Abstract
In this study, differences in the ruminal bacterial community between high-yield and low-yield lactating dairy cows under the same dietary conditions were investigated. Sixteen lactating dairy cows with similar parity and days in milk were divided into high-yield (HY) and low-yield (LY) groups based on their milk yield. On day 21, rumen content samples were collected, and their microbiota compositions were determined using high-throughput sequencing of the 16S rRNA gene by the Illumina MiSeq platform. During the study period, dry matter intake (DMI) and milk yield were measured daily, and milk composition was assessed 3 times per week. The results showed that the milk of the LY group tended to have higher fat (P = 0.08), protein (P = 0.01) and total solid contents (P = 0.04) than that of the HY group, while the HY group had higher ruminal propionate (P = 0.08) proportion and volatile fatty acid (VFA) (P = 0.02) concentrations. Principal coordinate analysis indicated significant differences in ruminal bacterial community compositions and structures between the HY group and LY group. The abundances of Ruminococcus 2, Lachnospiraceae and Eubacterium coprostanoligenes were significantly higher in the HY group than in the LY group. In addition, Bacteroides, Ruminococcus 2 and Candidatus-Saccharimonas were positively correlated with ruminal propionate proportion (r>0.4, P<0.05). These findings enhance the understanding of bacterial synthesis within the rumen and reveal an important mechanism underlying differences in milk production in dairy cows.
Introduction
A symbiotic relationship exists with regard to the rumen microbiota of cattle. The rumen is a highly specialized organ of ruminant animals that promotes a community of mutualistic microbial species while simultaneously absorbing nutrients derived from digestion of plant fibre and cellular material [1]. The rumen microbial community has a direct relationship with volatile fatty acid (VFA) and microbial protein biosynthesis, which play important roles in milk production efficiency [2]. In addition, the bacterial community also determines the production traits [3], production variables [4–7], and milk production and composition [8, 9] in dairy cows. Rumen microbial dynamics have been reported to involve both core and variable microbial components [10–12]. Similar to the microbial community in the gut of non-ruminants, the structure and function of the microbial community in the cow rumen are shaped by dynamic physical, chemical, and predatory environments [13, 14]. In turn, the microbial community regulates nutrient cycling to the host [15]. However, a more in-depth comparison is warranted to improve our understanding of differences in rumen bacterial community composition between high-yield and low-yield dairy cows.
Recent efforts to study the rumen microbiome have focused on identifying and quantifying ruminal microbial communities [8, 16]. As a powerful molecular approach for taxonomic analyses, the application of 16S rRNA gene sequencing technology has provided novel insight into the microbiome ecology of gastrointestinal tracts [17, 18]. Indeed, this technique has been widely used to study microbial diversity and the metabolic capabilities of microbiomes in different ecological niches [19], fermented food [20, 21], waste-water treatment facilities [22], and human and animal gastrointestinal tracts [23–25]. Recently, Paz et al. [26] reported on the compositions of various bacterial communities in different dairy breeds. Furthermore, some distinct rumen bacterial communities were significantly associated with the rumen fermentation parameters, which affect milk production [27]. Therefore, the objective of the present study was to examine differences in ruminal bacterial community compositions between high-yield and low-yield lactating cows under the same dietary conditions.
Materials and methods
Animals and experimental design
The experimental protocol was approved by the Institutional Animal Care and Use Committee at the Beijing University of Agriculture, in compliance with regulations for the administration of affairs concerning experimental animals (The State Science and Technology Commission of P. R. China, 1988). According to the principle of parity and lactation days, 16 Holstein lactating dairy cows of similar parity were used and assigned to a high-yield group (average production 31.90±1.76 kg/d, mean±SD) or a low-yield group (average production 19.30±1.76 kg/d), with 8 each. All the cows used in this experiment averaged 2.6±0.4 parity. At the beginning of the experiment, the average days in milk (DIM) was 114.6±7.5 days, the average body weight was 670±24 kg and the average dry mater intake was 24.2±2.7 kg/d. There was no initial difference between groups in terms of these parameters except the milk production. The cows had free access to water and were housed in a tie-stall barn. Cows were milked 3 times per day (0700,1400 and 2100 h). Feed intake and milk production data were recorded daily throughout the experiment. The experimental duration was 21 d, with an adaptation period of 14 d as control period and a sampling period of 7 d (D15-D21). These lactating dairy cows were the fed under the same dietary conditions, the composition of which is shown in Table 1.
Table 1. Ingredients and nutrient composition (% of DM) of the basal diet.
| Item | Content |
|---|---|
| Ingredient, % of DM | |
| Alfalfa hay | 6.90 |
| Corn silage | 46.32 |
| Oat grass | 2.40 |
| Ground corn | 9.88 |
| Soybean meal | 5.10 |
| Steam-flaked corn | 4.40 |
| DDGS1 | 4.40 |
| Corn bran | 3.70 |
| Extruded soybean | 3.00 |
| Barley | 2.66 |
| Wheat barn | 2.66 |
| Sodium cyclamate | 2.40 |
| Oat | 1.50 |
| Canola meal | 1.07 |
| Cottonseed meal | 1.07 |
| Magalac2 | 0.90 |
| NaHCO3 | 0.59 |
| Limestone | 0.48 |
| NaCl | 0.27 |
| Premix3 | 0.30 |
| Nutrient composition4 | |
| CP | 17.4 |
| NDF | 31.1 |
| ADF | 16.6 |
| Ether extract | 5.00 |
| Ca | 0.78 |
| P | 0.44 |
| NEL, Mcal/kg | 1.76 |
1DDGS = dried distillers’ grain with solubles.
2Church and Dwight Co. Inc., Princeton, NJ.
