Abstract
The cellular RNA pool in animals arises from two separate genomes stored in the nucleus and multiple mitochondria. Chemical methods to track nascent RNA synthesis are unable to distinguish between these two with stringency. Herein we report that spatially restricting bioorthogonal nucleoside biosynthesis enables, for the first time, selective metabolic labeling of the RNA transcribed in the mitochondria. We envision this approach could open the door for heretofore-impossible analyses of mitochondrial RNA. Beyond our results revealed herein, our approach provides a roadmap for researchers to begin to design strategies to examine biomolecules within subcellular compartments.
Graphical Abstract
Understanding the synthesis and lifetime of RNA molecules is key to elucidating how cells behave and control their biology1, 2 Recent approaches toward understanding the dynamics of transcription have relied on metabolic labeling of RNA.3–5 For example, alkynyl-6–8 and azido-containing9 modified nucleoside metabolic intermediates can be introduced into cells and nascent RNA can be tracked through bioorthogonal chemical reactions. Both Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) and strain-promoted alkyne-azide cycloaddition (SPAAC) can be utilized for imaging or biotinylation and streptavidin enrichment of labeled RNA.
RNAs are transcribed from two DNA repositories inside cells: the nucleus and the mitochondria. Both organelles have distinct genome sequences and thus can give rise to distinct RNA pools that perform diverse biological functions. A critical missing component in metabolic labeling of RNA (as well as other biomolecules) is the ability to distinguish with high stringency between RNA that arises from the two genomes. This represents a major challenge to overcome and may present a unique opportunity to expand the capabilities of chemical methods to study RNA molecules in living systems.
Mitochondria are the powerhouses of the cell and are vitally important for the normal physiology of every cell type. Changes in their function have been linked to many cancer types and neurological disorders.10, 11 Characterizing the molecular components of the mitochondria is critical for understanding its function. For mitochondrial analysis the traditional method is fractionation and enrichment.12–17 These approaches are extremely laborious and suffer from a very high false-positive rate.18 This is mostly attributed to the reportedly high RNA content on the outer membrane of the mitochondria.12, 13, 19, 20 Furthermore, fractionation methods suffer from the inability to track nascent RNA synthesis, due to mitochondria still being viable during centrifugation protocols.21–23 The canonical animal mitochondrial transcriptome consists of 13 mRNAs, 22 tRNAs, and 2 rRNAs, whose expression can be profiled by conventional RT-PCR;24 however, recent reports have suggested that the types of RNAs originating from the mitochondrial genome are much more complex than previously thought, surprisingly comprising miRNAs, antisense RNAs, and long non-coding RNAs.12, 13, 25–27 Moving beyond such a narrow view of mitochondrial RNA and identifying novel RNAs that arise from the mitochondrial genome is a major challenge in the field.12, 13 The lack of tools to stringently purify and identify new RNAs solely transcribed from the mitochondrial genome (while removing labeling of RNAs from the nuclear genome) prevents such analyses and discovery.
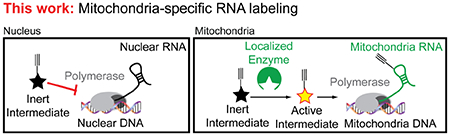
Within this Letter we report a novel approach toward addressing the many weaknesses described above. We demonstrate that spatially-restricting modified nucleotide biosynthesis within mitochondria, permits selective metabolic labeling of RNA that arises from the mitochondrial transcriptome (Figure 1). We also show that this approach can be used to specifically track mitochondrial RNA synthesis.
Figure 1.
Outline of mitochondria-specific metabolic labeling of RNA.
