There is controversy concerning the role of dihydroceramide desaturase (Degs1) in regulating cell survival, with studies showing that it can both promote and protect against apoptosis. We have therefore investigated the molecular basis for these opposing roles of Degs1.
KEYWORDS: dihydroceramide desaturase, sphingosine 1-phosphate, ER stress, proteasome, sphingosine kinase
ABSTRACT
There is controversy concerning the role of dihydroceramide desaturase (Degs1) in regulating cell survival, with studies showing that it can both promote and protect against apoptosis. We have therefore investigated the molecular basis for these opposing roles of Degs1. Treatment of HEK293T cells with the sphingosine kinase inhibitor SKi [2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole] or fenretinide, but not the Degs1 inhibitor GT11 {N-[(1R,2S)-2-hydroxy-1-hydroxymethyl-2-(2-tridecyl-1-cyclopropenyl)ethyl]octan-amide}, induced the polyubiquitination of Degs1 (Mr = 40 to 140 kDa) via a mechanism involving oxidative stress, p38 mitogen-activated protein kinase (MAPK), and Mdm2 (E3 ligase). The polyubiquitinated forms of Degs1 exhibit “gain of function” and activate prosurvival pathways, p38 MAPK, c-Jun N-terminal kinase (JNK), and X-box protein 1s (XBP-1s). In contrast, another sphingosine kinase inhibitor, ABC294640 [3-(4-chlorophenyl)-adamantane-1-carboxylic acid (pyridin-4-ylmethyl)amide], at concentrations of 25 to 50 μM failed to induce formation of the polyubiquitinated forms of Degs1. In contrast to SKi, ABC294640 (25 μM) promotes apoptosis of HEK293T cells via a Degs1-dependent mechanism that is associated with increased de novo synthesis of ceramide. These findings are the first to demonstrate that the polyubiquitination of Degs1 appears to change its function from proapoptotic to prosurvival. Thus, polyubiquitination of Degs1 might provide an explanation for the reported opposing functions of this enzyme in cell survival/apoptosis.
INTRODUCTION
De novo synthesis of ceramide involves a pathway in which serine and palmitoyl CoA undergo condensation to form 3-ketosphinganine, which is catalyzed by serine palmitoyltransferase and which is the rate-limiting step in ceramide biosynthesis (1). 3-Ketosphinganine is converted into sphinganine (dihydrosphingosine) by 3-ketosphinganine reductase, which is then acylated to dihydroceramide by ceramide synthase (CerS). Dihydroceramide desaturase (Degs1) introduces a trans 4,5 double bond to dihydroceramide, thereby forming ceramide de novo. Ceramide can also be formed by the sphingomyelinase-catalyzed hydrolysis of sphingomyelin in the so-called “salvage” pathway. Deacylation of ceramide by ceramidase produces sphingosine (Sph), which is then phosphorylated by sphingosine kinase 1 (SK1) or SK2 to form sphingosine 1-phosphate (S1P). Sphingosine kinase can also phosphorylate sphinganine to produce sphinganine 1-phosphate (dihydrosphingosine 1-phosphate). S1P is either cleaved by S1P lyase (to hexadecenal and phosphoethanolamine) or converted back to ceramide, via sphingosine, by S1P phosphatase and CerS. In general terms, ceramide and sphingosine promote senescence or apoptotic cell death (1, 2), while S1P promotes cell growth (3).
Although initially regarded as simply intermediates in de novo ceramide biosynthesis, dihydroceramides are now recognized to have distinct biological functions, including in relation to autophagy, cell proliferation, hypoxia, and several diseases (4). For example, dihydroceramides that accumulate in response to Degs1 inhibitors or the addition of exogenous dihydroceramides promotes autophagy (5–7) and endoplasmic reticulum (ER) stress (7). However, some effects of Degs1 inhibitors on autophagy are independent of dihydroceramides (8). Nonetheless, genetic evidence supports a role for dihydroceramides in autophagy, which is linked with cell survival. Thus, Degs1 deficiency produces an antiapoptotic effect via activation of AKT and autophagy (9). However, other studies have shown that inhibition of Degs1 promotes cell death, which may involve autophagy or apoptosis (7, 10). For example, apoptosis is induced by Degs1 inhibitors, including fenretinide and resveratrol (11–13), and this is prevented when enzymes upstream of Degs1 in the de novo ceramide synthesis pathway are inhibited (12). In contrast, inhibition of Degs1 or its genetic manipulation can produce resistance to apoptosis (7, 9, 14, 15).
One mechanism by which Degs1 might regulate cell survival is via ER stress, as lipid composition and particularly ceramide and dihydroceramide in the ER membrane activate ER stress responses (7, 16, 17). This can lead to an unfolded protein response (UPR), which is a survival process; however, a sustained UPR results in apoptosis (18). ER stress involves the dissociation of the ER chaperone binding immunoglobulin protein (BiP) from the lumenal domains of ER stress sensors, which are protein kinase R-like ER kinase (PERK), inositol-requiring kinase 1α (IRE1α), and activating transcription factor 6 (ATF6). PERK is a kinase which phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF2α) to inhibit protein synthesis. ER stress also initiates ER-associated protein degradation (ERAD) to remove misfolded proteins. Recent studies have demonstrated that the SK2 inhibitor ABC294640 [3-(4-chlorophenyl)-adamantane-1-carboxylic acid (pyridin-4-ylmethyl)amide] or K145 induces ER stress, with the latter increasing the expression of X-box protein 1s (XBP-1s) and p-eIF2α (19). Targeting SK2 with K145 also contributed to ER stress and UPR activation induced by bortezomib, as evidenced by stimulation of the IRE1, c-Jun N-terminal kinase (JNK), and p38 mitogen-activated protein kinase (MAPK) pathways, thereby resulting in potent synergistic apoptosis of myeloma cells in vitro (19). We have also shown that 2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole (SKi) induces ER stress/UPR in T-cell lymphoblastic leukemic (T-ALL) cells that results in protective cell survival autophagy (20). The effects of ABC294640 and SKi on Degs1 activity therefore raise the possibility for their involvement in both a “survival”-promoting UPR and an “apoptotic” UPR.
Given the current controversy surrounding the role of Degs1 in regulating cell survival versus apoptosis, we have investigated the molecular basis for these opposing roles of Degs1. We demonstrate that Degs1 is subject to regulation by polyubiquitination induced by the sphingosine kinase inhibitor SKi or fenretinide. Polyubiquitination of Degs1 promotes a “gain of function,” which enables the activation of prosurvival pathways, including p38 MAPK, JNK, and XBP-1s. In contrast, another sphingosine kinase inhibitor, ABC294640, failed to induce polyubiquitination of Degs1 at a concentration that increases de novo synthesis of ceramide and induces apoptosis via a Degs1-dependent mechanism. These findings are the first to suggest that the opposing functions of Degs1 in cell survival and apoptosis might be due to the polyubiquitination of Degs1.
RESULTS
Effects of ABC294640 and SKi on the polyubiquitination of Degs1.
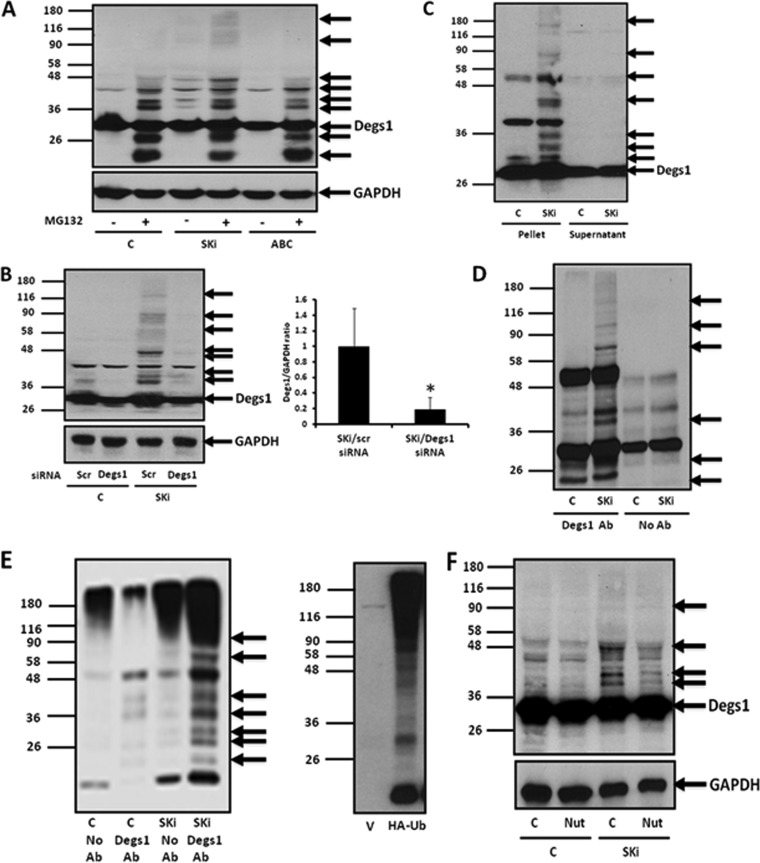
The sphingosine kinase inhibitors SKi and ABC294640 have previously been shown to reduce Degs1 activity, and this is associated with the proteasomal degradation of the enzyme in androgen-independent LNCaP-AI prostate cancer cells (21). We therefore assessed the effects of these SK inhibitors on Degs1 expression levels in HEK293T cells. Native Degs1 is expressed as a 32-kDa protein in HEK293T cells, detected with an anti-Degs1 antibody on Western blots (Fig. 1A). Treatment of HEK293T cells with SKi (10 μM for 24 h) induced the appearance of a ladder of higher-molecular-mass protein bands that immunoreacted with anti-Degs1 antibody (Fig. 1A), suggesting that SKi can stimulate the posttranslational modification of Degs1. Confirmation of their identity was established using small interfering RNA (siRNA) to knock down Degs1 expression, which reduced the immunoreactive intensity of the 32-kDa protein and that of the laddered protein bands (Fig. 1B), e.g., an 80% reduction in the expression of the 46-kDa posttranslationally modified form of Degs1 (Fig. 1B). In contrast to SKi, treatment of HEK293T cells with ABC294640 (25 μM for 24 h) did not induce the formation of the Degs1 ladder (Fig. 1A). Identical results were obtained with the parental HEK293 cell line (data not shown).
FIG 1.
