Abstract
The proteome is the study of the protein content of a definable component of an organism in biology. However, the tissue specific expression of proteins and the varied post translational modifications, splice variants and protein - protein complexes that may form, make the study of protein a challenging yet vital tool in answering many of the unanswered questions in medicine and biology to date. Indeed, the spatial, temporal and functional composition of proteins in the human body has proven difficult to elucidate for many years. Given the effect of microRNA and epigenetic regulation on silencing and enhancing gene transcription, the study of protein arguably provides more accurate information on homeostasis and perturbation in health and disease. There have been significant advances in the field of proteomics in recent years, with new technologies and platforms available to the research community. In this review, we briefly discuss some of these new technologies and developments in the context of respiratory disease. We also discuss the types of data science approaches to analyses and interpretation of the large volumes of data generated in proteomic studies. We discuss the application of these technologies with regard to respiratory disease and highlight the potential for proteomics in generating major advances in the understanding of respiratory pathophysiology into the future.
Keywords: proteomics, lung disease
1. Introduction
Proteomics is the study of “proteomes” or the study and characterization of the protein composition of a cell, organ or other definable compartment of living organisms. Proteins are compounds of one or more long chains of amino acids and are vital parts of all living organisms. Amino acids are compounds composed of both a carboxyl (—COOH) and an amino (—NH2) group and form the building blocks of proteins. These proteins are generated from translation of mRNA and provide the principal information on how cells or organs function1. The lung is a fascinating and complex arena for proteomic studies, with innate and adaptive immune systems, extracellular matrix/interstitium, resident and recruited leucocytes, and an epithelial lining that is constantly exposed to the external environment. Pulmonary diseases remain a major contributor to global morbidity and mortality and there are many difficult questions that remain unanswered in pulmonary pathophysiology2. Proteomics has the potential to address many of these shortcomings.
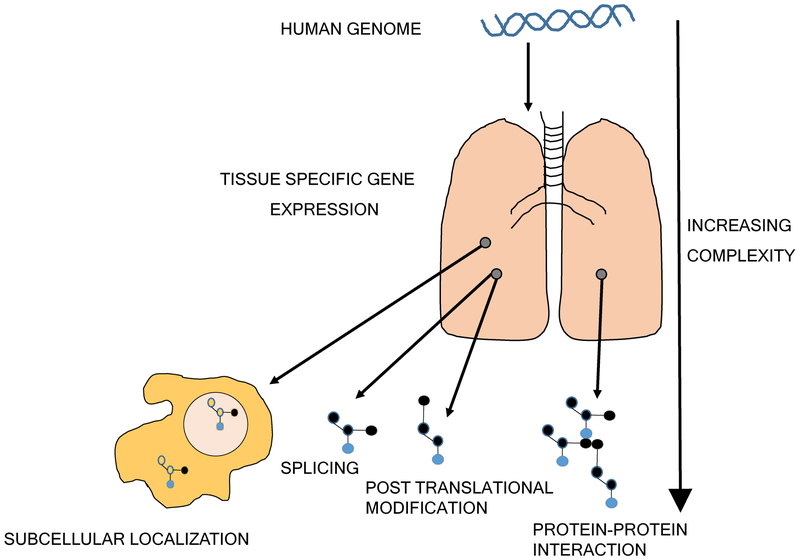
Protein is generated from translation of mRNA, yet flow of information from DNA to mRNA and then protein is confounded by epigenetic changes and microRNAs which can work to alter, amplify or dampen these genetic signals3,4. The human genome consists of approximately 31,000 protein coding genes5, and remains largely unchanged throughout life. Therefore, study of DNA and mRNA sequences does not account for changing environmental influences. Nucleic acid studies provide data on the potential for organ and cellular pathobiology and risk of disease and perturbation5,6. However, the human proteome adds incredible complexity to the human genome. The tissue specific expression of genes, translation of protein and subsequent splice variants, post translational modifications (PTMs) and protein-protein complexes/interactions7 and regulation of protein abundance largely at a translational level8 mean that interrogating the human proteome is arguably more challenging than interrogating the human genome (Fig.1).
Figure 1. The increasing complexity of the proteome.
The flow of information from DNA to mRNA and then protein is associated with ever increasing complexity. This is emphasized at the protein stage where subcellular localization, spatial transiency, multiple isoforms, large numbers of potential post translational modifications and protein-protein interactions lead to changes in expression and function.
Historically, technologies available to quantify protein in biological matrices were limited, costly and cumbersome. There has been considerable recent progress. The human proteome has been tentatively mapped using an integrative omics’ approach (transcriptomics and antibody based techniques) and represents a major step forward for proteomic research9. Central to this has been development of the human protein atlas, an invaluable research tool for protein localization and tissue expression, which includes a proteome map of normal human lung10,11. One of the major focuses of proteomic research to date has been identification of accurate disease biomarkers and targets for intervention12. Proteomics arguably has the most potential of the “omics” fields to provide new knowledge on disease pathogenesis, generate reliable biomarkers and facilitate discovery of new therapeutic strategies for human disease.
In this review, we discuss some of the recent advances in proteomic technology and describe current proteomic applications including mass spectrometry and aptamer approaches. We also detail several bioinformatics techniques and workflows to approach, analyze and interpret proteomic data. Finally, we highlight the application of proteomic technology to respiratory diseases and discuss some of the potential future uses of these technologies.
2. Proteomics applications and challenges
Herein, we provide a basic guide to some proteomic applications, namely mass spectrometry (MS) and aptamer approaches. An important consideration is that proteomic platforms are constantly evolving, have mixed versatility, difficulty and technical challenges. For instance, the field includes diverse projects from cell organelle protein expression profiling to human blood biomarker identification. Certain platforms may be better suited to addressing different scientific questions over others. All proteomic approaches will not be covered here and the interested reader is directed to a comprehensive review of proteomic applications and mass spectrometry elsewhere13.