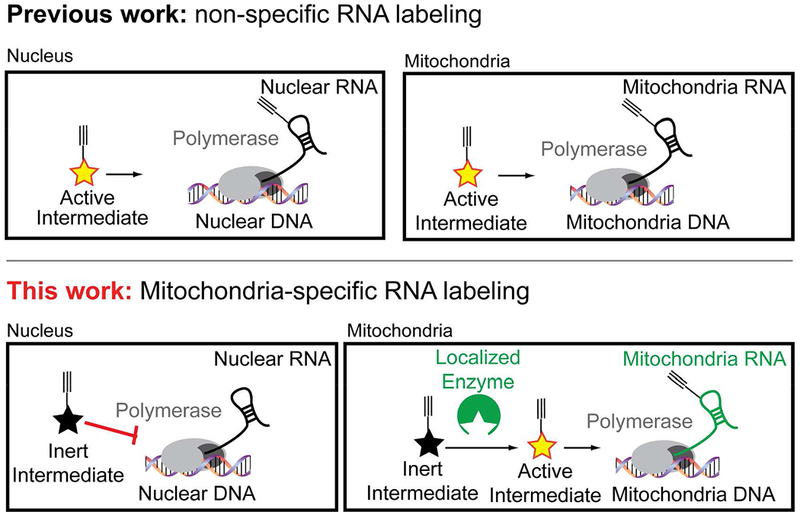
We recently demonstrated that nascent RNA synthesis can be tracked using 5-ethynyluracil (Inert Intermediate) with the enzyme uracil phosphoribosyltranferase (UPRT).28 UPRT converts uracil and phosphoribosyl pyrophosphate (PRPP) to 5’-phosphoro-5-ethynyluridine (Active Intermediate, 5’P-5EU), which is eventually incorporated into nascent RNA. Our original hypothesis was that we could spatially restrict UPRT to enable nascent RNA labeling within the mitochondria.
Consistent with this hypothesis PRPP has been observed within chloroplasts, which are plant organelles with their own genomes akin to mitochondria.29 Mitochondria are also known to have nucleoside kinase enzymes localized within their inner matrix, which would permit the eventual biosynthesis of a modified nucleoside triphosphate.30, 31 As such, we rationalized that localizing UPRT in the mitochondria may permit specific synthesis of 5’P-5EU (Figure 2, A) and eventual RNA labeling.
Figure 2. Design and localization of a UPRT enzyme construct to mitochondria.
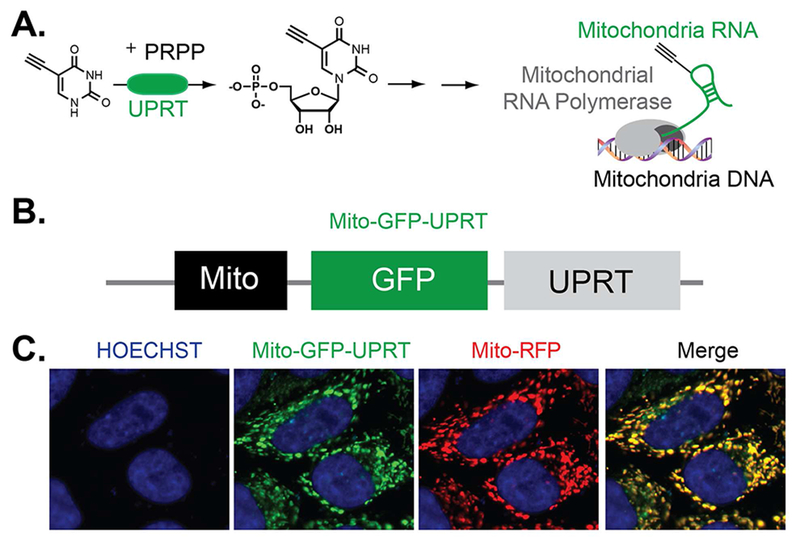
A. Schematic of proposed UPRT-mediated RNA synthesis in the mitochondria. B. Design of a mitochondria-targeted UPRT construct. C. Imaging of the mitochondria-targeted UPRT construct displays overlap with a Mito-RFP construct. Hoechst stains nuclear DNA.
