APOBEC enzymes are DNA cytosine deaminases that normally serve as virus restriction factors, but several members, including APOBEC3H, also contribute to cancer mutagenesis. Despite their importance in multiple fields, little is known about cellular processes that regulate these DNA mutating enzymes.
KEYWORDS: APOBEC3H, cancer mutagenesis, cytoplasmic retention, DNA deamination, nuclear import, retrovirus restriction, subcellular localization
ABSTRACT
APOBEC enzymes are DNA cytosine deaminases that normally serve as virus restriction factors, but several members, including APOBEC3H, also contribute to cancer mutagenesis. Despite their importance in multiple fields, little is known about cellular processes that regulate these DNA mutating enzymes. We show that APOBEC3H exists in two distinct subcellular compartments, cytoplasm and nucleolus, and that the structural determinants for each mechanism are genetically separable. First, native and fluorescently tagged APOBEC3Hs localize to these two compartments in multiple cell types. Second, a series of genetic, pharmacologic, and cell biological studies demonstrate active cytoplasmic and nucleolar retention mechanisms, whereas nuclear import and export occur through passive diffusion. Third, APOBEC3H cytoplasmic retention determinants relocalize APOBEC3A from a passive cell-wide state to the cytosol and, additionally, endow potent HIV-1 restriction activity. These results indicate that APOBEC3H has a structural zipcode for subcellular localization and selecting viral substrates for restriction.
INTRODUCTION
The APOBEC3 (A3) enzymes are comprised of either one (A3A, A3C, and A3H) or two (A3D, A3F, A3G, and A3B) zinc-coordinating domains that are grouped into three identified clades based on sequence homology (Z1, Z2, and Z3) (1–3). A3H is unique among the human A3 loci in that it is the most distantly related and is classified as the only Z3 domain (1, 4). A3H is also the most naturally variable A3, with at least seven different haplotypes and four unique splice variants that display differential enzymatic activity, stability, and subcellular distribution (5–10). The canonical function of the A3s is to restrict a wide variety of reverse-transcribing retroviruses and retroelements through both deaminase-dependent and -independent mechanisms (4, 11–14). A3D, A3F, A3G, and A3H incorporate into nascent HIV-1 particles and exert antiviral activity by mutating cytosines to uracils in newly synthesized viral cDNA (G-to-A mutation on the reverse strand) (8, 15–29). Interestingly, several lines of evidence have implicated A3H, A3B, and A3A as major sources of mutation in several different cancer types, including cervical, lung, head/neck, and bladder cancers (30–35). APOBEC mutational signatures are present in over half of all human cancers and often account for the majority of mutations in primary tumors, and in the case of breast cancer, they can account for greater than 50% of the mutations in metastatic tumors (36, 37).
Although DNA deamination is the hallmark A3 enzymatic activity, RNA has emerged as a significant regulator. Not only is RNA binding activity of restrictive A3s critical for encapsidation into viral particles and subsequent antiviral activity, but at least two family members, APOBEC1 (38) and A3A (39), exhibit RNA-editing capabilities. Additionally, RNA binding has also been implicated in maintaining proper subcellular localization for at least three different A3s: A3F, A3G, and recently A3H (5, 40–47). Furthermore, interactions between several A3s and cellular proteins can be disrupted by RNase A treatment, suggesting that RNA plays a pivotal role in regulating A3 activities. This is further evidenced by recent studies requiring RNase A treatment in order to purify high quantities of recombinant A3H protein from cell lysates (40, 41, 48).
Multiple groups recently succeeded in solving the X-ray crystal structure of A3H and discovered a surprising RNA-mediated dimerization mechanism (40, 48). In these structures, two A3H monomers dimerize around a short, 7- to 9-base-pair RNA duplex. Both the human and macaque A3H dimers were shown to interact extensively with the RNA strands of the duplex through contacts with loop 1, loop 7, and α-helix 6. Several separation-of-function mutants were identified within the RNA binding interface that retained DNA deaminase activity but lost RNA binding and dimerization, cytoplasmic localization, encapsidation, and anti-HIV activity (5, 40, 41). Curiously, disruption of the RNA-mediated dimer resulted in strong nuclear enrichment of A3H (5, 6, 30, 40, 41). Together, these findings further emphasize the importance of RNA in regulating A3 activities.
Here, we investigate the most common stably expressed and active A3H enzyme in global populations, the 183-amino-acid splice variant of haplotype II (7, 29). Initially, we sought to further characterize the role of the RNA binding interface in regulating A3H subcellular localization, but we were surprised by distinct nucleolar and cytoplasmic localization patterns for both native (untagged) and mCherry (mCh)-tagged enzymes. Further characterization of these properties identified structural determinants on the surface of A3H that regulate subcellular localization by genetically separable mechanisms, resulting in active cytoplasmic or nucleolar retention with passive nucleocytoplasmic diffusion. Finally, we show that A3H cytoplasmic retention determinants can relocalize A3A to the cytosol, which endows potent HIV-1 restriction activity. Overall, we propose a model in which A3H contains overlapping structural zipcodes that regulate steady-state subcellular localization.
RESULTS
Subcellular distribution of A3H.
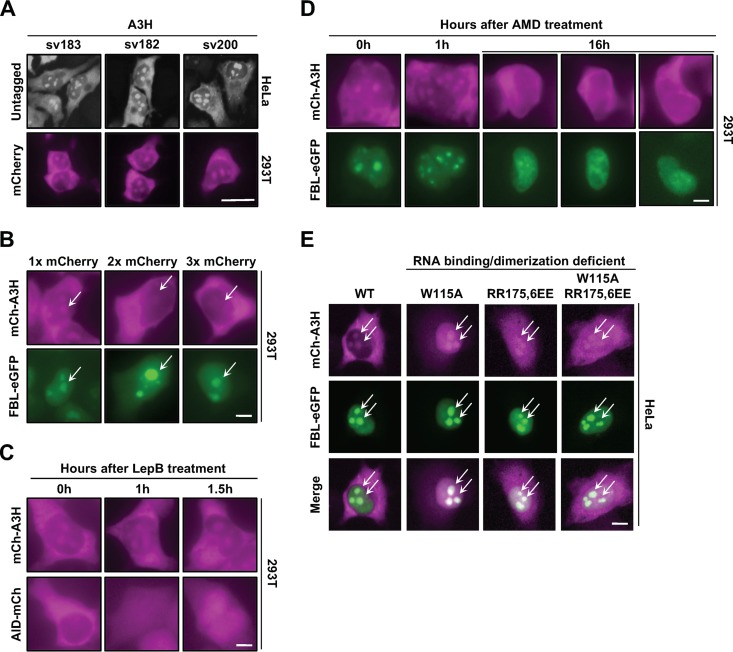
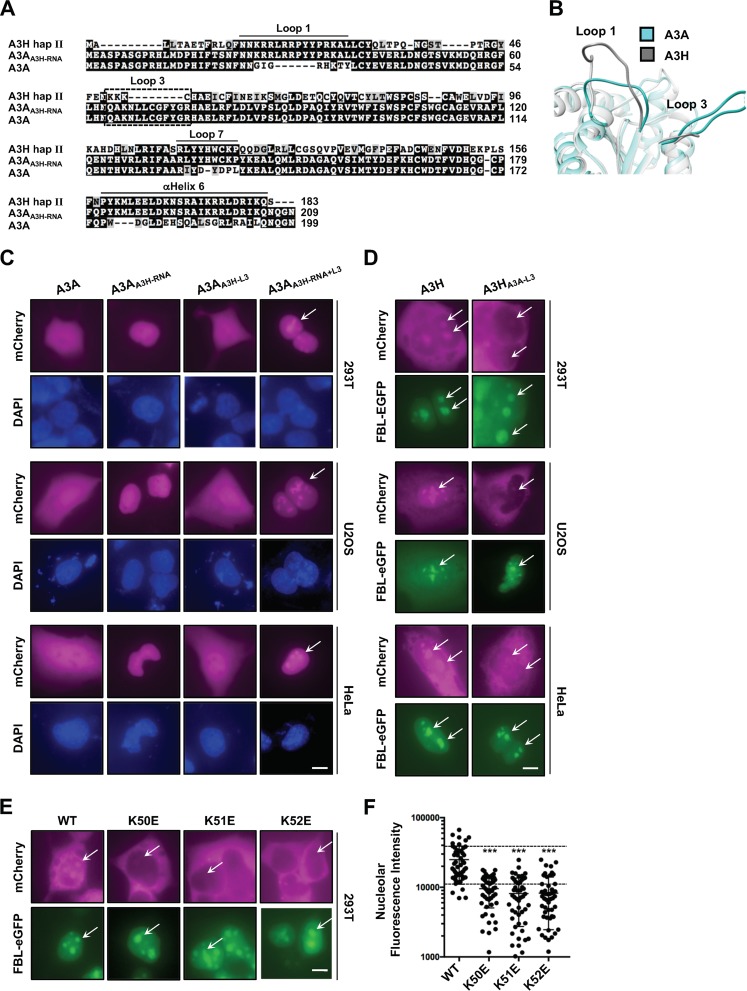
Previous studies have described A3H localization as predominantly cytoplasmic (6, 40, 49, 50), so we were surprised when immunofluorescence experiments using an A3H-specific polyclonal antibody detected two distinct subcellular pools of native A3H in HeLa cells (cytoplasmic and nuclear) (Fig. 1A, top). To confirm that this localization pattern was not due to an artifact of the immunofluorescence procedure, we added an N-terminal mCherry (mCh) fluorescence tag and still observed dual localization in 293T cells, indicating that this localization pattern is independent of detection method and cell type (Fig. 1A, bottom). In addition, two C-terminal splice variants of 182 and 200 residues in length had indistinguishable localization patterns, demonstrating that shared residues 1 to 182 govern both localization mechanisms (Fig. 1A, top and bottom).
