Abstract
It has been two decades since the lipid raft hypothesis was first presented. Even today, whether these nanoscale cholesterol-rich domains are present in cell membranes is not completely resolved. However, especially in the last few years, a rich body of literature has demonstrated both the presence and the importance of non-random distribution of biomolecules on the membrane, which is the focus of this review. These new developments have pushed the experimental limits of detection and have brought us closer to observing lipid domains in the plasma membrane of live cells. Characterization of biomolecules associated with lipid rafts has revealed a deep connection between biological regulation and function and membrane compositional heterogeneities. Finally, tantalizing new developments in the field have demonstrated that lipid domains might not just be associated with the plasma membrane of eukaryotes but could potentially be a ubiquitous membrane-organizing principle in several other biological systems.
Keywords: Lipid rafts, membrane domains, super-resolution fluorescence microscopy, immune receptor clustering, HIV, bacterial toxins
Introduction1
Spatiotemporal organization of molecules in cells is increasingly thought to play an important role in biological regulation [1]. A ubiquitous method by which cells utilize such spatiotemporal organization to bring about regulation is through clustering of biomolecules [2]. One of the best studied examples of how cells exploit clustering as a mechanism to functionally organize biomolecules is by patterning membrane proteins and lipids to form micro- and nano-domains in cell membranes [3–5]. It is now well recognized that cell membranes contain a variety of such domains and that these domains impose a local structure that prevents free mixing of the lipid and protein components of the membrane [6].
A major paradigm that has shaped much of our thinking about domains in cell membranes is the lipid (membrane) raft model [7]. Given the diversity in the interpretation of membrane rafts, we have utilized a traditional meaning of raft as cholesterol-rich, nanoscale heterogeneity in cell membranes formed as a consequence of liquid-liquid phase separation of lipids as the working definition in this review. This model, while recognized as over-simplified [8, 9], has provided a useful testbed for generating and testing hypothesis regarding the underlying mechanisms that control the size and shape of domains in the two-dimensional environment of cell membranes. Given the notorious difficulty in detecting raft domains in cells, the raft model has also driven advances in technology to develop techniques capable of imaging biomolecules with increasing spatial and temporal resolution, as well as new methodologies to label those molecules to enable their detection.
In this review, we highlight recent advances in the study of rafts as a pattern generation mechanism in cell membranes. First, we provide an overview of current optical microscopy-based methods and probes used to detect rafts in cells. Next, we describe new conceptual advances regarding the functional consequences of such patterning. Specific examples we discuss include how rafts function to compartmentalize cellular signaling, mechanisms responsible for targeting transmembrane proteins to rafts, and how pathogens exploit rafts to gain entry into cells. Finally, we discuss newly discovered sites where membrane domains form and areas for future exploration such as the relationship between membrane geometry and compositional heterogeneity.
Technical advances in optically detecting membrane nanodomains in cell membranes
Several important properties of rafts can be studied in model membrane systems using conventional microscopy-based approaches. For example, the phase preference of membrane proteins and lipids can be directly imaged in giant plasma membrane vesicles (GPMVs), which form micron-scale coexisting raft and non-raft domains [10]. However, current wisdom holds that in living cells rafts are highly dynamic and have sizes on the orders of tens of nanometers [11]. These spatial and temporal constraints have made it difficult to study them using conventional microscopy methods. In the last decade, there have been several developments in experimental techniques that have enabled a better understanding of native membrane organization by addressing both these bottlenecks.
Stochastic localization and single molecule techniques
The raft model predicts that raft-associated proteins and lipids should cluster together to form nanodomains that exclude non-raft proteins. However, since these domains are diffraction-limited, they cannot be resolved using regular fluorescence imaging methods. Techniques such as Förster Resonance Energy Transfer (FRET) have been used previously to probe the distribution of raft-associated proteins and lipids over distances of 100Å or less [12]. However, a limitation of FRET-based approaches is that they report on distances and not distributions of molecules per se, necessitating the use of theoretical modeling to interpret their meaning [12].
In recent years, super-resolution methods have advanced our understanding of several biological systems including plasma membranes organization by their ability to probe past the diffraction limit. One approach to super-resolution microscopy, the use of stochastic localization of fluorophores, has become a powerful tool to probe for the presence of different types of local heterogeneities in cell membranes (Figure 1). In these techniques, the different fluorophores which are spatially localized together within a diffraction limited spot are individually isolated temporally and various reconstruction methods are used to fit isolated emitters [13–15]. Typically, these methods can achieve localization to within tens of nanometers while imaging live cells [16]. These super-resolution methods have been used to describe aspects of several membrane organization including how raft associated immune-receptors cluster and signal [17–19], viral protein-membrane interaction [20, 21] and growth factor receptor organization [22, 23]. Importantly, several analysis methods are being simultaneously developed to understand the organization in a reconstructed image [24–27]. Clustering tools are now increasingly complex and can describe even the three-dimensional organization of molecules in a cell and not just in the membrane [28, 29]. Further, a number of analysis tools for super-resolution are now available free to users [28, 30–32]. Despite these advances in spatial prowess, a major drawback in these experiments is that the temporal resolution of the technique is still a few seconds, which tends to average many transient clustering events [24].
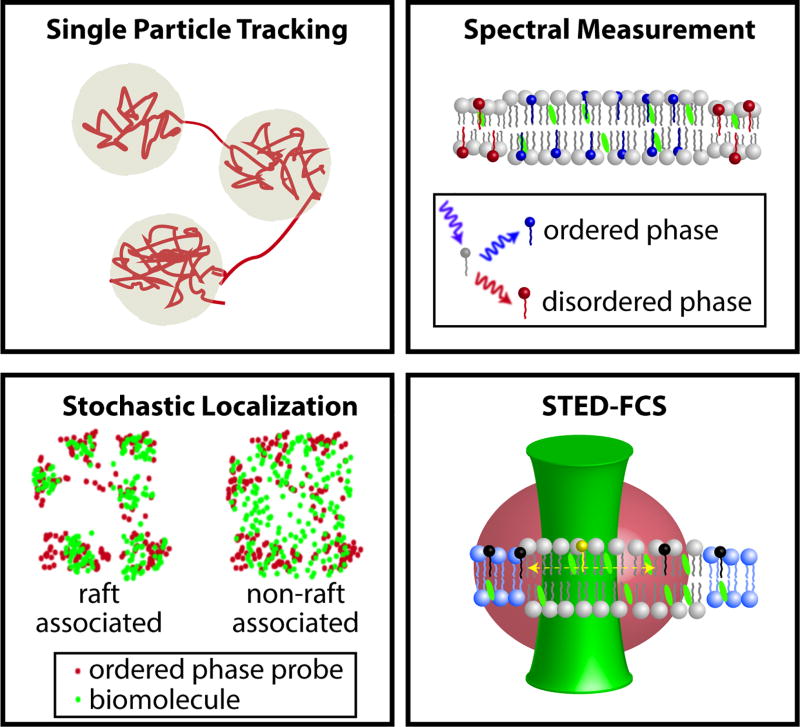
Figure 1. Examples of optical methods to probe membrane heterogeneity in cells.
(Upper left) High-speed single particle tracking is commonly used to detect regions in membrane where anomalous diffusion occurs. (Upper right) Spectral imaging of membrane order utilizes lipid probes whose fluorescence emission are sensitive to the local membrane environment. A typical spectral probe is red-shifted in the disordered phase and is blue-shifted in ordered phase. Thus, by spectrally delineating the emission spectra, it is possible to detect ordered raft-like region in cell membranes. (Bottom left) In stochastic localization methods, the membrane clustering of the biomolecule of interest is determined relative to the distribution of a known (dis)ordered phase preferring probe. If the molecule co-clusters with the ordered phase probe, it is raft associated. (Bottom right) STED-FCS measures the diffusion of membrane-associated proteins and lipids through a sub-diffraction focal volume. In the presence of membrane heterogeneity, their diffusion time through the beam varies as a function of the beam waist diameter. By determining this relationship, the underlying molecular architecture of the membrane can be deciphered.
Ameliorating these deficiencies in super-resolution methods are complementary developments in live cell single molecule methods. These high-speed experiments have been used to study of diffusion of lipid probes in membrane domains and confinement of molecules in membrane in the millisecond timescale [33–35]. These approaches allow for the interrogation of clustering events that occur over very fast timescales as well as the identification of underlying features of membrane architecture that slow or transiently halt the diffusion of proteins and lipids in the plane of the membrane (Figure 1). Advances in this area have been closely coupled to the development of fluorescent analogs of physiologically relevant lipids. This has represented a significant challenge to the field since the presence of fluorophores can modify the properties of lipids [36]. In recent years, the development of high-quality lipid probes has facilitated high spatiotemporal studies in lipid and protein dynamics [36–40]. These studies have provided information on the residency and co-diffusion timescales of different biomolecules in membrane. Further, the trajectories of single molecule measurements reveal the underlying characteristics of molecular motion. For instance, the trajectory of a molecule undergoing hop-diffusion is different from the trajectory of a molecule undergoing confined diffusion [41].
Of specific importance in understanding membrane organization has been the development of novel sphingomyelin and ganglioside probes used to measure lipid-lipid and lipid-protein interactions in diverse model systems from artificial membranes to high-speed live cell experiments [36, 37, 39]. These experiments have demonstrated evidence of transient recruitment of sphingomyelin, gangliosides to CD59-rich plasma membrane domains in cells. These studies have shown that the co-diffusion timescales are on the order of hundreds of milliseconds, demonstrating the importance of the sensitivity of the experimental technique. Understanding dynamics of lipids is still a nascent field and combining these single-molecule tracking experiments with other complementary super-resolution techniques will be a powerful tool to understand lipid dynamics and behavior.
Fluorescent correlation measurements
Another general approach for studying diffusion characteristics of fluorescent molecules and how membrane heterogeneities affect diffusion is fluorescence correlation spectroscopy. Fluorescence correlation measurements determine how the fluorescence in a diffraction limited spot or volume fluctuates [42, 43]. The fluctuations in the signal are dependent on the diffusion of fluorescent molecules in and out of the observation volume and are used to generate correlation functions which are then related to the diffusion of the molecule, usually through analytical or semi-analytical methods. While correlation experiments are relatively old techniques with fluorescent correlation spectroscopy (FCS) being developed close to half a century ago (reviewed in [44]), new developments in the field have enabled application of this technique to the study of lipid rafts in cells. The advantage compared to other techniques such as STORM is that the temporal resolution is in the milliseconds range. Though typically performed as a spectroscopic technique in a confocal volume, correlation measurements are often combined with other methods to extend the capabilities of the system. Variants of FCS techniques such as brightness analysis methods, Total internal Reflection FCS, scanning FCS, Fluorescence cross correlation spectroscopy, and inverse FCS, have all been developed on the backbone of traditional FCS methods and have extended its capabilities [45–49].
One of the most exciting development in this field is the development of spot variation FCS (svFCS) [50]. Here, diffusion coefficients are measured for different illumination volumes. While the principle of svFCS was established over 10 years ago [42, 51], the development of stimulated emission depletion (STED) sub-resolution illumination methods have now moved svFCS past the diffraction limit, thereby tremendously improving its use [52]. In STED, by combining two lasers, one to excite and other to deplete, and with clever spatial arrangement of the two laser pulses, it is possible to selectively excite a sub-diffraction volume. The variation of the diffusion coefficient as a function of beam diameter is characteristic of membrane confinement behavior [53]. Thus, using STED-FCS it is potentially possible to distinguish the underlying biological mechanism behind confinement of biomolecules (Figure 1). STED-FCS has been used to show that sphingolipids and GPI-anchored proteins do not diffuse freely in the plasma membrane, while phosphoglycerol lipids do [54–56]. Further, it has been used to demonstrate that both actin and lipid nanodomains contribute to the compartmentalization of lipids in the plasma membrane [57].
An important caution while interpreting FCS measurements is to understand the caveats in data interpretation. For instance, using Ising model simulations, Veatch and colleagues showed that incorrect assumptions in svFCS experiments can result in erroneous interpretation of diffusion behavior as anomalous diffusion when in reality it could still be Brownian [58]. Thus, it is important to reconcile the results of FCS with other methods, especially SPT [59], to ensure that complementary techniques demonstrate consistent results.
Development of novel chemical tools to study lipids in membranes
As previously noted, a critical component for the use of fluorescent microscopy in studying lipid domains is the development of lipid-specific fluorophore probes. The development of specific sensors for different lipids has received considerable attention. Click chemistry has been used to generate novel ceramide probes which can be used in super-resolution studies [38]. Naturally derived proteins and peptides with binding propensity for specific lipids have been retooled as lipid sensors. Perfringolysin O (PFO) secreted by Clostridium perfringens is the most studied of these and has been used for sensing cholesterol [60]. In a recent study, PFO was modified to probe the transbilayer distribution of cholesterol on membrane bilayers [61]. Other proteins have been isolated from different organisms that bind either selectively or non-selectively to different lipids. Lysenin, a protein isolated from the earthworm Eisenia foetida, binds to sphingomyelin with high specificity [62]. On the other end of the spectrum, Nakanori, a protein recently isolated from a non-edible mushroom, was found to bind to both cholesterol and sphingomyelin [63]. Many of these proteins can be fluorescently conjugated and thereby are thus extremely useful in studying various membrane properties.