Challenges in proteomic applications have been significant and have dampened enthusiasm for these platforms over the years. The spectrum of proteins that exist span a dynamic concentration range of at least 12 logs and this has hampered progress14,15. For instance, albumin, a large abundant protein in plasma is separated from the rarest measurable plasma proteins by 10 orders of magnitude15. The complexity of proteins involving splice variant and PTMs has also generated difficulties. The exact frequency of PTMs is unclear although the top 15 experimentally validated modifications represent the bulk of reported PTMs16. Common PTMs are listed in Table 1. Moreover, splice variants add further complexity. Indeed, fibronectin, an important component of the pulmonary interstitium, has more than 20 known isoforms17. Despite these hurdles, new advances have improved our technical abilities and these challenges have become less daunting.
Table 1.
Common post translational modifications
| Phosphoserine | 4-hydroxyproline |
| Phosphothreonine | Pyrrolidone carboxylic acid |
| N-linked glycosylation | N-acetylalanine |
| N-6 acetyllysine | O-linked glycosylation |
| Glycyl lysine isopeptide | Phosphotyrosine |
| Citrullination |
2.1. Mass Spectrometry (MS)
Significant advances in MS technology have accumulated in the last decade. These advances have improved the ability of these platforms to accurately measure thousands of proteins in a biological matrix.
Protein extraction from biological samples requires pre-formed knowledge of study design as different types of biological matrices and methods of extraction may induce bias and affect protein quantity and activity. For instance, blood within a tissue sample may give non representative falsely elevated results for certain proteins. Lysis and digestion of a biological matrix can generate peptide mixtures which need some degree of fractionation or enrichment to be compatible with proteomic applications. Fractionation can be achieved based on charge, isoelectric point or hydrophobicity properties of peptides and is typically achieved using gel electrophoresis, affinity chromatography or isoelectric focusing13. Specific subsets of peptides can be enriched by targeting PTMs (e.g. phosphorylation, acetylation) using affinity resins or antibody immunoprecipitation. Liquid chromatography (LC) is then applied to the reduced samples for further separation and sample reduction. MS is the next crucial analytical step as information garnered is then used to identify varied proteins. In brief, as MS measures the mass to charge ratio of ions (m/z) in gas phase, peptides must be transferred into the gas phase and then ionized. Once ionized, peptide precursor ions are submitted to the mass spectrometer where the m/z ratio is measured. Single precursor ions are selected then and subjected to tandem MS to generate characteristic fragment ions. This combination of precursor m/z ratio and its fragment ions is then matched to known peptide sequences from curated protein databases for protein identification. There are multiple technologies and methods for peptide fractionation, enrichment, ionization and types of mass spectrometers commercially available13. MS has been used widely in biomarker studies of respiratory disease to date including chronic obstructive pulmonary disease (COPD)18–20, acute respiratory distress syndrome (ARDS)21–23 and interstitial lung disease (ILD)24–26.
2.2. Aptamer based techniques
Aptamers are short single stranded RNA or DNA oligonucleotides that bind specific parts of a target molecule with high affinity and specificity27. Aptamer generation is less expensive and less arduous than antibody generation, and aptamers are not known to be toxic or immunogenic28. In recent years a new class of aptamer has been developed, termed slow off-rate modified aptamers (SOMAmers), which consist of single stranded DNA-based molecular recognition elements29,30. They are fully synthetic and developed in vitro using libraries of randomized sequences through modifications of the systematic evolution of ligands by exponential enrichment (SELEX) process. The selected SOMAmers have distinct recognizable nucleotide sequences and act as protein binding elements with defined shapes. The nucleotide sequences can be recognized by complimentary hybridization probes. The assay takes advantage of the slow dissociation rate between SOMAmers and their cognate proteins. Non-cognate interactions between SOMAmers and protein will dissociate rapidly. The cognate SOMAmers are hybridized to complementary probes on a standard DNA microarray. The SOMAmer data quantitatively represents the protein concentration in the sampled matrix. This is achieved by converting the assay signal in relative fluorescent units to protein concentration31. SOMAmers have been used to develop biomarker tools in several forms of respiratory disease including lung cancer32–34, pulmonary tuberculosis35,36 and idiopathic pulmonary fibrosis (IPF)37,38.
3. Bioinformatics Analysis of Proteomic Data
New proteomic experimental technologies generate large volumes of data, but a major challenge lies in analyzing these data to provide new biological insight. The fields of bioinformatics, computational biology and systems biology have developed techniques to facilitate curating, analysis and interpretation of “omics” data with many of these approaches described as either data-driven or knowledge-based39. Data-driven approaches rely only on protein data to identify proteins of interest in differentiating clinical or biological groups, and knowledge-based approaches rely on previously reported functions and pathways.
3.1. Data-driven Analysis
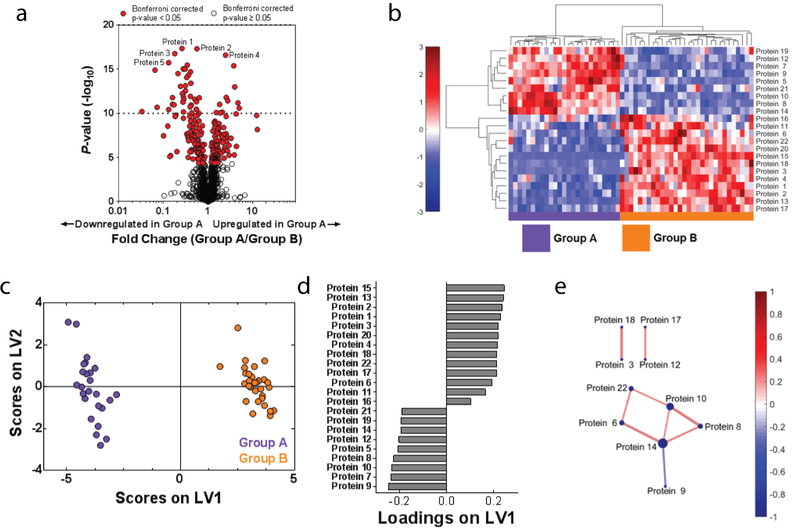
The goal of data-driven analysis is to use proteomic data to discover new proteins that are associated with certain experimental or clinical groups, without employing prior knowledge of these proteins’ functions. One way to begin analysis of a new proteomics dataset involves employing data-driven tools to enable visualization of the overall differences in protein expression data between clinical or biological groups. A common method used to visualize protein expression is a volcano plot, which displays information about each protein’s fold change in expression across groups on the x axis, vs. the significance of this change (determined by t test or other statistical analysis) on the y axis40 (Fig.2A). Since determining statistical significance in large proteomic data sets may involve performing many statistical tests, it is important to correct for multiple comparisons to control for the Type I error rate in order to reduce the number of false positive findings. The Bonferroni41 or Benjamini-Hochberg42 correction are common tests used in order to control for this, and can also be displayed on the volcano plot (Fig.2A).