FIG 1.
Evidence for cytoplasmic and nucleolar A3H. (A) Representative images of native A3H and mCherry-tagged (mCh) splice variants. Scale bar, 10 μm. (B) Representative images of mCh-A3H with 1, 2, or 3 copies of mCherry coexpressed with fibrillarin-eGFP (FBL-eGFP). Arrows indicate regions of interest (ROI). (C and D) Representative images of a time course treatment with either leptomycin B (LepB) (40 ng/ml) (C) or actinomycin D (AMD) (0.001 ng/ml) (D). (E) Representative images of the indicated mCh-A3H constructs coexpressed with FBL-eGFP in HeLa cells. ROI are indicated by arrows. Scale bars in panels B to E, 5 μm.
These results suggested that A3H maintains steady-state levels in two distinct subcellular compartments, which would be unique among the A3s since they typically exhibit a single localization pattern (46, 51–53). First, we wanted to determine where A3H localizes within the nucleus. The largest intranuclear organelles are the nucleoli, which are comprised primarily of actively transcribed ribosomal DNA (rDNA), nascently synthesized rRNA, and ribosome assembly proteins (54–59). To test for nucleolar localization, we coexpressed mCh-A3H with enhanced green fluorescent protein-tagged fibrillarin (FBL-eGFP), a well-studied nucleolar protein (60). As shown in Fig. 1B, the bulk of the nuclear A3H overlapped with FBL-eGFP in nucleolar structures.
Recently, we and others discovered that A3H forms RNA-mediated homodimers and that disruption of the dimer results in strong nuclear enrichment (40, 41). Because of this, we were curious if A3H contains a nuclear localization sequence (NLS), which would also account for the robust nucleolar localization observed in Fig. 1A. To test this idea, we generated 1×, 2×, and 3× mCh-A3H constructs and expressed them in 293T cells (Fig. 1B). The maximal size of a protein that can passively diffuse through the nuclear pore is 60 to 70 kDa (61–63); therefore, the addition of multiple copies of mCherry would render A3H too big to passively diffuse into the nuclear compartment. As demonstrated in Fig. 1B, the addition of 2× and 3× mCherry resulted in exclusively cytoplasmic localization even though FBL-eGFP was observed in nucleolar structures, suggesting that A3H import occurs through a passive mechanism.
Next, we asked whether A3H is exported actively from the nucleus to maintain steady-state levels in distinct subcellular compartments. The related family member AID has been shown to undergo rapid nucleocytoplasmic shuttling, with import dependent on CTNNBL1 and export on CRM1 (64–66). To determine if A3H also undergoes rapid nucleocytoplasmic shuttling, we used the CRM1 nuclear export inhibitor leptomycin B (LepB) (67, 68). As shown in Fig. 1C, treatment of 293T cells with LepB resulted in nuclear accumulation of AID-mCh over time. However, LepB treatment of cells expressing mCh-A3H did not display nuclear enrichment or increased nuclear accumulation, suggesting that A3H is not rapidly exported from the nucleus through a CRM1-mediated pathway. Taken together, these results indicated that movement of A3H into and out of the nuclear compartment is most likely due to passive diffusion followed by retention.
We next used actinomycin D (AMD) to better understand how A3H is retained within the nucleolus. AMD normally inhibits RNA transcription at low dosages for short treatment times (69, 70); however, increased dosage or treatment duration causes redistribution of nucleolar compartments (71–73). We hypothesized that accumulation of A3H within nucleoli occurs through binding to unknown nucleolar factors and that dispersion of the nucleolar compartments would result in loss of A3H within the nucleus. As expected, long-term treatment of 293T cells with AMD resulted in partial relocalization of FBL-eGFP from nucleolar to nuclear, indicating nucleolar disassembly (Fig. 1D, compare 0 h versus 16 h FBL-eGFP). Similarly, in support of our idea, AMD caused A3H to disappear from nucleolar foci and become predominantly cytoplasmic (Fig. 1D).
The loss of A3H from nucleoli following AMD treatment suggested that A3H may be actively retained in nucleolar compartments. Since nucleoli are comprised primarily of a dense concentration of rRNA and recent evidence suggests that a majority of A3H activities are mediated by RNA-based interactions, we reasoned that A3H is retained in nucleoli through interactions with rRNA. To test this, we examined the localization patterns of previously reported separation-of-function mutants that maintain potent DNA deamination activity but lose RNA binding and dimerization activities (5, 40). As shown in Fig. 1E, wild-type A3H and three separation-of-function mutants (W115A, RR175,6EE, and W115A/RR175,6EE) showed similar levels of nucleolar accumulation. We were surprised by these results because they suggested that nucleolar retention of A3H is not due to RNA binding activity but rather that A3H may be targeted specifically to this compartment by a yet-unknown nucleolar factor.
A3H loop 1 is required but not sufficient for nucleolar localization.
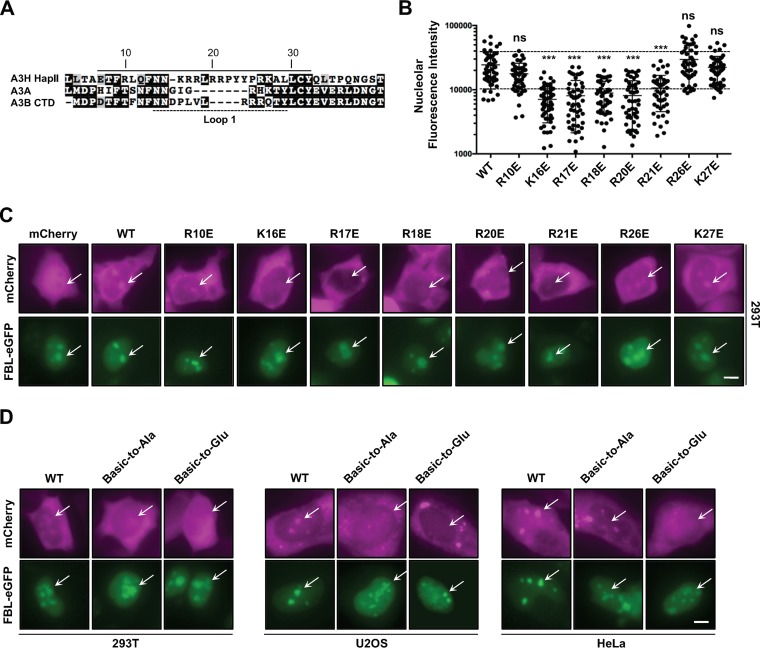
We next predicted that nucleolar retention of A3H would be due to specific cis-acting residues. Analysis of the A3H protein sequence using a prediction algorithm based on known nucleolar retention sequences (74, 75) suggested that a nucleolar targeting sequence may exist within loop 1, which encompasses a stretch of positively charged arginine and lysine residues (Fig. 2A). While no strict consensus motif has been defined for nucleolar targeting, numerous proteins have been characterized as having targeting sequences comprised of stretches of basic amino acids (76, 77). To determine if these residues have a role in nucleolar targeting, we changed each basic residue within loop 1 to glutamate and assessed the effect on localization. As quantified in Fig. 2B and depicted in representative images in Fig. 2C, single-amino-acid substitutions at positions K16, R17, R18, R20, and R21 significantly attenuated nucleolar localization compared to that for wild-type A3H and FBL-eGFP controls, while substitutions at positions R10, R26, and K27 had little effect. Additionally, several of these mutants (K16E, R18E, R26E, and K27E) have been reported to have potent DNA deamination activity and antiviral activity against HIV-1, and therefore, loss of nucleolar accumulation is not due to protein stability or misfolding (references 40 and 41 and data not shown). Furthermore, replacing all positive residues from position 16 to 21 with either alanine or glutamate resulted in abrogated nucleolar localization when expressed in 293T, U2OS, or HeLa cells (Fig. 2D). Taken together, these data indicated that loop 1 could be acting as a nucleolar targeting sequence.
FIG 2.
A predicted nucleolar targeting sequence in loop 1 is required for nucleolar localization. (A) Alignment of A3H, A3A, and A3B C-terminal domain amino acid sequences flanking loop 1 (dashed line). The predicted nucleolar localization sequence of A3H is indicated by the solid line. (B) Quantification of nucleolar fluorescence intensity of the indicated A3H constructs (n = 50 nucleoli). The dashed lines highlight ±SEM from wild-type quantification. ns, no significance; ***, P < 0.001 by the unpaired Student t test. (C and D) Representative images of the indicated mCh-A3H constructs coexpressed with FBL-eGFP. ROI are indicated by arrows. Basic-to-Ala/Glu refers to amino acids K16 to R21 replaced by either alanine or glutamate. All scale bars, 5 μm.
