Abstract
Sudden unexpected death is an upsetting event, which can remain unexplained even after post-mortem investigation. Internationally, molecular autopsies have shown to resolve up to 44% of unexplained cases; however, it is currently unclear how many of these were infants. This systematic literature review showed that significantly fewer infant cases were resolved (median: 4%) compared with cohorts of 1 to 45 years old (median: 32%). Further, no study involving indigenous African participants has yet been published. Overall, molecular autopsies hold immense value to living family members and is motivation to explore new avenues in infant cohorts.
Keywords: sudden infant death syndrome, negative autopsy, medico-legal investigation
Introduction
Sudden unexpected death of infants (SUDI) is a devastating occurrence worldwide. In most countries, such cases are referred for medico-legal autopsy to establish the cause of death; however, in many instances, the cause of death remains undetermined. By a matter of exclusion, these cases are categorized as sudden infant death syndrome (SIDS) and in developed countries, is the leading cause of death in infants. 1 2 While SUDI refers to the rapid and unexpected fatal episode of an infant less than 1 year old, SIDS refers specifically to those cases which remain unexplained following a full autopsy, clinical history review, and death scene investigation. 3
Molecular autopsies have been reported to be of value in cases of sudden unexpected death in the young (SUDY) (individuals younger than 40 years old), particularly in autopsy-negative cases. 4 5 A molecular autopsy involves the analysis of the deceased individuals' DNA to determine if they had a genetic mutation which contributed to and/or was likely to have caused their death. Most of the research has been focused on channelopathies, which are difficult to detect at a conventional autopsy (as the heart appears anatomically normal), but analysis of candidate genes implicated in channelopathy diseases (e.g., long QT syndrome and Brugada syndrome) can yield a molecular diagnosis. 5 Molecular autopsies have shown a resolution of up to 44% in previously unexplained cases (cohort aged 1–19 years old), which brings closure to family members, while also making it possible for blood relatives to be tested for these mutations. 6
Various case reports and studies have been published regarding the number of sudden unexpected death cases which have been potentially resolved by a molecular autopsy; 7 8 however, it is currently unclear how many of these cases were infants. Therefore, the aim of this study was to review the literature to assess the percentage of SUDI cases resolved by molecular autopsies. The objectives were to assess which diseases were most commonly inferred by molecular results and to compare the resolution rates between SUDI cases and older SUDY cohorts.
Methodology
The PubMed database was searched using the following search terms: ((Sudden[All Fields] AND unexpected[All Fields] AND (“death”[MeSH Terms] OR “death”[All Fields] OR “deaths”[All Fields]) AND (“infant”[MeSH Terms] OR “infant”[All Fields] OR “infants”[All Fields])) OR (“sudden infant death”[MeSH Terms] OR (“sudden”[All Fields] AND “infant”[All Fields] AND “death”[All Fields]) OR “sudden infant death”[All Fields] OR (“sudden”[All Fields] AND “infant”[All Fields] AND “death”[All Fields] AND “syndrome”[All Fields]) OR “sudden infant death syndrome”[All Fields])) AND (molecular[All Fields] AND (“autopsy”[MeSH Terms], OR “autopsy”[All Fields])). No search restrictions were applied on the dates of publication, and all articles up until December 31, 2017 were included.
All articles returned from the search were then scanned for relevance by reading the abstract and were included for further analysis if they met certain criteria, as follows: original research articles and case reports were included, but reviews and opinion articles were excluded. Articles pertaining to the development and/or optimization of molecular methods only were not included. Articles in a language other than English were excluded. Articles that described a cohort inclusive of 0 to 1-year-olds (but not necessarily limited to infants) were included, but the exact number of infants in their studies were also recorded and used in subsequent analysis. Articles whereby the cohort were all greater than 1-year-old were put aside for comparison purposes. It was also noted if a case report was subsequently included in a larger cohort study by the same research group. After all relevant articles were obtained, their reference lists were searched manually for other potential articles, which were subjected to the same criteria as described above.
The following variables were collected from each article: article details, including journal name and date; country of the population studied; total cohort size (including age range) as well as number of infants in the study; scope of molecular investigation (including the number of genes investigated); pathways/disease groups associated with the genes studied; and resolution of cases following molecular analysis. This information was collected for both infants, as well as older cohorts.
Descriptive statistics were applied to the data, and the non-parametric Mann–Whitney test was used to assess for significant differences in the percentages of resolved cases between infants and cohorts of more than 1-year-old, assuming a confidence interval of 95%. A Spearman rank sum test was used to assess for correlation between the number of genes included and the percentages of resolved cases. Graphs were created, and statistical tests were performed in GraphPad Prism 6.
Results
A total of 94 articles were returned, of which 55 were subsequently excluded. These were review articles ( n = 15), retrospective case reviews not involving molecular autopsies ( n = 8), viewpoint or recommendation articles ( n = 6), studies involving method optimization or molecular virus detection ( n = 10), or articles not relevant to the topic ( n = 16). Only one article was not in English, but was a review article, so was excluded anyway. Numerous articles were published relating to the Scripps Translational Science Institute study, 9 10 11 but only the most relevant article pertaining to this review was included, 11 to avoid overrepresentation of the cohort.