3Formulated to provide (per kg of DM) 4,560 mg of Cu, 3,000 mg of Fe, 12,100 mg of Zn, 4,590 mg of Mn, 60 mg of Co, 200 mg of Se, 270 mg of I, 10,000 IU of vitamin E, 450,000 IU of vitamin D, 2,000,000 IU of vitamin A, and 3,000 mg of nicotinic acid.
4Chemical composition is based on chemical analysis of the total mixed ration (TMR), as described.
Rumen fluid sampling and parameter measurement
Rumen fluid samples were collected from the oral cavity at 3–4 h after the morning feeding on day 21 (D21). The rumen contents were strained through 4 layers of cheesecloth with a mesh size of 250 μm. Ruminal pH was immediately measured using a portable pH metre (Testo 205, Testo AG, Germany). The filtered rumen fluid samples were centrifuged at 10,000 × g for 15 min at 4°C, aliquoted into 5-mL cryopreservation tubes, frozen in a liquid nitrogen tank and stored at -80°C until analysis of the ruminal bacterial community. A VFA analysis was conducted on 1 mL of each rumen fluid sample, which was preserved by adding 0.2 mL of 25% HPO3, by gas chromatography as reported by Mao et al. [28]. An ammonia-N analysis was performed using a colorimetric method consistent with AL and EP [29].
DNA extraction and polymerase chain reaction (PCR) amplification
Microbial DNA was extracted from rumen fluid samples using EZNA Bacterial DNA Kit (Omega Bio-Tek, Norcross, U.S.) according to the manufacturer’s protocols.
The yield and purity of the extracted DNA were assessed with a NanoDrop 1000 instrument (NanoDrop, Wilmington, DE).
16S rRNA analysis
The V3-V4 regions of the 16S ribosomal RNA gene were amplified by PCR. The reaction mixture contained 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM deoxyribonucleotide triphosphates (dNTPs), 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, 0.2 μL of bovine serum albumin (BSA) and 10 ng of the template DNA. The PCR protocol was set as follows: 95°C for 3 min, followed by 27 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 45 s and a final extension at 72°C for 10 min. The sequences of the primers used for PCR were as follows: 338F 5’-barcode-ACTCCTACGGGAGGCAGCAG)-3’ and 806R 5’- GGACTACHVGGGTWTCTAAT-3’. The reactions were performed in triplicate on 20-μL mixtures. Amplicons were excised from 2% agarose gels and purified using AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, U.S.) according to the manufacturer’s instructions and then quantified using a NanoDrop 2000 (Thermo, U.S.). Purified amplicons were pooled in equimolar ratios and pair-end sequenced (2 × 300) on the Illumina MiSeq platform according to standard protocols. The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP136923).
Statistical analysis
Data of dry matter intake, milk yield, milk composition, ruminal pH, VFA concentrations, and alpha diversity index were analyzed using PROC MIXED of SAS 9.4 (SAS Institute, Inc, Cary, NC) as shown in the following model: Yij = μ + Ti + ej, where Yij is the dependent variable, μ is the overall mean, Ti is the effect of treatment (LY or HY, considered fixed), and ej is the residual. A P<0.05 was considered statistically significant, and a trend was indicated by P<0.10. A principal coordinate analysis (PCoA) and Pearson correlations were carried out using R-3.2 with vegan package on the online Majorbio I-Sanger Cloud Platform (http://www.i-sanger.com). Pearson correlations were used to analyse the environmental factors and bacterial relationships using the pheatmap package. The resulting numerical matrix is visually displayed in a heat map diagram. The colour change reflects the data information in a two-dimensional matrix or table. The colour depth indicates the size of the data value (correlation value). The matrix directly shows the size of the data value with the defined colour. Asterisks indicate that the correlation coefficients (r) were >0.4 and the P values were <0.05.
Results
Dry matter intake, milk yield, and milk composition
Dry matter intake (DMI) was significantly greater in the high-yield (HY) group than in the low-yield (LY) group (P = 0.03). Milk production, 4% fat-corrected milk (FCM) and energy-corrected milk (ECM) were significantly lower in the LY group than in the HY group (Table 2). The milk fat content tended to be higher (P = 0.08), and the milk protein content was significantly higher in the LY group than in the HY group (P<0.01). No difference was observed in milk lactose content between the LY and HY groups (P = 0.21). Fat, protein and lactose yields were significantly greater in the HY group than in the LY group. However, somatic cell count (SCC) was not different between the HY and LY groups (P = 0.13; Table 2).
Table 2. Milk and ECM from high-yielding and low-yielding dairy cows during the entire sampling period1.
| Items | LY | HY | SEM | P-value |
|---|---|---|---|---|
| DMI (kg/d) | 23.4 | 25.6 | 0.43 | 0.0347 |
| Milk production (kg/d) | 19.3 | 31.9 | 1.76 | <0.0001 |
| 4%FCM production (kg/d)2 | 18.95 | 29.20 | 1.73 | <0.0001 |
| ECM production (kg/d)3 | 21.07 | 32.09 | 1.93 | <0.0001 |
| Milk composition | ||||
| Fat % | 4.02 | 3.48 | 0.28 | 0.0788 |
| Fat yield (kg/d) | 0.75 | 1.10 | 0.07 | 0.0004 |
| Protein % | 3.50 | 3.07 | 0.11 | 0.0023 |
| Protein yield (kg/d) | 0.66 | 0.98 | 0.06 | 0.0002 |
| Lactose % | 4.87 | 5.05 | 0.14 | 0.2127 |
| Lactose yield (kg/d) | 0.92 | 1.61 | 0.10 | <0.0001 |
| Total solid content % | 13.15 | 12.30 | 0.38 | 0.0417 |
| SCC (log10) | 2.46 | 3.10 | 2.78 | 0.1339 |
1Data are presented as least squares means.