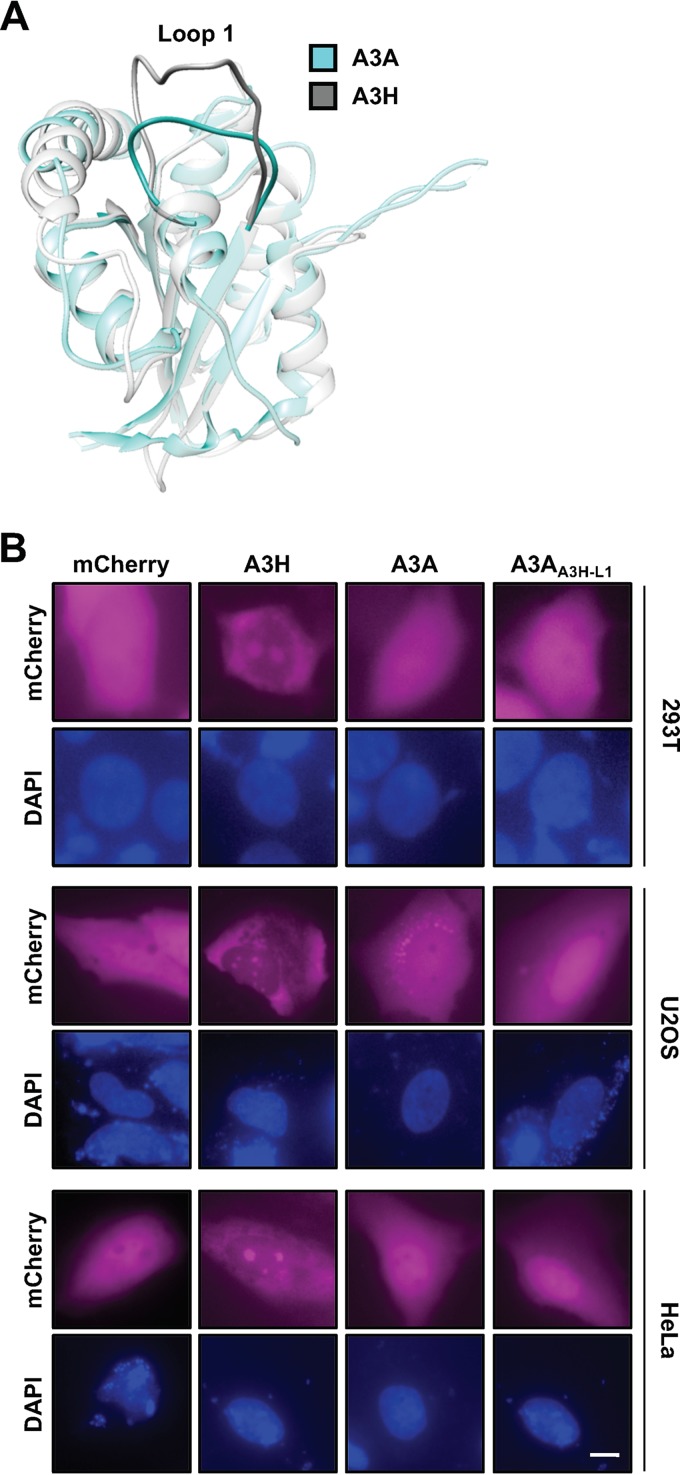
To determine if loop 1 is sufficient to drive nucleolar accumulation of a related single-domain enzyme, we exchanged loop 1 from A3H with loop 1 from A3A, which exhibits cell-wide localization when overexpressed (53) or cytoplasmic localization at endogenous expression levels (78). Importantly, the crystal structure of A3A has been determined (79–81) which allows for structure-guided analysis between A3A and A3H (40, 41) (structural overlay in Fig. 3A). Additionally, A3A is known to have very potent deaminase activity, and because of this, all initial studies using A3A as a molecular scaffolding were done with the catalytically inactive E72A mutant to avoid cytotoxicity (31, 82–84). As expected, overexpressed mCh-A3A displayed cell-wide localization; however, exchanging loop 1 from A3H had no discernible effect on localization when expressed in 293T, U2OS, or HeLa cells (Fig. 3B).
FIG 3.
A3H loop 1 is not sufficient for redistribution of A3A to the nucleolus. (A) Structural overlay of A3H (PDB 6B0B) and A3A (PBD 4XXO), highlighting the positioning of loop 1. (B) Representative images of A3H loop 1 swapped into mCh-A3A. Scale bar, 5 μm.
Chimeras exchanging loop 1 and either loop 7 or α-helix 6 relocalize A3A to the nucleus through an active import mechanism.
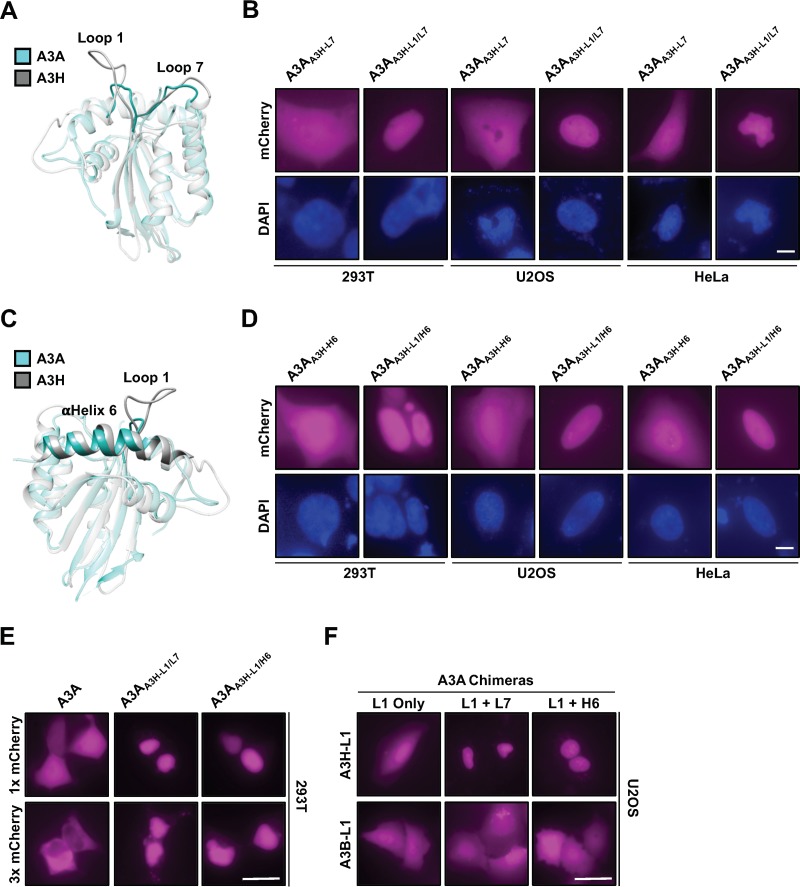
Since A3H and A3A are single-domain enzymes with nearly identical cores, we constructed additional chimeras to ask what surface residues are required to endow A3A with A3H properties. We first tested other components of the RNA binding interface because our work implicated loop 1 in nucleolar retention (see above) and structural studies showed select loop 1 residues together with loop 7 or α-helix 6 residues combining to bind duplex RNA (40, 41, 48). We therefore generated A3A chimeras that exchanged loop 7 or α-helix 6 alone or in combination with loop 1 and assessed localization (structural overlay in Fig. 4A and C). Neither loop 7 nor α-helix 6 alone had any effect on A3A localization (similar to exchange of loop 1 alone). However, when exchanged in combination with A3H loop 1, either loop 7 or α-helix 6 residues were sufficient to relocalize A3A to the nucleus in all cell lines tested (A3AA3H-L1/L7 and A3AA3H-L1/H6 in Fig. 4B and D).
FIG 4.
Loop 1 of A3H in combination with either loop 7 or α-helix 6 can relocalize A3A to the nucleus through an active import mechanism. (A and C) Structural representation of A3H overlaid with A3A to highlight the orientations of loop 1, α-helix 6, and loop 7 within the RNA binding interface. (B and D) Representative images of cells expressing the indicated mCh-A3A construct. Scale bars, 5 μm. (E) Representative images of the indicated construct with either 1× or 3× mCherry. (F) Representative images of A3A chimeras with loop 1 exchanged from either A3H or A3B alone or in combination with A3H loop 7 or α-helix 6. Scale bars, 10 μm.
It was odd that these A3A chimeras displayed such a strong nuclear localization phenotype given that neither wild-type A3A nor A3H displays this localization pattern normally. We wondered if these chimeras were being actively imported into the nuclear compartment through the putative nucleolar targeting motif in loop 1. Canonical nuclear import sequences are comprised of clusters of positively charged residues (85, 86), much like the composition of A3H loop 1, and there have been several examples of nuclear/nucleolar import sequences that have overlapping import functions (87–89). To test this idea, we generated A3A wild-type, A3AA3H-L1/L7, and A3AA3H-L1/H6 constructs with a 3× mCherry tag and assessed localization. As we would expect, 3× mCh-A3A displayed nuclear exclusion (fully cytoplasmic); however, both the A3AA3H-L1/L7 and A3AA3H-L1/H6 chimeras displayed strong nuclear localization even in the presence of 3× mCherry (Fig. 4E). To confirm that the relocalization of A3A chimeras shown in Fig. 4 was due to the presence of A3H loop 1 and not, for instance, due to the loss of protein determinants involved in regulating A3A localization, we tested the exchange of A3B C-terminal domain (CTD) loop 1 alone or in combination with loop 7 or α-helix 6 from A3H (a sequence alignment is depicted in Fig. 2A). As shown in Fig. 4F, when A3B CTD loop 1 was exchanged in combination with either loop 7 or α-helix 6 from A3H, the A3A chimeras failed to relocalize to the nuclear compartment in all cell lines tested (293T and HeLa cells also display cell-wide localization [data not shown]), further indicating that A3H loop 1 functions as a nuclear import/nucleolar retention sequence under certain conditions.