Figure 2. Data-driven analysis aids in proteome visualization.
(A) A volcano plot highlights significantly differences in expressed proteins between Groups A and B. Red indicates proteins that were significantly different (p < 0.05) between the two groups after correcting for multiple comparisons with the Bonferroni test. (B) Hierarchical clustering illustrates groupings of proteins that differ in expression between Group A and B. Color intensity indicates abundance, with increased expression in red, white unchanged, and decreased expression in blue compared to mean values (color bar to left of figure). Pearson’s correlation was used as the distance metric in this cluster. (C) A PLSDA scores plot illustrates distinct clustering between Groups A and B with loadings (D) indicating a distinct signature (determined using LASSO) of 22 proteins that best classified Groups A and B. (E) A protein correlation network based on protein expression in Group A. Each node is a protein, with lines indicating significant correlations (p < 0.05) to other proteins. Line thickness and color indicates Pearson’s correlation coefficient, with node size indicating the number of significant correlations. Significance was determined after correcting for the Type 1 error with the Bonferroni method.
Hierarchical clustering is another visualization technique that additionally highlights the presence of protein clusters that differentiate multiple groups of interest. The hierarchical clustering algorithm employs a distance metric (such as Pearson’s correlation coefficient, Euclidean distance, or others described by Jaskowiak et al.43) to cluster samples and proteins in terms of similarity. Identified clusters can then be displayed as dendrograms, with an associated heat map of color intensity to display changes in expression of each protein across groups of interest (Fig.2B).
Two other data-driven analytical approaches used to visualize differences between clinical or biological groups employ linear algebra: principal components analysis (PCA) and partial least squares discriminant analysis (PLSDA)44,45. PCA and PLSDA algorithms identify weighted linear combinations (or “patterns”) of measured proteins that capture variance across the samples. Each sample can then be plotted on these key combinations (called latent variables (LV) in PLSDA and principal components (PC) in PCA), generating an interpretable scores plot in which differences between groups of interest may be visualized. Although PCA and PLSDA create figures that can look similar, an important difference between them is that the PLSDA algorithm also receives information about patient groups and searches for variance that differentiates these groups, making it a “supervised” approach (Fig.2C, 2D). In contrast, PCA only evaluates overall variance (without information about groups), making it “unsupervised”.
Another approach for evaluating large proteomic datasets is correlation network analysis, which enables graphical visualization of significant correlations between protein pairs. Correlation networks are constructed by calculating Pearson or Spearman correlation coefficients between measured proteins. A map is then created indicating significant connections and the strength of each correlation (Fig.2E). These graphs allow quick identification of highly connected proteins that may be network regulators, and how these interactions change across groups. Again, multiple comparison tests should be used to reduce Type 1 Error.
In addition to visualization, data-driven approaches are also useful for eliminating proteins that are not relevant to a biological or clinical question of interest. In proteomics datasets, considerably large numbers of proteins may be unchanged between groups of interest, masking the important and differentially regulated proteins. In this case, quantitative feature selection techniques inherent to some data-driven approaches can be used to identify subsets, or “minimum signatures,” of proteins that best separate the groups of interest. Two such examples are the least absolute shrinkage and selection operator method (LASSO)46 and selection using variable importance in projection (VIP) scores in PLSDA47.
3.2. Knowledge-based Analysis
Knowledge-based bioinformatics tools take advantage of prior knowledge to analyze proteomics datasets in the context of known protein function and ontology. These tools enable identification of biological pathways that are both enriched in the dataset and known to be involved in specific functions and processes.
Knowledge-based analysis employs previously generated databases where proteins have been tagged with unique identifier labels. Uniprot IDs48 are the most commonly used protein identifiers, though gene IDs (with gene identifiers given by Ensembl49) or the Enzyme Commission numbering system50 are also often used in proteomics. The choice in which identifier to use depends on the type of proteins that are being measured, and on which identifiers a given knowledge-based database will accept. Once proteins are linked with unique identifiers, prior knowledge databases with annotated information about biological functions and pathways can be employed to identify associated processes. One such tool is Gene Ontology (GO)51. GO terms, which standardize the naming of genes and gene products, are used to report the specific “biological processes,” “molecular functions,” and “cellular compartments” annotations associated with measured genes and proteins. Similar to GO terms, the Kyoto Encyclopedia of Genes and Genomes (KEGG) links protein and gene names with their functions and chemical information52. KEGG differs from GO in that it is more focused on known protein interactions. KEGG’s mapped pathways include those describing metabolism, human disease, signal transduction, and many others. Other pathway databases include Reactome53, PANTHER pathways54, and WikiPathways55.