An important factor to be kept in mind in the use of lipid probes is that specificity does not imply uniqueness. Further, it is important to understand the effects of the probe on membrane rafts. A classic example is cholera toxin B (CTxB), a widely utilized lipid raft marker in cellular membranes. While CTxB is an indicator of ordered phase, it also induces phase separation and increases membrane order [64, 65]. Also, the phase partitioning of GM1 can influence CTxB binding to rafts [66]. Further, besides GM1, CTxB is capable of binding to glycoproteins [67]. Thus, it is important to understand the strengths and weaknesses of a particular lipid probe, especially when using single molecule and super-resolution methods. Also, no protein is completely raft associated or completely non-raft associated. Even model raft-associated proteins such as CTxB do not partition entirely into the ordered phase. Thus, a critical control for determining raft partitioning in cells is to use to two distinct control probes, one for the ordered phase and other for the disordered phase. A lipid raft probe should not only be raft-associated but also should be significantly depleted from the non-raft region. However, currently, there are not many good probes which partition into the ordered phase. Development of ordered phase preferring probes is an important area of future research [68].
Spectral Imaging
A prediction of raft hypothesis is the presence of nanoscale regions of altered membrane order in plasma membrane of cells. Microscopy can be used to detect the presence of increased membrane order in live cells the help of fluorescent probes whose emission spectra depend on the local environment (Figure 1). While most of the previously described optical techniques probe membrane heterogeneity through the lens of a few well-characterized membrane biomolecules, spectral microscopy has the ability to determine the properties of the membrane as a whole [69]. In spectral imaging, a lipid probe whose fluorescence spectrum is dependent on its local environment is added exogenously to the membrane system being studied so as to report the membrane order. The most well characterized of these dyes is laurdan, which absorbs in the UV range and has a blue-shifted spectral emission with increasing membrane order [70]. However, laurdan suffers from two limitations: a) the dye bleaches rapidly and b) the separation of two emission wavelengths corresponding to ordered and disordered phase (430–470 and 480–530) is not ideal for measurement in many confocal systems [71]. A derivative of laurdan known as C-laurdan with similar absorption/emission characteristics but less photobleaching has been developed [71, 72]. Another dye that is increasingly used is the commercially available ANEPPDHQ. This dye absorbs at 488nm and has emission spectra corresponding to 488 and Cy3 channels depending on membrane order. It thus can be used in most confocal microscopes with standard filters [73]. New probes are continuing to be developed with different spectral properties, enhanced photophysics and increased sensitivity to membrane order [74–76]. Currently, spectral imaging has been used to characterize gross membrane order changes [69]. It would be interesting to determine how membrane order changes when a specific protein binds. For these experiments it becomes imperative to use spectral probes along with fluorescent protein tags. C-Laurdan for instance can be used in conjunction with proteins conjugated with red tags such as mKate or mCherry to determine how binding of protein affects membrane order [25]. However, there is a need for developing environment sensitive red dyes which can be used in conjunction with GFP, for better simultaneous imaging of membrane order and protein binding.
Another bottleneck with spectral imaging had been the spatial resolution of the technique, which until recently was diffraction limited. More recently, spectral imaging has been combined with super-resolution methods to detect domains on the order of tens of nanometers in size [77, 78]. Using this technique, nanometer-scale spatially resolved domains in live cell membranes were observed. Spectral super-resolution imaging is still in its infancy and has the potential to directly visualize membrane rafts.
While we focus on the fluorescent experimental methods in this review, other non-fluorescent methods have also enabled substantial contributions in our understanding of membrane domain organization. Detergent Resistant Membrane (DRM), the traditional method of understanding of studying rafts, has largely been sidelined as the principal experimental method to determine whether a protein is targeted to ordered phase [7]. However, many biochemical experiments can now be combined with other experimental methods as microscopy or novel probes have been used to understand biological role for different lipids. For instance, biochemical methods were used to show that there are three pools of cholesterol that are important in regulating cholesterol levels in the plasma membrane [79]. This biochemical work has since been complemented using an emerging high resolution technique, NanoSIMS, where the authors have shown that accessible cholesterol is enriched in microvilli in Chinese hamster ovary cells [80]. Similarly, atomic force microscopy, a well-characterized experimental method to determine membrane properties has been combined with emerging methods such as STED-FCS, mass-spectrometry and X-ray techniques which has greatly expanded the applicability of each of the different methods [81, 82]. Other novel non-fluorescent methods such as high-resolution mass spectrometry, interferometric scattering microscopy, Raman microscopy have gained prominence in determining various membrane properties and detecting lipid clusters although given the specialized nature of these methods, wide use of these methods has currently been rather limited [83–88]. High resolution mass spectrometry methods have suggested for example that hemaggluthin, an influenza envelope protein which is known to cluster in plasma membrane, does not localize to cholesterol and sphingolipid enriched domains at a resolution of around 50 nm [85]. More recently, this technique has been expanded for three-dimensional imaging [89]. Other traditional methods such as EPR, NMR, x-ray and neutron scattering, though still not popular with biologists in the field are used extensively in determining various membrane properties including order in model membranes [90–92]. Classical approaches such as electron microscopy and its variants also remain useful to detect clustering of membrane-associated molecules [93–96]. Use of electron microscopy methods for studying lipid raft field has resurged in part due to its increasing use in studies of bacterial rafts [97–99].
Besides experimental methods, in-silico simulations have been used to study membrane properties inaccessible to experiments [100, 101]. For instance, molecular dynamics have been used look at chain lengths and how coupling between the bilayer affects membrane organization [102]. In recent years, several developments have emerged in computational approach for studying complex membranes, including development of better forcefields and increasing complexity of the lipids that are simulated [103–105]. Recently model membranes with more than 50 types of lipids have been simulated to look at domain formation [106, 107]. Finally, in recent years, in a growing number of studies computer simulations have been used to interpret and validate experimental results [58, 108]. Discussing all these are beyond the scope of this review and we would like to point an interested reader to several reviews that dwell more deeply into in silico methods for understanding membrane organization [109–113].
New insights into the role of rafts as spatial organizers in signaling and trafficking events
In addition to advances in technology that can be used to interrogate membrane architecture, our understanding of functional properties of raft-like domains has also continued to evolve over recent years. Lipid rafts have long been proposed to functionally segregate proteins and lipids within different local membrane compartments, thus regulating their interactions. Here we discuss recent advances in our understanding of rafts in compartmentalization of signaling and trafficking events.
Role of stabilized rafts in BCR signaling
A classic example of how clustering and crosslinking of proteins by rafts is linked to functional outcomes is the activation of immune receptors in cell membranes. Initial links between the activation of immune cell receptors and lipid rafts were established through the isolation of detergent-resistant membranes, cholesterol depletion, and biophysical approaches [114–116]. In recent years, single-molecule microscopy and super-resolution microscopy have shed detailed light on both the temporal and spatial activation of immune receptors in cells and how lipid domains play a role in these processes [29, 117, 118].
A recent example of the role of rafts in compartmentalizing immune cell signaling comes from studies of B-Cell Receptor (BCR) activation using super-resolution techniques [17]. To identify nanoscopic raft and non-raft domains in cells membranes, model peptides and proteins known to preferentially associate with either raft (ordered) or non-raft (disordered) domains in model membrane systems were used as markers. For instance, the well-established raft marker CTxB was found in distinct clusters which excluded a peptide with a polybasic sequence and geranylgeranyl modification that prefers the disordered phase. The size of the ordered phase clusters was found to be around 50nm, close to the resolution limit of the method. These findings suggest that phase-like domains exist in intact B-cell membranes.
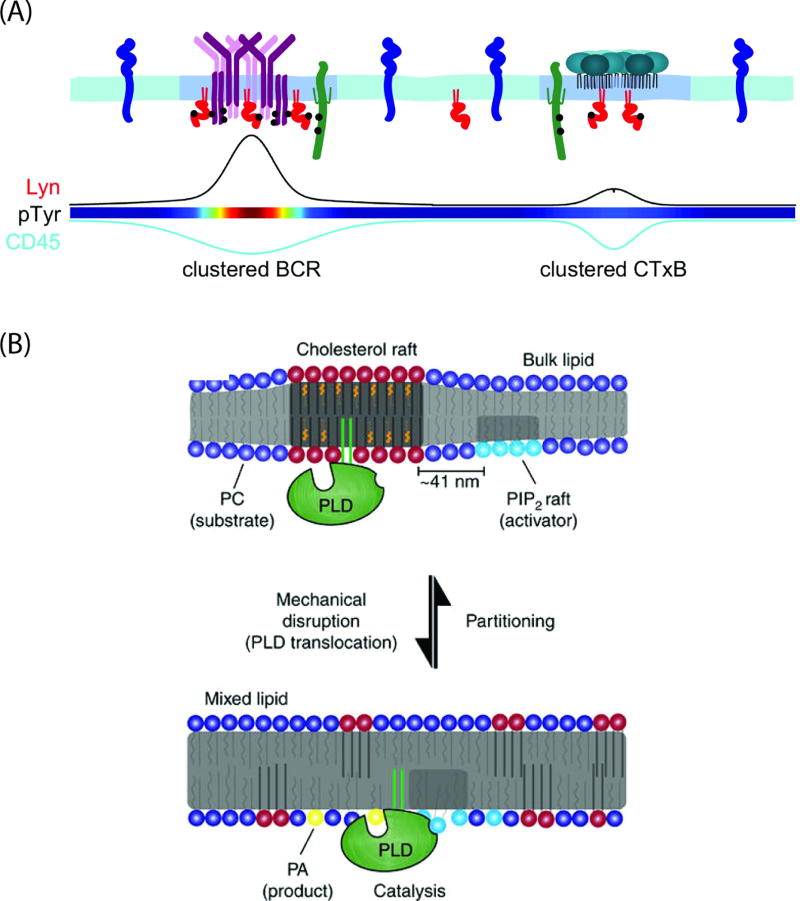
Using these markers to differentiate raft and non-raft domains, the authors next asked whether the BCR was enriched in pre-existing ordered domains. In the absence of stimulation, BCR was only weakly associated with ordered domains. However, upon clustering with an antigen, BCR clusters become enriched in ordered phase probes while excluding disordered phase probes. This suggests that BCR clustering stabilizes an ordered membrane domain rather than moving into existing rafts in response to crosslinking. Interestingly, the ordered domains extended beyond the BCR clusters themselves, indicating additional signaling complexes may be assembled close to the BCR clusters. Lyn kinase, a regulator of early BCR signaling, was recruited into the ordered domains stabilized by BCR signaling, whereas the phosphatase CD45, another important regulator of BCR activation, was excluded (Figure 2A). Computational modeling demonstrated the stabilization of a raft phase by BCR crosslinking and corresponding exclusion of non-raft signaling partners can give rise to local phosphorylation of proteins, a result that was also observed experimentally. These experiments demonstrate raft stabilization induced by receptor crosslinking can functionally sequester receptors and their effectors in cell membranes [17]. It will be important to determine whether this represents a general mechanism utilized by other raft-dependent signaling pathways in future studies.
Figure 2. Role of lipid domains in signaling.
(A) Model highlighting the role of ordered phase domains in downstream signaling of BCR. Clustered BCR behaves similar to clustered CTxB in its preference for partitioning with ordered phase probes in cell membranes. These clusters also recruit kinases such as Lyn which enable phosphorylation of effector molecules and exclude CD45, a phosphatase. The curves on the bottom illustrate the level of enrichment or depletion of the indicated proteins from the clustered BCR and clustered CTxB domains. Figure is adapted from reference [17] (Creative Commons Attribution License). (B) Model for how mechanical force induces PLD2 signaling. In resting cells, PLD2 is localized in rafts, which sequester it away from its substrate PC and activator PIP2. Mechanical disruption causes disruption of rafts, thereby increasing access of PLD2 to both PC and PIP2. Figure is adapted from reference [119] (Creative Commons Attribution 4.0 International License).
Impact of controlled disruption of rafts on mechanotransduction
If rafts function to compartmentalize signaling, then disruption of rafts should enable molecules that are otherwise segregated from one another to interact. An interesting example of how disruption of rafts regulate cellular events comes from a recent study using fast super-resolution imaging to investigate the role of rafts in mechanosensation [119]. The focus of this study was phospholipase D2 (PLD2), an enzyme whose activation is important for the transduction of force in muscle and cell growth. PLD2 is dually palmitoylated [120] and is predicted to associate with rafts. Consistent with this, PLD2 was shown to partially localize to domains enriched in the raft marker CTxB in a cholesterol-dependent manner. These domains were distinct from those occupied by phosphatidylinositol 4,5-bisphosphate (PIP2), which is known to activate PLD2 [121]. This leads to the interesting question of how PLD2 can interact either with PIP2 or its substrate phosphatidylcholine, which also presumably resides primarily outside of rafts. One mechanism by which this could be accomplished is through the dissolution of PLD2-containing rafts. Consistent with this, cholesterol depletion led to the release of PLD2 from CTxB-containing rafts and increased its proximity to PIP2. To test whether mechanical force might have a similar effect, PLD2 activation was monitored using a fluorescent product release assay. Strikingly, either mechanical shear or cholesterol depletion was found to lead to activation of PLD2, and combining the two treatments had a synergistic effect.
These findings suggest an intriguing mechanism by which mechanical forces are transduced in cell membranes [119]. In this model, shear force disrupts rafts, thereby liberating PLD2 from a raft-confined localization and allowing it access to both its substrate and activator (Figure 2B). Thus, this model proposes force-induced mixing of membrane domains ultimately underlies mechanotransduction. However, it not yet clear how much mechanical force is required to cause raft disruption or how long this effect would be expected to last. It is also unclear if cholesterol depletion may have additional effects beyond raft disruption that contribute to these signaling events. Thus, more work needs to be done to understand how these interesting effects of force on membranes are mediated. Finally, it is possible that the coupling of mechanosensing and rafts is not restricted to PLD2 but is a general paradigm for many other membrane-associated proteins.