Loop 3 in combination with the RNA binding interface governs A3H nucleolar localization.
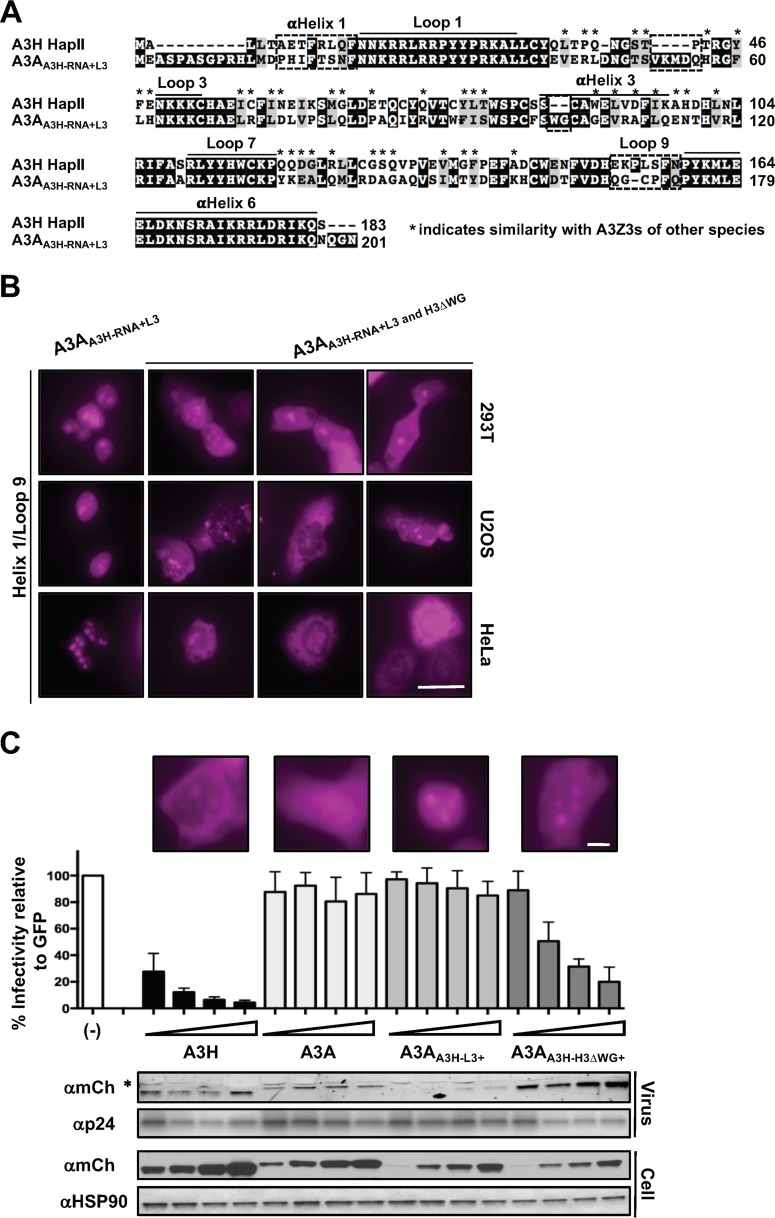
While relocalization of the A3A chimeras to the nucleus was interesting, it still did not explain what protein components are required for A3H nucleolar retention. The results in Fig. 1 and 4 suggested that components of the RNA binding interface may have overlapping functions as localization determinants, since partial reconstitution of the RNA binding interface in A3A could initiate active nuclear import. Therefore, we wondered whether exchanging the entire A3H RNA binding interface in A3A would result in an A3H-like nucleolus/cytoplasm localization pattern. We exchanged these components in A3A (referred to as A3AA3H-RNA) and still only observed nuclear localization, demonstrating that additional residues outside the RNA binding interface contribute to both A3H nucleolar and cytoplasmic retention mechanisms (Fig. 5C).
FIG 5.
Loop 3 of A3H contributes to nucleolar retention along with the RNA binding interface. (A) Alignment of wild-type A3H, A3A, and an A3AA3H-RNA chimera (A3AA3H-RNA has loop 1, loop 7, and α-helix 6 exchanged from A3H) amino acid sequences that highlight loop 3 (boxed sequence). (B) Structural representation of A3H overlaid with A3A to highlight the proximity of loop 3 to loop 1. (C to E) Representative images of the indicated A3A and A3H constructs. ROI are indicated by arrows. (F) Quantification of nucleolar fluorescence intensity of the indicated A3H constructs (n = 50 nucleoli). The dashed lines highlight ±SEM from wild-type quantification. ns, no significance; ***, P < 0.001 by the unpaired Student t test. All scale bars, 5 μm.
To gain further insight as to what these protein components might be, we examined the structures of A3H and A3A, looking for regions outside the RNA binding interface that are significantly different (the sequence alignment is shown in Fig. 5A, and the structural overlay is shown in Fig. 5B). One obvious region is loop 3, which is comprised of three consecutive lysine residues in A3H and is 10 residues longer with no concentrated positive charge in A3A. Because of this, we wondered if A3A loop 3 was sterically hindering relocalization of the chimeras tested for Fig. 4. As shown in Fig. 5C, exchanging loop 3 alone had no effect on A3A localization; however, exchanging loop 3 along with the RNA binding surface from A3H resulted in strong nucleolar accumulation (Fig. 5C). Additionally, A3H nucleolar accumulation was compromised in all cell lines tested by exchanging loop 3 with A3A loop 3 (Fig. 5D), and replacement of individual A3H loop 3 residues K50, K51, and K52 by glutamate also caused nucleolar accumulation to be strongly reduced (Fig. 5E and F). Taken together, these data identified loop 3 as part of the nucleolar localization mechanism and indicate that several cis elements on the surface of A3H, such as loop 1, loop7, and α-helix 6, work cohesively to promote nucleolar localization.
Relocalization of A3A to the cytoplasm endows potent HIV-1 restriction activity.
Closer examination of the A3A and A3H crystal structures led to the identification of both the RNA binding interface and loop 3 as determinants involved in regulating nucleolar localization of A3H. However, these chimeric constructs still failed to recapitulate A3H cytoplasmic localization even though we had reconstituted the full RNA binding interface. To better understand the cytoplasmic retention mechanism, we assessed the localization patterns of A3H homologs (A3Z3s) from related species. We tested mCherry-tagged rhesus macaque, cow, pig, sheep, and cat A3Z3 domains and confirmed cytoplasmic localization and, in some instances, also nucleolar accumulation, consistent with previous reports (references 6, 90, and 91 and data not shown). With this in mind, we compared amino acid sequences from these related A3Z3s to human A3H and A3AA3H-RNA+L3 to identify amino acid residues that were conserved among A3Z3s but differed from those in the A3AA3H-RNA+L3 chimera. We reasoned that excluding these residues from our search would reveal additional determinants involved in cytoplasmic retention that were not apparent in the cocrystal structure of A3H in complex with duplexed RNA (40, 41, 48). As shown in Fig. 6A, several regions were identified as potential candidates: α-helix 1, a four-amino-acid “insertion” at the end of β2, loop 9, and a WG “insertion” at the top of α-helix 3. We exchanged various combinations of these regions using the A3AA3H-RNA+L3 chimera as a scaffold and observed no discernible differences in localization patterns (data not shown). However, when we exchanged α-helix 1 in combination with loop 9 and the deletion of the “WG” sequence at the top of α-helix 3, the A3A chimera displayed a dual-localization pattern similar to that of wild-type A3H in all cell lines tested (Fig. 6B).
FIG 6.
Relocalization of A3A to the cytoplasm confers antiviral activity against Vif-deficient HIV-1. (A) Amino acid alignment of A3H and the A3AA3H-RNA+L3 chimera. Asterisks indicate amino acids in analogous A3Z3s that have similar biophysical properties. Dashed boxes indicate ROI. (B) Representative images of the indicated mCh-A3 chimeric proteins. Scale bar, 10 μm. (C) Top, Vif-deficient HIV-1 restriction assay with the indicated A3 constructs (“+” indicates that α-helix 1, loop 9, and the RNA binding interface are exchanged along with the indicated regions). A representative image of each enzyme is shown above the corresponding viral infectivity results. Bottom, immunoblots demonstrating expression of each construct in the cell lysate and the ability of the A3 protein to package into Vif-deficient HIV-1 particles. The asterisk indicates a nonspecific band in the viral particle blot. Scale bar, 5 μm.