In addition to biological annotation, knowledge-based analysis can be used to identify functions or pathways that are significantly enriched in datasets of interest. This involves comparing how many times a certain pathway is included in the protein set of interest with how many times it appears in a reference (control) set of proteins or genes, (such as that organism’s genome). A p-value can be calculated and used to determine if the pathway is significantly enriched in the proteomic dataset of interest. One such tool that both annotates proteins and performs functional enrichment analyses is the Database for Annotation, Visualization and Integrated Discovery (DAVID)56. DAVID’s strength is that it performs enrichment analyses on multiple annotation types (such as GO terms and KEGG pathways) and displays the results in both charts and clustered heat maps. Other knowledge-based enrichment analysis and visualization tools include Cytoscape57 and its ClueGO plug in58, EnrichNet59, and the commercial Ingenuity Pathway Analysis (IPA)60, as well as others as described by Laukens et al.61.
3.3. Combining Data-driven and Knowledge-based Analysis Techniques
Data-driven and knowledge-based proteomic analyses complement each other well when combined. In this process, data-driven tools can identify key minimum signatures of proteins that differentiate the groups of interest, with knowledge-based tools providing a deeper biological context for this smaller list of proteins. For example, a feature selection technique (LASSO or VIP scores) or a volcano plot can be used to narrow down the proteomics dataset into a list of the proteins that vary between the groups of interest. These identified significant proteins can subsequently be labeled with Uniprot IDs and input into DAVID to discover enriched pathways and biological processes, generating new hypotheses regarding mechanisms of action associated with disease.
4. Proteomic Applications in Respiratory Disease:
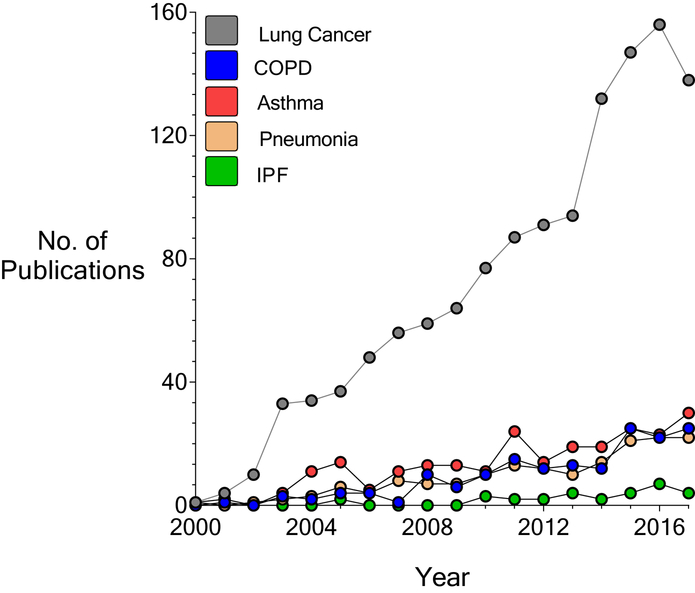
Diseases of the respiratory system remain a major source of global morbidity and mortality2. Proteomic discovery in lung is a rapidly evolving field, and currently much of the focus has been centered on the role of proteomics in lung cancer (Fig.3).
Figure 3. Proteomic studies in respiratory disease: increasing interest and number of publications.
The number of PubMed citations from the year 2000 to 2017 were recorded for each of the following: Lung Cancer, COPD, Asthma, Pneumonia, and IPF. MESH terms “proteomics” and “specific lung disease” (i.e IPF) were used as input. No filters were applied.
4.1. IPF
IPF is the most common form of ILD and is invariably fatal with a median survival of 2 to 3 years62. IPF etiology and pathogenesis are poorly understood63. The disease results in aberrant accumulation of extracellular matrix within the interstitium of the lung, promoting impaired gas exchange and respiratory failure37.
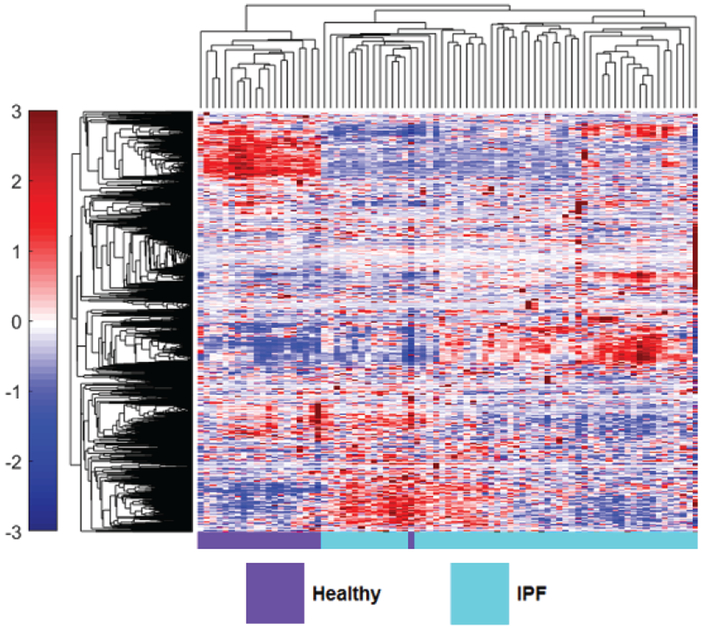
However, recent studies have started to explore differences in protein expression and profiling in IPF patients. Comparative proteomic analysis of lung tissue samples derived from IPF patients and human donor transplant lungs using 2D gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) MS demonstrated significant differences in protein expression25. Fifty-one proteins were upregulated and 38 down-regulated in IPF lung compared to normal. Proteins involved in unfolded protein response (UPR) were upregulated and immunohistochemistry confirmed induction of markers of UPR within type 2 pneumocytes. Furthermore, there was downregulation of anti-oxidants and structural epithelial proteins supporting epithelial cell injury as a key feature of IPF pathogenesis. The ability to differentiate between different types of ILD pathology using proteomic profiles would mark a major advancement in ILD management. Landi et al. employed bronchoalveolar lavage fluid (BALF) derived from IPF, sarcoidosis, Langerhans cell histiocytosis and scleroderma (SSc) associated ILD patients to examine differentially expressed protein profiles26. They reported novel findings supporting the regulation of ILD pathogenesis by factors in alternative complement activation, blood coagulation, protein folding and Slit-Robo signaling. The acquisition of BALF however may be challenging in chronic lung disease. Recent work by our group applied novel aptamer approaches to investigate the blood plasma proteome in IPF patients from the COMET study38. SOMAmers were measured in IPF patients and then analyzed to generate a panel of 6 plasma biomarkers to predict disease progression based on a composite disease progression index. IPF patients with high levels of inducible T cell costimulatory (ICOS) and trypsin 3 (TRY3) and low levels of ficolin-2 (FCN2), cathepsin-S (Cath-S), legumain (LGMN), and soluble vascular endothelial growth factor receptor 2 (VEGFsR2) predicted poorer progression-free survival. We next examined the differential expression of plasma proteins in healthy volunteers and IPF patients. In this recent proof of concept study, we employed hierarchical clustering of statistically significant differentially expressed proteins in IPF patients and healthy volunteers, demonstrating visually distinct plasma proteomes between healthy volunteers and IPF patients37 (Fig.4). This study highlights the potential use of proteomic profiles derived from easily accessible blood in the diagnostic workup of ILD patients. Foster and colleagues recently employed two different proteomic platforms to BALF from IPF patients and demonstrated the increased expression of osteopontin64. This work importantly validating previous studies and results across quantitative proteomic platforms65. Schiller et al. recently applied quantitative label free mass spectrometry to address common protein regulations across apparently heterogeneous lung fibrosis tissue from human patients (including IPF)66. They report a possible common regulator, MZB1 + plasma B cells, present at high prevalence in both fibrotic lung and skin tissue including IPF, hypersensitivity pneumonitis (HP), cryptogenic organizing pneumonia (COP), scleroderma associated ILD and unclassifiable ILD.
