS capillary cell and collected continuously, with a frame duration of 0.5 s and a dead time between frames of 0.5 s. Data reduction to absolute units, frame averaging and subtraction were done using FOXTROT60, a dedicated home-made application. All subsequent data processing and analysis steps were carried out with PRIMUS and other programs of the ATSAS suite61. The program GNOM62 was used to compute the pair-distance distribution functions, P(r).
Isothermal titration Calorimetry (ITC)
Interactions between Ku70/80 wild-type and Ku70/80cc and the different peptides containing the KBM and XLM motifs were determined by isothermal titration calorimetry (ITC) using a VP-ITC calorimeter (Malvern). Prior to measurements, all solutions were degassed under vacuum. The reaction cell of the ITC (volume 1.8 mL) was loaded with Ku heterodimers alone or complexed with DNA or peptides for competition experiments. Proteins were extensively dialyzed against buffer I (20 mM Tris, pH 8.0, 150 mM NaCl, and 5 mM β-mercaptoethanol). Peptides and DNA were prepared at high concentrations. The syringe (290 μL) was filled with the different peptides at concentration between 20 μM to 200 μM. The Ku heterodimer present in the cell was titrated by automatic injections of 6–10μL of the different peptides. Enthalpy ΔH (in kcal.mol−1), stoichiometry of the reaction N, and association constant Ka (in M−1) were obtained by nonlinear least-squares fitting of the experimental data using the single set of independent binding sites model of the Origin software provided with the instrument. The free energy of binding (ΔG) and the entropy (ΔS) were determined using the classical thermodynamic formula, ΔG = - RT ln(Ka) and ΔG = ΔH - TΔS. All binding experiments were performed in duplicate or triplicate at 25°C. Control experiments were performed with peptides injected into the buffer to evaluate the heat of the dilution.
EMSA:
Binding reactions (10 μL) were performed by incubating the annealed oligonucleotides (oligonucleotides used in this study are listed in Table Sxx and indicated in the figure legends) at a final concentration of 25 nM, with the indicated final concentrations of proteins in 75 mM KCl, 10 mM Tris (pH 7.5), 0.5 mM EDTA, 0.5 mM DTT, 0.5 mg/mL acetylated-BSA, and 5% glycerol. Reactions were incubated at room temperature for 1 hr and fractionated by 6% PAGE (29%/1% [w/v] Acrylamide:Bis-acrylamide) in 0.53 standard Tris-borate-EDTA (TBE) buffer at 80 V for 45 min to 1 hr. After electrophoresis, DNA was visualized using a ChemiDoc MP imaging system (Bio-Rad), either by direct detection of the fluorescently labeled DNA (FAM) or after staining with 0.2 mg/mL EtBr. Data were processed and quantified with the Image Lab software version 5.2.1 (Bio-Rad).
switchSENSE measurements:
All switchSENSE measurements were carried out on a DRX 2400 instrument, using a multi-purpose 48bp chip (both Dynamic Biosensors GmbH; Planegg, Germany). The sample and running buffer was Tris 10mM pH 7.4, 140mM NaCl, 0.05% Tween20, 50μM EDTA, 50μM EGTA. In all kinetics experiments, complementary DNA to the strand immobilized on the chip was first hybridized on the measurement electrodes. On top of the 48bp complementary to the chip, this DNA carried a 32bp-overhang hybridized with its 32bp complementary strand. Second, the Ku70/80 protein was associated at 100nM for 3min (reaching saturation) to the immobilized dsDNA. Finally, the association and dissociation of the XLF protein was measured using triplicates of each concentration, at a flow rate of 2mL/min. For each mutant, the surface was not regenerated in between concentrations – Ku does not dissociate in the timescale of the XLF measurement, and XLF fully dissociates for each concentration. The electrodes were only regenerated between the measurements of different mutants of XLF. Kinetics values were determined using Origin software.
Cell lines and cell culture
U2OS human osteosarcoma cells and immortalized BuS cells (derived from an XLF-deficient patient, gift from Jean-Pierre de Villartay, Institut Imagine, Paris, France) were grown in DMEM and RPMI, respectively. Media were supplemented with 10% fetal calf serum (Eurobio), 125 U/ml penicillin and 125 μg/ml streptomycin. Cells were maintained at 37°C in a 5% CO2 humidified incubator. Cell lines were tested negative for mycoplasma by PCR. All culture media and antibiotics were from Invitrogen. When necessary (conditional expression of shRNA against Ku80), doxycyclin (Sigma-Aldrich) was added to the medium at a 4 μg/ml final concentration.