Since the cytoplasmic retention of A3H has been linked to RNA binding, dimerization, and antiviral activity, we were curious about whether the A3A chimera that exhibited wild-type A3H-like localization could also restrict HIV-1. We tested the A3A chimeras that displayed either nucleolar/cytoplasmic localization or nuclear/nucleolar localization for their ability to restrict Vif-deficient HIV-1 particles. Vif is a well-characterized HIV-1 accessory gene product that counteracts the restrictive potential of the A3s by targeting these enzymes for proteasomal degradation in virus-producing cells (92). Because of this, we performed viral infectivity assays with Vif-deficient HIV-1 proviral plasmids to accurately assess antiviral activity of the A3A/H chimeras. As shown in Fig. 6C, the A3A chimera that displayed nucleolar/cytoplasmic localization could restrict HIV-1 particles in a dose-dependent manner, while the nucleolar/nuclear chimera failed to restrict virus at all concentrations tested (similar infectivity results were observed for untagged wild-type and mutant constructs [data not shown]). Additionally, the A3A chimera that displayed an A3H-like localization pattern and robust antiviral activity was observed at higher levels in viral particles than were wild-type and nuclear/nucleolar A3A proteins. This is the first instance in which the virus-targeting mechanism from one A3 enzyme has been transferred fully to a normally nonrestrictive A3 enzyme.
DISCUSSION
Human A3H is unique among the A3 family members because it has the most natural variation across global populations, and it is the only single-domain enzyme to display potent antiviral activity against HIV-1. Here, testing an extensive panel of A3 chimeras and amino acid substitutions in a variety of cell lines revealed that A3H is actively retained in two subcellular compartments, cytoplasm and nucleolus, by overlapping but genetically separable mechanisms. Moreover, the engraftment of cytoplasmic retention determinants from A3H into A3A proved sufficient to change A3A's “zipcode” to relocalize it from a cell-wide state to the cytosol predominantly and to endow it with antiviral activity against HIV-1. Although these studies provide multiple mechanistic insights and will help guide future work, it should be noted that a necessary requirement was A3 overexpression in model cell lines. Additional technologies and studies will be needed to extend these results to endogenous A3H in primary cell types.
A recent study implicated the RNA binding and dimerization activities of A3H in cytoplasmic retention (40). We expected to find the same mechanism for nucleolar retention; however, we directly tested this idea and found that three RNA binding/dimerization-defective A3H mutants still accumulated to levels comparable to that of the wild-type enzyme (Fig. 1E). When we further probed the nucleolar localization mechanism of A3H, we found that single-amino-acid substitutions in loop 1 or loop 3 significantly decreased nucleolar accumulation (Fig. 2 and 5D and E). Interestingly, when we asked which A3H surface residues are necessary to relocalize A3A from cell-wide to nucleolar localization, we discovered that loop 3 in combination with the RNA binding surface is required to relocalize A3A to nucleolar compartments (Fig. 4B and D and 5C and data not shown). Taken together, these results suggest that some determinants involved in the cytoplasmic retention/RNA binding mechanism are also involved in mediating nucleolar localization, resulting in partly overlapping structural zipcodes. Additionally, we demonstrate that nucleocytoplasmic shuttling of A3H occurs through a passive mechanism with active retention. This conclusion is similar to that of a previous study by Li and Emerman where the nuclear enrichment of A3H hap I was shown to occur through a passive mechanism and the cytoplasmic localization of A3H hap II through an active retention mechanism (6).
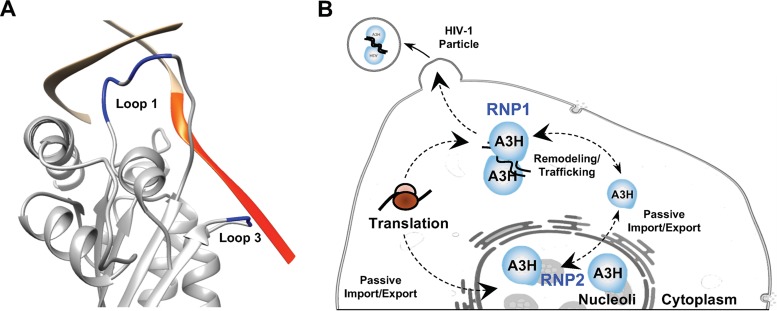
However, if our model of partially overlapping determinants were correct, one might expect loop 3 to be included as part of the RNA binding surface. Cocrystal structures between A3H and the duplex RNA resolve only 7 to 9 bp, which is too short to assess potential interactions between RNA and loop 3 residues. To overcome this lack of information, computational modeling was used to establish the lowest-energy conformation of an extended RNA chain to determine if loop 3 could contribute to the overall RNA binding mechanism. As shown in Fig. 7A, these computational studies suggested that the negatively charged phosphate backbone of the RNA will interact with the positively charged residues in loop 3 (50KKK52). Based on this model, the RNA chain would extend past the enzymatic active site and interact directly with loop 3, which is likely to sterically inhibit single-stranded DNA binding and deamination. This is consistent with observations that recombinant A3H lacks detectable enzymatic activity if purified without RNase A (40). Importantly, this model indicated that loop 3 has a dual role in mediating both cytoplasmic and nucleolar localization, which is supported by our reciprocal chimera data in Fig. 5C and D, as well as by the single-amino-acid substitutions shown in Fig. 5E and F.
FIG 7.
Computational modeling predicts that the duplexed RNA phosphate backbone interacts with loop 3 of A3H. (A) Structural model of the A3H/RNA cocrystal structure with 5 uracil nucleobases computationally added onto on the RNA chain that extends toward the enzymatic active site (additional residues are colored orange). Loop 1 and loop 3 are highlighted in blue. (B) Predicted model of how A3H maintains steady-state localization in two distinct subcellular compartments. RNP, ribonucleotide protein complex.
Although our results in Fig. 1E indicated that nucleolar localization is independent of RNA binding activity, additional data, including computational modeling, nucleolar A3A/H chimeras, and the A3H/A loop 3 chimera and loop 3 single-amino-acid substitutions, raised the possibility that A3H forms two distinct types of ribonucleoprotein (RNP) complexes to regulate subcellular localization (model in Fig. 7B). Nascently translated A3H is directed down one of two paths. In one instance, select loop 1 residues together with loop 7 or α-helix 6 residues combine to bind duplex RNA and heterodimerize in the cytoplasm (RNP1). In the other, monomeric A3H diffuses into the nuclear compartment and is actively retained in the nucleolus through a second RNP complex requiring select residues in loop 1 and loop 3 and potentially residues in loop 7 or α-helix 6 (RNP2). Homeostasis is maintained by remodeling of cytoplasmic RNAs to release the dimer or through cell cycle progression which dissolves nucleoli to release monomers (Fig. 7B). This model explains why nucleolar accumulation of A3A requires that the entire A3H RNA binding surface be exchanged in combination with loop 3 (Fig. 3 to 5), why mutations in the previously described RNA binding domain have no effect on nucleolar localization, and why substitutions in loops 1 and 3 do not disrupt cytoplasmic retention. This model also raises the interesting possibility of A3H regulation by a specific RNA(s) in each subcellular compartment.
An A3A/H chimera analysis enabled us to define the A3H cytoplasmic retention mechanism, and it indicated that α-helix 1, loop 9, and the “WG” sequence at the top of α-helix 3 are important (Fig. 6B and 7A). As determined by A3H crystal structures, α-helix 1 and loop 9 are in close proximity to each other and may act as a type of “molecular clasp” that stabilizes the RNA binding interface and locks the surface into the correct orientation for dimerization. Likewise, the “WG” sequence at the top of α-helix 3 is adjacent to loop 3 and may be involved with stabilizing the predicted loop 3/RNA backbone interaction (Fig. 7A). Interestingly, when A3A was modified to display an A3H-like localization pattern (Fig. 6B), it also gained the ability to package into and restrict the infectivity of Vif-deficient HIV-1 particles in a dose-dependent manner (Fig. 6C).
The data presented here and recent works (40, 48, 93) have linked A3H cytoplasmic retention and RNA binding to antiviral activity against HIV-1. This implies an intimate connection between subcellular localization and virus restriction and raises the question of nucleolar A3H function. Based on known biochemical activities of A3H (30, 41, 94), this enzyme may edit viral DNA cytosines or methylated cytosines as part of a nuclear virus restriction mechanism. Indeed, many different viral proteins are known to travel through the nucleoli from diverse viral families such as Herpesviridae, Coronaviridae, and Flaviviridae (95–101). An additional problem, however, is that most of these potential viral substrates have RNA-based genomes. Thus, we speculate that the larger active-site cavity of A3H required for methyl-cytosine editing may also be able to accommodate RNA cytosine bases and edit RNA cytosines to uracils and, therefore, that A3H may act as a guardian of the nucleolus, protecting this compartment from viruses that partially, or fully, require nucleolar translocation.
MATERIALS AND METHODS
Cell lines, culture conditions, and inhibitor treatments.