To localize UPRT into the mitochondria we constructed a Mito-GFP-UPRT fusion construct (Figure 2, B). Cellular imaging of the fusion demonstrated precise localization into the mitochondria, with strong overlap with a mitochondrial localized red fluorescent protein (Figure 2, C) and mitotracker fluorescent dye, which localizes into the mitochondrial matrix (Figure S1).
To demonstrate that the Mito-GFP-UPRT fusion converts 5EU to 5’P-5EU, with eventual incorporation into RNA, we performed a time-course dot blot analysis. 5EU was fed to cell media and RNA incorporation was assayed using dot blot analysis as shown in Figure 3, A and Figure S2. Comparison to whole-cell UPRT, there was markedly less RNA signal, which may be attributed to either weak activity or the extremely low percentage of total RNA that is comprised of mitochondrial RNA. Normal fractionation of mitochondria (inner matrix, outer membrane, and cytosolic RNA) demonstrated that the majority of signal is in mitochondrial-enriched RNA, with undetectable labeling of cytoplasmic RNA (Figure S3). Furthermore, to test for potential toxicity, we isolated mitochondria and performed a Cytochrome c oxidase assay, which reports on the activity and concentration of Cytochrome c at the mitochondrial membrane. As shown in Figure S4, there was no observed difference in Cytochrome c activity in the presence of 5EU or not, even after 8 hours of 5EU incubation. Overall, these data suggest that our approach can be utilized to metabolically label RNA in the mitochondrial matrix.
Figure 3. Mitochondria-targeted UPRT enables metabolic incorporation of 5EU into mitochondrial RNA.
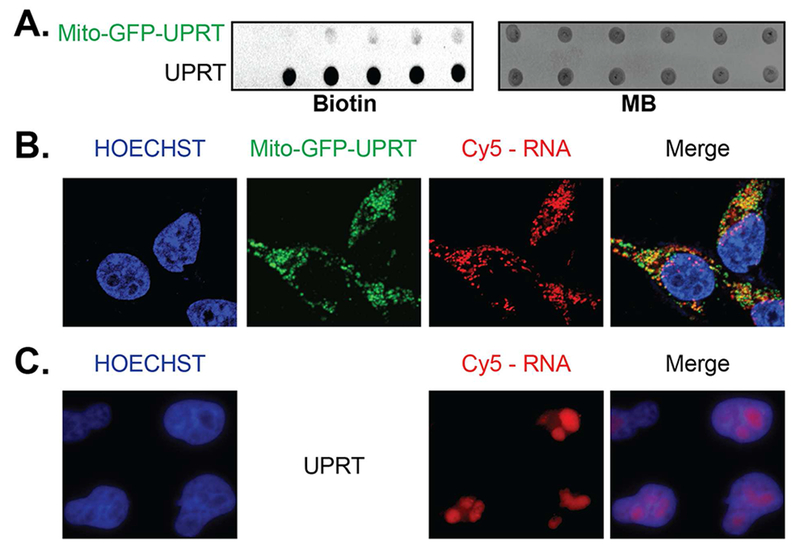
A. Dot blot analysis of equal amount of RNA derived from Mito-GFP-UPRT and spatially-unrestricted UPRT expressing cells in a time titration of 5EU treatment (left to right): 0, 0.5, 1, 2, 3 and 5h. MB: methylene blue stain-served as loading control B. Imaging of Mito-GFP-UPRT-transiently expressed cells and metabolically labeled RNA (Cy5-azide) show overlap in mitochondria. C. Imaging of spatially-unrestricted UPRT cells show RNA labeling in nucleolus, indicative of whole-cell RNA labeling. Hoechst stains nuclear DNA.