Figure 4. The peripheral blood proteome of IPF differs from healthy.
Hierarchical clustering of 1129 measured blood proteins in healthy and IPF patients illustrates visually distinct expression in the two groups. Proteomic abundance is displayed with color intensity, with red indicating overabundant proteins and blue indicating underabundant proteins compared to the mean expression level. Clustering was created using unsupervised average linkage with Pearson’s correlation as the distance metric.
4.2. Asthma
Asthma is a chronic inflammatory airway disorder characterized by variable airflow obstruction67. The disease is associated with exposure to aeroallergens which leads to immunological changes within the airway epithelium. To date, there are several studies that have examined the role of proteomic technologies in both development of biomarkers and improved understanding of asthma pathogenesis.
Initial studies using high performance liquid chromatography (HPLC) resulted in discovery of the chemokine CCL5 (RANTES) as a BALF biomarker of allergic inflammation and eosinophilic activation in asthma patients68. A further study of endobronchial biopsies in a small number of asthma patients and healthy volunteers using mass spectrometry also identified CCL5 as a biomarker. These authors used pathway analysis to identity biologically important functional pathways including acute phase response, cell-cell signaling and tissue development in asthmatic airways compared to controls69. Hamsten and colleagues also demonstrated alterations in CCL5 plasma protein levels with significantly lower levels reported in children with persistent asthma compared to controls70. Wu et al. used LC-MS/MS of BALF samples after allergen challenge in asthma patients to describe the complex biological pathways activated in the lung71. They found approximately 150 proteins that were upregulated in response to allergen exposure in BALF, and the upregulated proteins were associated with wide ranging functional pathways including proteolysis, inflammation, cell proliferation and signal transduction. Potentially interesting upregulated proteins included MMP9 and SERPINA3. MMP9 is a matrix metalloproteinase involved in lung remodeling that is generated in part by airway neutrophils72. Proteomic studies of sputum samples from asthma patients have also been employed to study asthma pathobiology. Gharib et al. examined airway sputum samples from 10 patients and reported 17 target proteins including alpha 1-antichymotrypsin73. Sputum samples are acquired by non-invasive means and therefore provide an advantage over other types of pulmonary sampling. Aptamer approaches have also been reported in studies of asthma. Loza et al. reported increases in serum CRP and IgE and reductions in serum carbonic anhydrase 6 and osteomodulin in severe asthma74.
4.3. COPD
COPD is a common disease with global impact and high related morbidity and mortality. COPD is characterized by airflow obstruction that is poorly reversible75. There is obstruction of small airways and destruction of distal alveolar structures resulting in air trapping, impaired gas exchange, cough, dyspnea and sputum production76. Proteomic approaches have been utilized in studies of COPD from BALF, tissue and blood for biomarker discovery. Nano-LC-MS techniques identified 76 differentially expressed proteins in BALF from COPD patients, and pathway analysis identified biological processes including inflammatory processes, glycolysis, and oxidation reduction77. Given the issues with dilution of epithelial lining fluid (ELF) on BALF acquisition, one investigative group obtained ELF directly from the airway using microprobes during bronchoscopy and then applied microfluidics based nano-LC-MS/MS to identify and quantify proteins in the ELF of COPD patients. They identified elevated levels of lactotransferrin, high-mobility group protein B1 (HGMB1) and alpha-1 antichymotrypsin in ELF from COPD patients compared to healthy controls78. Interestingly, alpha-1 antichymotrypsin encoded for by the SERPINA 3 gene is reportedly elevated in the sputum of asthma patients, possibly reflecting a shared mechanism in chronic inflammatory airway disorders71. Studies have also examined proteins in sputum to better understand COPD pathogenesis. Baraniuk and colleagues identified a higher abundance of mucin 5AC in sputum from COPD and healthy smokers. Patients with emphysema features had higher levels of defensins and protein components of neutrophil extracellular traps (NETS)79. Lee et al. employed MALDI-TOF-MS in tissue samples from COPD patients and healthy smokers19. They reported significant upregulation of MMP-13 mainly in alveolar macrophages and thioredoxin-like 2 (TXL2) in bronchial epithelium compared to healthy smokers. A further comprehensive study of tissue, plasma and sputum in COPD, IPF and alpha-1-antitrypsin deficiency patients identified the protein transglutaminase 2 (TGM2) as a COPD specific protein80. Tissue levels of TGM2 associated with disease severity, and sputum and plasma levels of TGM2 correlated with FEV1% predicted values.