A total of 20 original research studies 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 and seven case reports 31 32 33 34 35 36 37 thus met the inclusion criteria for the review, which included at least one infant greater than 1-year-old in their study ( Table 1 ). One of the seven case reports, however, 34 was also represented in one original research article. 13 The remaining articles ( n = 12) had cohorts of individuals older than 1 year old only.
Table 1. A total of 20 original articles and 7 case reports met the inclusion criteria for the systematic review.
| Date | Country | Number of infants (<1 year) in study cohort | Molecular autopsy approach | Number of genes/variants investigated | Sample used from deceased infant | Potential resolution in infant cases (%) | Ref. |
|---|---|---|---|---|---|---|---|
| Original articles | |||||||
| 2017 | United States | 8 | NGS, WES | Exome & mtDNA genome | Blood | 37.5 | 11 |
| 2017 | Canada | 182 | NGS, panel of genes | 71 genes | Frozen tissue | 6.04 | 12 |
| 2017 | Europe (multi-center) | 161 | NGS, WES | Focused on 192 genes | Tissue | 20.0 | 23 |
| 2016 | United States | 280 | NGS, panel of genes | 64 genes | Dried blood spot | 2.80 | 24 |
| 2015 | Germany | 38 | NGS, panel of genes | 86 genes | Frozen blood or tissue | 5.00 | 25 |
| 2015 | France | 6 | NGS, panel of genes | 23 genes | Frozen tissue | 33.3 | 26 |
| 2014 | United States | 141 | NGS, panel of genes | 6 genes | Dried blood spot or tissue | 13.5 | 27 |
| 2014 | New Zealand | 102 | Sanger sequencing | 6 genes | Blood, dried blood spot or tissue | 1.96 | 28 |
| 2014 | Netherlands | 138 | Various targeted genotyping methods | 25 variants in 11 genes | FFPE tissue | Not available | 29 |
| 2013 | Australia | 46 | Sanger sequencing | 2 genes | Blood or tissue | 4.00 | 30 |
| 2013 | Italy | 10 | Real-time PCR | 1 gene + gene expression | Tissue | 10.0 | 13 |
| 2011 | Japan | 30 | Sanger sequencing | 1 gene | Frozen blood | 3.33 | 14 |
| 2010 | United States | 292 | HPLC and Sanger sequencing | 4 genes | Blood or frozen tissue | 1.03 | 15 |
| 2010 | New Zealand | 1 | HPLC and Sanger sequencing | 5 genes | Dried blood spot | 0.00 | 16 |
| 2009 | France | 52 of which 32 were SIDS | HPLC and Sanger sequencing | 5 genes | Blood | 9.38 | 17 |
| 2007 | United States | 16 | Sanger sequencing | 2 genes | FFPE tissue | 12.5 | 18 |
| 2007 | United States | 221 | HPLC and Sanger sequencing | 1 gene | Blood | 1.35 | 19 |
| 2007 | Germany | 6 | Sanger sequencing | 2 genes | Blood | 0.00 | 20 |
| 2001 | United States | 93 | HPLC and Sanger sequencing | 1 gene | Frozen tissue | 2.15 | 21 |
| 1992 | United States | 67 | Restriction enzyme digestion | 1 variant | FFPE tissue | 0.00 | 22 |
| Mean = 8.62% Median = 4.00% |
|||||||
| Case studies | |||||||
| 2015 | United States | 1 | NGS, WES | Exome | Blood | Yes | 31 |
| 2014 | Japan | 1 | Sanger sequencing | 1 gene | Blood | Yes | 32 |
| 2014 | Spain | 1 | NGS, panel of genes | 104 genes | Blood | Yes | 33 |
| 2013 | Italy | 1 | PCR and agarose gel electrophoresis | 1 variant | Tissue | Yes | 34 |
| 2009 | United States | 1 | Sanger sequencing | 1 variant | Blood | Yes | 35 |
| 2001 | Germany | 2 | Sanger sequencing | 4 genes | Blood | No | 36 |
| 1997 | United States | 1 | Restriction enzyme digestion | 1 gene | Tissue | Yes | 37 |
Abbreviations: FFPE, formalin fixed paraffin embedded; HPLC, high-performance liquid chromatography; mtDNA, mitochondrial DNA; NGS, next generation sequencing; PCR, polymerase chain reaction; Ref., reference; USA, the United States of America; SIDS, sudden infant death syndrome; WES, whole exome sequencing.
Note: Details pertaining to these studies are summarized. Potential resolution is reported to three significant numbers.
Articles that met the inclusion criteria were published between 1992 and 2017, with 23 (85%) published in the past 11 years ( Fig. 1 ). The articles were published in 21 different journals; most journals published a single article, besides Forensic Science International ( n = 5), Archives of Disease in Childhood ( n = 2), and Heart Rhythm ( n = 2).
Fig. 1.