SCC = somatic cell count.
Ruminal pH and VFA concentrations
Rumen pH was not different between the HY group and the LY group (Table 3). NH3-N (mg/dL) was greater in the HY group than in the LY group (P<0.01), and there was an increase trend proportion of propionate (P = 0.08) and total VFA concentrations in the HY group relative to the LY group (P<0.05). In addition, the proportion of acetate had a tendency lower in the HY group than in the LY group (P = 0.06). A lower trend was observed for the acetate to propionate ratio in the HY group compared to the LY group (P = 0.06).
Table 3. Effects of differences between high-yielding and low-yielding dairy cows on metabolites in the rumen.
| Items | LY | HY | SEM | P-value |
|---|---|---|---|---|
| pH | 6.73 | 6.71 | 0.02 | 0.69 |
| NH3-N, mg/dL | 7.99 | 13.28* | 1.03 | 0.01 |
| Proportion | ||||
| Acetate | 62.38 | 60.72 | 0.45 | 0.06 |
| Propionate | 21.95 | 23.14 | 0.34 | 0.08 |
| Isobutyrate | 0.83 | 0.74 | 0.03 | 0.13 |
| Butyrate | 12.06 | 12.55 | 0.22 | 0.27 |
| Isovalerate | 1.21 | 1.31 | 0.05 | 0.34 |
| Valerate | 1.58 | 1.55 | 0.04 | 0.74 |
| Acetate:propionate ratio | 2.85 | 2.64 | 0.06 | 0.06 |
| Total VFA (mmol/L) | 99.76 | 113.63* | 3.14 | 0.02 |
*P<0. 05
Values within a sampling day followed by superscripted asterisks differ.
SEM = standard error of the mean.
Diversity and richness of microbial communities
In total, 2,382,338 merged sequences were acquired for the 16 samples from the dairy cows and 1,191,169 high-quality sequences, with an average read length of 440 bp, were classified as bacterial. On average, at least 54,144 sequences were obtained per sample, and greater than 99% depth coverage was achieved. The rarefaction curve generated tended to plateau, showing that the number of OTUs did not rise with an increasing volume of data. This finding showed that the data volume of sequencing was reasonable. The results of this study show that the sequencing data were reasonable and could reflect changes in most bacterial flora.
No significant differences were observed in alpha diversity index results between the HY and LY groups (P>0.05) (Table 4). However, the coverage of the HY group was significantly higher than that of the LY group (P<0.01), indicating greater community diversity in the HY group.
Table 4. Alpha diversity index of rumen bacteria.
| Item | LY | HY | SEM | P-value |
|---|---|---|---|---|
| Sobs | 1196.13 | 1214.88 | 17.21 | 0.60 |
| Shannon | 5.41 | 5.49 | 0.04 | 0.42 |
| Simpson | 0.01 | 0.01 | 0.00 | 0.74 |
| ACE | 1375.65 | 1360.65 | 18.75 | 0.70 |
| Chao | 1378.64 | 1376.82 | 20.10 | 0.97 |
| Coverage | 0.992 | 0.994** | 0.00 | 0.001 |
*P<0. 05
**P<0. 01:
Values within a sampling day followed by superscript asterisks differ. SEM = standard error of the mean
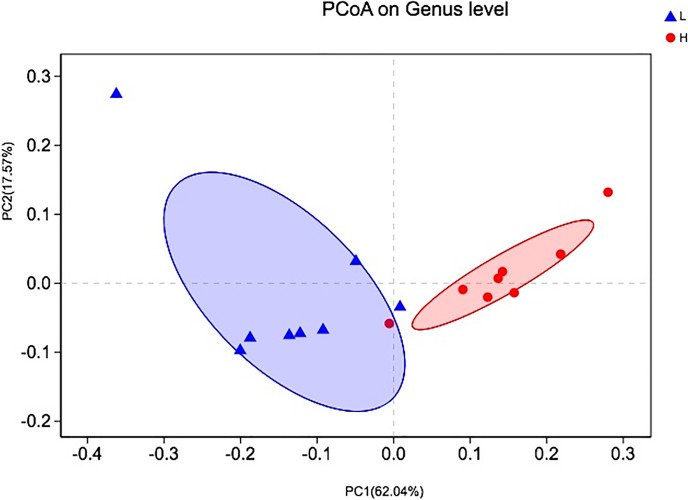
To understand the differences in the overall rumen bacterial community between high- and low-yield lactating dairy cows, a PCoA was used to analyse bacterial diversity, followed by the weighted UniFrac metrics (Fig 1). As shown in Fig 1, principal coordinate 1 accounted for 62.04% and principal coordinate 2 accounted for 17.57% of the total variation.
Fig 1. Principal coordinate analysis (PCoA) of bacterial community structures of the ruminal microbiota in the high-yielding group (red circles) and the low-yielding group (blue triangles).
PCoA plots were constructed using the unweighted UniFrac method.