Effect of ABC294640 or SKi on Degs1 protein laddering in HEK293T cells. Cells maintained in serum and grown to 70% confluence were treated with ABC294640 (ABC) (25 μM) or SKi (10 μM) for 24 h. In certain cases, cells were treated with scrambled (scr) or Degs1 siRNA (100 nM) for 48 h or with nutlin (Nut) (10 μM) or MG132 (10 μM) for 30 min prior to treatment with SKi or ABC294640. Where indicated, cells were transfected with a HA-ubiquitin (Ub) plasmid construct for 8 h prior to treatment with or without SKi. (A) Western blot probed with anti-Degs1 antibody showing the effect of SKi or ABC294640, in the presence or absence of MG132, on Degs1 laddering. (B) Western blot probed with anti-Degs1 antibody showing the effect of Degs1 siRNA on the formation of the Degs1 ladder in response to SKi. The bar graph demonstrates the reduction in polyubiquitinated Degs1 (Mr = 46 kDa) expression levels by Degs1 siRNA in the presence of SKi. Results are expressed as Degs1/GAPDH ratios (means ± standard errors of the means [SEM] for 3 independent experiments; *, P < 0.01 versus scrambled siRNA/SKi). (C) Western blot probed with anti-Degs1 antibody showing the localization of the Degs1 ladder in the pellet fraction. (D) Western blot probed with anti-Degs1 antibody showing the presence of the Degs1 ladder in anti-Degs1 immunoprecipitates. (E) Western blot showing the formation of HA-ubiquitinated forms of Degs1 in response to SKi, detected in anti-Degs1 immunoprecipitates with anti-HA antibody (Ab). Also shown is a Western blot of lysates from cells transfected with the vector or the HA-ubiquitin plasmid construct probed with anti-HA antibody. (F) Western blot probed with anti-Degs1 antibody showing the effect of nutlin on the formation of the Degs1 ladder in response to SKi. Blots in panels A, B, and F were reprobed for GAPDH using anti-GAPDH antibody to ensure comparable protein loading. Results are representative of data from at least 3 independent experiments. Lanes C, control.
We also sought a second independent method to confirm the identity of the proteins in the ladder. In this regard, the posttranslationally modified forms of Degs1 are present in a high-speed pellet fraction containing cell membranes (Fig. 1C) and can be released into a high-speed supernatant fraction using deoxycholate-containing buffer but not NP-40 (data not shown). By using this method, the posttranslationally modified forms of Degs1 were immunoprecipitated by the anti-Degs1 antibody (Fig. 1D).
Ubiquitination, which is catalyzed by E3 ligase, will typically promote posttranslational modification of proteins that can be resolved on SDS-PAGE gels as a ladder similar to that observed here for Degs1. We therefore transfected HEK293T cells with a plasmid construct encoding hemagglutinin (HA)-tagged ubiquitin and then treated these cells with SKi (10 μM for 24 h), after which Degs1 was immunoprecipitated with anti-Degs1 antibody and then probed with anti-HA antibody on Western blots. SKi promoted the formation of a ladder of HA-ubiquitinated Degs1 forms (Fig. 1E). The HA-ubiquitinated Degs1 laddering in immunoprecipitates was similar to that detected in immunoprecipitates probed with anti-Degs1 antibody on Western blots (Fig. 1D). Unpublished research from our laboratory has demonstrated that SKi promotes the ubiquitination of p53, which is catalyzed by Mdm2 (22). We therefore considered whether Mdm2 might be involved in regulating the ubiquitination of Degs1. Indeed, the formation of the Degs1 ladder in response to SKi was reduced when cells were pretreated with a specific inhibitor of Mdm2, nutlin (Fig. 1F).
Effect of the proteasome inhibitor MG132 on the polyubiquitination of Degs1.
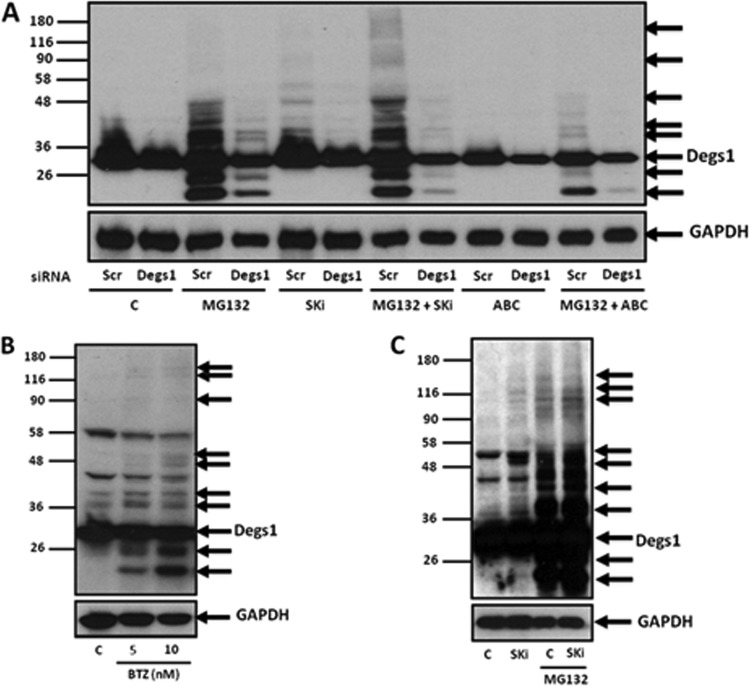
Given that the formation of the Degs1 ladder appears to reflect ongoing ubiquitin-proteasomal degradation, we assessed whether the proteasome inhibitor MG132 (carbobenzoxy-Leu-Leu-leucinal) induces the formation of the Degs1 ladder, and, indeed, this was the case (Fig. 1A and 2A). Interestingly, there were additional anti-Degs1 immunoreactive protein bands with molecular masses below 32 kDa that are not formed in response to SKi (Fig. 1A and 2A). These smaller Degs1 forms might represent proteasomal degradation products that have accumulated due to inhibition of the proteasome by MG132. The identity of the proteins formed in response to MG132 was confirmed using Degs1 siRNA (Fig. 2A). These findings suggest that Degs1 is subject to dynamic turnover in HEK293T cells.
FIG 2.
MG132- and bortezomib-induced formation of the Degs1 ladder. Cells maintained in serum and grown to 70% confluence were treated with ABC294640 (25 μM) or SKi (10 μM) for 24 h. In certain cases, cells were treated with scrambled siRNA or Degs1 siRNA (each at a final concentration of 100 nM) for 48 h or with MG132 (10 μM) for 30 min prior to treatment with ABC294640 or SKi. Cells were also treated with bortezomib (5 or 10 nM) for 24 h. (A) Western blot probed with anti-Degs1 antibody showing the effect of Degs1 siRNA on Degs1 laddering induced by MG132 in the presence or absence of SKi or ABC294640. (B) Western blot probed with anti-Degs1 antibody showing the effect of bortezomib (BTZ) on Degs1 laddering. (C) Western blot showing the formation of the Degs1 ladder in response to SKi or MG132 in PANC1 cells. Blots were reprobed for GAPDH using anti-GAPDH antibody to ensure comparable protein loading. Results are representative of data from at least 3 independent experiments. Lanes C, control.
We therefore assessed whether combined treatment of cells with SKi and MG132 had any effect on the formation of the Degs1 ladder. In this regard, combined treatment increased the formation of Degs1 forms with a Mr of >50 kDa (Fig. 2A), which represent polyubiquitinated forms of Degs1. Formation of the Degs1 ladder is a feature of proteasome inhibition, as a second proteasome inhibitor, bortezomib, which is chemically distinct from MG132, induced the same effect on Degs1 (Fig. 2B).
In order to establish whether Degs1 is subject to ubiquitin-proteasomal degradation in cancer cells, we treated PANC1 cancer cells with SKi and MG132. PANC1 cells express the native 32-kDa form of Degs1 (Fig. 2C), and SKi induced the appearance of higher-molecular-mass forms (53 kDa and 100 to 130 kDa) (Fig. 2C). Treatment of cells with MG132 also promotes the formation of a Degs1 ladder that is similar to that observed in HEK293T cells (Fig. 2C). These findings indicate that, as in HEK293T cells, Degs1 is subject to dynamic turnover via proteasomal degradation in PANC1 cells.
Therefore, we conclude that Degs1 is subject to polyubiquitination based on the following: (i) SKi induced the formation of HA-ubiquitinated Degs1 forms, which was detected in anti-Degs1 immunoprecipitates with anti-HA antibody on Western blots; (ii) the formation of the Degs1 ladder is reduced by pretreating cells with the Mdm2 inhibitor nutlin; (iii) the proteasome inhibitors MG132 and bortezomib induce the formation of the Degs1 ladder, consistent with the accumulation of polyubiquitinated forms of Degs1 that are trapped due to inhibition of the proteasome; and (iv) the formation of the Degs1 ladder is reduced by siRNA knockdown of Degs1.
Assessment of the roles of SK1 and SK2 in the polyubiquitination of Degs1.
We have previously shown that ABC294640 or SKi induces the proteasomal degradation of SK1 in cancer cells (21, 23). This was also the case in HEK293T cells, where ABC294640 (25 μM) or SKi (10 μM) treatment reduced SK1 expression by ∼80%, which was reversed by MG132 (Fig. 3A). The Degs1 ladder was not induced by siRNA knockdown of SK1 (Fig. 3B), which reduced SK1 protein expression by 70% (Fig. 3B). This suggests that SK1 is not involved in promoting the formation of the Degs1 ladder in response to SKi. This is supported by the fact that ABC294640 (25 μM) induces the proteasomal degradation of SK1 by 80% (Fig. 3A) in HEK293T cells yet fails to promote the formation of the Degs1 ladder at this concentration. Further confirmation for the lack of a role for SK1 was obtained using the SK1 inhibitor PF-543 (24), which exhibits a Ki of 4 nM for SK1 inhibition. In this regard, PF-543 failed to induce formation of the Degs1 ladder (Fig. 3C).
FIG 3.