4.4. Lung Cancer
The detection of lung cancer during the early phases of disease is crucial to providing optimal management strategies and potential cure, as it is often diagnosed at an advanced stage81. Therefore, the discovery of accurate and reliable biomarkers is an important goal. Extensive use of proteomic research applications has occurred in the lung cancer field but the proposed biomarkers have yet to be adopted for clinical applications82. Most lung cancer proteomic studies have been undertaken in diseased tissue samples, however some studies have been carried out on serum, blood, BALF, pleural fluid and saliva82.
Wu et al. studied plasma samples from lung adenocarcinoma (NSCLC) patients and age and gender matched healthy controls and reported nine candidate proteins that discriminated between cancer and health83. These proteins included gelsolin (GSN), galectin-1 (LGALS1) and actin cytoplasmic 1 (ACTB). It may be possible to use blood proteomics to stratify risk of developing lung cancer. One study applied proteomics to plasma from never, current or former smokers and reported a significant association with plasma apolipoprotein E (APOE) levels and the development of squamous metaplasia in the lungs, supporting the potential to develop proteomic plasma biomarkers capable of predicting pre-malignant and early forms of lung cancer84. However, lung tissue samples from cancer patients have received more extensive analysis. Numerous studies have analyzed proteomic changes within lung tissue samples. Pernemalm et al. used isobaric tags for relative and absolute quantitation (ITRAQ) based quantitative proteomics to compare lung cancer tissue samples associated with 2-year relapse and those without relapse. Using pathway analysis, they reported tumors associated with relapse had a higher dependence on glycolysis and higher hypoxia inducible factor (HIF) activity85. Kikuchi et al. pooled samples of lung adenocarcinoma (AC), squamous carcinoma (SCC) and control tissue and used shotgun proteomics to profile the lung tumor proteome. They found higher levels of Mapsin (SERPINAB5) in SCC tissue samples and identified for the first time, dysregulation of the p21-activated kinases in NSCLC86.
5. Translating Proteomic studies
To date, the results of many proteomic studies in medicine have been centered on the development of reliable biomarkers for disease and outcomes. For instance, the largest aptamer study of plasma proteins to date was employed to risk stratify patients with cardiovascular disease87. However, it is important to note that the study of proteins may have vast implications in medicine and science. Proteomics platforms may be used to generate hypothesis on disease pathophysiology, develop new therapies and novel strategies and assess for clinical efficacy and safety of new drugs88–91. Given the array of proteomic tools available including ELISA, aptamer and MS platforms, an important goal for the field is an improved understanding of accuracy and reproducibility across proteome specific platforms. These questions remain difficult to address without large scale cross platform studies in humans. We have previously shown significant correlations between protein measured by both ELISA and aptamer techniques within the same human cohort38. However, this is an area where further study is required. The future of proteomics is exciting and likely to yield major advances in medicine. Recent work has shown how proteomics may be integrated with genomic data to demonstrate overlap between quantitative gene, protein and disease associated loci, with evidence of causal links between specific proteins and disease92. These advances may lead to accurate mapping in real-time of disease states, biological pathways and therapeutic targets.
6. Conclusion
The last two decades have ushered in a timely revolution in proteomics. New technologies and modifications of old ones are facilitating studies of thousands of proteins in biological samples, allowing for an ever improved understanding of protein expression, function and dynamics. Leveraging the power of proteomics to provide an accurate estimate of immediate health or disease remains an achievable and vital goal. The continued evolution and expansion of proteomic technologies such as aptamer approaches and the parallel development of bioinformatics tools and applications will facilitate this goal. While challenges remain, evolving proteomic applications and the era of integrating genomic and proteomic human data in disease and health will alter the current architecture of how we understand, diagnose and manage human disease in the lung and elsewhere in the body.
Acknowledgements:
K.C.N. was supported by a Department of Education Graduate Assistance in Areas of National Need (GAANN) Fellowship awarded to the biomedical engineering department at the University of Michigan (PR Award Number: P200A150170). D.O.D was supported by NIH grant K99HL139996, B.B.M was supported by NIH grants AI117229 HL127805.