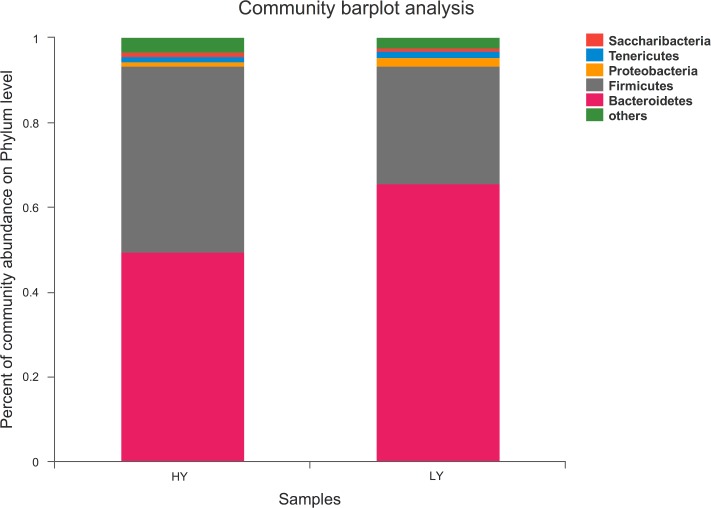
Twenty-one bacterial phyla were identified across all samples. Bacteroidetes, Firmicutes and Proteobacteria were the three dominant groups, representing 57.59%, 35.86%, and 1.53% of the total sequences, respectively (Fig 2). Thus, at the phylum level, Bacteroidetes and Firmicutes were particularly dominant. The HY group exhibited a greater abundance of Firmicutes and lower abundance of Bacteroidetes than did the LY group (P<0.01), whereas Proteobacteria was less abundant (P<0.05) (S1 Table).
Fig 2. Percent composition of predominant phyla in the rumen fluid.
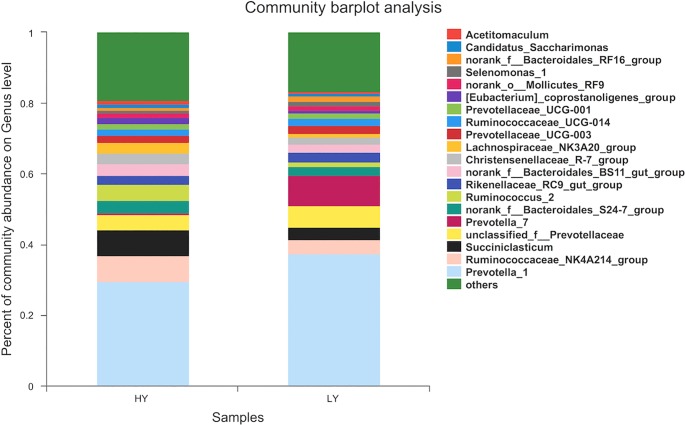
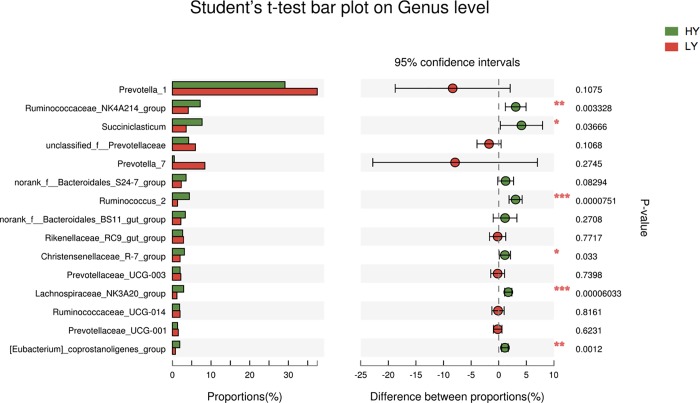
At the genus level, taxa with a relative abundance of ≥1% in at least one sample were further analysed, and the relevant genera are presented in Figs 3 and 4. Twenty-one genera were identified, 6 of which exhibited significantly different abundances between the groups. Specifically, 4 genera were more abundant in the HY group at P<0.01, including Ruminococcaceae-NK4A214-group, Ruminococcus 2, Lachnospiraceae-BS11-gut-group, and [Eubacterium]-coprostanoligenes-group, and 2 were more abundant in the HY group at P<0.05: Succiniclasticum and Christensenellaceae-R-7-group (S1 Table).
Fig 3. Percent composition of genera in the rumen fluid.
Fig 4. Percent composition and significance of genera in the rumen fluid.
Correlations between bacterial communities and ruminal variables
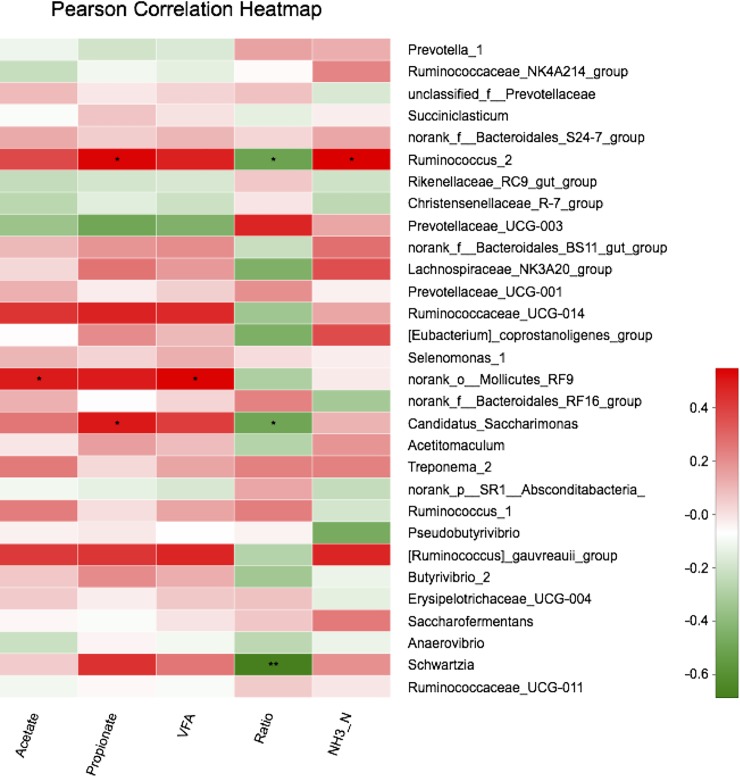
As shown in Fig 5, the relative abundances of the genera Bacteroides and Ruminococcus 2 were positively correlated with ruminal propionate and NH3-N concentrations (r>0.4, P<0.05) but negatively correlated with the ruminal ratio (acetate:propionate ratio) (r<-0.4, P<0.05). In addition, norank_o__Mollicutes_RF9 was positively correlated with ruminal acetate and VFA concentrations (r>0.4, P<0.05). Candidatus-Saccharimonas was positively correlated with the ruminal propionate concentration (r>0.4, P<0.05) but negatively correlated with the ruminal ratio (r<-0.4, P<0.05). Moreover, the ratio was negatively correlated with Schwartzia (r<-0.6, P<0.05).