Assessment of the roles of SK1 and SK2 in the formation of the Degs1 ladder. Cells maintained in serum and grown to 70% confluence were treated with ABC294640 (25 μM), SKi (10 μM), or PF-543 (100 nM) for 24 h. In certain cases, cells were treated with SK1 or SK2 siRNA (each at a final concentration of 100 nM) for 48 h or with MG132 (10 μM) for 30 min prior to treatment with SKi or ABC294640. (A) Western blot probed with anti-SK1 antibody showing the effect of ABC294640 or SKi on SK1 expression and reversal by MG132. The bar graph demonstrates the reduction in SK1 expression levels with SKi or ABC294640. Results are expressed as SK1/GAPDH ratios (means ± SEM for 6 independent experiments; *, P < 0.01 versus the control). (B) Western blot probed with anti-Degs1 antibody showing the lack of an effect of SK1 siRNA on the formation of the Degs1 ladder in response to SKi. Also shown are a Western blot and a bar graph demonstrating the reduction in SK1 protein expression with SK1 siRNA. Results are expressed as SK1/GAPDH ratios (means ± SEM for 5 independent experiments; *, P < 0.01 versus scrambled siRNA). (C) Western blot probed with anti-Degs1 antibody showing the lack of an effect of PF-543 on the formation of the Degs1 ladder. (D) Western blot probed with anti-Degs1 antibody showing the lack of an effect of SK2 siRNA on the formation of the Degs1 ladder in response to SKi. Also shown are data from RT-qPCR analysis demonstrating reductions in SK2 mRNA transcripts with SK2 siRNA. Results are expressed as percent decreases in the expression of SK2 (means ± SEM for 3 independent determinations; *, P < 0.01 versus scrambled siRNA). Blots were reprobed for GAPDH using anti-GAPDH antibody to ensure comparable protein loading, and results are representative of data from at least 3 independent experiments. Lanes C, control.
Both ABC294640 and SKi also inhibit SK2 activity (25). Therefore, it is unlikely that SK2 inhibition is involved in promoting the formation of the Degs1 ladder because ABC294640 (25 μM) failed to induce the Degs1 ladder (Fig. 1A). Moreover, siRNA knockdown of SK2 failed to induce formation of the Degs1 ladder (Fig. 3D). The efficiency of siRNA knockdown of the SK2 mRNA transcript was 50% (Fig. 3D). In addition, neither SK1 nor SK2 knockdown affected the formation of the Degs1 ladder in response to SKi (Fig. 3B and D).
Effects of SKi and ABC294640 on PARP cleavage and DNA synthesis.
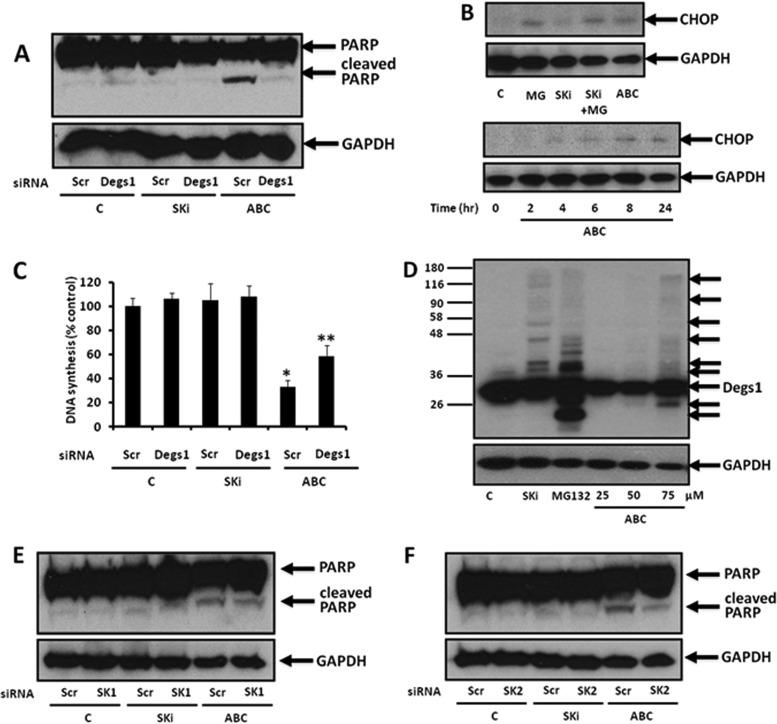
We next tested the effects of SKi and ABC294640 on the growth and apoptosis of HEK293T cells. For this purpose, we measured the cleavage of poly(ADP-ribose) polymerase (PARP) and the induction of CCAAT enhancer binding protein homologous protein (CHOP) expression as surrogate reporters for apoptosis (26), and we measured [3H]thymidine incorporation into DNA to measure growth (although this does not exclude DNA repair). Treatment of HEK293T cells with ABC294640 (25 μM for 24 h) but not SKi (10 μM for 24 h) induced PARP cleavage and CHOP expression and reduced DNA synthesis (Fig. 4A to C), indicating that ABC294640 promotes apoptosis. The effect of ABC294640 on PARP cleavage and DNA synthesis was reversed by siRNA knockdown of Degs1 expression (Fig. 4A and C), indicating that Degs1 is involved in regulating apoptosis and DNA synthesis in response to ABC294640. We also noted that a higher concentration of ABC294640 (75 μM for 24 h) produced a very weak Degs1 ladder (Fig. 4D), indicating that ABC294640 is less potent in inducing posttranslational modification of Degs1 than SKi (Fig. 4A and C). We can therefore formally exclude the polyubiquitinated forms of Degs1 from inducing apoptosis because SKi fails to promote this response.
FIG 4.
Effect of ABC294640 or SKi on PARP cleavage in HEK293T cells. Cells maintained in serum and grown to 70% confluence were treated with ABC294640 (25 to 75 μM) or SKi (10 μM) for 24 h or the times indicated. In certain cases, cells were treated with scrambled, Degs1, SK1, or SK2 siRNA (each at a final concentration of 100 nM) for 48 h prior to treatment with ABC294640 or SKi. (A) Western blot probed with anti-PARP antibody showing the effect of Degs1 siRNA, SKi, or ABC294640 (25 μM) on PARP cleavage. (B) Western blot probed with anti-CHOP antibody showing the effect of SKi (10 μM), ABC294640 (25 μM), or MG132 (MG) (10 μM) on CHOP expression. Also shown is the time-dependent increase in CHOP expression in response to ABC294640. (C) Bar graph showing the effect of Degs1 siRNA, SKi, or ABC294640 (25 μM) on DNA synthesis (*, P < 0.05 for ABC294640/scrambled siRNA versus control/scrambled siRNA; **, P < 0.05 for ABC294640/scrambled siRNA versus ABC294640/Degs1 siRNA [n = 3 independent experiments]). (D) Western blot probed with anti-Degs1 antibody showing the effect of ABC294640 (25, 50, or 75 μM) on Degs1 laddering. (E) Western blot probed with anti-PARP antibody showing the effect of SK1 siRNA, SKi, or ABC294640 (25 μM) on PARP cleavage. (F) Western blot probed with anti-PARP antibody showing the effect of SK2 siRNA, SKi, or ABC294640 (25 μM) on PARP cleavage. Blots were reprobed for GAPDH using anti-GAPDH antibody to ensure comparable protein loading. Results in panels A, B, and D to F are representative of data from at least 3 independent experiments. Lanes C, control.
Given the effects of ABC294640 on SK1 expression (Fig. 3A) and SK2 activity (25), it was necessary to evaluate the roles of these lipid kinases in regulating apoptosis in these cells. In this regard, SK1 knockdown with siRNA had no effect alone or on ABC294640-induced PARP cleavage (Fig. 4E), while SK2 knockdown (∼50%) reduced the cleavage of PARP in response to ABC294640 treatment (Fig. 4F). These findings suggest that SK2 and native Degs1 might collaborate to induce apoptosis in HEK293T cells in response to ABC294640.
Role of polyubiquitinated forms of Degs1 in regulating p38 MAPK and JNK activation.
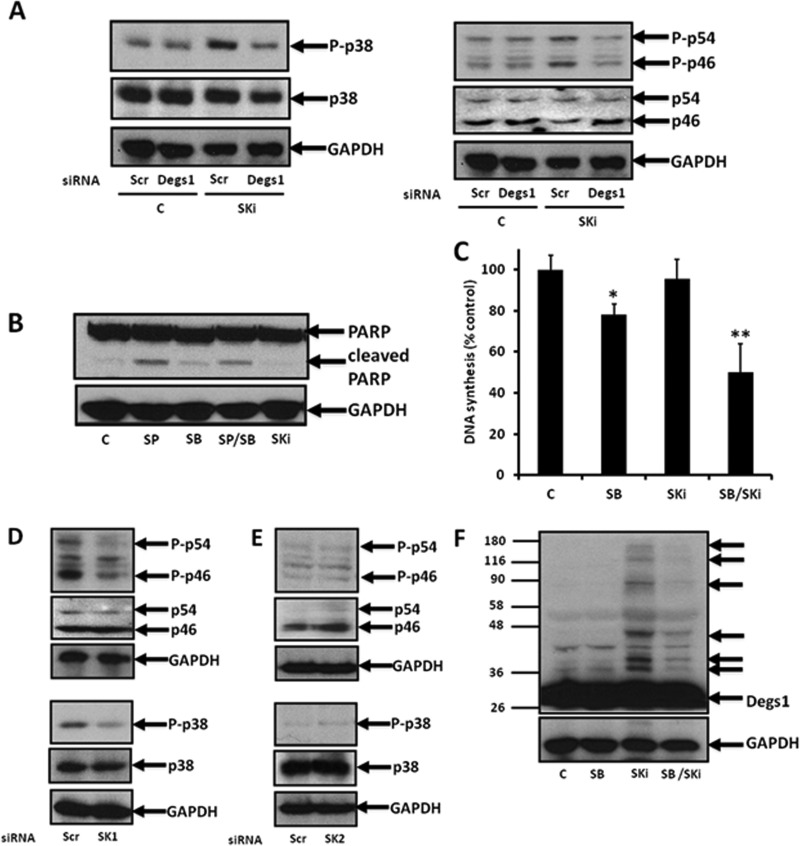
To further explore whether the polyubiquitinated forms of Degs1 are protective against apoptosis, we assessed whether SKi might activate prosurvival signaling pathways. In this regard, SKi induced an increase in the phosphorylation of p38 MAPK and JNK in HEK293T cells (Fig. 5A). These are known as “alarm signals” that might function to counter sustained UPR-induced cell death in this case. Indeed, treatment of cells with the JNK inhibitor SP600125 induced the formation of cleaved PARP (Fig. 5B), while the p38 MAPK inhibitor SB203580 reduced DNA synthesis (Fig. 5C), suggesting that JNK is protective against apoptosis, while p38 MAPK promotes DNA synthesis. In addition, while SKi alone had no effect on DNA synthesis, it enhanced the inhibitory effect of SB203580 (Fig. 5C). This might suggest that SKi is also capable of activating growth-inhibitory pathways but that these are suppressed when phosphorylated p38 MAPK levels are increased by SKi. Furthermore, treatment of HEK293T cells with Degs1 siRNA abolished the effect of SKi on the p38 MAPK and JNK pathways (Fig. 5A). In order for p38 MAPK/JNK signaling in response to SKi to be reduced by the siRNA knockdown of Degs1, the polyubiquitinated forms of Degs1 are likely to have acquired a gain of function.