Expression vectors
Cell transfection and transduction
Production of lentiviral particles in HEK-293T cells and transduction of U2OS and BuS cells were performed as previously described31. Transduced cells were used as populations without clonal selection, except when indicated otherwise.
Plasmid recircularization assay
U2OS cell populations expressing wild-type or mutated Ku80 protein were seeded to near confluence in 6-well plates and incubated overnight at 37°C. Cells were then transfected with 1 μg BamHI-linearized pEGFP-N1 plasmid (Clontech) and 1 μg pmCherry-C1–3NLS circular plasmid as a transfection control (gift from Dyche Mullins (Addgene #58476),63). Cells were split 24 h later, incubated at 37°C for two additional days and analyzed by flow cytometry on a Fortessa X-20 (BD Biosciences). For each cell population, the integrated GFP fluorescence signal was normalized to mCherry signal. End-joining activity was set to 100% for Ku80-WT expressing cells. Results were plotted as the mean values of four independent experiments ±s.d.
Distal End-Joining assay
U2OS-EJ5 cells (a kind gift from Jeremy Stark, City of Hope, Duarte, USA) containing one integrated copy of a GFP reporter cassette which allows to measure rejoining of two tandem I-SceI cut sites separated by a ~1.8-kb insert, were modified as follows. The cells were first transduced with lentiviruses produced from pLV-tTR-KRAB and pLV3-Tet-RFP ISceI-GRLBD to allow inducible expression and nuclear translocation of the I-SceI yeast meganuclease. The resulting cells were then transduced with lentiviruses prepared from pLVTHM2-shKu80. A positive clone enabling doxycyclin-dependent conditional knockdown of Ku80 expression and subsequent cell death was isolated. These cells were further transduced with lentiviruses produced from pLV3-HA-Ku80-shR-(WT or mutants) plasmids to replace, in the presence of doxycyclin, endogenous Ku80 expression by expression of the various Ku80 constructs described in the study. To perform Distal End-Joining assay, the different U2OS-EJ5 modified cell populations were seeded onto 6-well plates and incubated at 37°C for 24 h. Dexamethasone (Sigma-Aldrich) was then added to a final concentration of 250 nM. Cells were washed 24 h later, further incubated at 37°C for two to three days and analyzed by flow cytometry on a Fortessa X-20 analyzer (BD Biosciences). The fraction of GFP-positive cells was measured and normalized to 100% for Ku80-WT expressing cells. Results were plotted as the mean values of seven independent experiments ±s.d.
Ionizing irradiation and cell survival analysis
Three to six thousand cells per well were seeded in 6-well plates. Plated cells were exposed 24 h later to various doses of X-ray using a Faxitron RX-650 device (130 kV, 5 mA, dose rate 0.5 Gy.min−1). Six to seven days later, cells were washed with PBS, stained 10 min with crystal violet (0.1% aqueous solution). Stained cells were extensively washed with water and plates were air dried. Staining was dissolved with 10% acetic acid solution and absorption was measured at 570 nm (Ultrospec-3000 spectrophotometer, Pharmacia Biotech). Results were plotted as mean values of 5–10 independent experiments ±s.d. using Microsoft Excel software.
Live-cell microscopy and micro-irradiation
U2OS or BuS cells were seeded in 35-mm glass-bottom culture dishes (MatTek) two days prior laser irradiation. Experiments were carried out with a Zeiss LSM-710 confocal laser scanning microscope equipped with a coherent chameleon Vision-II tunable laser (690– 1080 nm), a 40X/1.3 oil immersion objective and a heated environmental chamber set at 37°C in 5% CO2 atmosphere. ECFP was excited using biphotonic laser at 800 nm (1.5% of maximum power). Confocal image series were recorded with a frame size of 512×512 pixels. Nuclei micro-irradiation was carried out at 800 nm at 20% of maximum power (mean max power was 3070 mW) in rectangle of 15 μm2 area during 50 μs pixel dwell time. Before and after micro-irradiation, confocal image series of one mid z-section were recorded at 1.94 s time interval (typically 9 pre-irradiation and 40–45 post-irradiation frames). For evaluation of the recruitment kinetics, fluorescence intensities of the irradiated region were corrected for total nuclear loss of fluorescence over the time course and normalized to the pre-irradiation value. Data from micro-irradiation of individual cells obtained in several independent experiments performed on different days were averaged, analyzed and displayed using PRISM software. p values at last time point were calculated using unpaired Student’s t-test.