Abbreviations:
- mRNA
messenger ribonucleic acid
- DNA
deoxyribonucleic acid
- PTMs
post translational modifications
- MS
Mass Spectrometry
- LC
liquid chromatography
- COPD
chronic obstructive pulmonary disease
- ARDS
acute respiratory distress syndrome
- ILD
interstitial lung disease
- SELEX
systematic evolution of ligands by exponential enrichment
- SOMAmers
slow off rate modified aptamers
- IPF
idiopathic pulmonary fibrosis
- PCA
principal component analysis
- PLSDA
partial least squares discriminant analysis
- LV
latent variables
- LASSO
least absolute shrinkage and selection operator
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- MALDI
matrix-assisted laser desorption/ionization
- TOF
time of flight
- UPR
unfolded protein response
- BALF
bronchoalveolar lavage fluid
- SSc
scleroderma
- ICOS
inducible T cell costimulatory
- TRY3
trypsin 3
- FCN2
ficolin-2
- Cath-S
cathepsin-S
- LGMN
legumain
- VEGFsR2
soluble vascular endothelial growth factor receptor 2
- HP
hypersensitivity pneumonitis
- COP
cryptogenic organizing pneumonia
- MZB
Marginal Zone B And B1 Cell Specific Protein
- HPLC
high performance liquid chromatography
- CCL5/RANTES
Regulated Upon Activation, Normally T-Expressed And Presumably Secreted
- MMP9
matrix metalloprotease 9
- SERPINA3
Serpin Family A Member 3
- 2D-DIGE
2 dimensional difference gel electrophoresis
- VEGF
vascular endothelial growth factor
- FABP5
fatty acid binding protein 5
- CRP
C reactive protein
- IgE
immunoglobulin E
- ELF
epithelial lining fluid
- HGMB1
high-mobility group protein
- NETS
neutrophil extracellular traps
- MMP-13
matrix metalloproteinase 13
- TXL2
thioredoxin-like 2
- TGM2
transglutaminase 2
- FEV1
forced expiratory volume in 1 second
- NSCLC
non-small cell lung carcinoma
- SCLC
small cell lung carcinoma
- GSN
gelsolin
- LGALS1
Galectin-1
- ACTB
actin cytoplasmic 1
- SERPINA4
Serpin Family A Member 4
- PON1
arylesterase 1
- APOE
Apolipoprotein E
- ITRAQ
Isobaric tags for relative and absolute quantitation
- HIF
hypoxia inducible factor
- AC
adenocarcinoma
- SCC
squamous carcinoma
- SERPINAB5
Mapsin
- PCA
principal components analysis
- PLSDA
partial least squares discriminant analysis
- LV
latent variable
- PC
principal component
- LASSO
least absolute shrinkage and selection operator
- VIP
variable importance in projection
- GO
gene ontology
- KEGG
Kyoto encyclopedia of genes and genomes
- DAVID
database for annotation, visualization and integrated discovery
- IPA
Ingenuity Pathway Analysis
The authors
Katy C. Norman is a graduate student in the Department of Biomedical Engineering at the University of Michigan, Ann Arbor. She is interested in using computational approaches to gain systems-level insight into immunological diseases. Her current work uses data-driven analysis techniques to identify key cytokine and cellular relationships involved in IPF and COPD.
Kelly B. Arnold is an Assistant Professor in the Department of Biomedical Engineering at the University of Michigan, Ann Arbor. Dr. Arnold uses experimental and computational techniques to understand and uncover the immune cell communication networks present in the inflammatory environment of disease states. Specifically, she investigates diseases affecting mucosal immunology, such as HIV, IPF, and COPD.
Beth B. Moore is a Professor in the Department of Internal Medicine, Division of Pulmonary and Critical Care Medicine and the Department of Microbiology and Immunology at the University of Michigan. Dr. Moore has a long history of research in the field of pulmonary fibrosis and has published extensively in IPF. Her recent work involves the use of omics including microbiome and proteomic analysis to identify novel host and microbiota related mechanisms involved in IPF disease progression.
David N. O’Dwyer is an Assistant Professor of Internal Medicine, Division of Pulmonary and Critical Care Medicine at the University of Michigan, Ann Arbor. He has specific interest in IPF and other forms of ILD. His research interests include host innate immune and pulmonary interactions in chronic lung injury and the use of proteomic biomarkers in the study of IPF pathogenesis and progression.
References
- 1.Cox J & Mann M Is proteomics the new genomics? Cell 130, 395–398, doi: 10.1016/j.cell.2007.07.032 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Ferkol T & Schraufnagel D The global burden of respiratory disease. Ann Am Thorac Soc 11, 404–406, doi: 10.1513/AnnalsATS.201311-405PS (2014). [DOI] [PubMed] [Google Scholar]
- 3.Allis CD & Jenuwein T The molecular hallmarks of epigenetic control. Nat Rev Genet 17, 487–500, doi: 10.1038/nrg.2016.59 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Ha M & Kim VN Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15, 509–524, doi: 10.1038/nrm3838 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Baltimore D Our genome unveiled. Nature 409, 814–816, doi: 10.1038/35057267 (2001). [DOI] [PubMed] [Google Scholar]
- 6.International Human Genome Sequencing, C. Initial sequencing and analysis of the human genome. Nature 409, 860, doi:10.1038/35057062 10.1038/35057062https://www.nature.com/articles/35057062#supplementary-informationhttps://www.nature.com/articles/35057062#supplementary-information (2001). [DOI] [PubMed] [Google Scholar]
- 7.Havugimana PC et al. A census of human soluble protein complexes. Cell 150, 1068–1081, doi: 10.1016/j.cell.2012.08.011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwanhausser B et al. Global quantification of mammalian gene expression control. Nature 473, 337–342, doi: 10.1038/nature10098 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Uhlen M et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419, doi: 10.1126/science.1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Thul PJ & Lindskog C The human protein atlas: A spatial map of the human proteome. Protein Sci 27, 233–244, doi: 10.1002/pro.3307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindskog C et al. The lung-specific proteome defined by integration of transcriptomics and antibody-based profiling. FASEB J 28, 5184–5196, doi: 10.1096/fj.14-254862 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Tambor V et al. Application of proteomics in biomarker discovery: a primer for the clinician. Physiol Res 59, 471–497 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Mallick P & Kuster B Proteomics: a pragmatic perspective. Nat Biotechnol 28, 695–709, doi: 10.1038/nbt.1658 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Wilkins MR et al. High-throughput mass spectrometric discovery of protein post-translational modifications. J Mol Biol 289, 645–657, doi: 10.1006/jmbi.1999.2794 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Anderson NL & Anderson NG The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics 1, 845–867 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Khoury GA, Baliban RC & Floudas CA Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep 1, doi: 10.1038/srep00090 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White