FIG 5.
p38 MAPK and JNK signaling. Cells maintained in serum and grown to 70% confluence were treated with SKi (10 μM) for 24 h. In certain cases, cells were treated with scrambled, Degs1, SK1, or SK2 siRNA (each at a final concentration of 100 nM) for 48 h or with SB203580 (10 μM) or SP600125 (20 μM) for 30 min prior to treatment with and without SKi. (A) Western blot probed with anti-phospho-p38 MAPK or anti-phospho-JNK antibodies showing the effect of Degs1 siRNA on the phosphorylation levels of p38 MAPK and JNK in response to SKi. (B) Western blot probed with anti-PARP antibody showing the effect of SB203580 (SB) or SP600125 (SP) on PARP cleavage. (C) Bar graph showing the effect of SKi with or without SB203580 on DNA synthesis (*, P < 0.05 for SB203580 versus the control; **, P < 0.05 for SB203580/SKi versus SB203580 alone [n = 3 independent experiments]). (D and E) Western blot probed with anti-phospho-p38 MAPK and anti-phospho-JNK antibodies showing no effect of SK1 siRNA (D) or SK2 siRNA (E) on the phosphorylation levels of p38 MAPK and JNK. (F) Western blot probed with anti-Degs1 antibody showing the effect of SB203580 on Degs1 laddering in response to SKi. Blots were reprobed for GAPDH using anti-GAPDH antibody, anti-p38 MAPK antibody, or anti-JNK antibody to ensure comparable protein loading. Results in panels A, B, and D to F are representative of data from at least 3 independent experiments. Lanes C, control.
Degs1 siRNA reduces the expression of both native and polyubiquitinated forms of Degs1 that accumulate in response to SKi (Fig. 2A). Therefore, the question arises as to which form(s) of Degs1 (native or polyubiquitinated forms) accounts for the increases in phosphorylated p38 MAPK and JNK levels. We can exclude native Degs1 because the levels of phosphorylated p38 MAPK and JNK are not significantly reduced in cells treated with Degs1 siRNA alone (Fig. 5A). To evaluate the role of SK1 or SK2 in regulating p38 MAPK and JNK signaling, we used SK1 siRNA or SK2 siRNA. However, neither siRNA recapitulated the effect of SKi on p38 MAPK and JNK phosphorylation levels (Fig. 5D and E).
For completeness, we evaluated the effects of SB203580 (p38 MAPK inhibitor) and SP600125 (JNK inhibitor) on the polyubiquitination of Degs1 in response to SKi. Surprisingly, pretreatment of cells with SB203580 (Fig. 5F) but not SP600125 (data not shown) decreased the formation of the Degs1 ladder in response to SKi. Thus, p38 MAPK might be involved in a positive-feedback loop that enhances the polyubiquitination of Degs1.
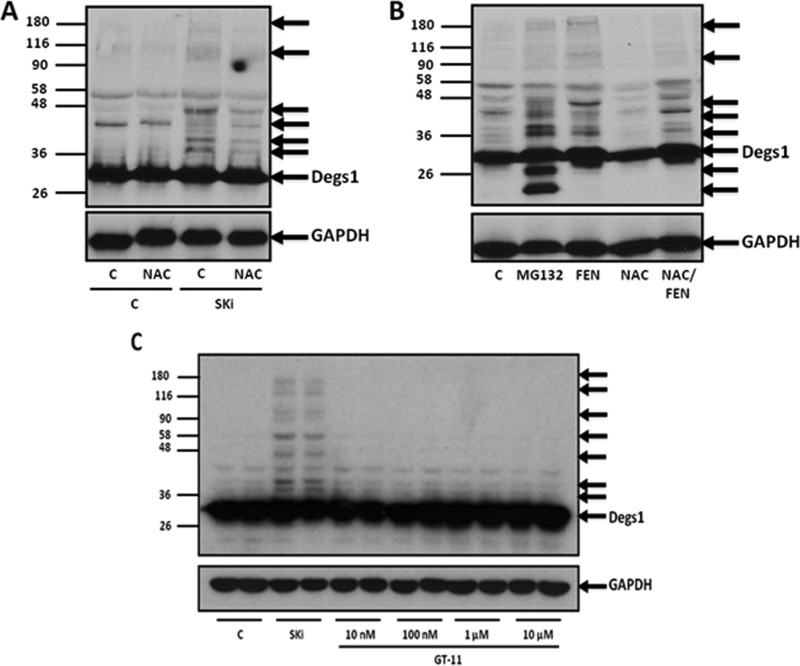
Effects of NAC, fenretinide, and GT11 on the polyubiquitination of Degs1.
Previous studies have proposed that SKi indirectly inhibits Degs1 activity via a mechanism involving an oxidative stress response and cytochrome b5 reductase (27). Similarly, fenretinide has been shown to inhibit Degs1 activity and induce oxidative stress (5, 28, 29). Therefore, we assessed whether the antioxidant N-acetyl-l-cysteine (NAC) and fenretinide can affect the posttranslational modification of Degs1. In this regard, the pretreatment of HEK293T cells with NAC reduced the formation of the Degs1 ladder in response to SKi (Fig. 6A). Fenretinide also induced the formation of posttranslationally modified forms of Degs1, and pretreatment with the antioxidant NAC reduced their formation (Fig. 6B). To more directly establish a link between compounds that can modulate Degs1 activity and formation of the Degs1 ladder, we used the competitive Degs1 inhibitor GT11 {N-[(1R,2S)-2-hydroxy-1-hydroxymethyl-2-(2-tridecyl-1-cyclopropenyl)ethyl]octanamide}. GT11 has been shown to competitively inhibit Degs1 with a Ki of 6 μM (30), and treatment of primary neurons for 24 h maximally inhibited the desaturation of ceramide at 100 nM (31). Higher concentrations of GT11 (10 μM) inhibit S1P lyase activity (31). Therefore, we treated HEK293T cells with GT11 (10 nM to 10 μM), which failed to induce the Degs1 ladder (Fig. 6C). Taken together, these findings suggest that it might be necessary for SKi and fenretinide to induce oxidative stress in order to indirectly stimulate the polyubiquitination of Degs1 and that the effects are not likely a consequence of direct binding to Degs1.
FIG 6.
Oxidative stress and polyubiquitination of Degs1. Cells maintained in serum and grown to 70% confluence were treated with SKi (10 μM), MG132 (10 μM), fenretinide (FEN) (1 μM), or GT11 (10 nM to 10 μM) for 24 h. In certain cases, cells were treated with NAC (10 mM) for 30 min prior to treatment with SKi, fenretinide, or GT11. (A and B) Western blot probed with anti-Degs1 antibody showing the effect of NAC on Degs1 laddering in response to SKi (A) or fenretinide (B). (C) Western blot showing the lack of an effect of GT11 on Degs1 laddering. Blots were reprobed for GAPDH using anti-GAPDH antibody to ensure comparable protein loading. Results are representative of data from at least 3 independent experiments. Lanes C, control.
Effects of ABC294640 and SKi on PERK and XBP-1s.
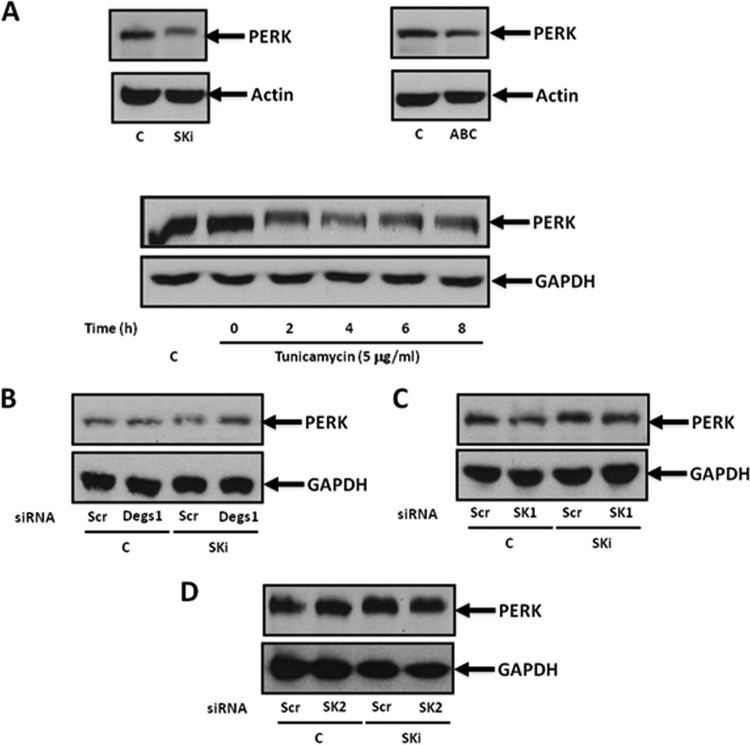
Changes in the lipid composition of the ER membrane, and particularly ceramide/dihydroceramide, are involved in ER stress. In addition, ER stress can contribute both prosurvival and apoptotic signals to the cell. Therefore, we considered whether the regulation of Degs1 by SKi and ABC294640 might involve this pathway. To evaluate this, we measured the levels of two ER stress effectors, namely, protein kinase R-like ER kinase (PERK) and X-box binding protein 1s (XBP-1s). Treatment of HEK293T cells with SKi induced a mobility shift in PERK on SDS-PAGE gels (Fig. 7A), which has previously been reported to be a consequence of phosphorylation (32). Tunicamycin (5 μg/ml), a bona fide inducer of ER stress, also promoted a mobility shift in PERK (Fig. 7A).
FIG 7.
PERK signaling. Cells maintained in serum and grown to 70% confluence were treated with ABC294640 (25 μM) or SKi (10 μM) for 24 h or with tunicamycin (5 μg/ml) for up to 8 h. In certain cases, cells were treated with scrambled, Degs1, SK1, or SK2 siRNA (each at a final concentration of 100 nM) for 48 h prior to treatment with SKi. Western blots probed with anti-PERK antibody show the effect of SKi, ABC294640, or tunicamycin (A), Degs1 siRNA (B), SK1 siRNA (C), or SK2 siRNA (D) on the mobility shift of PERK induced by SKi. Blots were reprobed for GAPDH or actin using anti-GAPDH or antiactin antibodies to ensure comparable protein loading. Results are representative of data from at least 3 independent experiments. Lanes C, control.