Raft targeting signals and their role in directing protein trafficking through the secretory pathway
Intertwined with the idea of rafts influencing biological functions is the question as to how certain proteins are targeted to lipid rafts. Several factors are thought to contribute to this process [122]. Lipid modifications such as palmitoylation and GPI anchors are well-known raft targeting motifs [123]. Further, certain proteins can bind to lipids which are enriched in the ordered phase. Crosslinking of membrane-associated biomolecules can also result in raft stabilization. Crosslinking of GM1 via CTxB binding to membranes close to a demixing point drives phase separation [65], and as discussed above, raft stabilization in response to receptor crosslinking is important in BCR signaling [17]. It should be noted however that this is not general scenario; immobilization of GPI-anchored proteins, for example, does not nucleate the formation of ordered domains [124].
Many transmembrane proteins are excluded from raft domains, and packing of transmembrane domains into an ordered raft phase has long been thought to be energetically unfavorable [125]. Nevertheless, a number of membrane proteins are associated with rafts, even independent of lipid modifications such as palmitoylation [126]. Until recently, the structural basis for raft targeting of these proteins was unclear. Recent work has now revealed that both the length and surface area of a transmembrane domain are critical determinants of raft partitioning [127, 128].
It is known from model membrane studies that lipid rafts are thicker than non-raft phase ordered phase [129]. Thus, a protein which has a longer transmembrane domain would theoretically partition into the ordered phase. This was recently experimentally tested using a panel of single-pass transmembrane domain proteins with varying lengths, and indeed, the length of the transmembrane domain was directly correlated with their partitioning into lipid rafts into GPMVs [127].
The solvent-accessible area of the transmembrane domain was also recently identified as an important determinant of raft partitioning [128]. In particular, proteins with a smaller solvent accessible area prefer to partition into raft phases, whereas the presence of bulky amino acid residues interferes with this process. These findings, together with information on whether a transmembrane protein contains a palmitoylation site, have been used to develop a model which predicts the affinity of single-pass transmembrane proteins for rafts [128]. Application of the model to the entire human proteome of single-pass transmembrane proteins revealed that compared to ER/Golgi associated proteins, plasma membrane proteins are more likely to be palmitoylated, have longer transmembrane domains, and a smaller solvent accessible area of their transmembrane domain. These findings strongly suggest raft partitioning is closely linked to the sorting and trafficking of single-pass transmembrane proteins to the plasma membrane. The mechanism by which this sorting is accomplished is unknown but appears to depend most importantly on the transmembrane domain. The mechanisms responsible for targeting multi-pass transmembrane proteins to ordered domains and the extent to which raft affinity governs their trafficking to the plasma membrane remain to be determined.
How and why pathogens target rafts
Pathogens interact with the plasma membrane of host cells during at least two stages in their life cycle, entry and egress. Rafts have long been thought to serve as a target that pathogens exploit to gain entry into cells as well as facilitate their spread [130, 131]. Here, we discuss three examples of how pathogens utilize rafts in different ways.
CTx and STx induce membrane reorganization to facilitate their uptake into cells
Association with lipid rafts is a common internalization strategy for many bacterial toxins and viruses [131, 132]. Perhaps the best-studied example of pathogens that target and exploit lipid rafts as a means to promote their internalization into host cells are the bacterial toxins cholera toxin and Shiga toxin, the causative agents of cholera and dysentery, respectively (reviewed in [133, 134]).
Cholera toxin and Shiga toxin are members of the AB5 class of bacterial toxins, consisting of an enzymatically active A subunit coupled to homopentameric membrane-binding B-subunits. Both toxins bind to multiple copies of their glycolipid receptors, ganglioside GM1 for the case of cholera toxin, and globotriaosylceramide Gb3 for the case of Shiga toxin. Membrane binding and trafficking of cholera toxin and Shiga toxin are driven primarily by their B-subunits (CTxB and STxB, respectively). CTxB and STxB are non-toxic and can be fluorescently labeled and added exogenously to cells. The endocytic mechanisms responsible for CTxB and STxB to enter cells have been extensively studied and are thought to consist of a combination of raft-dependent, clathrin-independent mechanisms and conventional clathrin-dependent endocytosis [131, 133, 134].
CTxB and STxB are not mere hijackers of rafts: they are instead thought to function as domain inducers that stabilize rafts. This activity has been especially well documented for the case of CTxB (reviewed in [135]) (Figure 3A). In vitro, CTxB binding induces the formation of a textured lipid phase [136]. The addition of CTxB to model membranes close to a phase boundary leads to the formation of large-scale liquid ordered (Lo) and liquid disordered (Ld) domains [65]. Similar effects have been observed in plasma membrane-derived vesicles membranes in response to CTxB binding, leading to the suggestion that cell membranes exist in a state where rafts can be readily coalesced [137]. CTxB binding is also correlated with an increase in the phase transition temperature of GPMVs, indicating rafts are stabilized in its presence [64, 138]. In cells, local ordering of the plasma membrane occurs in response to antibody-induced aggregation of CTxB which is correlated with an increase in actin filaments attached to the plasma membrane [139]. Additional evidence in support of the idea that CTxB binding induces membrane order comes from super-resolution spectral imaging, which revealed nanometer-scale domains in live cell plasma membranes form in response to CTxB binding [77].
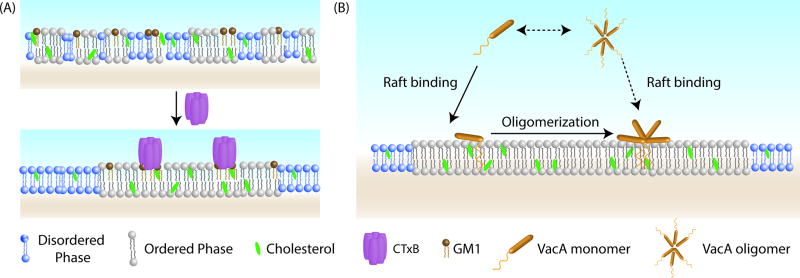
Figure 3. Role of oligomerization in bacterial toxin-raft association.
(A) CTxB is a homopentamer that binds up to 5 GM1 molecules. Binding of CTxB to multiple GM1’s stabilizes rafts and enhances toxin partitioning into the raft phase. (B) VacA is a pore-forming bacterial toxin secreted by Helicobacter pylori. Binding of VacA to lipid rafts is uncoupled from oligomerization, suggesting clustering is not essential for VacA to associated with rafts.
One factor that contributes importantly to the ability of both STx and CTx to build stabilized rafts is their ability to crosslink multiple copies of their glycolipid receptors. This has important consequences on the ability of the toxins to intoxify host cells. The use of CTx mutants with reduced ability to bind multiple copies of GM1 clearly indicate multivalent binding enhances intoxification of cells by CTx [140, 141]. A single GM1 binding site is sufficient to complete the intoxification pathway but is much less efficient than the holotoxin [140]. This could potentially in part reflect an inability of CTxB to stabilize rafts and a decreased ability to associate with rafts when only a single GM1 binding site is functional [64]. Multivalent binding is also important for induction of membrane curvature by STxB [142]. The mechanism of curvature generation is linked to the structure of the toxin, which upon binding to multiple copies of Gb3 imposes local curvature on membranes [143]. This curvature generation capability turn is closely linked to the ability of the toxin to stimulate its own endocytosis [142]. Clustering of toxin-induced domains also appears to enhance their uptake into cells [144]. Clustering of STxB requires tight binding of the toxin to membrane surfaces and has been hypothesized to result from an effective attraction between toxin molecules generated by a thermal Casimir force [144].
Although there is clear evidence that lipid binding by these bacterial toxins increases local membrane order, stabilizes rafts, and bends membranes, how these processes are linked to cellular events required to facilitate toxin trafficking remains unknown. One open question is the identity of the cellular machinery involved in the initial uptake of rafts via clathrin-independent endocytosis. Some insights have recently begun to emerge [145, 146], but much of the machinery required for raft-dependent clathrin-independent endocytosis is still unknown. How rafts enable cells to sense and respond to toxin binding also remains uncertain. Given that multiple pathogens utilize similar strategies to gain entrance into cells, efforts to answer these fundamental questions are likely to have widespread implications in developing directed therapeutic targeting strategies.
VacA, an example of a pore-forming toxin that targets rafts
Even though a large number of bacterial toxins are thought to associate with rafts, in many instances it is not yet clear how they target rafts or how rafts impact their function. One interesting example is Vacuolating toxin A (VacA), the major secreted exotoxin of the bacterium Helicobacter pylori (reviewed in [147]). Intoxification of host cells by VacA is initiated by binding of the toxin to the plasma membrane, followed by toxin oligomerization, membrane insertion, and pore formation [148]. Current models suggest that one or more of these events occur in lipid rafts.
Early studies demonstrating VacA associates with lipid rafts relied on biochemical approaches to isolate raft-enriched fractions and/or depleting cells of cholesterol to interfere with raft integrity and function [5, 149–151]. More recent work has now confirmed VacA’s raft association by showing it preferentially associates with the raft phase in GPMVs [152]. How VacA is targeted to lipid rafts is currently not entirely clear and may involve multiple mechanisms. Some studies indicate that sphingomyelin, one of the receptors of VacA, acts to recruit VacA to rafts [5], while others have shown that initial binding of VacA is to receptors in non-lipid raft microdomains and the raft partitioning of VacA occurs subsequently as a result of clustering [151]. Interestingly, unlike other bacterial toxins such as CTx that depend at least in part on multivalent binding to their receptor to facilitate raft targeting, VacA need not form oligomers in order to partition into rafts [152] (Figure 3B). Furthermore, the ability of the toxin to form pores is not required for it to associate with rafts per se [152].
Why then does VacA associate with rafts? One potential answer is that this is linked to VacA’s internalization mechanism: VacA enters cells via clathrin-independent endocytic pathways, which are typically raft-dependent [153]. However, how rafts influence VacA’s pore-forming activity is not yet known. For example, it is currently unclear whether the structure of pores formed by VacA differs in raft versus non-raft environments. This is an especially important question because there are multiple examples of pore-forming toxins that associate with rafts [154]. Future studies using VacA should help to provide insights into this question, as well as to better delineate raft targeting mechanisms for this interesting class of toxins.
HIV selectively binds and fuses at raft/non-raft boundaries
Not just bacteria, but also viruses are known to target lipid rafts. One important example is the association of HIV, an enveloped RNA virus, with membrane domains [155, 156]. Rafts are thought to play a role in multiple steps in HIV assembly and release. For example, cholesterol is important for viral fusion and infection of cells by HIV [157]. Furthermore, the host cell receptor for HIV, receptor CD4, has been identified as a raft-associated protein [158]. However, until recently, the exact mechanisms by which the virus targets rafts for entry into cells has remained enigmatic.
In a series of interesting studies from both a membrane biology and virology standpoint, HIV has been shown to selectively bind and fuse to the interface between liquid ordered (Lo) and liquid disordered (Ld) domains [159–161]. Initial evidence in support of this idea came from studies showing that reconstitution of the fusion peptide (FP) of HIV gp41 into liposomes mimicking the composition of HIV viral membranes facilitates their fusion to supported bilayers consisting of mixtures of Lo and Ld domains [160]. Strikingly, liposomes containing HIV FP preferentially accumulated at the boundary between Lo and Ld domains. Further, both phase separation and cholesterol were found to be required to facilitate fusion. This behavior was specific to the HIV FP because liposomes containing the influenza FP showed no preference for the boundary [160]. HIV-1 psuedoviruses also preferentially bound to the domain boundary, demonstrating this behavior is not limited to the isolated FP [160].
An interesting question raised by these findings is why HIV virions prefer to fuse at domain boundaries. Both lipid-driven and protein-mediated factors have been shown to be important in this process. One contributing factor that promotes fusion is the hydrophobic mismatch and line tension at the Lo/Ld domain interface [161]. The phase preference of the HIV receptor CD4 and co-receptor CCR5 also contributes to this process [159]. Studies in GPMVs revealed CD4 partitions primarily into the raft phase, consistent with previous reports that it is a raft-associated protein. Intriguingly, CCR5 instead preferentially localizes to the raft/ non-raft domain boundaries [159]. The presence of CCR5 at the phase boundary appears to plays an important role in recruiting HIV to this site as evidenced by studies of the effects of the HIV entry inhibitors maraviroc and enfuvirtide on the recruitment of virions to the raft/ non-raft interface. Maraviroc is known to prevent binding of HIV to CCR5 [162], while enfuvirtide inhibits a conformational change in gp41 required for fusion [163]. Remarkably, maraviroc treatment prevents binding of virions to the phase boundary, but enfuvirtide does not, further supporting the notion that targeting of the virions to the Lo/Ld domain boundary requires binding to CCR5.
Taken together, these findings suggest a model in which HIV initially binds to CD4 in raft domains (Figure 4A) [159]. It subsequently binds to nearby CCR5 molecules located at domain boundaries. Exposure of the gp41 FP then occurs. The FP also prefers to bind to the raft/non-raft interface, resulting in fusion at this site.