Protein extraction and immunoblotting
Sub-confluent cells from 60-mm culture dishes were harvested and washed with PBS. Pellets were resuspended in 100 μl of lysis buffer (50 mM HEPES.KOH pH 7.5, 450 mM NaCl, 1% Tritin-X100, 1 mM EDTA, 1 mM DTT, protease-phosphatase Halt Inhibitor cocktail (Pierce, Thermo Scientific)) and processed by four freeze/thaw cycles. After centrifugation at 14000 g for 10 min, protein concentration was measured in the supernatant with the Bradford assay (Bio-Rad). Proteins from 50 μg of cell extracts were separated in 4–15% Mini-Protean TGX precast polyacrylamide gels (Bio-Rad) and transferred to PVDF membrane (Millipore). Immunoblotting analysis was performed with the following antibodies: anti-Ku70 monoclonal antibody (clone N3H10 from NeoMarkers), anti-Ku80 monoclonal antibody (clone 111 from Thermo Fisher Scientific), anti-DNA-PKcs monoclonal antibody (clone 18.2 from Abcam), anti-XLF polyclonal antibody (Bethyl Laboratories), anti-APLF polyclonal antibody (SK3595, kind gift from K.W. Caldecott, University of Sussex, Brighton, UK, see25), anti-β-Actin monoclonal antibody (clone AC-15 from Ambion).
Cell culture and preparation for super-resolution imaging
BuS cells were grown in RPMI medium with 10% FBS and 100 U/mL Penicillin-Streptomysin. For super-resolution imaging, cells were cultured on glass coverslips for 24 hours, followed by serum starvation for 48 hours. The cells were then released into full medium for 4 hours so that most of the cells were in G1 phase.
The synchronized cells were then washed twice with PBS, and permeabilized with 0.5% Triton X-100 in CSK buffer (10 mM Hepes, 200 mM Sucrose, 100 mM NaCl, and 3 mM MgCl2, pH=7.4) for 10 minutes64. Cells were then fixed with paraformaldehyde (4%) for 20 mins, and blocked in block solution (2% glycine, 2% BSA, 0.2% gelatin, and 50 mM NH4Cl in PBS) overnight at 4°C.
Fixed cells were then immunostained with validated monoclonal antibodies: XLF was stained with XLF-antibody (3D6, NBP2–03275, NOVUS) for 1 hour at room temperature, followed by Alexa Fluor 647 conjugated goat-anti-mouse 2nd antibody staining (ab 150115, abcam) for 30 minutes at room temperature. Cells were then stained with Alexa Fluor conjugated anti-Ku80 antibody (EPR3467, ab202659, abcam) for 1 hour at room temperature. Cells were then mounted onto microscope glass slide, and imaged with freshly mixed imaging buffer (1 mg/mL glucose oxidase, 0.02 mg/mL catalase, 10% glucose, and 100 mM cycteanube (MEA)).
Microscope and Single-Molecule Localization imaging
Super-resolution imaging was performed on a custom-built optical imaging platform based on a Leica DMI 300 inverse microscope, equipped with a 488 nm (OBIS, Coherent) and a 639 nm laser line (MRL-FN-639–800, CNI). Laser lines were reflected into an HCX PL APO 63X NA=1.47 OIL CORR TIRF Objective (Zeiss) by a penta-edged dichroic beam splitter (FF408/504/581/667/762-Di01–22×29), and the emitted fluorescence was further extended by a 2X lens tube (Diagnostic Instruments), filtered by single-band filters (Semrock FF01–531/40 and FF01–676/37 for Alexa Fluor 488 and Alexa Fluor 647, respectively), and collected onto a sCMOS camera (Prim95B, Photometrics). A 405 nm Laser line (MDL-III-405–150, CNI) was also equipped to reactivate Alexa Fluor 647 fluorophores.