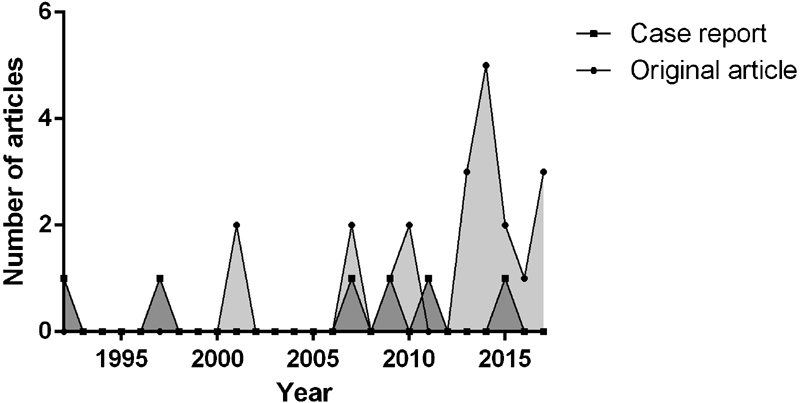
Number of original articles and case reports published per year. The majority of articles (85%) were published within the last 11 years.
Articles represented cohorts from 10 different countries, in addition to the one multi-center study in Europe ( Fig. 2 ). The United States of America had the most articles published (11/27 = 40.74%). No study was from an indigenous African population. Cohort sizes for infants ranged from n = 1 to 292 infants (median: n = 30, standard deviation: ± 31.5).
Fig. 2.
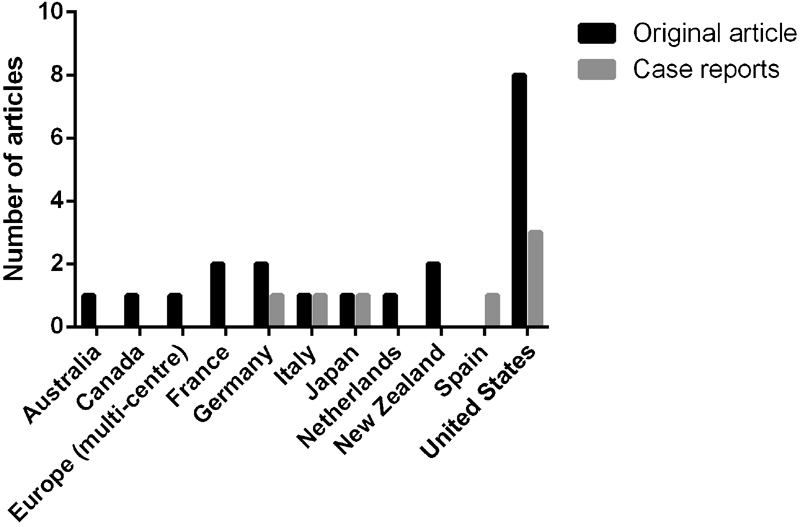
Number of articles published per country. A total of 41% of articles represented cohorts from the United States.
The scope of molecular analysis was between genotyping one variant to whole exome sequencing; however, the majority of studies (20/27 = 74%) researched 11 or fewer candidate genes by a variety of molecular methods. Two studies used whole exome sequencing (infants in cohort: 7–161), while four studies sequenced a panel of 23 genes or more using next generation sequencing technology (cohorts included 6 to 280 infants). In total, however, seven studies utilized next generation sequencing, and these were also the seven most recently published studies (2014–2017). When considering the studies that utilized next generation sequencing, a trend was seen in that—the smaller the cohort, the larger the percentage of potentially resolved cases ( R 2 = –0.7143); however, this was not statistically significant ( p = 0.0881).
When case reports were excluded, and the 20 “cohort” studies were analyzed, a mean of 8.6% (median: 4.0%) of sudden infant death cases were potentially resolved after a molecular autopsy. This was compared with the studies involving SUDY cohorts, over 1 year old, whereby a mean of 29.36% (median: 32.13%) of cases were potentially resolved following molecular autopsy ( Fig. 3 ). A Mann–Whitney test showed that older cohorts of SUDY (1–45 years old) were significantly more resolved by molecular autopsies compared with infants (0–1 years) ( p < 0.0001).
Fig. 3.