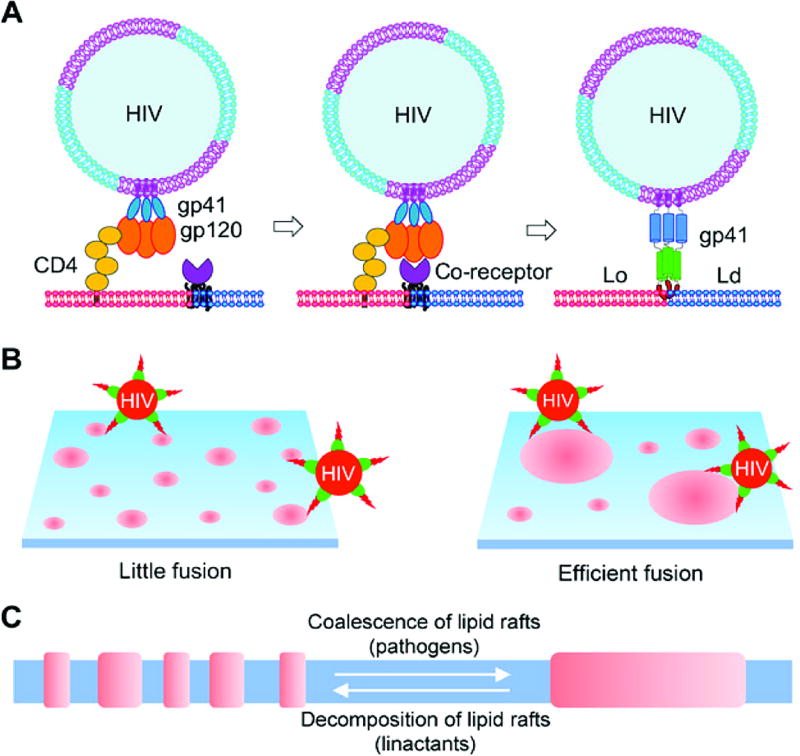
Figure 4. Role of membrane domains in HIV-plasma membrane interaction.
(A) The host binding partners of HIV are the raft associated CD4 and raft/non-raft boundary associated co-receptor, CCR5. HIV binds initially to CD4 in rafts. It is subsequently associated with CCR5 and thus becomes enriched at the Ld/Lo interface, where the gp41 FP inserts to mediate fusion. (B) Larger domains have increased line tension, enabling more efficient fusion of HIV compared to the case of smaller domains with lower line tension. (C) T-cell activation, which is thought to occur prior to the entry of HIV into cells, is thought to cause clustering of raft domains. Linactants break up raft domains, making them a potential therapeutic option to inhibit HIV infection. In all panels, raft domains are shown in red and non-raft domains in blue. Figure is adapted from reference [159] (Creative Commons Attribution Noncommercial License). 2017, AAAS.
The finding that HIV preferentially binds and fuses at raft/non-raft domains not only provides insights into the mechanism of action of existing therapies but also suggests new approaches to drug development. One new line of investigation is the potential use of linactants to inhibit fusion (Figure 4B, C). These types of compounds are thought to partition preferentially at the domain boundary and decrease the line tension. In support of this idea, vitamin E (α-tocopherol), which has been reported to serve as a linactant [164], decreases HIV FP-mediated membrane fusion [161]. This finding also offers a potential explanation for why vitamin E suppresses latent HIV activation in vitro [165]. Another potential therapeutic approach is to shift CCR5 away from the domain interface. A related idea was tested earlier in a work that deliberately targeted CD4 to non-raft domains [166]. These types of interventions would be greatly aided by a better understanding of mechanisms responsible for targeting CCR5 to raft/ non-raft boundaries.
In addition to defining the role of rafts in HIV entry, these findings also have several broader implications. First, they identify CCR5 as one of the few known examples of membrane proteins that preferentially localize to raft/non-raft interfaces. To date, only a handful of proteins have been shown to target the raft/non-raft boundary, including Ras and Rac [167, 168]. Ras and Rac contain multiple lipid modifications that are thought to be important to target them to domain interfaces. How CCR5, a G-protein coupled receptor containing seven transmembrane domains, is targeted to the domain interface is currently unknown. Thus, CCR5 would be an ideal experimental model system to study how phase preference is determined for multi-pass transmembrane proteins. Second, they reveal that domain boundaries can function as a preferred site for viral fusion. Line tension and lipid packing defects appear to make this region of the membrane particularly vulnerable to attack. This does not appear to be a universal mechanism, as the FP of influenza [160] and VSV-G virions [160] do not prefer the phase boundary. Whether this mechanism is exploited by viruses other than HIV remains to be determined. Finally, they illustrate that the boundary between raft and non-raft domains is functionally relevant. This is important because, under physiological conditions where domains are predicted to be small, boundaries between domains would be predicted to be abundant.
Newly discovered sites of domain formation
Much of the work on membrane organization and spatial patterning mechanisms has focused on the plasma membrane of mammalian cells, a site where lipid rafts are thought to be especially prevalent. Given that membranes are ubiquitous in cells and the diversity of composition of plasma membrane between different organisms, the paucity of research in understanding membrane organization in non-mammalian systems is surprising. Here, we discuss two examples of newly described membrane domains found in very different places: large-scale domains in yeast vacuoles, and nanoscale domains in bacteria.
Large-scale domains in yeast
As discussed above, most recent studies suggest that raft-like domains are nanoscale in dimensions in live cell membranes. An interesting exception to the general rule of nanoscale domains comes from the observation that yeast vacuoles contain micron-sized membrane domains when the cells enter stationary phase [169]. Initial evidence for this type of phase separation came as early as the 1980’s [170]. These domains can be readily visualized by fluorescence microscopy and also have been detected by freeze fracture electron microscopy [171]. While their biological functions are not completely understood, initial reports suggest vacuolar membrane domains are important for lipophagy and microautophagy [171, 172].
The dramatic appearance of the vacuolar domains (Figure 5) is reminiscent of large-scale domains observed in model membrane systems exhibiting Lo-Ld phase separation often used to model rafts. Consistent with this possibility, several lines of evidence suggest the domains form in a manner consistent with a membrane demixing mechanism, including the observations that they segregate vacuolar membrane proteins and lipid probes [169, 171] and that depletion of the major yeast sterol ergosterol inhibits domain formation [169, 172]. However, until recently, whether they actually represent bona fide coexisting liquid-liquid phases has not been rigorously tested. Recent studies now show that these domains fulfill two major criteria of a membrane that separates into two liquid phases: i) the domains coalesce over time and ii) they undergo reversible mixing and demixing at a constant transition temperature [173]. Thus, these micron-scale domains in live yeast consist of co-existing fluid phases formed by a phase separation mechanism, similar to that observed in model membrane systems [173].
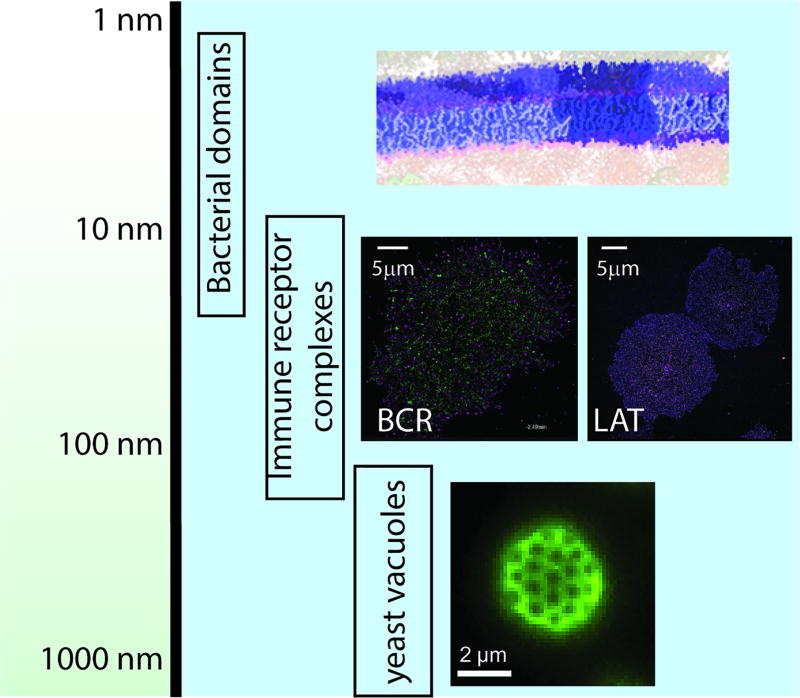
Figure 5. Scale of domains in cellular membranes.
Membrane domains span almost three orders in length. At the lower end of the scale, lipid domains found in bacteria are thought to be at the most tens of nanometers in length. The representation of bacterial domains was adapted from reference [177] (Creative Commons Attribution License). At the other end of spectrum, lipid domains reaching micron scale have been observed in yeast vacuoles. The yeast vacuole figure is reprinted from reference [173], Copyright (2017), with permission from Elsevier. For reference, typical immune receptor clusters are thought to be greater than tens of nanometers but smaller than the diffraction limit. The figure showing the BCR was taken from reference [17] and the figure showing a super-resolution image of LAT in T-cell was kindly provided by Dr. Dylan Owen and Dr. David Williamson.
These findings provide an important missing link between lipid-based domains in vitro and in vivo. It is not yet clear, however, why the domains in vacuolar membranes are so large while in other biological membranes domains tend to be much smaller in size. For instance, the role (or lack thereof) of actin in vacuolar membrane organization could be important in controlling the size of domains. Given that these domains occur in unperturbed cell membranes, in future studies, it would be interesting to use the yeast vacuole as a model to better understand the connection between membrane domain size, composition, and function.
Prokaryotic membrane domains
There is also now growing evidence that membrane domains, including raft-like domains, exist in prokaryotes [174, 175]. For example, the bacterium Borrelia burgdorferi, the causative agent of Lyme disease, contains raft-like membrane domains that contain cholesterol and cholesterol glycolipids [97, 99]. These domains can be isolated as detergent-resistant membrane fractions, and their formation requires sterols capable of forming ordered membrane domains. They can also be detected by FRET and transmission electron microscopy [97, 99]. There is also evidence that a bacterial homolog of a mammalian raft-associated membrane protein known as flotillin generates functional membrane domains in Staphylococcus aureus [176]. Disruption of these domains renders methicillin-resistant S. aureus (MRSA) susceptible to penicillin treatment [98]. These findings suggest bacterial domains are a potential therapeutic target.
Bacteria have also served as a testbed for the development of new methods to probe membrane organization in intact cells through a combination of metabolic labeling and neutron scattering approaches. Using the gram-positive bacterium Bacillus subtilis as a model, this approach was used to examine the overall structure of the bacterial membrane as well as to test for the presence of membrane domains [177]. A major advance put forward in this study was the use of a chemical biology-based approach to differentially label different cellular structures, thereby providing a contrast mechanism to separate out neutron scattering arising from the bacterial membranes. The bacterial membrane was shown to exhibit a characteristic lamellar structure expected for a membrane bilayer, with a hydrophobic acyl core thickness of ~24Å. Interestingly, this value is similar to that obtained from small-angle x-ray measurements of basolateral membranes purified from rat hepatocytes [178]. Even more remarkably, evidence of lateral demixing of lipids within the plane of the membrane over a length scale of 30–40 nm was observed (Figure 5). While the mechanisms underlying lipid demixing in this system are currently unknown, these results point to the presence of lipid domains in intact bacterial membranes.
Domains in bacterial systems are at the same time both complex (due to the presence of the cell wall) and simple (since there are no raft-targeting post-translational modifications such as palmitoylation). Further, most bacteria have neither cholesterol nor actin but instead possess analogs of both which are thought to be important in membrane organization [179, 180]. Thus, though rules governing bacterial rafts would have to be viewed differently compared to eukaryotic plasma membrane rafts, regulation by rafts could be evolutionarily ancient. Comparative analysis of the domains formed by these different systems would provide a comprehensive view of how lipid rafts are formed, how they function and how they have evolved.
Looking forward
The advent of newer, more sensitive experimental methods has made it possible to discern the structure and dynamics of membrane domains in biological membranes and probe the myriad physiological functions in which rafts play a role. Despite these advances, the field remains fraught with continued uncertainties about the nature of rafts and the exact mechanisms by which they control cellular functions [8]. In the final section of this review, we discuss several considerations and areas for future exploration that would advance our understanding of the role that rafts play in functionally patterning membranes.
With the continued development of many new experimental systems, it is important to bear in mind that different experimental methods report on distinct features of membranes. For instance, membrane order detected by spectral imaging is not necessarily linked to clustering of proteins in membranes reported by super resolution approaches. Further, different probes may also reflect different membrane properties. For the case of spectral measurements, the two commonly used dyes Laurdan and ANEPPDHQ do not actually measure packing of membranes the same way [181]. Thus, it is critical to probe membrane domains using a variety of experimental methods. By taking a multi-pronged approach, it is possible to develop a more nuanced understanding of the mosaic of data generated by different techniques.
It is also increasingly clear that cholesterol plays complex roles in biological membranes in addition to its proposed role in organizing rafts. A large number of proteins bind to cholesterol including PDZ domain-containing proteins [182, 183], and recent evidence suggests the transbilayer distribution of cholesterol is actively regulated [61]. It is thus not surprising that cholesterol depletion can have pleiotropic effects [184]. Despite this, much of the existing raft literature in cells relies on cholesterol depletion as a means to perturb raft-associated structures and events. In recent years, molecules that selectively tune membrane domains by either stabilizing or perturbing phase separation have been characterized [185–189]. However, to better differentiate between cholesterol-dependent and raft-dependent events, it would be helpful if new approaches to either perturb rafts or selectively move individual proteins in or out of domains were developed. The discovery of structural motifs that target single-pass transmembrane domain proteins to rafts [128] should enable the more selective testing of the role of raft association for specific proteins without disrupting rafts themselves.
The continued development of model membrane systems that recapitulate key features of biological membranes is also key to the advancement of the field. The properties of lipid domains have been extensively studied in well-controlled model systems such as the classic ternary lipid mixtures used to generate co-existing Lo and Ld domains [190, 191]. Over the past few years, efforts have been made to begin to incorporate increasing levels of complexity into these systems, such as the incorporation of actin cytoskeleton into model membranes to facilitate better understanding of the coupling between actin and domains [192–194]. This important goal merits further exploration given evidence that actin can influence the order of the outer plasma membrane leaflet, lipid domain formation, and the nanoclustering of GPI-anchored proteins in addition to its well-recognized role in compartmentalizing the plasma membrane via a picket fence mechanism (reviewed in [6, 195, 196]). Some efforts have also been made to develop asymmetric membrane models (reviewed in [195]). This also represents an area where further work is needed in order to enhance our understand how the lipid composition of each leaflet contributes to domain formation, as well as how coupling of domains between leaflets occurs [197].