293T cells were maintained in Dulbecco modified Eagle medium (DMEM) (HyClone, South Logan, UT) supplemented with 10% fetal bovine serum (FBS) (Gibco, Gaithersburg, MD) and 0.5% penicillin-streptomycin (50 units). HeLa cells were maintained in RPMI (HyClone) supplemented with 10% FBS (Gibco) and 0.5% penicillin-streptomycin (50 units). U2OS cells were maintained in McCoy's 5A medium (HyClone) supplemented with 10% FBS and 0.5% penicillin-streptomycin (50 units). 293T, HeLa, and U2OS cells were transfected with TransIT LTI (Mirus, Madison, WI) according to the manufacture's protocol. For inhibitor treatments, the indicated chemical was added to the culture medium for the indicated length of time and imaged. Leptomycin B (Sigma, St. Louis, MO) was used at a final concentration of 40 ng/ml, and actinomycin D (Sigma) was used at 0.001 ng/ml.
Plasmids and cloning.
All mCh-A3 expression plasmids used in this study were cloned into a pQCXIH retroviral expression vector. Point mutants were generated by PCR amplification using Phusion high-fidelity DNA polymerase (NEB, Ipswich, MA) and overlapping PCR to introduce the indicated mutation. A similar approach was taken to generate chimeric A3A/A3H constructs, except that the indicated A3s were used as individual templates and then overlapping PCR was used to combine the two amplicons. All mCh-A3 mutant amplicons were cloned in using AgeI and BsiWI restriction sites. The fibrillarin-eGFP was cloned into a pcDNA 4/TO construct using HindIII and NotI restriction sites.
Immunofluorescence.
Approximately 25,000 HeLa cells were plated on acid-washed and polylysine-treated 22- by 22- by 1.5-mm coverslips (12-541-B; Fisherbrand, Minneapolis, MN) in a 6-well plate and after 24 h were transfected with 500 ng of the indicated A3 plasmids, 1.5 μl TransIT-LT1 (2304; Mirus), and 100 μl serum-free RPMI 1640 (Corning, Corning, NY). After 48 h, medium was removed and coverslips were washed with phosphate-buffered saline (PBS), fixed in 4% methanol-free formaldehyde (28906; Thermo Scientific) for 15 min, and rinsed 3 times for 5 min each in PBS with gentle rocking. Cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min, washed three time for 5 min each in PBS, and then incubated in blocking buffer (0.0028 M KH2PO4, 0.0072 M K2HPO4, 5% goat serum [Gibco], 5% glycerol, 1% cold water fish gelatin [Sigma-Aldrich], 0.04% sodium azide, pH 7.2) for 1 h. Cells were washed in PBS and then incubated in rabbit anti-A3H primary antibody (NBP1-91682; Novus, Littleton, CO) at 1:200 diluted in blocking buffer overnight at 4°C. Cells were washed three times for 5 min each with PBS and then incubated in goat anti-rabbit secondary antibody–fluorescein isothiocyanate (FITC) (111095144; Jackson Laboratories, Bar Harbor, ME) at 1:1,000 dilution in blocking buffer for 1 h at room temperature. Coverslips were mounted on precleaned slides (Gold Seal Rite-On) using one drop (10 to 15 μl) of mounting medium (prepared by dissolving 1 g n-propyl gallate [Sigma] in 30 ml glycerol overnight, adding 0.35 ml 0.1 M KH2PO4, adjusting the pH to 8 to 8.5 with K2HPO4, and adding water to obtain a final volume of 50 ml). Slides were imaged on a Nikon inverted Ti-E deconvolution microscope and analyzed using NiS Elements including deconvolution of images.
Fluorescence microscopy experiments.
Approximately 10,000 293T, 6,000 HeLa, or 6,000 U2OS cells were plated into a 96-well CellBIND microplate (Corning) and allowed to adhere overnight. The next day, cells were transfected with 50 ng of the indicated mCh-A3 construct and 50 ng of FBL-eGFP, as indicated in appropriate figure panels, and imaged at 48 h posttransfection. For experiments without FBL-eGFP, 50 to 100 ng of the indicated mCh-A3 construct was transfected into the indicated cell lines and imaged 48 h later. For DAPI (4′,6′-diamidino-2-phenylindole) treatment, NucBlue live cell stain (Invitrogen) was added at the appropriate concentration to the culture medium, incubated at 37°C for 30 min, and then imaged. Images were collected at a magnification of ×40 using an EVOS FL color microscope (Thermo Fisher) or using a Nikon inverted Ti-E deconvolution microscope. Localization patterns for all mutant constructs and chemical treatments (i.e., each experimental condition) were examined from at least three independent fields of view and across three separate transfection experiments. At least 90% of the cells imaged for each experimental condition showed the same localization pattern, and one of these representative images was selected for presentation. Totals of at least 30 to 50 cells per mutant construct and at least 20 to 30 cells for each chemical treatment were analyzed. All images containing mCherry-tagged constructs have been pseudocolored with magenta instead of red to accommodate readers who may be affected by colorblindness.
Molecular modeling.
The coordinates of chains A, B, and C (which correspond to an A3H monomer and the two chains of the double-stranded RNA bound to it) in the A3H crystal structure PDB 6B0B were used for this study. In this structure, chain C corresponds to the RNA chain that is longer than the other chain of the double-stranded RNA and extends into the active site as a single RNA strand. Residue E52 was computationally mutated back to a lysine residue using MOE software (102). The coordinates of two RNA nucleotides (with a UU sequence) from chain C were saved as a template. This 2-nucleotide template was used to add one U nucleotide at the 5′ end of chain C by superimposing the 3′ nucleotide of the template onto the 5′ nucleotide of chain C. A3H bound to this extended RNA was then entirely minimized in MOE. Repetitively extending the RNA chain C by 1 nucleotide and minimizing, we obtained an A3H model with 5 more U nucleotides at the 5′ end of RNA chain C.
Viral infectivity.
Approximately 250,000 293T cells were plated into a 12-well culture plate and allowed to adhere overnight. The next day, cells were transfected with 100 ng of an HIV-1 vif-, vpr-, vpu-, env-, and nef-deficient eGFP reporter virus (where nef has been replaced with cytomegalovirus [CMV]-eGFP), 150 ng of a vesicular stomatitis virus (VSV) G expression construct, and increasing amounts of the indicated mCh-A3 expression constructs or an empty-vector control (25 to 100 ng in 25-ng increments). The medium was harvested at 48 h posttransfection and frozen for at least 8 h at −80°C, and then 100 μl of the viral medium was used to transduce 250,000 fresh 293T cells. After 48 h, eGFP-positive cells were analyzed using flow cytometry, and the percentage of infected cells was normalized to the empty-vector control.
Immunoblot analysis.
Cells transfected from the viral infectivity experiments described above were detached from plates using PBS-EDTA, collected into 1.5-ml Eppendorf tubes, and centrifuged at 500 × g for 10 min. Cell pellets were resuspended in 100 μl radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, and 140 mM NaCl), and 10% of the total cell lysate samples was combined with 2.5× SDS-PAGE loading buffer. Viral samples were pelleted through a 20% sucrose cushion for 2 h at 20,000 × g at 4°C. Residual medium and sucrose were aspirated from the pellet, and samples were resuspended in 2.5× SDS-PAGE loading buffer. Samples were separated on a 4 to 20% gradient SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF)-FL membranes (Millipore). The membranes were blocked in blocking solution (5% milk plus PBS supplemented with 0.1% Tween20) and then incubated with primary antibody diluted in blocking solution. Secondary antibodies were diluted in blocking solution plus 0.01% SDS. Membranes were imaged with a Li-Cor Odyssey instrument or film. Primary antibodies used in these experiments were anti-mCherry (ab167453; Abcam), anti-HSP90 (610418; BD Transduction Laboratories), and anti-p24 (183.H12.5C; NIH Reagent Program). Secondary antibodies used were anti-rabbit IRdye 800CW-conjugated (827-08365; Li-Cor), anti-mouse Alexa Fluor 680-conjugated (A-21057; Molecular Probes), anti-rabbit horseradish peroxidase (HRP)-conjugated (7074P2; Cell Signaling), and anti-mouse HRP-conjugated (7076P2; Cell Signaling) antibodies.
Statistical analysis.
Prism 6.0 (GraphPad) was used for statistical analysis. Quantitative data were represented as mean ± standard error of the mean (SEM). Statistical significance was determined by an unpaired Student t test. Nucleolar fluorescence intensity was determined by subtracting background fluorescence intensity from mCh-A3H fluorescence intensity within defined nucleolar compartments. The nucleolar compartments were defined by coexpression of FBL-eGFP with the indicated mCh-A3H mutants.
ACKNOWLEDGMENTS
D.J.S. received salary support from the University of Minnesota Craniofacial Research Training (MinnCResT) program (NIH T90DE022732). N.M.S. received salary support from NIGMS R01 (GM118000). This work was supported by NIAID R37 AI064046. R.S.H. is the Margaret Harvey Schering Land Grant Chair for Cancer Research, a Distinguished University McKnight Professor, and an Investigator of the Howard Hughes Medical Institute.
R.S.H. is a cofounder, shareholder, and consultant of ApoGen Biotechnologies Inc. R.E.A. is a cofounder of, is on the Scientific Advisory Board of, and has equity interest in Actavalon, Inc. The other authors declare no competing financial interests.
D.J.S. conceived and designed the studies. D.J.S. created all of the constructs. J.T.B. imaged RNA binding mutants. A.Z.C. performed immunofluorescence experiments using native A3H variants. J.L.M. performed immunoblot analyses. O.D. and R.E.A. generated the A3H model with extended RNA polymer. N.M.S. provided structural analysis. W.L.B. contributed to project management. D.J.S. and R.S.H. drafted the manuscript, and all authors contributed to revisions.