The mobility shift in PERK induced by SKi was not reversed by siRNA knockdown of Degs1 (Fig. 7B). Therefore, the SKi-induced phosphorylation of PERK likely does not involve Degs1. Alternatively, inhibition of SK1 or SK2 activity by SKi might affect PERK phosphorylation. However, neither SK1 nor SK2 knockdown with siRNA recapitulated or modified the effects of SKi on PERK (Fig. 7C and D), thereby also possibly excluding these kinases. This is consistent with the finding that ABC294640, which inhibits SK2 (25) and induces proteasomal degradation of SK1 in HEK293T cells (Fig. 3A), failed to induce a mobility shift in PERK (Fig. 7A).
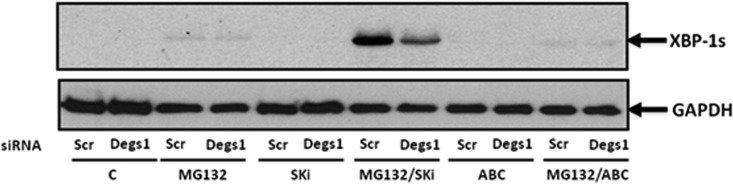
We next turned our attention to XBP-1s (Fig. 8). In this regard, neither ABC294640 nor SKi alone had any effect on XBP-1s expression. In contrast, MG132 induced a modest increase in the expression of XBP-1s. However, this was markedly enhanced by SKi (Fig. 8), which is significant because the combination of SKi and MG132 strongly stimulated the formation of the Degs1 ladder and particularly the accumulation of posttranslationally modified Degs1 forms with a Mr of >50 kDa (Fig. 2A). Moreover, the siRNA knockdown of Degs1 reduced the MG132/SKi-induced increase in XBP-1s expression (Fig. 8). In contrast, ABC294640 (25 μM) reduced the MG132-induced increases in XBP-1s levels (Fig. 8).
FIG 8.
Regulation of XBP-1s expression. Cells maintained in serum and grown to 70% confluence were treated with ABC294640 (25 μM) or SKi (10 μM) for 24 h. In certain cases, cells were treated with scrambled or Degs1 siRNA (final concentration of 100 nM) for 48 h or with MG132 (10 μM) for 30 min prior to treatment with SKi or ABC294640. A Western blot probed with anti-XBP-1s antibody shows the effect of Degs1 siRNA treatment on the induction of XBP-1s by MG132 in the presence and absence of SKi or ABC294640. Blots were reprobed for GAPDH using anti-GAPDH antibody to ensure comparable protein loading. Results are representative of data from at least 3 independent experiments. Lanes C, control.
Effects of SKi and ABC294640 on sphingolipid levels.
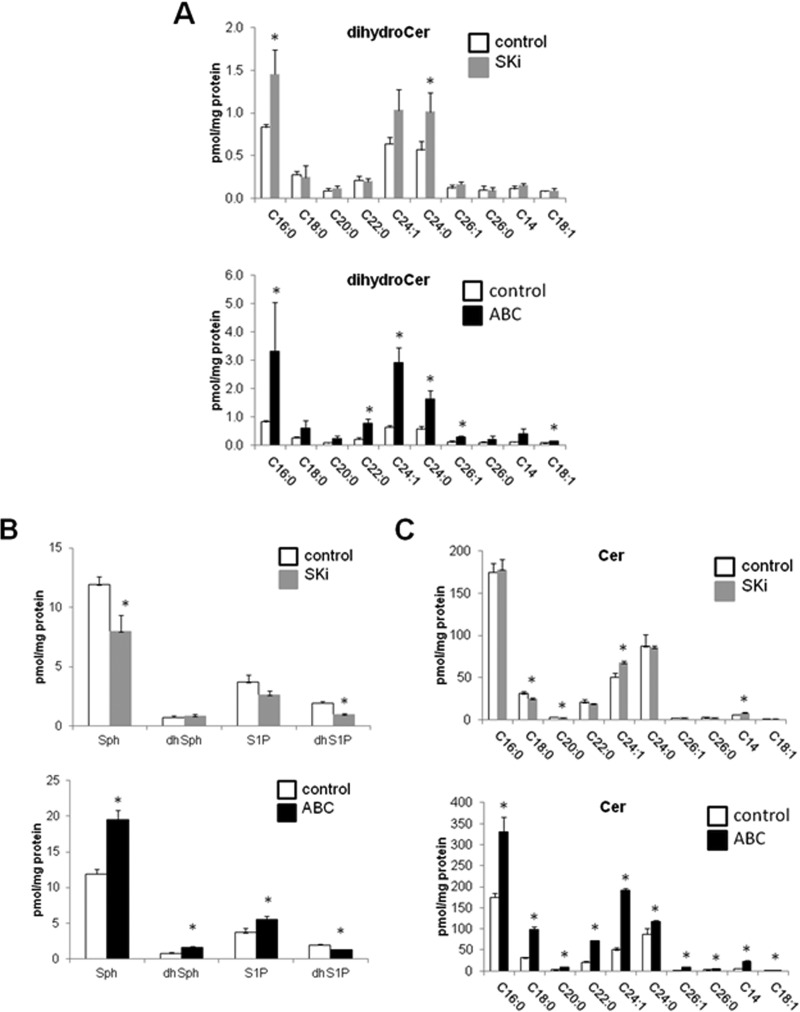
Since ABC294640 (25 μM) and SKi (10 μM) have differential effects on Degs1, we investigated whether these are reflected in changes in sphingolipids. First, we looked at dihydroceramide levels. In this regard, both compounds induced only modest increases in dihydroceramide levels. ABC294640 (25 μM) induced increases in C16:0, C18:1, C22:0, C24:1, C24:0, and C26:1 dihydroceramide levels, with C20:0 dihydroceramide levels approaching significance (Fig. 9A). SKi also induced increases in dihydroceramide levels, although, in contrast to ABC294640, only the levels of C16:0 and C24:0 dihydroceramides were elevated, while the increase in the C24:1 dihydroceramide level approached statistical significance (Fig. 9A). Hexosyl dihydroceramide levels were too low to produce a clear indication as to whether they were changed (data not shown).
FIG 9.
Effects of ABC294640 and SKi on dihydroceramide, sphingoid base, and ceramide levels. Cells maintained in serum and grown to 70% confluence were treated with ABC294640 (25 μM) or SKi (10 μM) for 24 h before snap-freezing. Lipid extracts were analyzed by LC-MS for different molecular species of dihydroceramide (A), sphingoid bases (B), and ceramides (C) (*, P < 0.05 for ABC294640 or SKi versus the control for 3 independent samples). Sphinganine and sphinganine 1-phosphate are denoted here dhSph and dhS1P, respectively. For panels A and C, the x axis annotates different N-acyl chain lengths and double-bond molecular species.
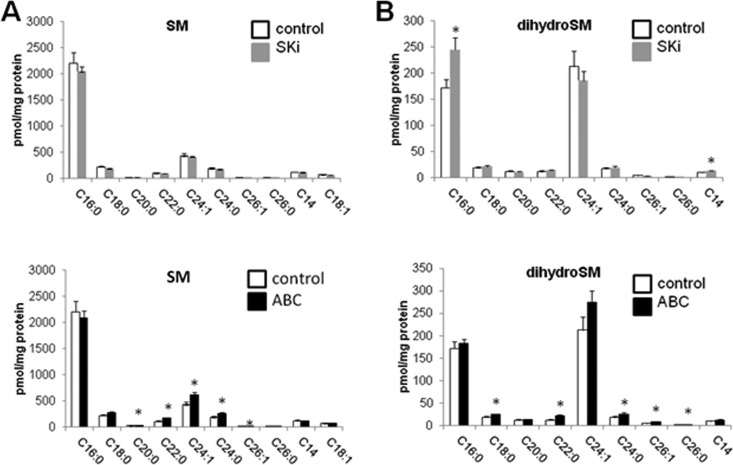
Both SKi and ABC294640 are SK1/SK2 inhibitors, and so we established their effects on long-chain sphingoid bases. Treatment of the cells with SKi induced a reduction in sphinganine 1-phosphate levels, with the decrease in S1P levels approaching statistical significance (P = 0.06) (Fig. 9B), consistent with its inhibitory effect on SK1 and SK2 activity. This was accompanied by a lack of an effect on sphinganine levels but a reduction in sphingosine levels. In contrast, ABC294640 induced an increase in sphingosine, sphinganine, and S1P levels (Fig. 9B), which might reflect increased de novo synthesis and/or uptake while reducing sphinganine 1-phosphate levels (Fig. 9B); the latter effect is consistent with SK inhibition. In line with the elevation of sphingosine and sphinganine levels, ABC294640 induced an increase in ceramide levels (Fig. 9C), and this might also reflect an induction of de novo biosynthesis and/or uptake from the medium. In this regard, ABC294640 induced an increase in the levels of various ceramide molecular species (including C14:0, C16:0, C18:0, C18:1, C20:0, C22:0, C24:1, C24:0, C26:0, and C26:1) (Fig. 9C), which is consistent with the ability of this compound to induce apoptosis. In contrast, SKi induced a decrease in the levels of C18:0 and C20:0 ceramides, while it induced increased levels of C24:1 and C14:0 ceramides (Fig. 9C). SKi had little effect on total ceramides, suggesting that it probably fails to raise the levels of “apoptotic” ceramides, thereby accounting for its lack of an effect on PARP and CHOP (Fig. 4A and B). In terms of sphingomyelin levels, ABC294640 induced an increase in the levels of C20:0, C22:0, C24:0, C24:1, and C26:1 species, with the increase for C18:0 approaching statistical significance, while SKi had no effect (Fig. 10A). ABC294640 also increased the levels of dihydrosphingomyelin (including C18:0, C22:0, C24:0, and C26:1, with C24:1 approaching statistical significance [P = 0.054]), while SKi increased the levels of C16:0 dihydrosphingomyelin (Fig. 10B).
FIG 10.
Effects of ABC294640 and SKi on sphingomyelin and dihydrosphingomyelin levels. Cells maintained in serum and grown to 70% confluence were treated with ABC294640 (25 μM) or SKi (10 μM) for 24 h before snap-freezing. Lipid extracts were analyzed by LC-MS for different molecular species of sphingomyelin (SM) (A) and dihydrosphingomyelin (B) (*, P < 0.05 for ABC294630 or SKi versus the control for 3 independent samples). The x axis annotates different N-acyl chain lengths and double-bond molecular species.