More work also needs to be done to understand how the architecture of cell membranes regulates domain organization. One important goal in this direction is to continue to catalog the varied mechanisms that compartmentalize membranes into subdomains, generate protein nanoclusters, and make connections with other organelles [9, 198–200]. It is important to understand that in addition to lipids and membranous proteins, associated proteins also sculpt the membrane. For instance, both the physical basis and the consequence of coupling of actin and microtubules to plasma membrane order, while thought to be essential, are still not understood. Thus, while there is no single membrane clustering mechanism, a critical area of research is to understand the relationship between the different mechanisms of membrane organization. It is likely that the relative importance of each of these different processes is dependent on the probes and model systems that are being used for these studies.
Finally, in cells, besides compositional heterogeneity, cellular membranes have structural organization. Currently, the relationship between the complex topology and geometry of cellular membranes and lipid domains is not obvious. From a practical perspective, membrane topology impacts the interpretation of several microscopy-based approaches used to study the properties of domains [201]. There is considerable evidence in model systems to show that lipid phase separation is coupled to curvature in membranes [202], and geometrical constraints in membrane can alter diffusion and thereby protein sorting [203–208]. Thus, the interplay between the two could form the cornerstone of many membrane-associated biological processes [209].
Our understanding of organization of membranes arises from a patchwork of different experimental techniques performed on diverse model systems to probe a fragmented set of membrane properties using an incomplete set of molecular players. Despite this, we now have a framework in hand that has enabled us to apply principles of membrane organization to several biological systems. The next level of abstraction should start with the coordination between multiple systems and look at membrane organization holistically at different scales, both in space and time.
Acknowledgments
We thank Drs. Dylan Owen and Dave Williamson for providing the super-resolution image of LAT.
Funding: Supported by R01 GM106720 and R01 AG056147 from the National Institutes of Health and a Pilot and Feasibility Award from Vanderbilt University Medical Center’s Digestive Disease Research Center supported by NIH grant P30DK058404. The funding sources had no role in writing the article or the decision to submit the paper for publication.
Footnotes
Abbreviations used in this article: BCR, B-cell receptor; CTxB, cholera toxin B-subunit; GPMV, giant plasma membrane-derived vesicle; FCS, fluorescence correlation spectroscopy; FRET, Förster Resonance Energy Transfer; FP, fusion peptide; Ld, liquid disordered; Lo, liquid ordered; MRSA, methicillin-resistant S. aureus; PFO, Perfringolysin O; PIP2, phosphatidylinositol 4,5-bisphosphate; PLD2, phospholipase D2; STORM, stochastic optical reconstruction microscopy; STxB, Shiga toxin B-subunit; svFCS, spot variation FCS; STED, Stimulated emission depletion; VacA, Vacuolating toxin A
References
- 1.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nussinov R, Jang H, Tsai CJ. Oligomerization and nanocluster organization render specificity. Biol Rev Camb Philos Soc. 2015;90:587–598. doi: 10.1111/brv.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernardino de la Serna J, Schutz GJ, Eggeling C, Cebecauer M. There Is No Simple Model of the Plasma Membrane Organization. Front Cell Dev Biol. 2016;4:106. doi: 10.3389/fcell.2016.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 5.Gupta VR, Patel HK, Kostolansky SS, Ballivian RA, Eichberg J, Blanke SR. Sphingomyelin functions as a novel receptor for Helicobacter pylori VacA. PLoS Pathog. 2008;4:e1000073. doi: 10.1371/journal.ppat.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KG. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson’s fluid-mosaic model. Annu Rev Cell Dev Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- 7.Sezgin E, Levental I, Mayor S, Eggeling C. The mystery of membrane organization: composition regulation and roles of lipid rafts. Nat Rev Mol Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sevcsik E, Schutz GJ. With or without rafts? Alternative views on cell membranes. Bioessays. 2016;38:129–139. doi: 10.1002/bies.201500150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu SM, Fairn GD. Mesoscale organization of domains in the plasma membrane - beyond the lipid raft. Crit Rev Biochem Mol Biol. 2018:1–16. doi: 10.1080/10409238.2018.1436515. [DOI] [PubMed] [Google Scholar]
- 10.Sezgin E, Kaiser HJ, Baumgart T, Schwille P, Simons K, Levental I. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat Protoc. 2012;7:1042–1051. doi: 10.1038/nprot.2012.059. [DOI] [PubMed] [Google Scholar]
- 11.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 12.Rao M, Mayor S. Use of Forster’s resonance energy transfer microscopy to study lipid rafts. Biochim Biophys Acta. 2005;1746:221–233. doi: 10.1016/j.bbamcr.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 14.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung BO, Chou KC. Review of super-resolution fluorescence microscopy for biology. Appl Spectrosc. 2011;65:967–980. doi: 10.1366/11-06398. [DOI] [PubMed] [Google Scholar]
- 17.Stone MB, Shelby SA, Nunez MF, Wisser K, Veatch SL. Protein sorting by lipid phase-like domains supports emergent signaling function in B lymphocyte plasma membranes. Elife. 2017;6 doi: 10.7554/eLife.19891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu CJ, Baumgart T. Spatial association of signaling proteins and F-actin effects on cluster assembly analyzed via photoactivation localization microscopy in T cells. PLoS One. 2011;6:e23586. doi: 10.1371/journal.pone.0023586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prescher J, Baumgartel V, Ivanchenko S, Torrano AA, Brauchle C, Muller B, Lamb DC. Super-resolution imaging of ESCRT-proteins at HIV-1 assembly sites. PLoS Pathog. 2015;11:e1004677. doi: 10.1371/journal.ppat.1004677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bobone S, Hilsch M, Storm J, Dunsing V, Herrmann A, Chiantia S. Phosphatidylserine Lateral Organization Influences the Interaction of Influenza Virus Matrix Protein 1 with Lipid Membranes. J Virol. 2017;91 doi: 10.1128/JVI.00267-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Needham SR, Hirsch M, Rolfe DJ, Clarke DT, Zanetti-Domingues LC, Wareham R, Martin-Fernandez ML. Measuring EGFR separations on cells with ~10 nm resolution via fluorophore localization imaging with photobleaching. PLoS One. 2013;8:e62331. doi: 10.1371/journal.pone.0062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Gao J, Guo X, Tong T, Shi X, Li L, Qi M, Wang Y, Cai M, Jiang J, Xu C, Ji H, Wang H. Regulation of EGFR nanocluster formation by ionic protein-lipid interaction. Cell Res. 2014;24:959–976. doi: 10.1038/cr.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone MB, Veatch SL. Steady-state cross-correlations for live two-colour super-resolution localization data sets. Nat Commun. 2015;6:7347. doi: 10.1038/ncomms8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owen DM, Williamson DJ, Magenau A, Gaus K. Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nat Commun. 2012;3:1256. doi: 10.1038/ncomms2273. [DOI] [PubMed] [Google Scholar]
- 26.Rossy J, Cohen E, Gaus K, Owen DM. Method for co-cluster analysis in multichannel single-molecule localisation data. Histochem Cell Biol. 2014;141:605–612. doi: 10.1007/s00418-014-1208-z. [DOI] [PubMed] [Google Scholar]
- 27.Veatch SL, Machta BB, Shelby SA, Chiang EN, Holowka DA, Baird BA. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS ONE. 2012;7:e31457. doi: 10.1371/journal.pone.0031457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levet F, Hosy E, Kechkar A, Butler C, Beghin A, Choquet D, Sibarita JB. SR-Tesseler: a method to segment and quantify localization-based super-resolution microscopy data. Nat Methods. 2015;12:1065–1071. doi: 10.1038/nmeth.3579. [DOI] [PubMed] [Google Scholar]
- 29.Griffie J, Shlomovich L, Williamson DJ, Shannon M, Aaron J, Khuon S, G LB, Boelen L, Peters R, Cope AP, Cohen EAK, Rubin-Delanchy P, Owen DM. 3D Bayesian cluster analysis of super-resolution data reveals LAT recruitment to the T cell synapse. Sci Rep. 2017;7:4077. doi: 10.1038/s41598-017-04450-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andronov L, Lutz Y, Vonesch JL, Klaholz BP. SharpViSu: integrated analysis and segmentation of super-resolution microscopy data. Bioinformatics. 2016;32:2239–2241. doi: 10.1093/bioinformatics/btw123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ovesny M, Krizek P, Borkovec J, Svindrych Z, Hagen GM. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malkusch S, Heilemann M. Extracting quantitative information from single-molecule super-resolution imaging data with LAMA - LocAlization Microscopy Analyzer. Sci Rep. 2016;6:34486. doi: 10.1038/srep34486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujiwara TK, Iwasawa K, Kalay Z, Tsunoyama TA, Watanabe Y, Umemura YM, Murakoshi H, Suzuki KG, Nemoto YL, Morone N, Kusumi A. Confined diffusion of transmembrane proteins and lipids induced by the same actin meshwork lining the plasma membrane. Mol Biol Cell. 2016;27:1101–1119. doi: 10.1091/mbc.E15-04-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liebel M, Hugall JT, van Hulst NF. Ultrasensitive Label-Free Nanosensing and High-Speed Tracking of Single Proteins. Nano Lett. 2017;17:1277–1281. doi: 10.1021/acs.nanolett.6b05040. [DOI] [PubMed] [Google Scholar]
- 35.Wu HM, Lin YH, Yen TC, Hsieh CL. Nanoscopic substructures of raft-mimetic liquid-ordered membrane domains revealed by high-speed single-particle tracking. Sci Rep. 2016;6:20542. doi: 10.1038/srep20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki KGN, Ando H, Komura N, Fujiwara TK, Kiso M, Kusumi A. Development of new ganglioside probes and unraveling of raft domain structure by single-molecule imaging. Biochim Biophys Acta. 2017;1861:2494–2506. doi: 10.1016/j.bbagen.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 37.Kinoshita M, Suzuki KG, Matsumori N, Takada M, Ano H, Morigaki K, Abe M, Makino A, Kobayashi T, Hirosawa KM, Fujiwara TK, Kusumi A, Murata M. Raft-based sphingomyelin interactions revealed by new fluorescent sphingomyelin analogs. J Cell Biol. 2017;216:1183–1204. doi: 10.1083/jcb.201607086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter T, Schlegel J, Burgert A, Kurz A, Seibel J, Sauer M. Incorporation studies of clickable ceramides in Jurkat cell plasma membranes. Chem Commun (Camb) 2017;53:6836–6839. doi: 10.1039/c7cc01220a. [DOI] [PubMed] [Google Scholar]
- 39.Komura N, Suzuki KGN, Ando H, Konishi M, Imamura A, Ishida H, Kusumi A, Kiso M. Syntheses of Fluorescent Gangliosides for the Studies of Raft Domains. Methods Enzymol. 2017;597:239–263. doi: 10.1016/bs.mie.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Sezgin E, Levental I, Grzybek M, Schwarzmann G, Mueller V, Honigmann A, Belov VN, Eggeling C, Coskun U, Simons K, Schwille P. Partitioning diffusion and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochim Biophys Acta. 2012;1818:1777–1784. doi: 10.1016/j.bbamem.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Ritchie K, Shan XY, Kondo J, Iwasawa K, Fujiwara T, Kusumi A. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys J. 2005;88:2266–2277. doi: 10.1529/biophysj.104.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wawrezinieck L, Rigneault H, Marguet D, Lenne PF. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys J. 2005;89:4029–4042. doi: 10.1529/biophysj.105.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2. Kluwer Academic/Plenum Press; 1999. Place Published. [Google Scholar]
- 44.Elson EL. Quick tour of fluorescence correlation spectroscopy from its inception. J Biomed Opt. 2004;9:857–864. doi: 10.1117/1.1779234. [DOI] [PubMed] [Google Scholar]
- 45.Rigler R, Elson ES, editors. Fluorescence Correlation Spectroscopy: Theory and Applications. Springer; 2001. Place Published. [Google Scholar]
- 46.Hassler K, Leutenegger M, Rigler P, Rao R, Rigler R, Gosch M, Lasser T. Total internal reflection fluorescence correlation spectroscopy (TIR-FCS) with low background and high count-rate per molecule. Opt Express. 2005;13:7415–7423. doi: 10.1364/opex.13.007415. [DOI] [PubMed] [Google Scholar]
- 47.Jiang Y, Pryse KM, Melnykov A, Genin GM, Elson EL. Investigation of Nanoscopic Phase Separations in Lipid Membranes Using Inverse FCS. Biophys J. 2017;112:2367–2376. doi: 10.1016/j.bpj.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez SA, Gratton E. Lipid--protein interactions revealed by two-photon microscopy and fluorescence correlation spectroscopy. Acc Chem Res. 2005;38:469–477. doi: 10.1021/ar040026l. [DOI] [PubMed] [Google Scholar]