REFERENCES
- 1.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol 22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 2.Harris RS, Liddament MT. 2004. Retroviral restriction by APOBEC proteins. Nat Rev Immunol 4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 3.LaRue RS, Andrésdóttir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jónsson SR, Landau NR, Löchelt M, Malik HS, Malim MH, Münk C, O'Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol 83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Refsland EW, Harris RS. 2013. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol 371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhen A, Du J, Zhou X, Xiong Y, Yu XF. 2012. Reduced APOBEC3H variant anti-viral activities are associated with altered RNA binding activities. PLoS One 7:e38771. doi: 10.1371/journal.pone.0038771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li MM, Emerman M. 2011. Polymorphism in human APOBEC3H affects a phenotype dominant for subcellular localization and antiviral activity. J Virol 85:8197–8207. doi: 10.1128/JVI.00624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. 2008. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. 2011. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol 85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harari A, Ooms M, Mulder LC, Simon V. 2009. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol 83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebrahimi D, Richards CM, Carpenter MA, Wang J, Ikeda T, Cheng AZ, Becker JT, McCann JL, Shaban NM, Salamango DJ, Starrett GJ, Lingappa JR, Yong J, Brown WL, Harris RS. Genetic and mechanistic basis for APOBEC3H alternative splicing, differential retrovirus restriction, and counteraction by HIV-1 protease. Nat Commun, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albin JS, Harris RS. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med 12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol 14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 13.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog 4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A 103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apolonia L, Schulz R, Curk T, Rocha P, Swanson CM, Schaller T, Ule J, Malim MH. 2015. Promiscuous RNA binding ensures effective encapsidation of APOBEC3 proteins by HIV-1. PLoS Pathog 11:e1004609. doi: 10.1371/journal.ppat.1004609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogerd HP, Cullen BR. 2008. Single-stranded RNA facilitates nucleocapsid: APOBEC3G complex formation. RNA 14:1228–1236. doi: 10.1261/rna.964708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. 2004. The interaction between HIV-1 Gag and APOBEC3G. J Biol Chem 279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- 18.Huthoff H, Malim MH. 2007. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and virion encapsidation. J Virol 81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan MA, Kao S, Miyagi E, Takeuchi H, Goila-Gaur R, Opi S, Gipson CL, Parslow TG, Ly H, Strebel K. 2005. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J Virol 79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schäfer A, Bogerd HP, Cullen BR. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, Freed EO, Hu WS, Pathak VK. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem 279:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 22.Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 2007. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol 81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.York A, Kutluay SB, Errando M, Bieniasz PD. 2016. The RNA binding specificity of human APOBEC3 proteins resembles that of HIV-1 nucleocapsid. PLoS Pathog 12:e1005833. doi: 10.1371/journal.ppat.1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zennou V, Perez-Caballero D, Göttlinger H, Bieniasz PD. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol 78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Refsland EW, Hultquist JF, Harris RS. 2012. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog 8:e1002800. doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, Zhang W, Tian C, Liu B, Yu Y, Ding L, Spearman P, Yu XF. 2008. Distinct viral determinants for the packaging of human cytidine deaminases APOBEC3G and APOBEC3C. Virology 377:71–79. doi: 10.1016/j.virol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang Y, Wang X, Esselman WJ, Zheng YH. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol 80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. 2011. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol 85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809. doi: 10.1016/S0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 30.Starrett GJ, Luengas EM, McCann JL, Ebrahimi D, Temiz NA, Love RP, Feng Y, Adolph MB, Chelico L, Law EK, Carpenter MA, Harris RS. 2016. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat Commun 7:12918. doi: 10.1038/ncomms12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS. 2013. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns MB, Temiz NA, Harris RS. 2013. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin DA. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjörd JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Imielinsk M, Jäger N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, López-Otín C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdés-Mas R, van Buuren MM, van 't Veer L, Vincent-Salomon A, Waddell N, Yates LR, Australian Pancreatic Cancer Genome Initiative, ICGC Breast Cancer Consortium, ICGC MMML-Seq Consortium, ICGC PedBrain, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. 2013. Signatures of mutational processes in human cancer. Nature 500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, McBride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, McLaren S, Butler AP, Teague JW, Jönsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerød A, Tutt A, Martens JW, Aparicio SA, Borg Å Salomon AV, Thomas G, Børresen-Dale AL, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR, Breast Cancer Working Group of the International Cancer Genome Consortium . 2012. Mutational processes molding the genomes of 21 breast cancers. Cell 149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullah I, Karthik GM, Alkodsi A, Kjällquist U, Stålhammar G, Lövrot J, Martinez NF, Lagergren J, Hautaniemi S, Hartman J, Bergh J. 2018. Evolutionary history of metastatic breast cancer reveals minimal seeding from axillary lymph nodes. J Clin Invest 128:1355–1370. doi: 10.1172/JCI96149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefebvre C, Bachelot T, Filleron T, Pedrero M, Campone M, Soria JC, Massard C, Lévy C, Arnedos M, Lacroix-Triki M, Garrabey J, Boursin Y, Deloger M, Fu Y, Commo F, Scott V, Lacroix L, Dieci MV, Kamal M, Diéras V, Gonçalves A, Ferrerro JM, Romieu G, Vanlemmens L, Mouret Reynier MA, Théry JC, Le Du F, Guiu S, Dalenc F, Clapisson G, Bonnefoi H, Jimenez M, Le Tourneau C, André F. 2016. Mutational profile of metastatic breast cancers: a retrospective analysis. PLoS Med 13:e1002201. doi: 10.1371/journal.pmed.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng B, Burant CF, Davidson NO. 1993. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 39.Sharma S, Patnaik SK, Taggart RT, Kannisto ED, Enriquez SM, Gollnick P, Baysal BE. 2015. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat Commun 6:6881. doi: 10.1038/ncomms7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaban NM, Shi K, Lauer KV, Carpenter MA, Richards CM, Salamango D, Wang J, Lopresti MW, Banerjee S, Levin-Klein R, Brown WL, Aihara H, Harris RS. 2018. The antiviral and cancer genomic DNA deaminase APOBEC3H is regulated by an RNA-mediated dimerization mechanism. Mol Cell 69:75–86.e9. doi: 10.1016/j.molcel.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito F, Yang H, Xiao X, Li SX, Wolfe A, Zirkle B, Arutiunian V, Chen XS. 2018. Understanding the structure, multimerization, subcellular localization and mC selectivity of a genomic mutator and anti-HIV factor APOBEC3H. Sci Rep 8:3763. doi: 10.1038/s41598-018-21955-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallois-Montbrun S, Holmes RK, Swanson CM, Fernández-Ocaña M, Byers HL, Ward MA, Malim MH. 2008. Comparison of cellular ribonucleoprotein complexes associated with the APOBEC3F and APOBEC3G antiviral proteins. J Virol 82:5636–5642. doi: 10.1128/JVI.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izumi T, Burdick R, Shigemi M, Plisov S, Hu WS, Pathak VK. 2013. Mov10 and APOBEC3G localization to processing bodies is not required for virion incorporation and antiviral activity. J Virol 87:11047–11062. doi: 10.1128/JVI.02070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. 2006. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J Biol Chem 281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- 45.Phalora PK, Sherer NM, Wolinsky SM, Swanson CM, Malim MH. 2012. HIV-1 replication and APOBEC3 antiviral activity are not regulated by P bodies. J Virol 86:11712–11724. doi: 10.1128/JVI.00595-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stenglein MD, Matsuo H, Harris RS. 2008. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J Virol 82:9591–9599. doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wichroski MJ, Robb GB, Rana TM. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog 2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bohn JA, Thummar K, York A, Raymond A, Brown WC, Bieniasz PD, Hatziioannou T, Smith JL. 2017. APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA. Nat Commun 8:1021. doi: 10.1038/s41467-017-01309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, Sata T, Tokunaga K. 2007. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res 35:2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Löwer J, Cichutek K, Flory E, Schumann GG, Münk C. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem 281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 51.Salamango DJ, McCann JL, Demir Ö Brown WL, Amaro RE, Harris RS. 2018. APOBEC3B nuclear localization requires two distinct N-terminal domain surfaces. J Mol Biol 430:2695–2708. doi: 10.1016/j.jmb.2018.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lackey L, Demorest Z, Land A, Hultquist J, Brown W, Harris R. 2012. APOBEC3B and AID have similar nuclear import mechanisms. J Mol Biol 419:301–314. doi: 10.1016/j.jmb.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lackey L, Law EK, Brown WL, Harris RS. 2013. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle 12:762–772. doi: 10.4161/cc.23713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pederson T. 2011. The nucleolus. Cold Spring Harb Perspect Biol 3:a000638. doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grummt I, Längst G. 2013. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim Biophys Acta 1829:393–404. doi: 10.1016/j.bbagrm.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 56.Ferreira-Cerca S, Pöll G, Gleizes PE, Tschochner H, Milkereit P. 2005. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell 20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 57.Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. 2007. The multifunctional nucleolus. Nat Rev Mol Cell Biol 8:574–585. [DOI] [PubMed] [Google Scholar]