Fig 5. Correlation analyses between the relative abundances of bacteria genera and ruminal fermentation parameters.
Only genera with abundances significantly associated with ruminal VFA, propionate and acetate concentrations are presented. Green represents a negative correlation between the abundance of the species and the ratio (r<−0.4), and red represents a positive correlation (r>0.4, 0.01<P< = 0.05 *; 0.001<P≤0.01 **; P≤0.001 ***; Values with a significant correlation followed by superscripted asterisks differ).
Discussion
Changes in rumen fermentation parameters and milk composition
Higher volatile fatty acid concentration and milk composition yield were found in the HY group compared to the LY group. Recent studies suggested an important relationship between VFAs and milk components [2, 30, 31]. Specifically, of the three principle VFAs, acetate and butyrate are substrates for oxidation and are precursors of lipids [32, 33], moreover propionate is the only glucogenic VFA, accounting for 65–80% of the net glucose supply in lactating dairy cows [34, 35]. In the present study, the proportion of propionate had an increasing trend in the HY group than that in the LY group, which may also be explained the mechanism of different milk composition and milk production. In line with the reported that milk yield was most highly related to rumen concentrations of butyrate and propionate [36].
Milk fat, protein and lactose yields were significantly greater in the HY group than in the LY group, which is consistent with previous research that milk production and VFA-producing bacterium have the positive correlation[37]. Weimer et al. [14] also showed that high-efficiency Holstein cows have greater VFA and propionate molar percentages compared with low-efficiency Holstein cows, which was apparently caused by differences in the ruminal bacterial community. Furthermore, it has been reported that even under the same dietary condition the bacterial communities also different between Holstein and Jersey cows [26]. Therefore, these findings suggested that probably due to host–microbiota interactions, different milk production dairy cows may harbour different microbial species compositions, which are probably closely related to distinct differences in rumen fermentation parameters and milk composition.
The NH3-N concentration in rumen fluid can reflect the balance of protein degradation and synthesis under varying feed conditions. Our results showed that the NH3-N concentration was within the normal range, though that in the HY group was significantly higher than that in the LY group. It is well known that NH3-N is an intermediate product of feed protein, non-protein nitrogen degradation and microbial protein synthesis, and it is mainly affected by feed protein degradation, rumen wall absorption, microorganism utilization and rumen chyme outflow rate [38–40]. Yang et al. [41] reported that the concentration of NH3-N should be higher than 5 mg/dL; otherwise, it will influence the "uncoupling" effect of ruminal fermentation and reduce the efficiency of microbial protein synthesis. Corroborating all these results indicate that rumen microbes promote protein degradation in high-yield dairy cows, providing a better understanding of the difference in milk proteins between the two groups.
Differences in rumen microbial composition between HY and LY groups
No differences were observed in bacterial community richness and diversity between the groups in our study. Three phyla predominated in both groups, which was consistent with previous studies reporting that the principal phyla of microbes in the rumen are Bacteroidetes, Firmicutes, and Proteobacteria [38]. The proportions of these three phyla account for approximately 94% of the total [40, 42, 43]. Interestingly, our results showed that the abundance of Firmicutes in the HY group was higher than that in the LY group, though the abundances of the two other dominant phyla were lower in the former than in the latter. In consistent with previous reported by Pan et al. [44], cows fed a high proportion of grain have a higher abundance of Firmicutes and a lower abundance of Proteobacteria than control cows, and other studies have shown that feeding a high amount of grain can promote milk production [45, 46]. Thus, our study provides a better understanding of why cows fed the same dietary condition can have different milk production. The present findings further demonstrate that Firmicutes plays an important role in milk production.
In agreement with other research results [12, 47], our study showed that Prevotella was the most abundant genus in all samples. Although Prevotella was more abundant in the LY group than in the HY group, the difference was not significant. In contrast, Ruminococcaceae-NK4A214-group, Ruminococcus 2, Lachnospiraceae-BS11-gut-group and [Eubacterium]-coprostanoligenes-group were significantly different between the two groups, with higher abundances in the HY group than in the LY group. Jiang et al. [48] reported that the increase in the relative abundance of Ruminococcus partly explains why adding live yeast to the diet increases the in vivo digestibility of DM and NDF and the performance of cows. Thus, this result illustrates that high-performance cows have higher abundances of Ruminococcus in the rumen fluid, which is consistent with the present research results. Besides, Ruminococcus spp. as major bacteria plays an important role in acetate production [49]. However, our results found that although the Ruminococcus spp. had significantly higher abundances in the HY group than in the LY group, the proportion of acetate was trend to lower in HY group compared with LY group.