- 49.Levi V, Ruan Q, Kis-Petikova K, Gratton E. Scanning FCS a novel method for three-dimensional particle tracking. Biochem Soc Trans. 2003;31:997–1000. doi: 10.1042/bst0310997. [DOI] [PubMed] [Google Scholar]
- 50.Mailfert S, Hamon Y, Bertaux N, He HT, Marguet D. A user’s guide for characterizing plasma membrane subdomains in living cells by spot variation fluorescence correlation spectroscopy. Methods Cell Biol. 2017;139:1–22. doi: 10.1016/bs.mcb.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 51.Lenne PF, Wawrezinieck L, Conchonaud F, Wurtz O, Boned A, Guo XJ, Rigneault H, He HT, Marguet D. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 2006;25:3245–3256. doi: 10.1038/sj.emboj.7601214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kastrup L, Blom H, Eggeling C, Hell SW. Fluorescence fluctuation spectroscopy in subdiffraction focal volumes. Phys Rev Lett. 2005;94:178104. doi: 10.1103/PhysRevLett.94.178104. [DOI] [PubMed] [Google Scholar]
- 53.Mueller V, Honigmann A, Ringemann C, Medda R, Schwarzmann G, Eggeling C. FCS in STED microscopy: studying the nanoscale of lipid membrane dynamics. Methods Enzymol. 2013;519:1–38. doi: 10.1016/B978-0-12-405539-1.00001-4. [DOI] [PubMed] [Google Scholar]
- 54.Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, Belov VN, Hein B, von Middendorff C, Schonle A, Hell SW. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 55.Honigmann A, Mueller V, Ta H, Schoenle A, Sezgin E, Hell SW, Eggeling C. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nat Commun. 2014;5:5412. doi: 10.1038/ncomms6412. [DOI] [PubMed] [Google Scholar]
- 56.Mueller V, Ringemann C, Honigmann A, Schwarzmann G, Medda R, Leutenegger M, Polyakova S, Belov VN, Hell SW, Eggeling C. STED nanoscopy reveals molecular details of cholesterol- and cytoskeleton-modulated lipid interactions in living cells. Biophys J. 2011;101:1651–1660. doi: 10.1016/j.bpj.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider F, Waithe D, Clausen MP, Galiani S, Koller T, Ozhan G, Eggeling C, Sezgin E. Diffusion of lipids and GPI-anchored proteins in actin-free plasma membrane vesicles measured by STED-FCS. Mol Biol Cell. 2017;28:1507–1518. doi: 10.1091/mbc.E16-07-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burns MC, Nouri M, Veatch SL. Spot size variation FCS in simulations of the 2D Ising model. J Phys D Appl Phys. 2016;49 doi: 10.1088/0022-3727/49/21/214001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lagerholm BC, Andrade DM, Clausen MP, Eggeling C. Convergence of lateral dynamic measurements in the plasma membrane of live cells from single particle tracking and STED-FCS. J Phys D Appl Phys. 2017;50:063001. doi: 10.1088/1361-6463/aa519e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maekawa M, Yang Y, Fairn GD. Perfringolysin O Theta Toxin as a Tool to Monitor the Distribution and Inhomogeneity of Cholesterol in Cellular Membranes. Toxins (Basel) 2016;8 doi: 10.3390/toxins8030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu SL, Sheng R, Jung JH, Wang L, Stec E, O’Connor MJ, Song S, Bikkavilli RK, Winn RA, Lee D, Baek K, Ueda K, Levitan I, Kim KP, Cho W. Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat Chem Biol. 2017;13:268–274. doi: 10.1038/nchembio.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamaji A, Sekizawa Y, Emoto K, Sakuraba H, Inoue K, Kobayashi H, Umeda M. Lysenin a novel sphingomyelin-specific binding protein. J Biol Chem. 1998;273:5300–5306. doi: 10.1074/jbc.273.9.5300. [DOI] [PubMed] [Google Scholar]
- 63.Makino A, Abe M, Ishitsuka R, Murate M, Kishimoto T, Sakai S, Hullin-Matsuda F, Shimada Y, Inaba T, Miyatake H, Tanaka H, Kurahashi A, Pack CG, Kasai RS, Kubo S, Schieber NL, Dohmae N, Tochio N, Hagiwara K, Sasaki Y, Aida Y, Fujimori F, Kigawa T, Nishibori K, Parton RG, Kusumi A, Sako Y, Anderluh G, Yamashita M, Kobayashi T, Greimel P, Kobayashi T. A novel sphingomyelin/cholesterol domain-specific probe reveals the dynamics of the membrane domains during virus release and in Niemann-Pick type C. FASEB J. 2017;31:1301–1322. doi: 10.1096/fj.201500075R. [DOI] [PubMed] [Google Scholar]
- 64.Raghunathan K, Wong TH, Chinnapen DJ, Lencer WI, Jobling MG, Kenworthy AK. Glycolipid crosslinking is required for cholera toxin to partition into and stabilize ordered domains. Biophys J. 2016;111:2547–2550. doi: 10.1016/j.bpj.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hammond AT, Heberle FA, Baumgart T, Holowka D, Baird B, Feigenson GW. Crosslinking a lipid raft component triggers liquid ordered-liquid disordered phase separation in model plasma membranes. Proc Natl Acad Sci U S A. 2005;102:6320–6325. doi: 10.1073/pnas.0405654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rissanen S, Grzybek M, Orlowski A, Rog T, Cramariuc O, Levental I, Eggeling C, Sezgin E, Vattulainen I. Phase Partitioning of GM1 and Its Bodipy-Labeled Analog Determine Their Different Binding to Cholera Toxin. Front Physiol. 2017;8:252. doi: 10.3389/fphys.2017.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wands AM, Fujita A, McCombs JE, Cervin J, Dedic B, Rodriguez AC, Nischan N, Bond MR, Mettlen M, Trudgian DC, Lemoff A, Quiding-Jarbrink M, Gustavsson B, Steentoft C, Clausen H, Mirzaei H, Teneberg S, Yrlid U, Kohler JJ. Fucosylation and protein glycosylation create functional receptors for cholera toxin. Elife. 2015;4:e09545. doi: 10.7554/eLife.09545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klymchenko AS, Kreder R. Fluorescent probes for lipid rafts: from model membranes to living cells. Chem Biol. 2014;21:97–113. doi: 10.1016/j.chembiol.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 69.Sezgin E, Waithe D, Bernardino de la Serna J, Eggeling C. Spectral imaging to measure heterogeneity in membrane lipid packing. Chemphyschem. 2015;16:1387–1394. doi: 10.1002/cphc.201402794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parasassi T, Gratton E. Membrane lipid domains and dynamics as detected by Laurdan fluorescence. J Fluoresc. 1995;5:59–69. doi: 10.1007/BF00718783. [DOI] [PubMed] [Google Scholar]
- 71.Barucha-Kraszewska J, Kraszewski S, Ramseyer C. Will C-Laurdan dethrone Laurdan in fluorescent solvent relaxation techniques for lipid membrane studies? Langmuir. 2013;29:1174–1182. doi: 10.1021/la304235r. [DOI] [PubMed] [Google Scholar]
- 72.Kim HM, Choo HJ, Jung SY, Ko YG, Park WH, Jeon SJ, Kim CH, Joo T, Cho BR. A two-photon fluorescent probe for lipid raft imaging: C-laurdan. Chembiochem. 2007;8:553–559. doi: 10.1002/cbic.200700003. [DOI] [PubMed] [Google Scholar]
- 73.Owen DM, Rentero C, Magenau A, Abu-Siniyeh A, Gaus K. Quantitative imaging of membrane lipid order in cells and organisms. Nat Protoc. 2012;7:24–35. doi: 10.1038/nprot.2011.419. [DOI] [PubMed] [Google Scholar]
- 74.Mazeres S, Joly E, Lopez A, Tardin C. Characterization of M-laurdan a versatile probe to explore order in lipid membranes. F1000Res. 2014;3:172. doi: 10.12688/f1000research.4805.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kreder R, Pyrshev KA, Darwich Z, Kucherak OA, Mely Y, Klymchenko AS. Solvatochromic Nile Red probes with FRET quencher reveal lipid order heterogeneity in living and apoptotic cells. ACS Chem Biol. 2015;10:1435–1442. doi: 10.1021/cb500922m. [DOI] [PubMed] [Google Scholar]
- 76.Karpenko IA, Niko Y, Yakubovskyi VP, Gerasov AO, Bonnet D, Kovtun YP, Klymchenko AS. Push-pull dioxaborine as fluorescent molecular rotor: far-red fluorogenic probe for ligand-receptor interactions. J Mater Chem C Mater. 2016;4:3002–3009. doi: 10.1039/C5TC03411F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moon S, Yan R, Kenny SJ, Shyu Y, Xiang L, Li W, Xu K. Spectrally Resolved, Functional Super-Resolution Microscopy Reveals Nanoscale Compositional Heterogeneity in Live-Cell Membranes. J Am Chem Soc. 2017;139:10944–10947. doi: 10.1021/jacs.7b03846. [DOI] [PubMed] [Google Scholar]
- 78.Sezgin E, Schneider F, Zilles V, Urbancic I, Garcia E, Waithe D, Klymchenko AS, Eggeling C. Polarity-Sensitive Probes for Superresolution Stimulated Emission Depletion Microscopy. Biophys J. 2017;113:1321–1330. doi: 10.1016/j.bpj.2017.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Das A, Brown MS, Anderson DD, Goldstein JL, Radhakrishnan A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. Elife. 2014;3 doi: 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He C, Hu X, Jung RS, Weston TA, Sandoval NP, Tontonoz P, Kilburn MR, Fong LG, Young SG, Jiang H. High-resolution imaging and quantification of plasma membrane cholesterol by NanoSIMS. Proc Natl Acad Sci U S A. 2017;114:2000–2005. doi: 10.1073/pnas.1621432114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Naulin PA, Alveal NA, Barrera NP. Toward atomic force microscopy and mass spectrometry to visualize and identify lipid rafts in plasmodesmata. Front Plant Sci. 2014;5:234. doi: 10.3389/fpls.2014.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gumi-Audenis B, Costa L, Carla F, Comin F, Sanz F, Giannotti MI. Structure and Nanomechanics of Model Membranes by Atomic Force Microscopy and Spectroscopy: Insights into the Role of Cholesterol and Sphingolipids. Membranes (Basel) 2016;6 doi: 10.3390/membranes6040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mashaghi A, Mashaghi S, Reviakine I, Heeren RM, Sandoghdar V, Bonn M. Label-free characterization of biomembranes: from structure to dynamics. Chem Soc Rev. 2014;43:887–900. doi: 10.1039/c3cs60243e. [DOI] [PubMed] [Google Scholar]
- 84.Spindler S, Ehrig J, König K, Nowak T, Piliarik M, Stein HE, Taylor RW, Garanger E, Lecommandoux S, Alves ID, Sandoghdar V. Visualization of lipids and proteins at high spatial and temporal resolution via interferometric scattering (iSCAT) microscopy. Journal of Physics D: Applied Physics. 2016;49:274002. [Google Scholar]
- 85.Wilson RL, Frisz JF, Klitzing HA, Zimmerberg J, Weber PK, Kraft ML. Hemagglutinin clusters in the plasma membrane are not enriched with cholesterol and sphingolipids. Biophys J. 2015;108:1652–1659. doi: 10.1016/j.bpj.2015.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ando J, Palonpon AF, Sodeoka M, Fujita K. High-speed Raman imaging of cellular processes. Curr Opin Chem Biol. 2016;33:16–24. doi: 10.1016/j.cbpa.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 87.Shirota K, Yagi K, Inaba T, Li PC, Murata M, Sugita Y, Kobayashi T. Detection of Sphingomyelin Clusters by Raman Spectroscopy. Biophys J. 2016;111:999–1007. doi: 10.1016/j.bpj.2016.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Donaldson SH, Jr, de Aguiar HB. Molecular Imaging of Cholesterol and Lipid Distributions in Model Membranes. J Phys Chem Lett. 2018;9:1528–1533. doi: 10.1021/acs.jpclett.8b00235. [DOI] [PubMed] [Google Scholar]
- 89.Yeager AN, Weber PK, Kraft ML. Three-dimensional imaging of cholesterol and sphingolipids within a Madin-Darby canine kidney cell. Biointerphases. 2016;11:02A309. doi: 10.1116/1.4939681. [DOI] [PubMed] [Google Scholar]
- 90.Loser L, Saalwachter K, Mendes Ferreira T. Liquid-liquid phase coexistence in lipid membranes observed by natural abundance (1)H-(13)C solid-state NMR. Phys Chem Chem Phys. 2018;20:9751–9754. doi: 10.1039/c8cp01012a. [DOI] [PubMed] [Google Scholar]
- 91.Ionova IV, Livshits VA, Marsh D. Phase diagram of ternary cholesterol/palmitoylsphingomyelin/palmitoyloleoyl-phosphatidylcholine mixtures: spin-label EPR study of lipid-raft formation. Biophys J. 2012;102:1856–1865. doi: 10.1016/j.bpj.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marquardt D, Heberle FA, Nickels JD, Pabst G, Katsaras J. On scattered waves and lipid domains: detecting membrane rafts with X-rays and neutrons. Soft Matter. 2015;11:9055–9072. doi: 10.1039/c5sm01807b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krager KJ, Sarkar M, Twait EC, Lill NL, Koland JG. A novel biotinylated lipid raft reporter for electron microscopic imaging of plasma membrane microdomains. J Lipid Res. 2012;53:2214–2225. doi: 10.1194/jlr.D026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Papahadjopoulos-Sternberg B. Freeze-Fracture Electron Microscopy on Domains in Lipid Mono- and Bilayer on Nano-Resolution Scale. Methods Mol Biol. 2017;1522:55–72. doi: 10.1007/978-1-4939-6591-5_5. [DOI] [PubMed] [Google Scholar]