To identify RNA synthesis within the mitochondria with higher stringency we used confocal imaging. After treatment with 5EU, cells were fixed, permeabilized, and stringently washed. RNA was imaged using CuAAC-mediated reaction to append Cy5-azide at 5’ethynyl uracil residues. As shown in Figure 3, B, cells expressing Mito-GFP-UPRT had RNA signal (Cy5-N3) that overlapped with Mito-GFP-UPRT localization. In contrast, cells expressing spatially-unrestricted UPRT had the large majority of their signal in the nucleus, which is consistent with our previous results with significant metabolic labeling of rRNA within the nucleolus (Figure 3, C).32 RNase treatment of labeled Mito-GFP-UPRT cells or Mito-GFP-UPRT cells lacking 5EU showed loss of signal in mitochondria, further demonstrating that CuAAC-mediated staining is specific for RNA (Figure S5).
To explore the selectivity of RNA incorporation we assayed RNA enrichment by RT-PCR. We chose five RNAs to compare: (1) GAPDH mRNA, which is one of the most highly abundant RNAs transcribed from the nuclear genome, (2) 18S rRNA a highly abundant RNA from the cytoplasmic ribosome, (3) U6 snRNA, an abundant splicing RNA that is transcribed in the nucleus, (4) 12S mitochondrial rRNA, and (5) MT-CO2 mRNA (cytochrome-c oxidase 2), mitochondrial mRNA.
Briefly, Mito-GFP-UPRT-containing cells were incubated with 5EU for 5 hours, and total RNA isolated. CuAAC-mediated reaction appended biotin to total RNA. Biotin-appended RNA was enriched and profiled by RT-PCR (Supplementary Information). As shown in Figure 4, A, enrichment RT-PCR clearly demonstrates enrichment of all assayed RNAs in cells expressing UPRT. RNAs transcribed from the mitochondrial genome are selectively enriched and those from the nuclear genome are not in Mito-GFP-UPRT expressing cells. This result is especially impressive because the raw copy numbers of the RNAs coming from the nuclear genome (such as 18S rRNA) are much higher than those of mitochondria, and as such any incorporation of 5EU into these RNAs would be robust. These results nicely demonstrate that mitochondrial RNA is selectively labeled over RNA transcribed from the nuclear genome.
Figure 4. Selectively labeled mitochondrial transcripts in Mito-GFP-UPRT expressing cells.
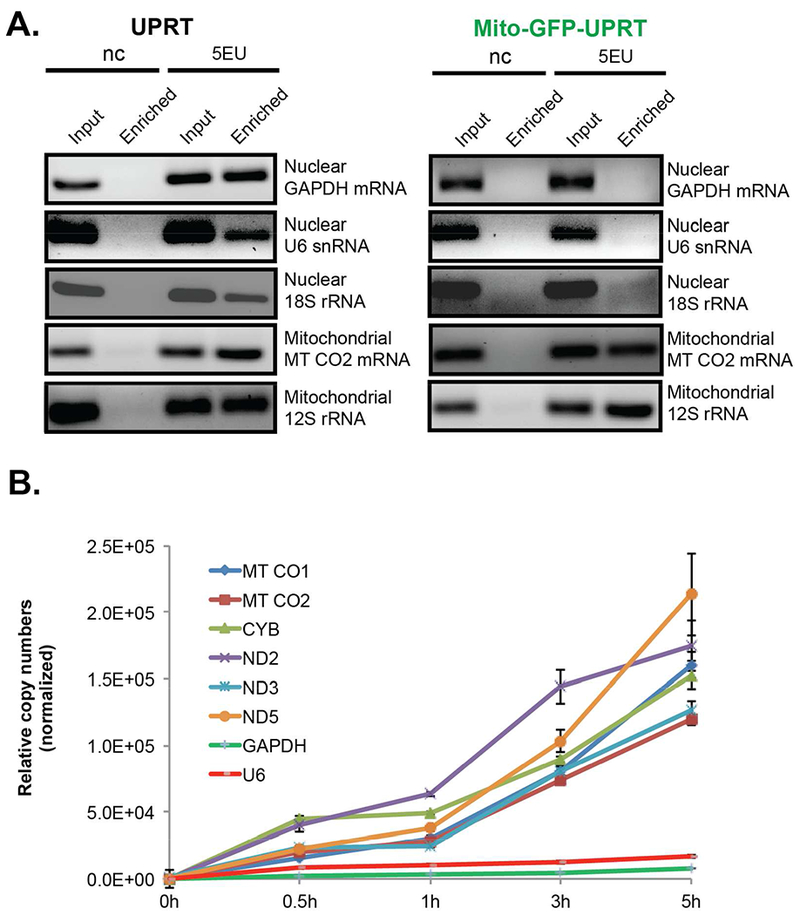
A. Enriched cDNA templates derived from RNA of spatially-unrestricted UPRT vs. Mito-GFP-UPRT expressing cells were assayed by PCR with specific primer pairs for nuclear or mitochondrial transcripts (nc: negative control = DMSO; 5EU = 5’Ethynyl uracil). B Assessment of six mitochondrial and two nuclear transcripts (served as negative controls) from RNA of transiently transfected Mito-GFP-UPRT cells exposed to 5EU in a time course by SYBR Green RT-qPCR analysis (n = two technical duplicates)