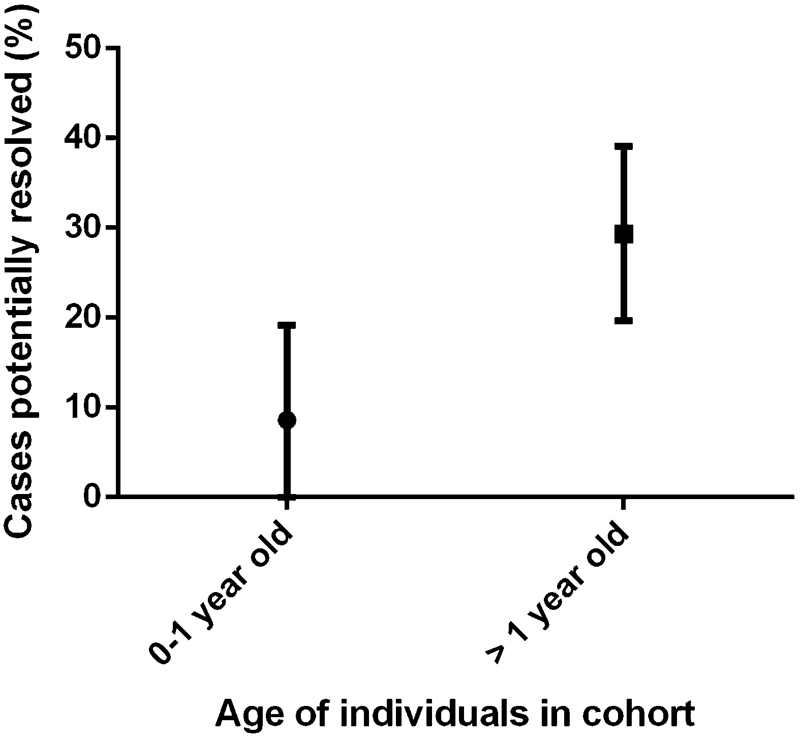
Percentages of cases potentially resolved following molecular autopsies, which was significantly different between infants (0–1 years old) and cohorts of “sudden unexpected death in the young” (1 to ∼45 years old).
Out of the seven case reports, one included the analysis of two infants, 36 while the remaining six each reported on one infant only. Researchers were successful at identifying pathogenic mutations, which probably contributed toward death, in the six single case reports. These mutations were involved in channelopathies (two case reports), metabolic disorders (three case reports), and autonomic nervous control (one case report). The case report including two infants was unsuccessful at identifying pathogenic mutations in four genes involved in channelopathies. 36
Of the 20 original articles, 12 identified putative pathogenic mutations in genes associated with channelopathies, 5 in cardiomyopathies, 4 in metabolic disorders, 2 in autonomic nervous control disorders, and 1 in mitochondrial DNA (mtDNA) disorders. The category of diseases with the highest “yield” of potentially resolved cases was that of channelopathies (6.7%) followed by metabolic disorders (5.08%), with the remaining three categories having a return of less than 1.6% resolution, each. Lastly, within these 20 articles, the number of genes investigated was significantly associated with the percentage of potentially resolved cases ( p = 0.0179) ( Fig. 4 ).
Fig. 4.
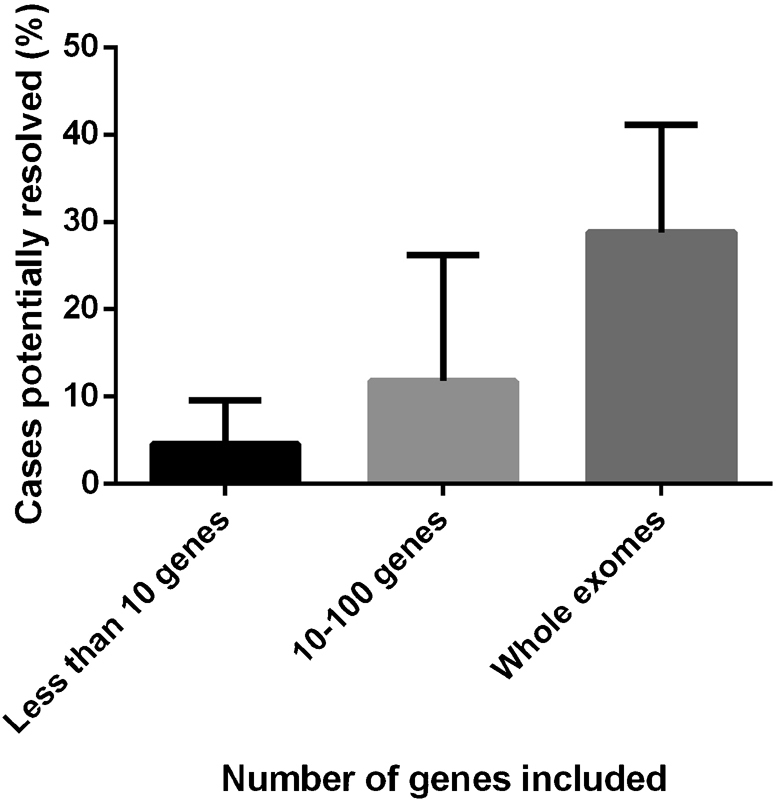
The percentage of cases potentially resolved by molecular autopsies increased as the number of genes analyzed increased. This is represented in this figure by means of categorizing the data; however, a Spearman rank sum test on the numerical data ( Table 1 ) showed that this association was statistically significant ( p = 0.0179).
Discussion
The role of genetic factors in human disease especially involving metabolic and cardiovascular diseases is well recognized. 38 The investigation of genetic factors to explain mortality is less well established; however, this trend is growing for metabolic and cardiovascular diseases. Some of these diseases do not have obvious physical/phenotypic manifestations, particularly in very young infants and, therefore, may go undetected at autopsy. However, such diseases can potentially be revealed with genetic analysis of the deceased individuals' DNA.