DISCUSSION
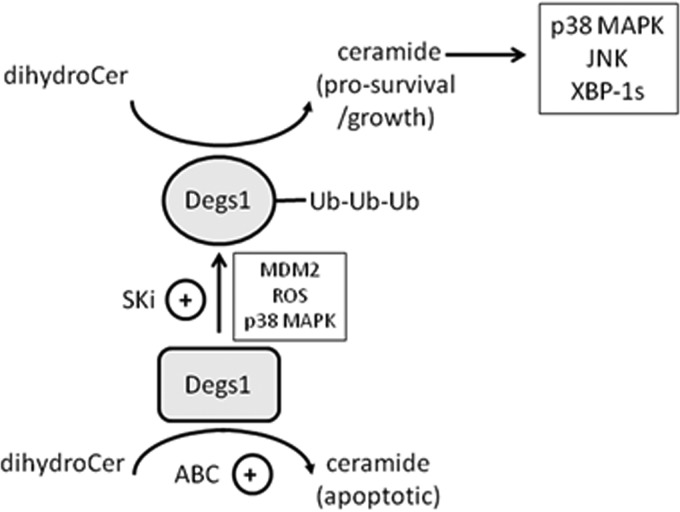
The major finding of this study is that Degs1 is regulated by polyubiquitination. Moreover, we demonstrate here that the native and polyubiquitinated forms of Degs1 exert opposing functions on cellular fate. Thus, the polyubiquitinated forms of Degs1 are linked with the activation of prosurvival and growth signaling pathways, e.g., JNK, XBP-1s, and p38 MAPK (Fig. 11). The polyubiquitinated forms of Degs1 appear to be in transit to proteasomal degradation, as the accumulation of the forms is increased by combined treatment of the cells with the proteasome inhibitor MG132 and SKi. Therefore, it is possible that some compounds remove Degs1 from cells via the ubiquitin-proteasomal degradation route, and this might account for cell death induced by these compounds, which has been reported in the literature (7, 10–13). However, the polyubiquitinated forms themselves are endowed with a prosurvival function, and therefore, if the degradation rate is low, then these forms might predominate and thereby account for the cell survival function of Degs1 (5–7, 9, 14, 15).
FIG 11.
Schematic showing the different roles of native and polyubiquitinated Degs1 forms in regulating prosurvival/growth and apoptotic pathways in HEK293T cells. Native Degs1 is proposed to regulate the formation of apoptotic ceramide species, while polyubiquitinated Degs1 forms are proposed to regulate the formation of prosurvival/growth ceramide species. ROS, reactive oxygen species.
We have also demonstrated a role for oxidative stress, p38 MAPK (functioning in a positive-feedback mechanism of regulation), and Mdm2 in promoting the polyubiquitination of Degs1. The mechanism by which p38 MAPK regulates the polyubiquitination of Degs1 remains to be determined. However, previous studies have shown that tumor necrosis factor alpha (TNF-α) uses the p38 MAPK pathway to stimulate the expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle (33). There is additional evidence for the link between p38 MAPK and Mdm2. Thus, hypoxia has been shown to induce downregulation of Mdm2, and this can be reduced by p38 MAPK inhibitors and by a dominant-interfering mutant of the p38-activating kinase mitogen-activated protein kinase kinase 3 (34).
In contrast to the polyubiquitinated forms of Degs1, the native form of Degs1 appears to be linked with the induction of apoptosis (Fig. 11). In this regard, ABC294640 (25 μM) fails to induce the polyubiquitination of Degs1 but promotes apoptosis. A higher concentration of ABC294640 (75 μM) stimulates the formation of the Degs1 ladder, suggesting that it is significantly less potent than SKi.
We have also established the effects of SKi and ABC294640 on sphingolipid levels, which provides information concerning the effect of modulating Degs1, SK1, and SK2 activity on the survival of HEK293T cells. Treatment of cells with SKi increased C16:0 and C24:0 dihydroceramide levels. The increase in the levels of these species is relatively modest and can be accounted for by the loss of some Degs1 via proteasomal degradation. However, the selective effect on these particular dihydroceramide species is somewhat surprising because this enzyme can catalyze the desaturation of many different molecular species of dihydroceramide. The dihydroceramide profile might, in fact, be an average of some loss of Degs1 via proteasomal degradation and some gain of function acquired by the polyubiquitinated forms. Second, the levels of C14:0 and C24:1 ceramides were shown to be elevated in response to treatment of cells with SKi, and it is tempting to speculate that these species might be formed from dihydroceramide by the polyubiquitinated Degs1 forms. In this regard, these molecular species might represent “prosurvival” ceramides linked to the activation of the p38 MAPK/JNK and XBP-1s pathways. The gain of function acquired by the polyubiquitinated forms of Degs1 might require redistribution to a specific functional compartment with effectors of the survival pathways, where local regulation of discrete pools of specific dihydroceramide molecular species can take place. Indeed, Beauchamp et al. (35) reported that Degs1 is N-myristoylated and targeted to the mitochondria, suggesting that there are two pools of Degs1, one in the ER and one in the mitochondria. This requires further investigation, as immunofluorescence analysis with anti-Degs1 antibody did not reveal any obvious change in the distribution of Degs1 with SKi (data not shown), although the polyubiquitinated forms of Degs1 might be resident in the ER because of the link with the activation of the ER stress-responsive effector JNK. The modest effect of SKi on dihydroceramide levels might also suggest that the rates of proteasomal degradation of Degs1 and, therefore, inactivation are rather low in HEK293T cells, thereby allowing the intermediate polyubiquitinated forms to accumulate and to therefore instigate a persistent cell survival program.
The treatment of HEK293T cells with ABC294640 increased both dihydroceramide and ceramide levels. This suggests an overall increase in flux through the de novo ceramide pathway or increased cellular uptake. The increased levels of dihydroceramide might effectively endow native Degs1 with a gain of function by a substrate induction mechanism to promote the formation of ceramide species that promote apoptosis. The changes in dihydroceramide levels with SKi and ABC294640 are consistent with previous studies that reported that these compounds promote senescence of androgen-independent LNCaP-AI prostate cancer cells associated with the proteasomal degradation of SK1 and Degs1 (21) and increased p53 and p21 expression (21). The inhibitory effects of SKi and ABC294640 on Degs1 activity result in increased dihydroceramide levels in prostate cancer cells (23, 36). SKi has been proposed to also indirectly inhibit Degs1 via a cytochrome b5 reductase-dependent mechanism (27) and also increased dihydroceramide levels in ovarian cancer cells (37).
The treatment of cells with ABC294640 also increased both sphingosine and S1P levels, which is rather surprising given its effect on inducing the proteasomal degradation of SK1 in HEK293T cells and its characterization as an SK2 inhibitor. Nevertheless, ABC294640 is a rather a weak inhibitor of SK2 (50% inhibitory concentration [IC50] = 50 μM) (38). Therefore, there might be sufficient SK2 activity remaining in the presence of ABC294640 to drive S1P formation as a consequence of elevated levels of sphingosine that accumulates in response to ABC294640. ABC294640 also reduced sphinganine 1-phosphate levels, which is consistent with its ability to induce the proteasomal degradation of SK1 in HEK293T cells. Therefore, these findings suggest that SK1 might be more important than SK2 in catalyzing the formation of sphinganine 1-phosphate in these cells. Indeed, a similar reduction in sphinganine 1-phosphate levels is observed with SKi, which also induces the proteasomal degradation of SK1. SKi also reduces S1P levels albeit moderately. A possible explanation for the difference in the actions of ABC294640 and SKi on S1P levels, then, is that SKi is a much stronger inhibitor of SK2 activity (IC50 = 2 μM) (our unpublished data) than ABC294640, and therefore, S1P levels might be predominantly regulated by SK2 in HEK293T cells.
The findings presented in this paper reveal that Degs1 is a very versatile enzyme, and polyubiquitination might convert the enzyme from being proapoptotic to being prosurvival. This might account for various conflicting reports on the role of Degs1 in promoting apoptosis or cell survival mediated by, for instance, autophagy (for a review, see reference 4). There is evidence for similar regulation of CerS1 by the ubiquitin-proteasome degradation pathway. For instance, stress induces the translocation of CerS1 from the ER to the Golgi apparatus and promotes its proteasomal degradation in HEK293 cells (39). Moreover, overexpression of CerS1 increases the activation of p38 MAPK by cisplatin in HEK293 cells (40). These findings are intriguing and suggest that CerS1 and Degs1 might operate through a similar linked ubiquitin-proteasomal degradation mechanism in response to chemical stress that results in the activation of the p38 MAPK pathway.
The prosurvival function of Degs1 is likely to be dictated by the lifetime of the polyubiquitinated forms of Degs1 in various different cellular systems. These findings might have important consequences when evaluating Degs1 as a target for therapeutic treatment of cancer. Our findings reveal possible modes of action by which drugs targeting Degs1 might act. First, drugs that prevent polyubiquitination of Degs1 might promote cell death if they also increase de novo biosynthesis of apoptotic ceramide. Second, drugs that accelerate the proteasomal degradation of Degs1 might remove the prosurvival function of the polyubiquitinated forms to potentially kill cancer cells via apoptotic/senescent pathways.
MATERIALS AND METHODS
Materials.
All general biochemicals were from Sigma (Poole, UK). ABC294640 (catalogue number S7174) was obtained from Stratech Scientific (UK). SKi (catalogue number 567731) was obtained from Merck Biosciences (Nottingham, UK). MG132 (catalogue number C2211), N-acetyl-l-cysteine (catalogue number A7250), nutlin (catalogue number N6287), and protein A-Sepharose 4B Fast Flow (catalogue number P9424) were obtained from Sigma (Poole, UK). SP600125 (catalogue number BML-EI305) and SB203580 (catalogue number BML-EI286) were obtained from Enzo Life Sciences (UK). Bortezomib (catalogue number sc-217785) and fenretinide (catalogue number HY-15373) were obtained from Insight Biotechnology Ltd. (UK). Anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (catalogue number sc-47724) antibody was obtained from Insight Biotechnology Ltd. (UK); anti-Degs1 (catalogue number ab185237) antibody was obtained from Abcam; and anti-phospho-JNK (catalogue number 4671), anti-phospho-p38 MAPK (catalogue number 9211), anti-PARP (catalogue number 9542), anti-PERK (catalogue number 5683), anti-CHOP (catalogue number 5554), and anti-XBP-1s (catalogue number 12782) antibodies were obtained from New England BioLabs Ltd. (Hitchin, UK). Anti-SK1 antibody was custom-made by Abcam. DharmaFECT reagent; ON-TARGETplus SMARTpool; and Degs1, SK1, and SK2 siRNAs were obtained from Dharmacon (Cromlington, UK). Scrambled siRNA (Allstars negative control) was obtained from Qiagen (Crawley, UK). [methyl-3H]thymidine (25 Ci/mmol; 37 MBq/ml) (catalogue number NET027A) was obtained from PerkinElmer (UK). The internal standard mix for quantitation of the sphingolipids by liquid chromatography (LC)-tandem mass spectrometry (MS) (catalogue number LM-6002) was obtained from Avanti Polar Lipids (Alabaster, AL). GT11 was obtained from Avanti Polar Lipids.