Members of the family Lachnospiraceae are gram-positive obligate anaerobes that are mostly non-spore-forming bacteria [50, 51]. Huws et al. [52] showed that Ruminococcaceae and Lachnospiraceae play predominant roles in biohydrogenation pathways within the rumen. Furthermore, as the primary succinate-utilizing bacterial taxon, Succiniclasticum accounted for 7.45% of the total bacterial community in the HY group, with significantly greater abundance than in the LY group. A higher level of Succiniclasticum has been associated with greater production of succinate from starch degradation [53], which could also explain the higher proportion of propionate in HY group compared with LY group. Moreover, the abundances of Christensenellaceae and Ruminococcaceae NK4A214 in the HY group were significantly higher than in the LY group, though little information about these two genera has been reported in the literature. The reasons for the altered status of genera in cows with different milk production are need further studies.
Conclusion
In summary, high-yield dairy cows have better ruminal fermentation patterns than do low-yield cows, which was partially attributed to the greater abundances of Bacteroides, Ruminococcus 2, Ruminococcaceae NK4A214, Lachnospiraceae, Succiniclasticum, Eubacterium and Christensenella in the former. Furthermore, rumen fermentation in high-yield cows exhibited higher VFA levels than that found in low-yield cows. The rumen microbial compositions of high-yield and low-yield dairy cows are different, and microbial species diversity and distribution contribute to production-related phenotypes. Overall, our findings enhance our understanding of rumen bacteria in cows with different milk yields and provide new strategies for improving dairy cow production performance.
Supporting information
(XLSX)
(XLS)
Acknowledgments
This study was financially supported by the Project of National Nature Science Foundation of China (Grant No. 31772629 and No. 31702302), Beijing Municipal Education Commission Project (SQKM201710020011), Open Project Program of Beijing Key Laboratory of Dairy Cow Nutrition, the National Key Research and Development Plan (2016YFD0700205, 2016YFD0700201, 2017YFD0701604). Hua Zhang thanks the Research Fund for Young Scientists of BUA. Jinjin Tong also thanks the Research Fund for Young Scientists of BUA, the foundation supporting the China postdoctoral foundation and the Beijing postdoctoral foundation.
Data Availability
The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP136923).
Funding Statement
This study was financially supported by the Project of National Nature Science Foundation of China (Grant No. 31772629 and No. 31702302), Beijing Municipal Education Commission Project (SQKM201710020011), Open Project Program of Beijing Key Laboratory of Dairy Cow Nutrition, the National Key Research and Development Plan (2016YFD0700205, 2016YFD0700201, 2017YFD0701604). Hua Zhang thanks the Research Fund for Young Scientists of BUA. Jinjin Tong also thanks the Research Fund for Young Scientists of BUA, the foundation supporting the China postdoctoral foundation and the Beijing postdoctoral foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Delong EF, Stephen L, Stackebrandt E, Rosenberg E. Prokaryotic biology and symbiotic associations: Springer; 2013. [Google Scholar]
- 2.Krehbiel CR. INVITED REVIEW: Applied nutrition of ruminants: Fermentation and digestive physiology 1. Professional Animal Scientist. 2014;30(2):129–39. [Google Scholar]
- 3.Schären M, Frahm J, Kersten S, Meyer U, Hummel J, Breves G, et al. Interrelations between the rumen microbiota and production, behavioral, rumen-fermentation, metabolic, and immunological attributes of dairy cows. Journal of Dairy Science. 2018. [DOI] [PubMed] [Google Scholar]
- 4.Guan LL, Nkrumah JD, Basarab JA, Moore SS. Linkage of microbial ecology to phenotype: correlation of rumen microbial ecology to cattle's feed efficiency. Fems Microbiology Letters. 2008;288(1):85–91. 10.1111/j.1574-6968.2008.01343.x [DOI] [PubMed] [Google Scholar]
- 5.Li F, Guan LL. Metatranscriptomic Profiling Reveals Linkages between the Active Rumen Microbiome and Feed Efficiency in Beef Cattle. Applied & Environmental Microbiology. 2017;83(9):AEM.00061-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myer PR, Smith TPL, Wells JE, Kuehn LA, Freetly HC. Rumen Microbiome from Steers Differing in Feed Efficiency. Plos One. 2015;10(6):e0129174 10.1371/journal.pone.0129174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shabat SK, Sasson G, Doronfaigenboim A, Durman T, Yaacoby S, Berg Miller ME, et al. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. Isme Journal. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jami E, White BA, Mizrahi I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. Plos One. 2014;9(1):e85423 10.1371/journal.pone.0085423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohammed R. Changes in ruminal bacterial community composition following feeding of alfalfa silage inoculated with a commercial silage inoculant. Journal of Dairy Science. 2012;95(1):328–39. 10.3168/jds.2011-4492 [DOI] [PubMed] [Google Scholar]