To date, many genes and variants have been identified as candidates for molecular autopsies as reviewed by various authors. 38 39 40 Some studies have targeted specific mutations in candidate genes, sometimes on a case-by-case basis, while other studies have screened cohorts for these mutations ( Table 1 ). In comparison, and more recently, a next generation sequencing approach has been used to identify new mutations which could underlie and perhaps explain sudden unexpected death cases.
One of the challenges in these larger scale studies, as with most next generation data, is the interpretation of rare variants, particularly those that are putatively pathogenic according to in silico analysis but lack functional and co-segregation studies to support the predicted pathogenicity of the variant. The value of co-segregation analysis was demonstrated by Campuzano et al 33 whereby the presence of rare variants in asymptomatic family members aided the exclusion of some variants as being causative of the infant's death and ultimately led to the identification of the variant likely related to the infant's sudden death. As Glengarry et al 28 noted, however, co-segregation studies are usually challenging to perform especially when the proband is an infant, due to difficulties in tracking families. The implication of this is two-fold; first, if a pathogenic variant is found which explains death, the information cannot be given to the family who is lost to follow up; and second, the lack of co-segregation analysis, together with the absence of functional studies, results in many variants being classified as “variants of unknown significance” (VUS), and their role within the sudden unexpected death cases remains speculative. Until functional data becomes available, the interpretation of the variant is purely hypothetical and does not warrant clinical testing in living relatives—that is, if family members could eventually be tracked, and they are willing to engage. For the studies that used next-generation sequencing in this review, only one out of seven studies performed co-segregation analysis for at least some of the cases. 11 Further, their reported percentage of “potentially resolved cases” was based on putatively pathogenic mutations, which means that the true percentage of actually resolved cases by molecular autopsy is likely to be lower than reported here.
A remarkable finding in this review was that significantly more cases were potentially resolved in older cohorts of SUDY compared with infants. This may have been a consequence of the specific mutations/genes investigated in the respective cohorts and suggests that there may be different mechanisms contributing toward death in the different age groups. For example, genes associated with channelopathies were investigated in the majority of studies in all age groups; however, cardiovascular diseases are probably more advanced and therefore of greater significance in older individuals, whereas other pathways may be more significant in infants, but were just not targeted during the molecular autopsy. Davis et al 41 also proposed that the interplay of genetic variants in long QT syndrome in SIDS cases is part of the vulnerable infant spectrum, and a paradigm shift is needed to fully appreciate the complex interplay of environmental factors. Therefore, while this review demonstrates the potential of molecular autopsies, particularly in the older cohorts, it also suggests that genes associated with other disease categories should possibly be targeted in infants, and environmental factors should also be considered. This notion was supported by Rueda et al 11 who showed that it might to worthwhile exploring variants in mtDNA, as four out of eight SIDS cases had an increased ratio of heteroplasmic variants. Out of the cohort studies included in this review, only the Scripps study investigated mtDNA variants and suggested that the systematic collation of data across multiple studies may provide the means to investigate this hypothesis with a larger cohort.
Tang et al 42 noted that although they were not the only cause of sudden unexpected death, cardiac channelopathies do play a significant role in such cases. Since 2008, the Molecular Genetics Laboratory of the New York City Office of the Chief Medical Examiner has been screening six candidate genes associated with cardiac channelopathies during molecular autopsy of cases of sudden unexplained death. Between 2008 and 2012, 274 sudden unexpected death cases with negative post-mortem investigation were screened for mutations in these candidate genes. A total of 46 genetic variants were detected (of which 24 were novel) in infants (13.5%) and non-infants (19.5%), most of which were in the sodium voltage-gated channel α subunit 5 ( SCN5A ) gene (which encodes for a part of the channel that allows sodium ions into cells). The cohort comprised individuals from diverse ethnic backgrounds, and the authors observed that the group with the highest risk of sudden unexpected death were African-American infants. 27 As observed in this review, however, there is a stark gap in forensic molecular autopsy data pertaining to indigenous Africans.
The finding in this review that significantly more infant cases were potentially resolved with the more genes investigated was not unexpected. Rueda et al 11 reviewed the data generated from the Scripps study ( n = 50; cohort 0–45 years) and analyzed the number of variants obtained following the bioinformatics pipeline on (1) the whole exome sequencing data and (2) a subset of the data, which was a simulated cardiac panel of 233 markers. They showed that there was a median of 354 variants per sample (range: 235–739) from the exome data, compared with a median of 7 per sample (range: 1–18) for the “panel” subset. This vast reduction of variants to consider within the case context is important for time and resource considerations; however, while time may be initially saved, there is also a chance that less cases would be resolved with a panel approach. The advantage of whole exome sequencing in this regard is that the filtering of subsets of genes from the larger dataset is possible, which may be the first step in data analysis; however, this is not true for data from a panel; as such, additional sequencing may need to be performed on molecular autopsy-negative cases. At this exploratory stage, especially in developing countries, it might make sense to strategically consider whole exome sequencing (ensuring good coverage), taking into account that one could resort to different modes of analysis from such data, i.e., looking at just component panels (e.g., channelopathies), while ultimately also having the capability to look for deleterious mutations across the exome. However, one must also consider that a molecular autopsy in SIDS cohorts is not yet a mature test, and relatively, whole exome sequencing is an expensive method to pursue. Recent case–control studies in 2018 (thus not included in systematic search) which used whole exome sequencing on large SIDS cohorts have also shown valuable but very slight differences between cases and controls. 43 44 Thus, whole exome sequencing should be reserved for large case–control cohorts or cases where families are available for co-segregation analysis.