Cell culture.
HEK293T and PANC1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM)-GlutaMAX supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% (vol/vol) fetal bovine serum at 37°C with 5% CO2 before treatment with compounds.
[3H]thymidine incorporation.
HEK293T cells (approximately 70% confluent) in 24-well plates were incubated with compounds or the vehicle (dimethyl sulfoxide [DMSO]; 0.1% [vol/vol] final concentration), as detailed in the figure legends, for 20 h prior to the addition of [3H]thymidine (9.25 kBq per well) for a further 5 h. Incubations were terminated by removing the medium and immediately adding 1 ml of ice-cold 10% (wt/vol) trichloroacetic acid, and the mixture was placed on ice for 10 min. This was replaced with a further 1 ml of ice-cold 10% (wt/vol) trichloroacetic acid for 10 min, and this step was repeated once more. Residual nuclear material was dissolved in 0.25 ml of 0.1% SDS–0.3 M NaOH. [3H]thymidine uptake was quantified by liquid scintillation counting. Radiometric values (means ± standard deviations [SD]) were obtained from 3 or more independent experiments.
siRNA transfection.
HEK293T cells were transiently transfected with siRNA constructs or scrambled siRNA (as a negative control) at a final concentration of 100 nM. Cells were transfected at approximately 50 to 60% confluence and maintained in the transfection mixture for 48 h.
Immunoprecipitation.
Cells were placed in ice-cold lysis buffer (500 μl/3 wells) containing 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 1% (wt/vol) Na deoxycholate, 10% (vol/vol) glycerol, 20 mM Tris base, 1 mg/ml bovine serum albumin (BSA), 0.5 mM Na3VO4, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), and leupeptin and aprotinin (both at 10 μg/ml) (pH 8.0). The samples were homogenized using a 0.24-mm-gauge needle and syringe and left to shake at 4°C for 60 min. After centrifugation to remove cell debris, 250 μl of the supernatant was precleared with 20 μl of protein A-Sepharose beads (1:1 with lysis buffer for 20 min at 4°C) before subsequent immunoprecipitation by adding 2 μl of the anti-Degs1 antibody and 20 μl protein A-Sepharose beads (1:1 with lysis buffer for 2 h at 4°C). Immunoprecipitates were collected by centrifugation and washed three times with buffer A (10 mM HEPES, 100 mM NaCl, 0.5% [vol/vol] NP-40, and 0.2 mM PMSF [pH 7.0]) and three times with buffer B (buffer A without NP-40). The beads were then sedimented by centrifugation, the supernatant was removed, and 20 μl of Laemmli buffer (0.125 M Tris-HCl, 10% [vol/vol] 2-mercaptoethanol, 20% [vol/vol] glycerol, 4% [wt/vol] SDS, and 0.004% [wt/vol] bromophenol blue [pH 6.7]) was added to the beads, which were then heated at 100°C for 3 min. The samples were then subjected to SDS-PAGE.
Western blotting.
Upon treatment, HEK293T cells were lysed in sample buffer containing 62.5 mM Tris-HCl (pH 6.7), 0.5 M sodium pyrophosphate, 1.25 mM EDTA, 1.25% (wt/vol) sodium dodecyl sulfate, 0.06% (wt/vol) bromophenol blue, 12.5% (vol/vol) glycerol, and 50 mM dithiothreitol. Proteins were separated on a 10% (vol/vol) acrylamide-bisacrylamide gel and transferred to a nitrocellulose Hybond membrane (GE Healthcare). Membranes were blocked in 5% (wt/vol) BSA (Fisher) in TBST buffer containing 20 mM Tris-HCl (pH 7.5), 48 mM NaCl, and 0.1% (vol/vol) Tween 20 for 1 h at room temperature prior to incubation with primary antibody (diluted in blocking buffer) overnight at 4°C. Following three washes in TBST, membranes were incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG secondary antibody, as required, for 1 h at room temperature. Immunoreactive protein bands were visualized using enhanced chemiluminescence.
RNA extraction.
Total RNAs from the cell samples were isolated using an RNeasy Plus minikit (Qiagen, Manchester, UK) according to the manufacturer's instructions. Spectrophotometric analysis with a Nanodrop 2000C instrument (ThermoFisher Scientific, Renfrew, UK) was performed to obtain the RNA concentrations, and all of the RNAs had A260/A280 ratios of approximately 2.1. The RNAs were stored at −80°C until required.
Gene expression analysis by quantitative real-time PCR.
RNAs from each cell sample were reverse transcribed to cDNA with a Tetro cDNA synthesis kit (Bioline, London, UK), along with aliquots processed without reverse transcriptase to function as no-template controls for real-time quantitative PCR (RT-qPCR), and were stored at −20°C. RT-qPCR was conducted by using PrimeTime qPCR probe assays for SPHK2 (assay Hs.PT.58.3726704) and GAPDH (assay Hs.PT.39a.22214836) as the reference gene with PrimeTime gene expression master mix (Integrated DNA Technologies, Leuven, Belgium). Thermal cycling was carried out on an ABI StepOnePlus real-time PCR system (Applied Biosystems, Warrington, UK) under the following protocol: polymerase activation at 95°C for 3 min followed by 40 cycles of denaturation at 95°C for 5 s and annealing/extension at 60°C for 30 s. Relative quantification of gene expression between the samples treated with the control scrambled siRNA and those treated with SPHK2 siRNA was performed by using the 2−ΔΔCT method to determine the percent knockdown of SPHK2 expression (41).
Lipidomics.
HEK293T cells were treated with the vehicle (DMSO; 0.1% [vol/vol] final concentration), ABC294640 (25 μM), or SKi (10 μM) for 24 h and then carefully rinsed twice with 1 ml ice-cold phosphate-buffered saline (PBS) before being scraped into ice-cold PBS. Cells were pelleted by centrifugation (180 × g at 4°C for 3 min), and the supernatant was carefully removed. The cell pellet was snap-frozen in liquid nitrogen for 5 s before being stored at −80°C for sphingolipid analysis, which was conducted as described previously (42, 43), using liquid chromatography, electrospray ionization-tandem mass spectrometry, and multiple-reaction monitoring for quantitation.
Densitometry.
Quantification of immunoreactive bands was performed using ImageJ.
ACKNOWLEDGMENTS
Funds for S.L.K. and A.H.M. were from the Smithgall Institute Endowed Chair in Molecular Cell Biology at Georgia Tech. M.A. thanks the Kuwait Government for Ph.D. sponsorship (reference number 1590235272).
REFERENCES
- 1.Hannun YA, Obeid LM. 2008. Principles of bioactive lipid signaling: lessons from sphingolipids. Nat Rev Mol Cell Biol 9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 2.Hannun YA, Obeid LM. 2011. Many ceramides. J Biol Chem 286:27855–27862. doi: 10.1074/jbc.R111.254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spiegel S, Milstien S. 2011. The ‘outs and the ins’ of sphingosine-1-phosphate in immunity. Nat Rev Immunol 11:403–415. doi: 10.1038/nri2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddique MM, Li Y, Chaurasia B, Kaddai VA, Summers SA. 2015. Dihydroceramides: from bit players to lead actors. J Biol Chem 290:15371–15379. doi: 10.1074/jbc.R115.653204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, Kelly S, Allegood JC, Liu Y, Peng Q, Ramaraju H, Sullards MC, Cabot M, Merrill AH Jr.. 2006. Ceramides and other bioactive sphingolipid backbones in health and disease: lipidomic analysis, metabolism and roles in membrane structure, dynamics, signaling and autophagy. Biochim Biophys Acta 1758:1864–1884. doi: 10.1016/j.bbamem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Signorelli P, Munoz-Olaya JM, Gagliostro V, Casas J, Ghidoni R, Fabriàs G. 2009. Dihydroceramide intracellular increase in response to resveratrol treatment mediates autophagy in gastric cancer cells. Cancer Lett 282:238–243. doi: 10.1016/j.canlet.2009.03.020. [DOI] [PubMed] [Google Scholar]
- 7.Gagliostro V, Casas J, Caretti A, Abad JL, Tagliavacca L, Ghidoni R, Fabrias G, Signorelli P. 2012. Dihydroceramide delays cell cycle G1/S transition via activation of ER stress and induction of autophagy. Int J Biochem Cell Biol 44:2135–2143. doi: 10.1016/j.biocel.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 8.Casasampere M, Ordóñez YF, Casas J, Fabrias G. 2017. Dihydroceramide desaturase inhibitors induce autophagy via dihydroceramide-dependent and independent mechanisms. Biochim Biophys Acta 1861:264–275. doi: 10.1016/j.bbagen.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 9.Siddique MM, Li Y, Wang L, Ching J, Mal M, Ilkayeva O, Wu YJ, Bay BH, Summers SA. 2013. Ablation of dihydroceramide desaturase 1, a therapeutic target for the treatment of metabolic diseases, simultaneously stimulates anabolic and catabolic signaling. Mol Cell Biol 33:2353–2369. doi: 10.1128/MCB.00226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernández-Tiedra S, Fabriàs G, Dávila D, Salanueva ÍJ, Casas J, Montes LR, Antón Z, García-Taboada E, Salazar-Roa M, Lorente M, Nylandsted J, Armstrong J, López-Valero I, McKee CS, Serrano-Puebla A, García-López R, González-Martínez J, Abad JL, Hanada K, Boya P, Goñi F, Guzmán M, Lovat P, Jäättelä M, Alonso A, Velasco G. 2016. Dihydroceramide accumulation mediates cytotoxic autophagy of cancer cells via autolysosome destabilization. Autophagy 12:2213–2229. doi: 10.1080/15548627.2016.1213927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu JM, DiPietrantonio AM, Hsieh TC. 2001. Mechanism of fenretinide (4-HPR)-induced cell death. Apoptosis 6:377–388. doi: 10.1023/A:1011342220621. [DOI] [PubMed] [Google Scholar]