- 95.Fujita A, Cheng J, Hirakawa M, Furukawa K, Kusunoki S, Fujimoto T. Gangliosides GM1 and GM3 in the living cell membrane form clusters susceptible to cholesterol depletion and chilling. Mol Biol Cell. 2007;18:2112–2122. doi: 10.1091/mbc.E07-01-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou Y, Hancock JF. Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim Biophys Acta. 2015;1853:841–849. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 97.LaRocca TJ, Pathak P, Chiantia S, Toledo A, Silvius JR, Benach JL, London E. Proving lipid rafts exist: membrane domains in the prokaryote Borrelia burgdorferi have the same properties as eukaryotic lipid rafts. PLoS Pathog. 2013;9:e1003353. doi: 10.1371/journal.ppat.1003353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garcia-Fernandez E, Koch G, Wagner RM, Fekete A, Stengel ST, Schneider J, Mielich-Suss B, Geibel S, Markert SM, Stigloher C, Lopez D. Membrane Microdomain Disassembly Inhibits MRSA Antibiotic Resistance. Cell. 2017;171:1354–1367. e1320. doi: 10.1016/j.cell.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.LaRocca TJ, Crowley JT, Cusack BJ, Pathak P, Benach J, London E, Garcia-Monco JC, Benach JL. Cholesterol lipids of Borrelia burgdorferi form lipid rafts and are required for the bactericidal activity of a complement-independent antibody. Cell Host Microbe. 2010;8:331–342. doi: 10.1016/j.chom.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Metzler R, Jeon JH, Cherstvy AG. Non-Brownian diffusion in lipid membranes: Experiments and simulations. Biochim Biophys Acta. 2016;1858:2451–2467. doi: 10.1016/j.bbamem.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 101.Rog T, Vattulainen I. Cholesterol sphingolipids and glycolipids: what do we know about their role in raft-like membranes? Chem Phys Lipids. 2014;184:82–104. doi: 10.1016/j.chemphyslip.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 102.Paz Ramos A, Lague P, Lamoureux G, Lafleur M. Effect of Saturated Very Long-Chain Fatty Acids on the Organization of Lipid Membranes: A Study Combining (2)H NMR Spectroscopy and Molecular Dynamics Simulations. J Phys Chem B. 2016;120:6951–6960. doi: 10.1021/acs.jpcb.6b04958. [DOI] [PubMed] [Google Scholar]
- 103.Wu EL, Cheng X, Jo S, Rui H, Song KC, Davila-Contreras EM, Qi Y, Lee J, Monje-Galvan V, Venable RM, Klauda JB, Im W. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J Comput Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hong C, Tieleman DP, Wang Y. Microsecond molecular dynamics simulations of lipid mixing. Langmuir. 2014;30:11993–12001. doi: 10.1021/la502363b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khakbaz P, Klauda JB. Probing the importance of lipid diversity in cell membranes via molecular simulation. Chem Phys Lipids. 2015;192:12–22. doi: 10.1016/j.chemphyslip.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 106.Ingolfsson HI, Melo MN, van Eerden FJ, Arnarez C, Lopez CA, Wassenaar TA, Periole X, de Vries AH, Tieleman DP, Marrink SJ. Lipid organization of the plasma membrane. J Am Chem Soc. 2014;136:14554–14559. doi: 10.1021/ja507832e. [DOI] [PubMed] [Google Scholar]
- 107.Ingolfsson HI, Carpenter TS, Bhatia H, Bremer PT, Marrink SJ, Lightstone FC. Computational Lipidomics of the Neuronal Plasma Membrane. Biophys J. 2017;113:2271–2280. doi: 10.1016/j.bpj.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Angiolini J, Plachta N, Mocskos E, Levi V. Exploring the dynamics of cell processes through simulations of fluorescence microscopy experiments. Biophys J. 2015;108:2613–2618. doi: 10.1016/j.bpj.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Poger D, Caron B, Mark AE. Validating lipid force fields against experimental data: Progress, challenges and perspectives. Biochim Biophys Acta. 2016;1858:1556–1565. doi: 10.1016/j.bbamem.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 110.Lyubartsev AP, Rabinovich AL. Force Field Development for Lipid Membrane Simulations. Biochim Biophys Acta. 2016;1858:2483–2497. doi: 10.1016/j.bbamem.2015.12.033. [DOI] [PubMed] [Google Scholar]
- 111.Ingolfsson HI, Arnarez C, Periole X, Marrink SJ. Computational 'microscopy' of cellular membranes. J Cell Sci. 2016;129:257–268. doi: 10.1242/jcs.176040. [DOI] [PubMed] [Google Scholar]
- 112.Chavent M, Duncan AL, Sansom MS. Molecular dynamics simulations of membrane proteins and their interactions: from nanoscale to mesoscale. Curr Opin Struct Biol. 2016;40:8–16. doi: 10.1016/j.sbi.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Perilla JR, Goh BC, Cassidy CK, Liu B, Bernardi RC, Rudack T, Yu H, Wu Z, Schulten K. Molecular dynamics simulations of large macromolecular complexes. Curr Opin Struct Biol. 2015;31:64–74. doi: 10.1016/j.sbi.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.He HT, Lellouch A, Marguet D. Lipid rafts and the initiation of T cell receptor signaling. Semin Immunol. 2005;17:23–33. doi: 10.1016/j.smim.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 115.Holowka D, Gosse JA, Hammond AT, Han X, Sengupta P, Smith NL, Wagenknecht-Wiesner A, Wu M, Young RM, Baird B. Lipid segregation and IgE receptor signaling: a decade of progress. Biochim Biophys Acta. 2005;1746:252–259. doi: 10.1016/j.bbamcr.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 116.Gupta N, DeFranco AL. Lipid rafts and B cell signaling. Semin Cell Dev Biol. 2007;18:616–626. doi: 10.1016/j.semcdb.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sherman E, Barr V, Manley S, Patterson G, Balagopalan L, Akpan I, Regan CK, Merrill RK, Sommers CL, Lippincott-Schwartz J, Samelson LE. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity. 2011;35:705–720. doi: 10.1016/j.immuni.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Williamson DJ, Owen DM, Rossy J, Magenau A, Wehrmann M, Gooding JJ, Gaus K. Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat Immunol. 2011;12:655–662. doi: 10.1038/ni.2049. [DOI] [PubMed] [Google Scholar]
- 119.Petersen EN, Chung HW, Nayebosadri A, Hansen SB. Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D. Nat Commun. 2016;7:13873. doi: 10.1038/ncomms13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xie Z, Ho WT, Exton JH. Functional implications of post-translational modifications of phospholipases D1 and D2. Biochim Biophys Acta. 2002;1580:9–21. doi: 10.1016/s1388-1981(01)00168-8. [DOI] [PubMed] [Google Scholar]
- 121.Lopez I, Arnold RS, Lambeth JD. Cloning and initial characterization of a human phospholipase D2 (hPLD2). ADP-ribosylation factor regulates hPLD2. J Biol Chem. 1998;273:12846–12852. doi: 10.1074/jbc.273.21.12846. [DOI] [PubMed] [Google Scholar]
- 122.Lorent JH, Levental I. Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem Phys Lipids. 2015 doi: 10.1016/j.chemphyslip.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 123.Levental I, Grzybek M, Simons K. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry. 2010;49:6305–6316. doi: 10.1021/bi100882y. [DOI] [PubMed] [Google Scholar]
- 124.Sevcsik E, Brameshuber M, Folser M, Weghuber J, Honigmann A, Schutz GJ. GPI-anchored proteins do not reside in ordered domains in the live cell plasma membrane. Nat Commun. 2015;6:6969. doi: 10.1038/ncomms7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shogomori H, Hammond AT, Ostermeyer-Fay AG, Barr DJ, Feigenson GW, London E, Brown DA. Palmitoylation and intracellular domain interactions both contribute to raft targeting of linker for activation of T cells. J Biol Chem. 2005;280:18931–18942. doi: 10.1074/jbc.M500247200. [DOI] [PubMed] [Google Scholar]
- 126.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci U S A. 2010;107:22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Diaz-Rohrer BB, Levental KR, Simons K, Levental I. Membrane raft association is a determinant of plasma membrane localization. Proc Natl Acad Sci U S A. 2014;111:8500–8505. doi: 10.1073/pnas.1404582111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lorent JH, Diaz-Rohrer B, Lin X, Spring K, Gorfe AA, Levental KR, Levental I. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat Commun. 2017;8:1219. doi: 10.1038/s41467-017-01328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gandhavadi M, Allende D, Vidal A, Simon SA, McIntosh TJ. Structure, composition, and peptide binding properties of detergent soluble bilayers and detergent resistant rafts. Biophys. J. 2002;82:1469–1482. doi: 10.1016/S0006-3495(02)75501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Manes S, del Real G, Lacalle RA, Lucas P, Gomez-Mouton C, Sanchez-Palomino S, Delgado R, Alcami J, Mira E, Martinez AC. Membrane raft microdomains mediate lateral assemblies required for HIV-1 infection. EMBO Rep. 2000;1:190–196. doi: 10.1093/embo-reports/kvd025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Aigal S, Claudinon J, Romer W. Plasma membrane reorganization: A glycolipid gateway for microbes. Biochim Biophys Acta. 2015;1853:858–871. doi: 10.1016/j.bbamcr.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 132.Chinnapen DJ, Chinnapen H, Saslowsky D, Lencer WI. Rafting with cholera toxin: endocytosis and trafficking from plasma membrane to ER. FEMS Microbiol Lett. 2007;266:129–137. doi: 10.1111/j.1574-6968.2006.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sandvig K, Bergan J, Kavaliauskiene S, Skotland T. Lipid requirements for entry of protein toxins into cells. Prog Lipid Res. 2014;54:1–13. doi: 10.1016/j.plipres.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 134.Johannes L. Shiga Toxin-A Model for Glycolipid-Dependent and Lectin-Driven Endocytosis. Toxins (Basel) 2017;9 doi: 10.3390/toxins9110340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Day CA, Kenworthy AK. Functions of cholera toxin B-subunit as a raft cross-linker. Essays Biochem. 2015;57:135–145. doi: 10.1042/bse0570135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Watkins EB, Miller CE, Majewski J, Kuhl TL. Membrane texture induced by specific protein binding and receptor clustering: active roles for lipids in cellular function. Proc Natl Acad Sci U S A. 2011;108:6975–6980. doi: 10.1073/pnas.1014579108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lingwood D, Ries J, Schwille P, Simons K. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc Natl Acad Sci U S A. 2008;105:10005–10010. doi: 10.1073/pnas.0804374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Johnson SA, Stinson BM, Go MS, Carmona LM, Reminick JI, Fang X, Baumgart T. Temperature-dependent phase behavior and protein partitioning in giant plasma membrane vesicles. Biochim Biophys Acta. 2010;1798:1427–1435. doi: 10.1016/j.bbamem.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dinic J, Ashrafzadeh P, Parmryd I. Actin filaments attachment at the plasma membrane in live cells cause the formation of ordered lipid domains. Biochim Biophys Acta. 2013;1828:1102–1111. doi: 10.1016/j.bbamem.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 140.Jobling MG, Yang Z, Kam WR, Lencer WI, Holmes RK. A single native ganglioside GM1-binding site is sufficient for cholera toxin to bind to cells and complete the intoxication pathway. MBio. 2012;3 doi: 10.1128/mBio.00401-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wolf AA, Jobling MG, Saslowsky DE, Kern E, Drake KR, Kenworthy AK, Holmes RK, Lencer WI. Attenuated endocytosis and toxicity of a mutant cholera toxin with decreased ability to cluster GM1. Infect Immun. 2008;76:1476–1484. doi: 10.1128/IAI.01286-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Römer W, Berland L, Chambon V, Gaus K, Windschiegl B, Tenza D, Aly MR, Fraisier V, Florent JC, Perrais D, Lamaze C, Raposo G, Steinem C, Sens P, Bassereau P, Johannes L. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 2007;450:670–675. doi: 10.1038/nature05996. [DOI] [PubMed] [Google Scholar]
- 143.Pezeshkian W, Hansen AG, Johannes L, Khandelia H, Shillcock JC, Kumar PB, Ipsen JH. Membrane invagination induced by Shiga toxin B-subunit: from molecular structure to tube formation. Soft Matter. 2016;12:5164–5171. doi: 10.1039/c6sm00464d. [DOI] [PubMed] [Google Scholar]
- 144.Pezeshkian W, Gao H, Arumugam S, Becken U, Bassereau P, Florent JC, Ipsen JH, Johannes L, Shillcock JC. Mechanism of Shiga Toxin Clustering on Membranes. ACS Nano. 2017;11:314–324. doi: 10.1021/acsnano.6b05706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Johannes L, Parton RG, Bassereau P, Mayor S. Building endocytic pits without clathrin. Nat Rev Mol Cell Biol. 2015;16:311–321. doi: 10.1038/nrm3968. [DOI] [PubMed] [Google Scholar]
- 146.Mayor S, Parton RG, Donaldson JG. Clathrin-independent pathways of endocytosis. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cover TL, Blanke SR. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat Rev Microbiol. 2005;3:320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- 148.Foegeding NJ, Caston RR, McClain MS, Ohi MD, Cover TL. An Overview of Helicobacter pylori VacA Toxin Biology. Toxins (Basel) 2016;8 doi: 10.3390/toxins8060173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ricci V, Galmiche A, Doye A, Necchi V, Solcia E, Boquet P. High cell sensitivity to Helicobacter pylori VacA toxin depends on a GPI-anchored protein and is not blocked by inhibition of the clathrin-mediated pathway of endocytosis. Mol Biol Cell. 2000;11:3897–3909. doi: 10.1091/mbc.11.11.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Patel HK, Willhite DC, Patel RM, Ye D, Williams CL, Torres EM, Marty KB, MacDonald RA, Blanke SR. Plasma membrane cholesterol modulates cellular vacuolation induced by the Helicobacter pylori vacuolating cytotoxin. Infect Immun. 2002;70:4112–4123. doi: 10.1128/IAI.70.8.4112-4123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nakayama M, Hisatsune J, Yamasaki E, Nishi Y, Wada A, Kurazono H, Sap J, Yahiro K, Moss J, Hirayama T. Clustering of Helicobacter pylori VacA in lipid rafts mediated by its receptor, receptor-like protein tyrosine phosphatase beta, is required for intoxication in AZ-521 Cells. Infect Immun. 2006;74:6571–6580. doi: 10.1128/IAI.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Raghunathan K, Foegeding NJ, Campbell AM, Cover TL, Ohi MD, Kenworthy AK. Determinants of Raft Partitioning of the Helicobacter pylori Pore-Forming Toxin VacA. Infect Immun. 2018;86 doi: 10.1128/IAI.00872-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gauthier NC, Monzo P, Kaddai V, Doye A, Ricci V, Boquet P. Helicobacter pylori VacA cytotoxin: a probe for a clathrin-independent and Cdc42-dependent pinocytic pathway routed to late endosomes. Mol Biol Cell. 2005;16:4852–4866. doi: 10.1091/mbc.E05-05-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Skocaj M, Bakrac B, Krizaj I, Macek P, Anderluh G, Sepcic K. The sensing of membrane microdomains based on pore-forming toxins. Curr Med Chem. 2013;20:491–501. doi: 10.2174/0929867311320040002. [DOI] [PubMed] [Google Scholar]