Nascent tracking of RNA synthesis is an important experimental approach toward characterizing RNA expression dynamics. Although there has been considerable analysis of nuclear RNA expression through pulse-labeling of nucleoside analogs, the approach has yet to be demonstrated in an approach only labeling mitochondrial RNA. We sought to test if our strategy could be used to track RNA synthesis for mitochondrial RNA. For this, we tracked several mitochondrial RNAs: MT-CO1/MT-CO2 mRNAs (cytochrome c oxidase subunit I & II enzymes, also known as COX1 and COX2), CYB mRNA (cytochrome B), ND2/ND3/ND5 mRNAs (NADH dehydrogenase subunits 2, 3 and 5), 33, 34 and along with nuclear transcribed RNA such as GAPDH and U6. Mito-GFP-UPRT-containing cells were treated with 5EU and total RNA isolated at different time points. RNA was enriched and quantified using RT-qPCR. As shown in Figure 4, B, we were able to enrich and track the nascent transcription of the mitochondrial RNAs, but not RNAs from the nuclear genome. Overall, these results demonstrate the ability to stringently label mitochondrial RNA (and not nuclear RNA), they also show the power of our approach in being able to track nascent RNA synthesis and dynamics.
Herein we have developed a method for spatially-restricted tracking of RNA biosynthesis in cells, through the controlled localization of an enzyme controlling bioorthogonal nucleotide biosynthesis. We have demonstrated, through imaging, that our Mito-specific UPRT-mediated analog and nascent RNA labeling with a bioorthogonal handle co-localize inside cells. Furthermore, we have demonstrated distinct populations of the controlled labeling of mitochondrial RNA, and undetectable labeling of RNA arising from the nuclear genome. This approach is well-positioned to enable controlled and high-stringency analysis of the dynamics of mitochondrial RNA synthesis, as demonstrated in Figure 4. This approach can be married with more unbiased analyses, such as RNA sequencing, to preferentially uncover novel RNA molecules produced from the mitochondrial genome. Furthermore, as the analogs and enzymes used herein have already been demonstrated to work in animals we anticipate our approach may also be used to study the dynamics of mitochondrial RNA expression in living animals,7 even perhaps in complex environments such as the central nervous system.35 Future efforts will be focused on extending these findings into complex cellular environments, in vivo.