- 58.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. 2005. Nucleolar proteome dynamics. Nature 433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 59.Scherl A, Couté Y, Déon C, Callé A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. 2002. Functional proteomic analysis of human nucleolus. Mol Biol Cell 13:4100–4109. doi: 10.1091/mbc.e02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baran V, Veselá J, Rehák P, Koppel J, Fléchon JE. 1995. Localization of fibrillarin and nucleolin in nucleoli of mouse preimplantation embryos. Mol Reprod Dev 40:305–310. doi: 10.1002/mrd.1080400306. [DOI] [PubMed] [Google Scholar]
- 61.Görlich D, Kutay U. 1999. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 62.Görlich D. 1998. Transport into and out of the cell nucleus. EMBO J 17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nigg EA. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 64.Patenaude AM, Orthwein A, Hu Y, Campo VA, Kavli B, Buschiazzo A, Di Noia JM. 2009. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Mol Biol 16:517–527. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- 65.Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, Nakata M, Honjo T. 2004. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A 101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ganesh K, Adam S, Taylor B, Simpson P, Rada C, Neuberger M. 2011. CTNNBL1 is a novel nuclear localization sequence-binding protein that recognizes RNA-splicing factors CDC5L and Prp31. J Biol Chem 286:17091–17102. doi: 10.1074/jbc.M110.208769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci U S A 96:9112–9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun Q, Carrasco YP, Hu Y, Guo X, Mirzaei H, Macmillan J, Chook YM. 2013. Nuclear export inhibition through covalent conjugation and hydrolysis of leptomycin B by CRM1. Proc Natl Acad Sci U S A 110:1303–1308. doi: 10.1073/pnas.1217203110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perry RP, Kelley DE. 1970. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J Cell Physiol 76:127–139. doi: 10.1002/jcp.1040760202. [DOI] [PubMed] [Google Scholar]
- 70.Hollstein U. 1974. Actinomycin chemistry and mechanism of action. Chem Rev 74:625–652. doi: 10.1021/cr60292a002. [DOI] [Google Scholar]
- 71.Recher L, Briggs LG, Parry NT. 1971. A reevaluation of nuclear and nucleolar changes induced in vitro by actinomycin D Cancer Res 31:140–151. [PubMed] [Google Scholar]
- 72.Schoefl G. 1964. The effect of actinomycin D on the fine structure of the nucleolus. J Ultrastruct Res 10:224–243. doi: 10.1016/S0022-5320(64)80007-1. [DOI] [PubMed] [Google Scholar]
- 73.Suter ER, Salomon JC. 1966. The effect of actinomycin D on the fine structure of the nucleolus of hepatocytes of rats treated with thioacetamide. Exp Cell Res 43:248–251. doi: 10.1016/0014-4827(66)90405-8. [DOI] [PubMed] [Google Scholar]
- 74.Scott MS, Troshin PV, Barton GJ. 2011. NoD: a nucleolar localization sequence detector for eukaryotic and viral proteins. BMC Bioinformatics 12:317. doi: 10.1186/1471-2105-12-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scott MS, Boisvert FM, McDowall MD, Lamond AI, Barton GJ. 2010. Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res 38:7388–7399. doi: 10.1093/nar/gkq653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Emmott E, Hiscox JA. 2009. Nucleolar targeting: the hub of the matter. EMBO Rep 10:231–238. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martin RM, Ter-Avetisyan G, Herce HD, Ludwig AK, Lättig-Tünnemann G, Cardoso MC. 2015. Principles of protein targeting to the nucleolus. Nucleus 6:314–325. doi: 10.1080/19491034.2015.1079680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Land AM, Law EK, Carpenter MA, Lackey L, Brown WL, Harris RS. 2013. Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and nongenotoxic. J Biol Chem 288:17253–17260. doi: 10.1074/jbc.M113.458661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bohn MF, Shandilya SMD, Silvas TV, Nalivaika EA, Kouno T, Kelch BA, Ryder SP, Kurt-Yilmaz N, Somasundaran M, Schiffer CA. 2015. The ssDNA mutator APOBEC3A is regulated by cooperative dimerization. Structure 23:903–911. doi: 10.1016/j.str.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi K, Carpenter MA, Banerjee S, Shaban NM, Kurahashi K, Salamango DJ, McCann JL, Starrett GJ, Duffy JV, Demir Ö Amaro RE, Harki DA, Harris RS, Aihara H. 2017. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat Struct Mol Biol 24:131–139. doi: 10.1038/nsmb.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kouno T, Silvas TV, Hilbert BJ, Shandilya SMD, Bohn MF, Kelch BA, Royer WE, Somasundaran M, Kurt Yilmaz N, Matsuo H, Schiffer CA. 2017. Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity. Nat Commun 8:15024. doi: 10.1038/ncomms15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. 2011. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep 12:444–450. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suspène R, Aynaud MM, Guétard D, Henry M, Eckhoff G, Marchio A, Pineau P, Dejean A, Vartanian JP, Wain-Hobson S. 2011. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc Natl Acad Sci U S A 108:4858–4863. doi: 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aynaud MM, Suspène R, Vidalain PO, Mussil B, Guétard D, Tangy F, Wain-Hobson S, Vartanian JP. 2012. Human Tribbles 3 protects nuclear DNA from cytidine deamination by APOBEC3A. J Biol Chem 287:39182–39192. doi: 10.1074/jbc.M112.372722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dingwall C, Sharnick S, Laskey R. 1982. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell 30:449–458. doi: 10.1016/0092-8674(82)90242-2. [DOI] [PubMed] [Google Scholar]
- 86.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. 2007. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem 282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Earley LF, Kawano Y, Adachi K, Sun XX, Dai MS, Nakai H. 2015. Identification and characterization of nuclear and nucleolar localization signals in the adeno-associated virus serotype 2 assembly-activating protein. J Virol 89:3038–3048. doi: 10.1128/JVI.03125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu J, Du X, Ke Y. 2006. Mapping nucleolar localization sequences of 1A6/DRIM. FEBS Lett 580:1405–1410. doi: 10.1016/j.febslet.2006.01.064. [DOI] [PubMed] [Google Scholar]
- 89.Sheng Z, Lewis JA, Chirico WJ. 2004. Nuclear and nucleolar localization of 18-kDa fibroblast growth factor-2 is controlled by C-terminal signals. J Biol Chem 279:40153–40160. doi: 10.1074/jbc.M400123200. [DOI] [PubMed] [Google Scholar]
- 90.LaRue RS, Jónsson SR, Silverstein KA, Lajoie M, Bertrand D, El-Mabrouk N, Hötzel I, Andrésdóttir V, Smith TP, Harris RS. 2008. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol 9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schmitt K, Guo K, Algaier M, Ruiz A, Cheng F, Qiu J, Wissing S, Santiago ML, Stephens EB. 2011. Differential virus restriction patterns of rhesus macaque and human APOBEC3A: implications for lentivirus evolution. Virology 419:24–42. doi: 10.1016/j.virol.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 93.Bieniasz PD, Kutluay SB. 2018. CLIP-related methodologies and their application to retrovirology. Retrovirology 15:35. doi: 10.1186/s12977-018-0417-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ito F, Fu Y, Kao SA, Yang H, Chen XS. 2017. Family-wide comparative analysis of cytidine and methylcytidine deamination by eleven human APOBEC proteins. J Mol Biol 429:1787–1799. doi: 10.1016/j.jmb.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bertrand L, Leiva-Torres GA, Hyjazie H, Pearson A. 2010. Conserved residues in the UL24 protein of herpes simplex virus 1 are important for dispersal of the nucleolar protein nucleolin. J Virol 84:109–118. doi: 10.1128/JVI.01428-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Salsman J, Zimmerman N, Chen T, Domagala M, Frappier L. 2008. Genome-wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS Pathog 4:e1000100. doi: 10.1371/journal.ppat.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Szekely L, Jiang WQ, Pokrovskaja K, Wiman KG, Klein G, Ringertz N. 1995. Reversible nucleolar translocation of Epstein-Barr virus-encoded EBNA-5 and hsp70 proteins after exposure to heat shock or cell density congestion. J Gen Virol 76:2423–2432. doi: 10.1099/0022-1317-76-10-2423. [DOI] [PubMed] [Google Scholar]
- 98.Wurm T, Chen H, Hodgson T, Britton P, Brooks G, Hiscox JA. 2001. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J Virol 75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yuan X, Yao Z, Shan Y, Chen B, Yang Z, Wu J, Zhao Z, Chen J, Cong Y. 2005. Nucleolar localization of non-structural protein 3b, a protein specifically encoded by the severe acute respiratory syndrome coronavirus. Virus Res 114:70–79. doi: 10.1016/j.virusres.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Melén K, Kinnunen L, Fagerlund R, Ikonen N, Twu KY, Krug RM, Julkunen I. 2007. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J Virol 81:5995–6006. doi: 10.1128/JVI.01714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fraser JE, Rawlinson SM, Heaton SM, Jans DA. 2016. Dynamic nucleolar targeting of dengue virus polymerase NS5 in response to extracellular pH. J Virol 90:5797–5807. doi: 10.1128/JVI.02727-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chemical Computing Group. 2018. Molecular Operating Environment (MOE) 2014.09. Chemical Computing Group ULC, Montreal, Quebec, Canada. [Google Scholar]