- 155.Freed EO. HIV-1 assembly, release and maturation. Nat Rev Microbiol. 2015;13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Graham DR, Chertova E, Hilburn JM, Arthur LO, Hildreth JE. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with beta-cyclodextrin inactivates and permeabilizes the virions: evidence for virion-associated lipid rafts. J Virol. 2003;77:8237–8248. doi: 10.1128/JVI.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Popik W, Alce TM, Au WC. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4(+) T cells. J Virol. 2002;76:4709–4722. doi: 10.1128/JVI.76.10.4709-4722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang ST, Kreutzberger AJB, Kiessling V, Ganser-Pornillos BK, White JM, Tamm LK. HIV virions sense plasma membrane heterogeneity for cell entry. Sci Adv. 2017;3:e1700338. doi: 10.1126/sciadv.1700338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yang ST, Kiessling V, Simmons JA, White JM, Tamm LK. HIV gp41-mediated membrane fusion occurs at edges of cholesterol-rich lipid domains. Nat Chem Biol. 2015;11:424–431. doi: 10.1038/nchembio.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang ST, Kiessling V, Tamm LK. Line tension at lipid phase boundaries as driving force for HIV fusion peptide-mediated fusion. Nat Commun. 2016;7:11401. doi: 10.1038/ncomms11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tan Q, Zhu Y, Li J, Chen Z, Han GW, Kufareva I, Li T, Ma L, Fenalti G, Li J, Zhang W, Xie X, Yang H, Jiang H, Cherezov V, Liu H, Stevens RC, Zhao Q, Wu B. Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex. Science. 2013;341:1387–1390. doi: 10.1126/science.1241475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 164.Muddana HS, Chiang HH, Butler PJ. Tuning membrane phase separation using nonlipid amphiphiles. Biophys J. 2012;102:489–497. doi: 10.1016/j.bpj.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Heredia A, Davis C, Amoroso A, Taylor G, Le N, Bamba D, Redfield RR. In vitro suppression of latent HIV-1 activation by vitamin E: potential clinical implications. AIDS. 2005;19:836–837. doi: 10.1097/01.aids.0000168981.82337.10. [DOI] [PubMed] [Google Scholar]
- 166.Del Real G, Jimenez-Baranda S, Lacalle RA, Mira E, Lucas P, Gomez-Mouton C, Carrera AC, Martinez AC, Manes S. Blocking of HIV-1 infection by targeting CD4 to nonraft membrane domains. J Exp Med. 2002;196:293–301. doi: 10.1084/jem.20020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nicolini C, Baranski J, Schlummer S, Palomo J, Lumbierres-Burgues M, Kahms M, Kuhlmann J, Sanchez S, Gratton E, Waldmann H, Winter R. Visualizing association of N-ras in lipid microdomains: influence of domain structure and interfacial adsorption. J Am Chem Soc. 2006;128:192–201. doi: 10.1021/ja055779x. [DOI] [PubMed] [Google Scholar]
- 168.Moissoglu K, Kiessling V, Wan C, Hoffman BD, Norambuena A, Tamm LK, Schwartz MA. Regulation of Rac1 translocation and activation by membrane domains and their boundaries. J Cell Sci. 2014;127:2565–2576. doi: 10.1242/jcs.149088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Toulmay A, Prinz WA. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J Cell Biol. 2013;202:35–44. doi: 10.1083/jcb.201301039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Moeller CH, Mudd JB, Thomson WW. Lipid phase separations and intramembranous particle movements in the yeast tonoplast. Biochim Biophys Acta. 1981;643:376–386. doi: 10.1016/0005-2736(81)90082-1. [DOI] [PubMed] [Google Scholar]
- 171.Tsuji T, Fujimoto M, Tatematsu T, Cheng J, Orii M, Takatori S, Fujimoto T. Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. Elife. 2017;6 doi: 10.7554/eLife.25960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang CW, Miao YH, Chang YS. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J Cell Biol. 2014;206:357–366. doi: 10.1083/jcb.201404115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rayermann SP, Rayermann GE, Cornell CE, Merz AJ, Keller SL. Hallmarks of Reversible Separation of Living, Unperturbed Cell Membranes into Two Liquid Phases. Biophys J. 2017;113:2425–2432. doi: 10.1016/j.bpj.2017.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bramkamp M, Lopez D. Exploring the existence of lipid rafts in bacteria. Microbiol Mol Biol Rev. 2015;79:81–100. doi: 10.1128/MMBR.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lopez D, Koch G. Exploring functional membrane microdomains in bacteria: an overview. Curr Opin Microbiol. 2017;36:76–84. doi: 10.1016/j.mib.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bach JN, Bramkamp M. Flotillins functionally organize the bacterial membrane. Mol Microbiol. 2013;88:1205–1217. doi: 10.1111/mmi.12252. [DOI] [PubMed] [Google Scholar]
- 177.Nickels JD, Chatterjee S, Stanley CB, Qian S, Cheng X, Myles DAA, Standaert RF, Elkins JG, Katsaras J. The in vivo structure of biological membranes and evidence for lipid domains. PLoS Biol. 2017;15:e2002214. doi: 10.1371/journal.pbio.2002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc Natl Acad Sci U S A. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Strahl H, Burmann F, Hamoen LW. The actin homologue MreB organizes the bacterial cell membrane. Nat Commun. 2014;5:3442. doi: 10.1038/ncomms4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Saenz JP, Grosser D, Bradley AS, Lagny TJ, Lavrynenko O, Broda M, Simons K. Hopanoids as functional analogues of cholesterol in bacterial membranes. Proc Natl Acad Sci U S A. 2015;112:11971–11976. doi: 10.1073/pnas.1515607112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Amaro M, Reina F, Hof M, Eggeling C, Sezgin E. Laurdan and Di-4-ANEPPDHQ probe different properties of the membrane. J Phys D Appl Phys. 2017;50:134004. doi: 10.1088/1361-6463/aa5dbc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hulce JJ, Cognetta AB, Niphakis MJ, Tully SE, Cravatt BF. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat Methods. 2013;10:259–264. doi: 10.1038/nmeth.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sheng R, Chen Y, Yung Gee H, Stec E, Melowic HR, Blatner NR, Tun MP, Kim Y, Kallberg M, Fujiwara TK, Hye Hong J, Pyo Kim K, Lu H, Kusumi A, Goo Lee M, Cho W. Cholesterol modulates cell signaling and protein networking by specifically interacting with PDZ domain-containing scaffold proteins. Nat Commun. 2012;3:1249. doi: 10.1038/ncomms2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zidovetzki R, Levitan I. Use of cyclodextrins to manipulate plasma membrane cholesterol content: Evidence, misconceptions and control strategies. Biochim Biophys Acta. 2007;1768:1311–1324. doi: 10.1016/j.bbamem.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Levental I, Veatch S. The Continuing Mystery of Lipid Rafts. J Mol Biol. 2016;428:4749–4764. doi: 10.1016/j.jmb.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhou Y, Maxwell KN, Sezgin E, Lu M, Liang H, Hancock JF, Dial EJ, Lichtenberger LM, Levental I. Bile acids modulate signaling by functional perturbation of plasma membrane domains. J Biol Chem. 2013;288:35660–35670. doi: 10.1074/jbc.M113.519116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Raghunathan K, Ahsan A, Ray D, Nyati MK, Veatch SL. Membrane Transition Temperature Determines Cisplatin Response. PLoS One. 2015;10:e0140925. doi: 10.1371/journal.pone.0140925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gray E, Karslake J, Machta BB, Veatch SL. Liquid general anesthetics lower critical temperatures in plasma membrane vesicles. Biophys J. 2013;105:2751–2759. doi: 10.1016/j.bpj.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Levental KR, Lorent JH, Lin X, Skinkle AD, Surma MA, Stockenbojer EA, Gorfe AA, Levental I. Polyunsaturated Lipids Regulate Membrane Domain Stability by Tuning Membrane Order. Biophys J. 2016;110:1800–1810. doi: 10.1016/j.bpj.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Veatch SL, Keller SL. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Heberle FA, Wu J, Goh SL, Petruzielo RS, Feigenson GW. Comparison of three ternary lipid bilayer mixtures: FRET and ESR reveal nanodomains. Biophys J. 2010;99:3309–3318. doi: 10.1016/j.bpj.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Honigmann A, Sadeghi S, Keller J, Hell SW, Eggeling C, Vink R. A lipid bound actin meshwork organizes liquid phase separation in model membranes. Elife. 2014;3:e01671. doi: 10.7554/eLife.01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Vogel SK, Greiss F, Khmelinskaia A, Schwille P. Control of lipid domain organization by a biomimetic contractile actomyosin cortex. Elife. 2017;6 doi: 10.7554/eLife.24350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Koster DV, Husain K, Iljazi E, Bhat A, Bieling P, Mullins RD, Rao M, Mayor S. Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer. Proc Natl Acad Sci U S A. 2016;113:E1645–1654. doi: 10.1073/pnas.1514030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Fujimoto T, Parmryd I. Interleaflet Coupling Pinning and Leaflet Asymmetry-Major Players in Plasma Membrane Nanodomain Formation. Front Cell Dev Biol. 2016;4:155. doi: 10.3389/fcell.2016.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Koster DV, Mayor S. Cortical actin and the plasma membrane: inextricably intertwined. Curr Opin Cell Biol. 2016;38:81–89. doi: 10.1016/j.ceb.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 197.Cheng HT, Megha, London E. Preparation and properties of asymmetric vesicles that mimic cell membranes: effect upon lipid raft formation and transmembrane helix orientation. J Biol Chem. 2009;284:6079–6092. doi: 10.1074/jbc.M806077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Krapf D. Compartmentalization of the plasma membrane. Curr Opin Cell Biol. 2018;53:15–21. doi: 10.1016/j.ceb.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 199.Goyette J, Gaus K. Mechanisms of protein nanoscale clustering. Curr Opin Cell Biol. 2017;44:86–92. doi: 10.1016/j.ceb.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 200.Johannes L, Pezeshkian W, Ipsen JH, Shillcock JC. Clustering on Membranes: Fluctuations and More. Trends Cell Biol. 2018;28:405–415. doi: 10.1016/j.tcb.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 201.Parmryd I, Onfelt B. Consequences of membrane topography. FEBS J. 2013;280:2775–2784. doi: 10.1111/febs.12209. [DOI] [PubMed] [Google Scholar]
- 202.Heinrich M, Tian A, Esposito C, Baumgart T. Dynamic sorting of lipids and proteins in membrane tubes with a moving phase boundary. Proc Natl Acad Sci U S A. 2010;107:7208–7213. doi: 10.1073/pnas.0913997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cheney PP, Weisgerber AW, Feuerbach AM, Knowles MK. Single Lipid Molecule Dynamics on Supported Lipid Bilayers with Membrane Curvature. Membranes (Basel) 2017;7 doi: 10.3390/membranes7010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Klaus CJ, Raghunathan K, DiBenedetto E, Kenworthy AK. Analysis of diffusion in curved surfaces and its application to tubular membranes. Mol Biol Cell. 2016;27:3937–3946. doi: 10.1091/mbc.E16-06-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Callan-Jones A, Sorre B, Bassereau P. Curvature-driven lipid sorting in biomembranes. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Roux A, Cuvelier D, Nassoy P, Prost J, Bassereau P, Goud B. Role of curvature and phase transition in lipid sorting and fission of membrane tubules. Embo J. 2005;24:1537–1545. doi: 10.1038/sj.emboj.7600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kusters R, Kapitein LC, Hoogenraad CC, Storm C. Shape-induced asymmetric diffusion in dendritic spines allows efficient synaptic AMPA receptor trapping. Biophys J. 2013;105:2743–2750. doi: 10.1016/j.bpj.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, Klumperman J, McMahon HT, Cullen PJ. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 209.von Erlach TC, Bertazzo S, Wozniak MA, Horejs CM, Maynard SA, Attwood S, Robinson BK, Autefage H, Kallepitis C, Del Rio Hernandez A, Chen CS, Goldoni S, Stevens MM. Cell-geometry-dependent changes in plasma membrane order direct stem cell signalling and fate. Nat Mater. 2018;17:237–242. doi: 10.1038/s41563-017-0014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]