Supplementary Material
ACKNOWLEDGMENT
We thank members of the Spitale lab for their careful reading and critique of the manuscript. We thank M. Cleary (UC Merced) for his generous gift of the UPRT plasmid used. RNA research in the Spitale lab is supported by start up funds from the University of California, Irvine, and the NIH Director’s New Innovator Award (1DP2GM119164 and 1R21MH113062 RCS). RCS is a Pew Biomedical Scholar.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Experimental methods, synthetic schemes and spectra, for all compounds are available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Chappell J, Takahashi MK, Meyer S, Loughrey D, Watters KE, and Lucks J (2013) The centrality of RNA for engineering gene expression, Biotechnology Journal 8, 1379–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharp PA (2009) The centrality of RNA, Cell 136, 577–580. [DOI] [PubMed] [Google Scholar]
- 3.Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, Amit I, and Regev A (2011) Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells, Nature Biotechnology 29, 436–U237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duffy EE, Rutenberg-Schoenberg M, Stark CD, Kitchen RR, Gerstein MB, and Simon MD (2015) Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry, Mol Cell 59, 858–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curanovic D, Cohen M, Singh I, Slagle CE, Leslie CS, and Jaffrey SR (2013) Global profiling of stimulus-induced polyadenylation in cells using a poly(A) trap, Nat Chem Biol 9, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Y, and Beal PA (2016) Synthesis and evaluation of an alkyne-modified ATP analog for enzymatic incorporation into RNA, Bioorg Med Chem Lett 26, 1799–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jao CY, and Salic A (2008) Exploring RNA transcription and turnover in vivo by using click chemistry, Proc Natl Acad Sci U S A 105, 15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curanovic D, Cohen M, Singh I, Slagle CE, Leslie CS, and Jaffrey SR (2013) Global profiling of stimulus-induced polyadenylation in cells using a poly(A) trap, Nat Chem Biol 9, 671–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nainar S, Beasley S, Fazio M, Kubota M, Dai N, Correa IR Jr., and Spitale RC (2016) Metabolic Incorporation of Azide Functionality into Cellular RNA, Chembiochem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vyas S, Zaganjor E, and Haigis MC (2016) Mitochondria and Cancer, Cell 166, 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattson MP, Gleichmann M, and Cheng A (2008) Mitochondria in Neuroplasticity and Neurological Disorders, Neuron 60, 748–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu GQ, Mercer TR, Shearwood AMJ, Siira SJ, Hibbs ME, Mattick JS, Rackham O, and Filipovska A (2013) Mapping of Mitochondrial RNA-Protein Interactions by Digital RNase Footprinting, Cell Rep 5, 839–848. [DOI] [PubMed] [Google Scholar]
- 13.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AMJ, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, and Mattick JS (2011) The Human Mitochondrial Transcriptome, Cell 146, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma H, Folmes CDL, Wu J, Morey R, Mora-Castilla S, Ocampo A, Ma L, Poulton J, Wang XJ, Ahmed R, Kang EJ, Lee Y, Hayama T, Li Y, Van Dyken C, Gutierrez NM, Tippner-Hedges R, Koski A, Mitalipov N, Amato P, Wolf DP, Huang TS, Terzic A, Laurent LC, Belmonte JCI, and Mitalipov S (2015) Metabolic rescue in pluripotent cells from patients with mtDNA disease, Nature 524, 234-+. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Vizarra E, Lopez-Perez MJ and Enriquez JA (2002) Isolation of biogenetically competent mitochondria from mammalian tissues and cultured cells, Methods 26, 292–297. [DOI] [PubMed] [Google Scholar]
- 16.Pallotti F, and Lenaz G (2007) Isolation and subfractionation of mitochondria from animal cells and tissue culture lines, Method Cell Biol 80, 3–44. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt S, Eberhagen C, Weber S, Aichler M, and Zischka H (2015) Isolation of Mitochondria from Cultured Cells and Liver Tissue Biopsies for Molecular and Biochemical Analyses, Proteomic Profiling: Methods and Protocols 1295, 87–97. [DOI] [PubMed] [Google Scholar]