As noted previously, molecular autopsies may not always have a high diagnostic yield in sudden expected death cases; 45 DNA from 59 Australian SUD cases underwent genetic testing of two genes encoding ion channels, and 23 cases showed at least one variation in these genes. However, none of them was deemed to be pathogenic. The authors suggested that a more selected approach should be adopted, and these genes should only be screened for those individuals who had a family history of cardiac channelopathy disorders. 45
This was demonstrated again in a combined “retrospective and prospective” study in New Zealand, where they too showed that a molecular autopsy of genes involved in long QT syndrome does not always yield significant results. 28 A total of 102 SIDS cases were included in the study, where 71 cases were “unselected,” and 31 cases were specially selected for a molecular autopsy subsequent to screening by a cardiac genetic service. Long QT syndrome genes were sequenced in these infants, and mutations were observed in 4% of unselected cases compared with 16% in selected cases. Their study showed that there was a significantly lower diagnostic yield among unselected SIDS cases compared with selected cases ( p < 0.05), and this may be attributed to other common risk factors in the unselected group, such as co-sleeping. 28
Overall, the role of molecular autopsies has important consequences for family members of the deceased victim. Apart from providing some sort of answer to the question of “what led to the death of my child?” the results of a molecular diagnosis have clinical value for relatives who may carry the same mutations; some of which are already clinically actionable. 6 12 This review has collated the results from previous molecular autopsy related studies and showed that a median of 4% of previous SIDS cases can potentially be resolved by a molecular autopsy. While this was significantly lower than the yield from older cohorts of SUDY, these results would have been of particular value to the respective family members. This review has also shown that channelopathies and metabolic disorders yielded the most positive results thus far in this emerging field, but other avenues, such as mtDNA, may be worthwhile to explore. It must be noted though, that a limitation of this study was that not all molecular autopsy studies might have been found using the systematic search methods described here.
While there is a growing momentum of genomics research in large cohorts of adults with various communicable and non-communicable diseases in Africa, 46 a special effort needs to be made toward developing large cohorts sudden unexpected deaths which remain unexplained. With the anticipated decrease in costs of genomic research and increased capacity in big data analysis and learning from the efforts in the developed world, we will be well-placed to investigate the genetics/genomics that potentially underlies sudden unexplained death. Moreover, we hope this will lead to the effective implementation of a molecular autopsy that will be locally or regionally relevant.
Footnotes
Conflict of Interest None.
References
- 1.Hauck F R, Tanabe K O. International trends in sudden infant death syndrome: stabilization of rates requires further action. Pediatrics. 2008;122(03):660–666. doi: 10.1542/peds.2007-0135. [DOI] [PubMed] [Google Scholar]
- 2.Byard R W, Krous H F. Sudden infant death syndrome: overview and update. Pediatr Dev Pathol. 2003;6(02):112–127. doi: 10.1007/s10024-002-0205-8. [DOI] [PubMed] [Google Scholar]
- 3.Krous H F, Beckwith J B, Byard R W et al. Sudden infant death syndrome and unclassified sudden infant deaths: a definitional and diagnostic approach. Pediatrics. 2004;114(01):234–238. doi: 10.1542/peds.114.1.234. [DOI] [PubMed] [Google Scholar]
- 4.Mazzanti A, Priori S G. Molecular autopsy for sudden unexplained death? Time to discuss pros and cons. J Cardiovasc Electrophysiol. 2012;23(10):1099–1102. doi: 10.1111/j.1540-8167.2012.02430.x. [DOI] [PubMed] [Google Scholar]
- 5.Baruteau A E, Tester D J, Kapplinger J D, Ackerman M J, Behr E R. Sudden infant death syndrome and inherited cardiac conditions. Nat Rev Cardiol. 2017;14(12):715–726. doi: 10.1038/nrcardio.2017.129. [DOI] [PubMed] [Google Scholar]
- 6.Anderson J H, Tester D J, Will M L, Ackerman M J. Whole-exome molecular autopsy after exertion-related sudden unexplained death in the young. Circ Cardiovasc Genet. 2016;9(03):259–265. doi: 10.1161/CIRCGENETICS.115.001370. [DOI] [PubMed] [Google Scholar]
- 7.Hertz C L, Christiansen S L, Ferrero-Miliani L et al. Next-generation sequencing of 34 genes in sudden unexplained death victims in forensics and in patients with channelopathic cardiac diseases. Int J Legal Med. 2015;129(04):793–800. doi: 10.1007/s00414-014-1105-y. [DOI] [PubMed] [Google Scholar]