For super-resolution imaging, the 488 and 639 laser lines were adjusted to ~ 1.0 and 1.5 kW/cm2, and a Highly Inclined and Laminated Optical sheet (HILO) illumination mode for sample excitation. Alexa Fluor 488 and Alexa Fluor 647 were sequentially exited and their emitted fluorescence was also sequentially collected by switching the single-band filters in a filter wheel. The emitted photons were collected onto a sCMOS camera (Photometrics Prime 95B), and a minimum of 2000 frames at 33 Hz were recorded for each image stack.
Reconstruction from Single-Molecule Localization image to super-resolution image
Each image of the image stack was first of all box-filtered with the box size of 4 times of the FWHM of a 2D gaussian point spread function (PSF). Considering the patterned noise for sCMOS camera construction, each pixel was weighted by the inverse of its noise variance during filtering. The low-pass filtered image was then extracted as the background from the raw image. The local maximums of the image were then recognized and segmented into single PSFs for 2D gaussian fit.
2D gaussian fit of each individual PSF was performed by GPU using the Maximum Likelihood Estimation (MLE) algorithm. Briefly, the likelihood function of each pixel was constructed by convolving the Poisson distribution of the shot noise governed by the photons emitted from fluorophores nearby, and the gaussian distribution of the readout noise of the camera itself65. The offsets, variance, and analog-to-digital conversion factor of each pixel of the camera was calibrated beforehand. The position, amplitude, sigma, and background of each individual 2D gaussian PSF were addressed by maximizing the likelihood function of each pixel given its readout digital value. The fitting accuracy was estimated by Cramér-Rao lower bound (CRLB) and the accuracy of Alexa Fluor 488 and Alexa Fluor 647 in this work are ~ 17 and 13 nm, respectively.
Alignment of images of different colors
Mapping of the two colors was performed using a polynomial mapping algorithm. Briefly, a spatially separated and broad-spectrum fluorescent bead (TetraSpec, ThermoFisher) slide was imaged on both color channels. Mass centers of each same bead but in different channels were recorded pairwise. The coefficient of each term in a 2nd order polynomial function was optimized by training the polynomial regression using all the recorded bead’s centers. The optimized polynomial function was then used for two color channels alignment. We note that choosing the proper order of polynomial function for optimization depends on the number of training beads, and higher order but not enough trainees would lead to overfit.
Pair-Correlation analysis
Coordinates localized within ~2.5 times of the averaged localization uncertainty, and from consecutive frames were considered as artificial blinking and grouped as one coordinate. This coordinate was calculated by taking the 1/var weighted average of all the coordinates within the group, where var is the localization uncertainty of each coordinate. The grouped coordinates were then rendered onto a pixelized image of 5 nm/pixel, and the pair-correlation was performed on this image by series 2D Fourier and inverse Fourier transfers66. The correlation was then fitted into two correlation terms: the correlation among coordinates within localization uncertainty, and that among the coordinates that form a cluster/molecular assemble/filament. Considering the size of XLF ‘clusters’ is not randomly distributed, we fitted the second term into a normal distribution and interpret the fitted sigma as the apparent radius of the averaged XLF ‘cluster’ radius (Figure XC) across the image.
Data availability
Crystal structures are deposited at the pdb with the following codes Ku-DNA-pAPLF (6ERF), Ku-DNA-pXLF (6ERH) and Ku-DNA-pXLFshort (6ERG) Source data for figures 3, 4 and 5 are available with the paper online Other data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
J-B.C is supported by ARC program (SLS220120605310), ANR (ANR-12-SVSE8–012), INCA DomRep (PLBIO 2012–280), CEFIPRA grant 5203C and by the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INBS-05. Work in E.R’s laboratory is supported by National Institutes of Health Grants CA187612, GM108119, and the American Cancer Society RSG DMC-16–241-01-DMC. P.C’s team is supported by the Ligue Nationale Contre Le Cancer (Equipe labellisée 2013 and 2018) and Electricité de France (EDF, Conseil de Radioprotection). J-B.C, M.M and P.C are supported by ANR (CE12 2017 NHEJLIG4 grant). We thank K.W. Caldecott (University of Sussex, Brighton, UK) for the gift of anti-APLF antibody and J.M. Stark (City of Hope, Duarte, USA) for the gift of U2OS cells engineered with the integrated distal-end-joining reporter. We would like to thank the Imaging Core Facility TRI-IPBS, in particular S. Mazeres and R. Poincloux for maintenance of the live-cell microscopy equipment and E. Näser for the maintenance of the flow-cytometers. Flow-cytometry equipment was with the financial support from ITMO Cancer Aviesan (Alliance Nationale Pour les Sciences de la Vie et de la Santé, National Alliance for Life Science and Health) within the framework of Cancer Plan. We thank J-P. de Villartay and F. Theillet for careful reading of the manuscript.