- 18.Havugimana PC, Hart GT, Nepusz T, Yang HX, Turinsky AL, Li ZH, Wang PI, Boutz DR, Fong V, Phanse S, Babu M, Craig SA, Hu PZ, Wan CH, Vlasblom J, Dar VUN, Bezginov A, Clark GW, Wu GC, Wodak SJ, Tillier ERM, Paccanaro A, Marcotte EM, and Emili A (2012) A Census of Human Soluble Protein Complexes, Cell 150, 1068–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaltimbacher V, Bonnet C, Lecoeuvre G, Forster V, Sahel JA, and Corral-Debrinski M (2006) mRNA localization to the mitochondrial surface allows the efficient translocation inside the organelle of a nuclear recoded ATP6 protein, Rna-a Publication of the Rna Society 12, 1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadir N, Haim-Vilmovsky L, Kraut-Cohen J, and Gerst JE (2011) Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae, Rna-a Publication of the Rna Society 17, 1551–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frezza C, Cipolat S, and Scorrano L (2007) Organelle isolation: functional mitochondria from mouse liver, muscle and cultured filroblasts, Nat Protoc 2, 287–295. [DOI] [PubMed] [Google Scholar]
- 22.Lampl T, Crum JA, Davis TA, Milligan C, and Moore VD (2015) Isolation and Functional Analysis of Mitochondria from Cultured Cells and Mouse Tissue, Jove-J Vis Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pallotti F, and Lenaz G (2001) Isolation and subfractionation of mitochondria from animal cells and tissue culture lines, Method Cell Biol 65, 1–35. [DOI] [PubMed] [Google Scholar]
- 24.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, and Mattick JS (2011) The human mitochondrial transcriptome, Cell 146, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forini F, D’aurizio R, Kusmic C, Nicolini G, Baumgart M, Groth M, Ucciferri N, Iervasi G, and Pitto L (2016) A high-throghput approach unveils putative miRNA-mediated mitochondria-targeted cardioprotective circuits activated by T3 in the post ischemia reperfusion setting, Cardiovasc Res 111, S95–S95. [Google Scholar]
- 26.Sripada L, Tomar D, Prajapati P, Singh R, Singh AK, and Singh R (2012) Systematic Analysis of Small RNAs Associated with Human Mitochondria by Deep Sequencing: Detailed Analysis of Mitochondrial Associated miRNA, PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rackham O, Shearwood AMJ, Mercer TR, Davies SMK, Mattick JS, and Filipovska A (2011) Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins, Rna-a Publication of the Rna Society 17, 2085–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen K, Fazio M, Kubota M, Nainar S, Feng C, Li X, Atwood SX, Bredy TW, and Spitale RC (2017) Cell-Selective Bioorthogonal Metabolic Labeling of RNA, J Am Chem Soc 139, 2148–2151. [DOI] [PubMed] [Google Scholar]
- 29.Krath BN, and Hove-Jensen B (1999) Organellar and cytosolic localization of four phosphoribosyl diphosphate synthase isozymes in spinach, Plant Physiol 119, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Von Ohlen T, Luce-Fedrow A, Ortega MT, Ganta RR, and Chapes SK (2012) Identification of critical host mitochondrion-associated genes during Ehrlichia chaffeensis infections, Infect Immun 80, 3576–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenberg N, Schumm DE, and Webb TE (1977) Uridine kinase activities and pyrimidine nucleoside phosphorylation in fluoropyrimidine-sensitive and -resistant cell lines of the Novikoff hepatoma, Biochem J 164, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen K, Fazio M, Kubota M, Nainar S, Feng C, Li X, Atwood SX, Bredy TW, and Spitale RC (2017) Cell-Selective Bioorthogonal Metabolic Labeling of RNA, Journal of the American Chemical Society 139, 2148–2151. [DOI] [PubMed] [Google Scholar]
- 33.Chen H, and Chan DC (2017) Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells, Cell Metab [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khacho M, and Slack RS (2017) Mitochondrial dynamics in the regulation of neurogenesis: From development to the adult brain, Dev Dyn [DOI] [PubMed] [Google Scholar]
- 35.Gay L, Miller MR, Ventura PB, Devasthali V, Vue Z, Thompson HL, Temple S, Zong H, Clear MD, Stankunas K, and Doe CQ (2013) Mouse TU tagging: a chemical/genetic intersectional method for purifying cell type-specific nascent RNA, Genes Dev 27, 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