- 12.Erdreich-Epstein A, Tran LB, Bowman NN, Wang H, Cabot MC, Durden DL, Vlckova J, Reynolds CP, Stins MF, Groshen S, Millard M. 2002. Ceramide signaling in fenretinide-induced endothelial cell apoptosis. J Biol Chem 277:49531–49537. doi: 10.1074/jbc.M209962200. [DOI] [PubMed] [Google Scholar]
- 13.Hail N Jr, Kim HJ, Lotan R. 2006. Mechanisms of fenretinide-induced apoptosis. Apoptosis 11:1677–1694. doi: 10.1007/s10495-006-9289-3. [DOI] [PubMed] [Google Scholar]
- 14.Siddique MM, Bikman BT, Wang L, Ying L, Reinhardt E, Shui G, Wenk MR, Summers SA. 2012. Ablation of dihydroceramide desaturase confers resistance to etoposide-induced apoptosis in vitro. PLoS One 7:e44042. doi: 10.1371/journal.pone.0044042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breen P, Joseph N, Thompson K, Kraveka JM, Gudz TI, Li L, Rahmaniyan M, Bielawski J, Pierce JS, Van Buren E, Bhatti G, Separovic D. 2013. Dihydroceramide desaturase knockdown impacts sphingolipids and apoptosis after photodamage in human head and neck squamous carcinoma cells. Anticancer Res 33:77–84. [PMC free article] [PubMed] [Google Scholar]
- 16.Spassieva SD, Mullen TD, Townsend DM, Obeid LM. 2009. Disruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem J 424:273–283. doi: 10.1042/BJ20090699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volmer R, Ron D. 2015. Lipid-dependent regulation of the unfolded protein response. Curr Opin Cell Biol 33:67–73. doi: 10.1016/j.ceb.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White-Gilbertson S, Hua Y, Liu B. 2013. The role of endoplasmic reticulum stress in maintaining and targeting multiple myeloma: a double-edged sword of adaptation and apoptosis. Front Genet 4:109. doi: 10.3389/fgene.2013.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallington-Beddoe CT, Bennett MK, Vandyke K, Davies L, Zebol JR, Moretti PAB, Pitman MR, Hewett DR, Zannettino ACW, Pitson SM. 2017. Sphingosine kinase 2 inhibition synergises with bortezomib to target myeloma by enhancing endoplasmic reticulum stress. Oncotarget 8:43602–43616. doi: 10.18632/oncotarget.17115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evangelisti C, Evangelisti C, Teti G, Chiarini F, Falconi M, Melchionda F, Pession A, Bertaina A, Locatelli F, McCubrey JA, Beak DJ, Bittman R, Pyne S, Pyne NJ, Martelli AM. 2014. Assessment of the effect of sphingosine kinase inhibitors on apoptosis, unfolded protein response and autophagy of T-cell acute lymphoblastic leukemia cells; indications for novel therapeutics. Oncotarget 5:7886–7901. doi: 10.18632/oncotarget.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNaughton M, Pitman M, Pitson SM, Pyne NJ, Pyne S. 2016. Proteasomal degradation of sphingosine kinase 1 and inhibition of dihydroceramide desaturase by the sphingosine kinase inhibitors, SKi or ABC294640, induces growth arrest in androgen-independent LNCaP-AI prostate cancer cells. Oncotarget 7:16663–16675. doi: 10.18632/oncotarget.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moll UM, Petrenko O. 2003. The MDM2-p53 interaction. Mol Cancer Res 1:1001–1008. [PubMed] [Google Scholar]
- 23.Loveridge C, Tonelli F, Leclercq T, Lim KG, Long JS, Berdyshev E, Tate RJ, Natarajan V, Pitson SM, Pyne NJ, Pyne S. 2010. The sphingosine kinase 1 inhibitor 2-(p-hydroxyanilino)-4-(p-chlorophenyl)thiazole induces proteasomal degradation of sphingosine kinase 1 in mammalian cells. J Biol Chem 285:38841–38852. doi: 10.1074/jbc.M110.127993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnute ME, McReynolds MD, Kasten T, Yates M, Jerome G, Rains JW, Hall T, Chrencik J, Kraus M, Cronin CN, Saabye M, Highkin MK, Broadus R, Ogawa S, Cukyne K, Zawadzke LE, Peterkin V, Iyanar K, Scholten JA, Wendling J, Fujiwara H, Nemirovskiy O, Wittwer AJ, Nagiec MM. 2012. Modulation of cellular S1P levels with a novel, potent and specific inhibitor of sphingosine kinase-1. Biochem J 444:79–88. doi: 10.1042/BJ20111929. [DOI] [PubMed] [Google Scholar]
- 25.Gao P, Peterson YK, Smith RA, Smith CD. 2012. Characterization of isoenzyme-selective inhibitors of human sphingosine kinases. PLoS One 7:e44543. doi: 10.1371/journal.pone.0044543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishitoh H. 2012. CHOP is a multifunctional transcription factor in ER stress. J Biochem 151:217–219. doi: 10.1093/jb/mvr143. [DOI] [PubMed] [Google Scholar]
- 27.Cingolani F, Casasampere M, Sanllehí P, Casas J, Bujons J, Fabrias G. 2014. Inhibition of dihydroceramide desaturase activity by the sphingosine kinase inhibitor SKI II. J Lipid Res 55:1711–1720. doi: 10.1194/jlr.M049759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Maurer BJ, Liu YY, Wang E, Allegood JC, Kelly S, Symolon H, Liu Y, Merrill AH Jr, Gouazé-Andersson V, Yu JY, Giuliano AE, Cabot MC. 2008. N-(4-Hydroxyphenyl)retinamide increases dihydroceramide and synergizes with dimethylsphingosine to enhance cancer cell killing. Mol Cancer Ther 7:2967–2976. doi: 10.1158/1535-7163.MCT-08-0549. [DOI] [PubMed] [Google Scholar]
- 29.Rahmaniyan M, Curley RW Jr, Obeid LM, Hannun YA, Kraveka JM. 2011. Identification of dihydroceramide desaturase as a direct in vitro target for fenretinide. J Biol Chem 286:24754–24764. doi: 10.1074/jbc.M111.250779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Triola G, Fabriàs G, Casas J, Llebaria A. 2003. Synthesis of cyclopropene analogues of ceramide and their effect on dihydroceramide desaturase. J Org Chem 68:9924–9932. doi: 10.1021/jo030141u. [DOI] [PubMed] [Google Scholar]
- 31.Triola G, Fabrias G, Dragusin M, Niederhausen L, Broere R, Llebaria A, van Echten-Deckert G. 2004. Specificity of the dihydroceramide desaturase inhibitor N-[(1R,2S)-2-hydroxy-1-hydroxymethyl-2-(2-tridecyl-1-cyclopropenyl)ethyl]octanamide (GT11) in primary cultured cerebellar neurons. Mol Pharmacol 66:1671–1678. doi: 10.1124/mol.104.003681. [DOI] [PubMed] [Google Scholar]
- 32.Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG. 2002. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol 22:7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li YP, Chen Y, John J, Moylan J, Jin B, Mann DL, Reid MB. 2005. TNF-α acts via p38 MAPK to stimulate expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle. FASEB J 19:362–370. doi: 10.1096/fj.04-2364com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y, Mao XO, Sun Y, Xia Z, Greenberg DA. 2002. p38 mitogen-activated protein kinase mediates hypoxic regulation of Mdm2 and p53 in neurons. J Biol Chem 277:22909–22914. doi: 10.1074/jbc.M200042200. [DOI] [PubMed] [Google Scholar]
- 35.Beauchamp E, Tekpli X, Marteil G, Lagadic-Gossmann D, Legrand P, Rioux V. 2009. N-Myristoylation targets dihydroceramide Delta4-desaturase 1 to mitochondria: partial involvement in the apoptotic effect of myristic acid. Biochimie 91:1411–1419. doi: 10.1016/j.biochi.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 36.Venant H, Rahmaniyan M, Jones EE, Lu P, Lilly MB, Garrett-Mayer E, Drake RR, Kraveka JM, Smith CD, Voelkel-Johnson C. 2015. The sphingosine kinase 2 inhibitor ABC294640 reduces the growth of prostate cancer cells and results in accumulation of dihydroceramides in vitro and in vivo. Mol Cancer Ther 14:2744–2752. doi: 10.1158/1535-7163.MCT-15-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Illuzzi G, Bernacchioni C, Aureli M, Prioni S, Frera G, Donati C, Valsecchi M, Chigorno V, Bruni P, Sonnino S, Prinetti A. 2010. Sphingosine kinase mediates resistance to the synthetic retinoid N-(4-hydroxyphenyl) retinamide in human ovarian cancer cells. J Biol Chem 285:18594–18602. doi: 10.1074/jbc.M109.072801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.French KJ, Zhuang Y, Maines LW, Gao P, Wang W, Beljanski V, Upson JJ, Green CL, Keller SN, Smith CD. 2010. Pharmacology and antitumor activity of ABC294640, a selective inhibitor of sphingosine kinase-2. J Pharmacol Exp Ther 333:129–139. doi: 10.1124/jpet.109.163444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sridevi P, Alexander H, Laviad EL, Min J, Mesika A, Hannink M, Futerman AH, Alexander S. 2010. Stress-induced ER to Golgi translocation of ceramide synthase 1 is dependent on proteasomal processing. Exp Cell Res 316:78–91. doi: 10.1016/j.yexcr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Min J, Mesika A, Sivaguru M, Van Veldhoven PP, Alexander H, Futerman AH, Alexander S. 2007. (Dihydro)ceramide synthase 1 regulated sensitivity to cisplatin is associated with the activation of p38 mitogen-activated protein kinase and is abrogated by sphingosine kinase 1. Mol Cancer Res 5:801–812. doi: 10.1158/1541-7786.MCR-07-0100. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Shaner RL, Allegood JC, Park H, Wang E, Kelly S, Haynes CA, Sullards MC, Merrill AH Jr.. 2009. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J Lipid Res 50:1692–1707. doi: 10.1194/jlr.D800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullards MC, Liu Y, Chen Y, Merrill AH Jr.. 2011. Analysis of mammalian sphingolipids by liquid chromatography tandem mass spectrometry (LC-MS/MS) and tissue imaging mass spectrometry (TIMS). Biochim Biophys Acta 1811:838–853. doi: 10.1016/j.bbalip.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]