Footnotes
ACCESSION CODES. Crystal structures are deposited at the pdb with the following codes Ku-DNA-pAPLF (6ERF), Ku-DNA-pXLF (6ERH) and Ku-DNA-pXLFshort (6ERG)
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Lieber MR The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End-Joining Pathway. Annu Rev Biochem 79, 181–211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang HHY, Pannunzio NR, Adachi N & Lieber MR Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 18, 495–506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker JR, Corpina RA & Goldberg J Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412, 607–14 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Grundy GJ, Moulding HA, Caldecott KW & Rulten SL One ring to bring them all-The role of Ku in mammalian non-homologous end joining. DNA Repair (Amst) 17, 30–8 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Rulten SL & Grundy GJ Non-homologous end joining: Common interaction sites and exchange of multiple factors in the DNA repair process. Bioessays 39(2017). [DOI] [PubMed] [Google Scholar]
- 6.Costantini S, Woodbine L, Andreoli L, Jeggo PA & Vindigni A Interaction of the Ku heterodimer with the DNA ligase IV/Xrcc4 complex and its regulation by DNA-PK. DNA Repair (Amst) 6, 712–22 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Nick McElhinny SA, Snowden CM, McCarville J & Ramsden DA Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol 20, 2996–3003 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yano KI, Morotomi-Yano K, Lee KJ & Chen DJ Functional significance of the interaction with Ku in DNA double-strand break recognition of XLF. FEBS Lett 585, 841–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundy GJ et al. APLF promotes the assembly and activity of non-homologous end joining protein complexes. Embo J 32, 112–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ropars V et al. Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining. Proc Natl Acad Sci U S A 108, 12663–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammel M et al. XRCC4 interactions with XRCC4-like factor (XLF) create an extended grooved scaffold for DNA ligation and double-strand break repair. J Biol Chem 286, 32638–50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andres SN et al. A human XRCC4-XLF complex bridges DNA. Nucleic Acids Res 40, 1868–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Q et al. Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions. Biochem Soc Trans 39, 1387–92 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Reid DA et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc Natl Acad Sci U S A 112, E2575–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahel I et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451, 81–5 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Rulten SL, Cortes-Ledesma F, Guo L, Iles NJ & Caldecott KW APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol Cell Biol 28, 4620–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanno S et al. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. Embo J 26, 2094–103 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirodkar P, Fenton AL, Meng L & Koch CA Identification and functional characterization of a Ku-binding motif in Aprataxin Polynucleotide kinase/phosphatase-Like Factor (APLF). J Biol Chem 288, 19604–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammel M et al. An Intrinsically Disordered APLF Links Ku, DNA-PKcs, and XRCC4-DNA Ligase IV in an Extended Flexible Non-homologous End Joining Complex. J Biol Chem 291, 26987–27006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnoult N et al. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549, 548–552 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossi ML, Ghosh AK & Bohr VA Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst) 9, 331–44 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tadi SK et al. PAXX Is an Accessory c-NHEJ Factor that Associates with Ku70 and Has Overlapping Functions with XLF. Cell Rep 17, 541–555 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Ochi T et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 347, 185–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bekker-Jensen S et al. Human Xip1 (C2ORF13) is a novel regulator of cellular responses to DNA strand breaks. J Biol Chem 282, 19638–43 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Iles N, Rulten S, El-Khamisy SF & Caldecott KW APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol Cell Biol 27, 3793–803 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macrae CJ, McCulloch RD, Ylanko J, Durocher D & Koch CA APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair (Amst) 7, 292–302 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Malivert L et al. The C-terminal domain of Cernunnos/XLF is dispensable for DNA repair in vivo. Mol Cell Biol 29, 1116–22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brosey CA, Ahmed Z, Lees-Miller SP & Tainer JA What Combined Measurements From Structures and Imaging Tell Us About DNA Damage Responses. Methods Enzymol 592, 417–455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blier PR, Griffith AJ, Craft J & Hardin JA Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem 268, 7594–601 (1993). [PubMed] [Google Scholar]