- 8.Ackerman M J. State of postmortem genetic testing known as the cardiac channel molecular autopsy in the forensic evaluation of unexplained sudden cardiac death in the young. Pacing Clin Electrophysiol. 2009;32 02:S86–S89. doi: 10.1111/j.1540-8159.2009.02393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloss C S, Zeeland A A, Topol S E et al. A genome sequencing program for novel undiagnosed diseases. Genet Med. 2015;17(12):995–1001. doi: 10.1038/gim.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torkamani A, Muse E D, Spencer E G et al. Molecular autopsy for sudden unexpected death. JAMA. 2016;316(14):1492–1494. doi: 10.1001/jama.2016.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rueda M, Wagner J L, Phillips T C et al. Molecular autopsy for sudden death in the young: is data aggregation the key? Front Cardiovasc Med. 2017;4:1–16. doi: 10.3389/fcvm.2017.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewar L J, Alcaide M, Fornika D et al. Investigating the genetic causes of sudden unexpected death in children through targeted next-generation sequencing analysis. Circ Cardiovasc Genet. 2017;10(04):1–9. doi: 10.1161/CIRCGENETICS.116.001738. [DOI] [PubMed] [Google Scholar]
- 13.Casale V, Oneda R, Matturri L, Lavezzi A M. Investigation of 5-HTT expression using quantitative real-time PCR in the human brain in SIDS Italian cases. Exp Mol Pathol. 2013;94(01):239–242. doi: 10.1016/j.yexmp.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto T, Tanaka H, Kobayashi H et al. Retrospective review of Japanese sudden unexpected death in infancy: the importance of metabolic autopsy and expanded newborn screening. Mol Genet Metab. 2011;102(04):399–406. doi: 10.1016/j.ymgme.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Tan B H, Pundi K N, Van Norstrand D W et al. Sudden infant death syndrome-associated mutations in the sodium channel beta subunits. Heart Rhythm. 2010;7(06):771–778. doi: 10.1016/j.hrthm.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gladding P A, Evans C A, Crawford J et al. Posthumous diagnosis of long QT syndrome from neonatal screening cards. Heart Rhythm. 2010;7(04):481–486. doi: 10.1016/j.hrthm.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 17.Millat G, Kugener B, Chevalier P et al. Contribution of long-QT syndrome genetic variants in sudden infant death syndrome. Pediatr Cardiol. 2009;30(04):502–509. doi: 10.1007/s00246-009-9417-2. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Lantz P E, Ibdah J A. Post-mortem analysis for two prevalent beta-oxidation mutations in sudden infant death. Pediatr Int. 2007;49(06):883–887. doi: 10.1111/j.1442-200X.2007.02478.x. [DOI] [PubMed] [Google Scholar]
- 19.Van Norstrand D W, Valdivia C R, Tester D J et al. Molecular and functional characterization of novel glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) mutations in sudden infant death syndrome. Circulation. 2007;116(20):2253–2259. doi: 10.1161/CIRCULATIONAHA.107.704627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiehne N, Kauferstein S. Mutations in the SCN5A gene: evidence for a link between long QT syndrome and sudden death? Forensic Sci Int Genet. 2007;1(02):170–174. doi: 10.1016/j.fsigen.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Ackerman M J, Siu B L, Sturner W Q et al. Postmortem molecular analysis of SCN5A defects in sudden infant death syndrome. JAMA. 2001;286(18):2264–2269. doi: 10.1001/jama.286.18.2264. [DOI] [PubMed] [Google Scholar]
- 22.Miller M E, Brooks J G, Forbes N, Insel R.Frequency of medium-chain acyl-CoA dehydrogenase deficiency G-985 mutation in sudden infant death syndrome Pediatr Res 199231(4 Pt 1):305–307. [DOI] [PubMed] [Google Scholar]
- 23.Neubauer J, Lecca M R, Russo G et al. Post-mortem whole-exome analysis in a large sudden infant death syndrome cohort with a focus on cardiovascular and metabolic genetic diseases. Eur J Hum Genet. 2017;25(04):404–409. doi: 10.1038/ejhg.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Methner D NR, Scherer S E, Welch K et al. Postmortem genetic screening for the identification, verification, and reporting of genetic variants contributing to the sudden death of the young. Genome Res. 2016;26(09):1170–1177. doi: 10.1101/gr.195800.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santori M, Blanco-Verea A, Gil R et al. Broad-based molecular autopsy: a potential tool to investigate the involvement of subtle cardiac conditions in sudden unexpected death in infancy and early childhood. Arch Dis Child. 2015;100(10):952–956. doi: 10.1136/archdischild-2015-308200. [DOI] [PubMed] [Google Scholar]