- 10.Creevey CJ, Kelly WJ, Henderson G, Leahy SC. Determining the culturability of the rumen bacterial microbiome. Microbial Biotechnology. 2014;7(5):467–79. 10.1111/1751-7915.12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson G, Cox F, Ganesh S, Jonker A, Young W, Collaborators GRC, et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Scientific Reports. 2016;6:14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu S, Baldwin RL, Li W, Li C, Connor EE, Li RW. The Bacterial Community Composition of the Bovine Rumen Detected Using Pyrosequencing of 16S rRNA Genes. Metagenomics. 2012;1(1):b1–11. [Google Scholar]
- 13.Newbold CJ, Gabriel DLF, Alejandro B, Eva RM, Mcewan NR. The Role of Ciliate Protozoa in the Rumen. Frontiers in Microbiology. 2015;6(Suppl 1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weimer PJ, Cox MS, Vieira dPT, Lin M, Hall MB, Suen G. Transient changes in milk production efficiency and bacterial community composition resulting from near-total exchange of ruminal contents between high- and low-efficiency Holstein cows. Journal of Dairy Science. 2017;100(9):7165–82. 10.3168/jds.2017-12746 [DOI] [PubMed] [Google Scholar]
- 15.Church DC. The ruminant animal. Digestive physiology and nutrition. 1988. [Google Scholar]
- 16.Mcsweeney C, Mackie R. Commission on Genetic Resources for Food and Agriculture. Micro-organisms and ruminant digestion: State of knowledge, trends and future prospects Background Study Paper; 2012. [Google Scholar]
- 17.Dowd SE, Callaway TR, Wolcott RD, Sun Y, Mckeehan T, Hagevoort RG, et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). Bmc Microbiology. 2008;8(1):125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards JE, Mcewan NR, Travis AJ, Wallace RJ. 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek. 2004;86(3):263–81. 10.1023/B:ANTO.0000047942.69033.24 [DOI] [PubMed] [Google Scholar]
- 19.Bucbe M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S, et al. 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytologist. 2009;184(2):449–56. 10.1111/j.1469-8137.2009.03003.x [DOI] [PubMed] [Google Scholar]
- 20.Hamberger A, Horn MA, Dumont MG, Murrell JC, Drake HL. Anaerobic consumers of monosaccharides in a moderately acidic fen. Applied & Environmental Microbiology. 2008;74(10):3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humblot C, Guyot JP. Pyrosequencing of Tagged 16S rRNA Gene Amplicons for Rapid Deciphering of the Microbiomes of Fermented Foods Such as Pearl Millet Slurries. Applied & Environmental Microbiology. 2009;75(13):4354–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanapareddy N, Hamp TJ, Gonzalez LC, Hilger HA, Fodor AA, Clinton SM. Molecular Diversity of a North Carolina Wastewater Treatment Plant as Revealed by Pyrosequencing. Applied & Environmental Microbiology. 2009;75(6):1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao SY, Huo WJ, Zhu WY. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environmental Microbiology. 2016;18(2):525 10.1111/1462-2920.12724 [DOI] [PubMed] [Google Scholar]
- 24.Gonzalo MF, Denman SE, Yang C, Jane C, Makoto M, Mcsweeney CS. Methane Inhibition Alters the Microbial Community, Hydrogen Flow, and Fermentation Response in the Rumen of Cattle. Frontiers in Microbiology. 2016;7(1087). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danielsson R, Dicksved J, Sun L, Gonda H, Müller B, Schnürer A, et al. Methane Production in Dairy Cows Correlates with Rumen Methanogenic and Bacterial Community Structure. Frontiers in Microbiology. 2017;8:226 10.3389/fmicb.2017.00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paz HA, Anderson CL, Muller MJ, Kononoff PJ, Fernando SC. Rumen Bacterial Community Composition in Holstein and Jersey Cows Is Different under Same Dietary Condition and Is Not Affected by Sampling Method. Frontiers in Microbiology. 2016;7(e0116704). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golder HM, Denman SE, Mcsweeney C, Wales WJ, Auldist MJ, Wright MM, et al. Effects of partial mixed rations and supplement amounts on milk production and composition, ruminal fermentation, bacterial communities, and ruminal acidosis. Journal of Dairy Science. 2014;97(9):5763–85. 10.3168/jds.2014-8049 [DOI] [PubMed] [Google Scholar]
- 28.Mao SY, Zhang G, Zhu WY. Effect of disodium fumarate on ruminal metabolism and rumen bacterial communities as revealed by denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA. Animal Feed Science & Technology. 2008;140(3–4):293–306. [Google Scholar]
- 29.AL C, EP M. Modified reagents for determination of urea and ammonia. Clinical chemistry. 1962;8(8):130–2. [PubMed] [Google Scholar]
- 30.Armentano LE. Ruminant hepatic metabolism of volatile fatty acids, lactate and pyruvate. Journal of Nutrition. 1992;122(3 Suppl):838–42. 10.1093/jn/122.suppl_3.838 [DOI] [PubMed] [Google Scholar]
- 31.Tang C, Zhang K, Zhan T, Zhao Q, Zhang J. Metabolic Characterization of Dairy Cows Treated with Gossypol by Blood Biochemistry and Body Fluid Untargeted Metabolome Analyses. Journal of Agricultural & Food Chemistry. 2017. [DOI] [PubMed] [Google Scholar]
- 32.Bauman DE, Griinari JM. NUTRITIONAL REGULATION OF MILK FAT SYNTHESIS. Annual Review of Nutrition. 2003;23(1):203–27. [DOI] [PubMed] [Google Scholar]
- 33.Dijkstra J, Boer H, Van BJ, Bruining M, Tamminga S. Absorption of volatile fatty acids from the rumen of lactating dairy cows as influenced by volatile fatty acid concentration, pH and rumen liquid volume. British Journal Nutrition. 1993;69(2):385–96. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds CK, Aikman PC, Lupoli B, Humphries DJ, Beever DE. Splanchnic metabolism of dairy cows during the transition from late gestation through early lactation. Journal of Dairy Science. 2003;86(4):1201–17. 10.3168/jds.S0022-0302(03)73704-7 [DOI] [PubMed] [Google Scholar]