- 30.Dolinsky TJ et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res 35, W522–5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Q et al. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Res 39, 9605–9619 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langer A et al. Protein analysis by time-resolved measurements with an electro-switchable DNA chip. Nat Commun 4, 2099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buck D et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124, 287–99 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Yano KI & Chen DJ Live cell imaging of XLF and XRCC4 reveals a novel view of protein assembly in the non-homologous end-joining pathway. Cell Cycle 7, 1321–5 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Hammel M, Yu Y, Fang S, Lees-Miller SP & Tainer JA XLF Regulates Filament Architecture of the XRCC4.Ligase IV Complex. Structure 18, 1431–42 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malivert L et al. Delineation of the XRCC4 interacting region in the globular head domain of cernunnos/XLF. J Biol Chem 285, 26475–83 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennardo N, Cheng A, Huang N & Stark JM Alternative-NHEJ Is a Mechanistically Distinct Pathway of Mammalian Chromosome Break Repair. PLoS Genet 4, e1000110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennardo N, Gunn A, Cheng A, Hasty P & Stark JM Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet 5, e1000683 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gunn A, Bennardo N, Cheng A & Stark JM Correct end use during end joining of multiple chromosomal double-strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. J Biol Chem 286, 42470–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grundy GJ et al. The Ku-binding motif is a conserved module for recruitment and stimulation of non-homologous end-joining proteins. Nat Commun 7, 11242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brouwer I et al. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature 535, 566–9 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Andres SN, Modesti M, Tsai CJ, Chu G & Junop MS Crystal Structure of Human XLF: A Twist in Nonhomologous DNA End-Joining. Mol Cell 28, 1093–101 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Lu H, Pannicke U, Schwarz K & Lieber MR Length-dependent binding of human XLF to DNA and stimulation of XRCC4: DNA ligase IV activity. J Biol Chem 282, 11155–62 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Wu PY et al. Interplay between cernunnos-XLF and NHEJ proteins at DNA ends in the cell. J Biol Chem 282, 31937–43 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Mari PO et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A 103, 18597–602 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu HL, Yannone SM & Chen DJ Defining interactions between DNA-PK and ligase IV/XRCC4. DNA Repair (Amst) 1, 225–35 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Leber R, Wise TW, Mizuta R & Meek K The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase. J Biol Chem 273, 1794–801 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Wang YG, Nnakwe C, Lane WS, Modesti M & Frank KM Phosphorylation and regulation of DNA ligase IV stability by DNA-dependent protein kinase. J Biol Chem 279, 37282–90 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Cottarel J et al. A noncatalytic function of the ligation complex during nonhomologous end joining. J Cell Biol 200, 173–86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graham TG, Walter JC & Loparo JJ Two-Stage Synapsis of DNA Ends during Non-homologous End Joining. Mol Cell 61, 850–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menon V & Povirk LF XLF/Cernunnos: An important but puzzling participant in the nonhomologous end joining DNA repair pathway. DNA Repair (Amst) 58, 29–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jspeert H et al. XLF deficiency results in reduced N-nucleotide addition during V(D)J recombination. Blood 128, 650–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY-REFERENCES
- 53.Bieniossek C, Imasaki T, Takagi Y & Berger I MultiBac: expanding the research toolbox for multiprotein complexes. Trends Biochem Sci 37, 49–57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kabsch W XDS. Acta Crystallogr D Biol Crystallogr 66, 125–32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Winn MD et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smart OS et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr 68, 368–80 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez J & Nishino Y Advances in X-ray scattering: from solution SAXS to achievements with coherent beams. Curr Opin Struct Biol 22, 670–8 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Petoukhov MV et al. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr 45, 342–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Svergun DI Small-angle X-ray and neutron scattering as a tool for structural systems biology. Biol Chem 391, 737–43 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Belin BJ, Lee T & Mullins RD DNA damage induces nuclear actin filament assembly by Formin −2 and Spire-(1/2) that promotes efficient DNA repair. Elife 4, e07735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Britton S, Coates J & Jackson SP A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol 202, 579–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang F et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat Methods 10, 653–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Veatch SL et al. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS ONE 7, e31457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structures are deposited at the pdb with the following codes Ku-DNA-pAPLF (6ERF), Ku-DNA-pXLF (6ERH) and Ku-DNA-pXLFshort (6ERG) Source data for figures 3, 4 and 5 are available with the paper online Other data that support the findings of this study are available from the corresponding author upon reasonable request.