- 26.Farrugia A, Keyser C, Hollard C, Raul J S, Muller J, Ludes B. Targeted next generation sequencing application in cardiac channelopathies: analysis of a cohort of autopsy-negative sudden unexplained deaths. Forensic Sci Int. 2015;254:5–11. doi: 10.1016/j.forsciint.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 27.Wang D, Shah K R, Um S Y et al. Cardiac channelopathy testing in 274 ethnically diverse sudden unexplained deaths. Forensic Sci Int. 2014;237:90–99. doi: 10.1016/j.forsciint.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Glengarry J M, Crawford J, Morrow P L, Stables S R, Love D R, Skinner J R. Long QT molecular autopsy in sudden infant death syndrome. Arch Dis Child. 2014;99(07):635–640. doi: 10.1136/archdischild-2013-305331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liebrechts-Akkerman G, Liu F, Lao O et al. PHOX2B polyalanine repeat length is associated with sudden infant death syndrome and unclassified sudden infant death in the Dutch population. Int J Legal Med. 2014;128(04):621–629. doi: 10.1007/s00414-013-0962-0. [DOI] [PubMed] [Google Scholar]
- 30.Evans A, Bagnall R D, Duflou J, Semsarian C. Postmortem review and genetic analysis in sudden infant death syndrome: an 11-year review. Hum Pathol. 2013;44(09):1730–1736. doi: 10.1016/j.humpath.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 31.Lopez H U, Haverfield E, Chung W K. Whole-exome sequencing reveals CLCNKB mutations in a case of sudden unexpected infant death. Pediatr Dev Pathol. 2015;18(04):324–326. doi: 10.2350/14-08-1543-CR.1. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi Y, Sano R, Nakajima T et al. Combination of postmortem mass spectrometry imaging and genetic analysis reveals very long-chain acyl-CoA dehydrogenase deficiency in a case of infant death with liver steatosis. Forensic Sci Int. 2014;244:e34–e37. doi: 10.1016/j.forsciint.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 33.Campuzano O, Allegue C, Sarquella-Brugada G et al. The role of clinical, genetic and segregation evaluation in sudden infant death. Forensic Sci Int. 2014;242:9–15. doi: 10.1016/j.forsciint.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Mecchia D, Casale V, Oneda R, Matturri L, Lavezzi A M. Sudden death of an infant with cardiac, nervous system and genetic involvement--a case report. Diagn Pathol. 2013;8(159):159. doi: 10.1186/1746-1596-8-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manoukian A A, Ha C E, Seaver L H, Bhagavan N V. A neonatal death due to medium-chain acyl-CoA dehydrogenase deficiency: utilization of the neonatal metabolic screen in a functional approach to sudden unexplained infant death. Am J Forensic Med Pathol. 2009;30(03):284–286. doi: 10.1097/PAF.0b013e318187e09b. [DOI] [PubMed] [Google Scholar]
- 36.Bajanowski T, Rossi L, Biondo Bet al. Prolonged QT interval and sudden infant death--report of two cases Forensic Sci Int 2001115(1-2):147–153. [DOI] [PubMed] [Google Scholar]
- 37.el-Schahawi M, Bruno C, Tsujino S et al. Sudden infant death syndrome (SIDS) in a family with myophosphorylase deficiency. Neuromuscul Disord. 1997;7(02):81–83. doi: 10.1016/s0960-8966(97)00424-0. [DOI] [PubMed] [Google Scholar]
- 38.Opdal S H, Rognum T O. The sudden infant death syndrome gene: does it exist? Pediatrics. 2004;114(04):e506–e512. doi: 10.1542/peds.2004-0683. [DOI] [PubMed] [Google Scholar]
- 39.Opdal S H, Rognum T O. Gene variants predisposing to SIDS: current knowledge. Forensic Sci Med Pathol. 2011;7(01):26–36. doi: 10.1007/s12024-010-9182-9. [DOI] [PubMed] [Google Scholar]
- 40.Sarquella-Brugada G, Campuzano O, Cesar S et al. Sudden infant death syndrome caused by cardiac arrhythmias: only a matter of genes encoding ion channels? Int J Legal Med. 2016;130(02):415–420. doi: 10.1007/s00414-016-1330-7. [DOI] [PubMed] [Google Scholar]
- 41.Davis A M, Glengarry J, Skinner J R. Sudden infant death: QT or not QT? That is no longer the question. Circ Arrhythm Electrophysiol. 2016;9(06):1–6. doi: 10.1161/CIRCEP.115.003859. [DOI] [PubMed] [Google Scholar]
- 42.Tang Y, Stahl-Herz J, Sampson B A. Molecular diagnostics of cardiovascular diseases in sudden unexplained death. Cardiovasc Pathol. 2014;23(01):1–4. doi: 10.1016/j.carpath.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Männikkö R, Wong L, Tester D Jet al. Dysfunction of NaV1.4, a skeletal muscle voltage-gated sodium channel, in sudden infant death syndrome: a case-control study Lancet 2018391(10129):1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tester D J, Wong L CH, Chanana P et al. Cardiac genetic predisposition in sudden infant death syndrome. J Am Coll Cardiol. 2018;71(11):1217–1227. doi: 10.1016/j.jacc.2018.01.030. [DOI] [PubMed] [Google Scholar]
- 45.Doolan A, Langlois N, Chiu C, Ingles J, Lind J M, Semsarian C. Postmortem molecular analysis of KCNQ1 and SCN5A genes in sudden unexplained death in young Australians. Int J Cardiol. 2008;127(01):138–141. doi: 10.1016/j.ijcard.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Rotimi C, Abayomi A, Abimiku Aet al. Research capacity. Enabling the genomic revolution in Africa Science 2014344(6190):1346–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]