- 35.Zarrin M, Grossen-Rösti L, Bruckmaier RM, Gross JJ. Elevation of blood β-hydroxybutyrate concentration affects glucose metabolism in dairy cows before and after parturition. Journal of Dairy Science. 2017;100(3):2323–33. 10.3168/jds.2016-11714 [DOI] [PubMed] [Google Scholar]
- 36.Seymour WM, Campbell DR, Johnson ZB. Relationships between rumen volatile fatty acid concentrations and milk production in dairy cows: a literature study. Animal Feed Science & Technology. 2005;119(1):155–69. [Google Scholar]
- 37.Sofyan A, Mitsumori M, Ohmori H, Uyeno Y, Hasunuma T, Akiyama K, et al. Differences in rumen fermentation characteristics between low‐yield and high‐yield dairy cows in early lactation. Animal Science Journal. 2017;88(7). [DOI] [PubMed] [Google Scholar]
- 38.Colmenero JJO, Broderick GA. Effect of Dietary Crude Protein Concentration on Milk Production and Nitrogen Utilization in Lactating Dairy Cows. Journal of Dairy Science. 2006;89(5):1704 10.3168/jds.S0022-0302(06)72238-X [DOI] [PubMed] [Google Scholar]
- 39.Mutsvangwa T, Davies KL, Mckinnon JJ, Christensen DA. Effects of dietary crude protein and rumen-degradable protein concentrations on urea recycling, nitrogen balance, omasal nutrient flow, and milk production in dairy cows. Journal of Dairy Science. 2016;99(8):6298–310. 10.3168/jds.2016-10917 [DOI] [PubMed] [Google Scholar]
- 40.Jin D, Zhao S, Wang P, Zheng N, Bu D, Yves B, et al. Insights into Abundant Rumen Ureolytic Bacterial Community Using Rumen Simulation System. Frontiers in Microbiology. 2016;7(887). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang WZ, Beauchemin KA, Rode LM. Effects of Grain Processing, Forage to Concentrate Ratio, and Forage Particle Size on Rumen pH and Digestion by Dairy Cows. Journal of Dairy Science. 2001;84(10):2203–16. 10.3168/jds.S0022-0302(01)74667-X [DOI] [PubMed] [Google Scholar]
- 42.Khafipour E, Krause DO, Plaizier JC. A grain-based subacute ruminal acidosis challenge causes translocation of lipopolysaccharide and triggers inflammation. Journal of Dairy Science. 2009;92(3):1060–70. 10.3168/jds.2008-1389 [DOI] [PubMed] [Google Scholar]
- 43.Khafipour E, Li S, Plaizier JC, Krause DO. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Applied & Environmental Microbiology. 2009;75(22):7115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan X, Xue F, Nan X, Tang Z, Wang K, Beckers Y, et al. Illumina Sequencing Approach to Characterize Thiamine Metabolism Related Bacteria and the Impacts of Thiamine Supplementation on Ruminal Microbiota in Dairy Cows Fed High-Grain Diets. Frontiers in Microbiology. 2017;8:1818 10.3389/fmicb.2017.01818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rugoho I, Cheng L, Aizimu W, Bryant RH, Edwards GR. Effects of post‐grazing herbage height and concentrate feeding on milk production and major milk fatty acids of dairy cows in mid‐lactation. Grass & Forage Science. 2017;72(2):211–9. [Google Scholar]
- 46.Ueda K, Mitani T, Kondo S. Effect of timing and type of supplementary grain on herbage intake, nitrogen utilization and milk production in dairy cows grazed on perennial ryegrass pasture from evening to morning. Animal Science Journal. 2017;88(1):107–18. 10.1111/asj.12605 [DOI] [PubMed] [Google Scholar]
- 47.Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Applied Microbiology & Biotechnology. 2007;75(1):165. [DOI] [PubMed] [Google Scholar]
- 48.Jiang Y, Ogunade IM, Qi S, Hackmann TJ, Staples C, Adesogan AT. Effects of the dose and viability of Saccharomyces cerevisiae. 2. Diversity of ruminal microbes as analyzed by Illumina MiSeq sequencing and quantitative PCR. Journal of Dairy Science. 2016;100(1):325 10.3168/jds.2016-11263 [DOI] [PubMed] [Google Scholar]
- 49.Jeyanathan J, Martin C, Morgavi DP. The use of direct-fed microbials for mitigation of ruminant methane emissions: a review. Animal. 2014;8(2):250–61. 10.1017/S1751731113002085 [DOI] [PubMed] [Google Scholar]
- 50.Cotta M, Forster R. The Family Lachnospiraceae, Including the Genera Butyrivibrio, Lachnospira and Roseburia: Springer US; 2006.: 727–35. p. [Google Scholar]
- 51.Hedberg ME, Moore ER, Svenssonstadler L, Hörstedt P, Baranov V, Hernell O, et al. Lachnoanaerobaculum gen. nov., a new genus in the Lachnospiraceae: characterization of Lachnoanaerobaculum umeaense gen. nov., sp. nov., isolated from the human small intestine, and Lachnoanaerobaculum orale sp. nov., isolated from saliva, and reclassifica. International Journal of Systematic & Evolutionary Microbiology. 2012;62(Pt 11):2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huws SA, Kim EJ, Lee MRF, Scott MB, Tweed JKS, Pinloche E, et al. As yet uncultured bacteria phylogenetically classified as Prevotella, Lachnospiraceae incertae sedis and unclassified Bacteroidales, Clostridiales and Ruminococcaceae may play a predominant role in ruminal biohydrogenation. Environmental Microbiology. 2011;13(6):1500–12. 10.1111/j.1462-2920.2011.02452.x [DOI] [PubMed] [Google Scholar]
- 53.Hook SE, Steele MA, Northwood KS, Dijkstra J, France J, Wright AD, et al. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. Fems Microbiology Ecology. 2011;78(2):275–84. 10.1111/j.1574-6941.2011.01154.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLS)
Data Availability Statement
The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP136923).