ES & Muro AF Fibronectin splice variants: understanding their multiple roles in health and disease using engineered mouse models. IUBMB Life 63, 538–546, doi: 10.1002/iub.493 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Ohlmeier S et al. Proteomic studies on receptor for advanced glycation end product variants in idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease. Proteomics Clin Appl 4, 97–105, doi: 10.1002/prca.200900128 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Lee EJ et al. Proteomic analysis in lung tissue of smokers and COPD patients. Chest 135, 344–352, doi: 10.1378/chest.08-1583 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Merali S et al. Analysis of the plasma proteome in COPD: Novel low abundance proteins reflect the severity of lung remodeling. COPD 11, 177–189, doi: 10.3109/15412555.2013.831063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowler RP et al. Proteomic analysis of pulmonary edema fluid and plasma in patients with acute lung injury. Am J Physiol Lung Cell Mol Physiol 286, L1095–1104, doi: 10.1152/ajplung.00304.2003 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Ren S et al. Deleted in malignant brain tumors 1 protein is a potential biomarker of acute respiratory distress syndrome induced by pneumonia. Biochem Biophys Res Commun 478, 1344–1349, doi: 10.1016/j.bbrc.2016.08.125 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Bhargava M et al. Proteomic profiles in acute respiratory distress syndrome differentiates survivors from non-survivors. PLoS One 9, e109713, doi: 10.1371/journal.pone.0109713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bargagli E et al. Calgranulin B (S100A9) levels in bronchoalveolar lavage fluid of patients with interstitial lung diseases. Inflammation 31, 351–354, doi: 10.1007/s10753-008-9085-z (2008). [DOI] [PubMed] [Google Scholar]
- 25.Korfei M et al. Comparative proteomic analysis of lung tissue from patients with idiopathic pulmonary fibrosis (IPF) and lung transplant donor lungs. J Proteome Res 10, 2185–2205, doi: 10.1021/pr1009355 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Landi C et al. Towards a functional proteomics approach to the comprehension of idiopathic pulmonary fibrosis, sarcoidosis, systemic sclerosis and pulmonary Langerhans cell histiocytosis. J Proteomics 83, 60–75, doi: 10.1016/j.jprot.2013.03.006 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Lakhin AV, Tarantul VZ & Gening LV Aptamers: problems, solutions and prospects. Acta Naturae 5, 34–43 (2013). [PMC free article] [PubMed] [Google Scholar]
- 28.Bouchard PR, Hutabarat RM & Thompson KM Discovery and development of therapeutic aptamers. Annu Rev Pharmacol Toxicol 50, 237–257, doi: 10.1146/annurev.pharmtox.010909.105547 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Gold L, Walker JJ, Wilcox SK & Williams S Advances in human proteomics at high scale with the SOMAscan proteomics platform. N Biotechnol 29, 543–549, doi: 10.1016/j.nbt.2011.11.016 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Wilson R High-content aptamer-based proteomics. J Proteomics 74, 1852–1854 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Kraemer S et al. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay. PLoS One 6, e26332, doi: 10.1371/journal.pone.0026332 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehan MR et al. Validation of a blood protein signature for non-small cell lung cancer. Clin Proteomics 11, 32, doi: 10.1186/1559-0275-11-32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehan MR et al. Protein signature of lung cancer tissues. PLoS One 7, e35157, doi: 10.1371/journal.pone.0035157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostroff RM et al. Early detection of malignant pleural mesothelioma in asbestos-exposed individuals with a noninvasive proteomics-based surveillance tool. PLoS One 7, e46091, doi: 10.1371/journal.pone.0046091 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Groote MA et al. Elucidating novel serum biomarkers associated with pulmonary tuberculosis treatment. PLoS One 8, e61002, doi: 10.1371/journal.pone.0061002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell TM et al. Potential of High-Affinity, Slow Off-Rate Modified Aptamer Reagents for Mycobacterium tuberculosis Proteins as Tools for Infection Models and Diagnostic Applications. J Clin Microbiol 55, 3072–3088, doi: 10.1128/JCM.00469-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Dwyer DN et al. The peripheral blood proteome signature of idiopathic pulmonary fibrosis is distinct from normal and is associated with novel immunological processes. Sci Rep 7, 46560, doi: 10.1038/srep46560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashley SL et al. Six-SOMAmer Index Relating to Immune, Protease and Angiogenic Functions Predicts Progression in IPF. PLoS One 11, e0159878, doi: 10.1371/journal.pone.0159878 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benedict KF & Lauffenburger DA Insights into proteomic immune cell signaling and communication via data-driven modeling. Curr Top Microbiol Immunol 363, 201–233, doi: 10.1007/82_2012_249 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Cui X & Churchill GA Statistical tests for differential expression in cDNA microarray experiments. Genome Biol 4, 210 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonferroni CE Teoria statistica delle classi e calcolo delle probabilitá. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze 8, 3–62, doi:citeulike-article-id:1778138 (1936). [Google Scholar]
- 42.Benjamini Y & Hochberg Y Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met 57, 289–300 (1995). [Google Scholar]
- 43.Jaskowiak PA, Campello RJ & Costa IG On the selection of appropriate distances for gene expression data clustering. BMC Bioinformatics 15 Suppl 2, S2, doi: 10.1186/1471-2105-15-S2-S2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janes KA & Yaffe MB Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol 7, 820–828, doi: 10.1038/nrm2041 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Martens H & Martens M Multivariate analysis of quality: an introduction. (John Wiley & Songs, Ltd., 2001). [Google Scholar]
- 46.Tibshirani R Regression shrinkage and selection via the Lasso. J Roy Stat Soc B Met 58, 267–288 (1996). [Google Scholar]
- 47.Wold S, Johansson E & Cocchi M PLS-partial least squares projections to latent structures. 3D QSAR in drug design 1, 523–550 (1993). [Google Scholar]
- 48.The UniProt C UniProt: the universal protein knowledgebase. Nucleic Acids Res 45, D158–D169, doi: 10.1093/nar/gkw1099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zerbino DR et al. Ensembl 2018. Nucleic Acids Res 46, D754–D761, doi: 10.1093/nar/gkx1098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.International Union of Biochemistry and Molecular Biology, Nomenclature Committee & Webb, E. C. Enzyme nomenclature 1992 : recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the nomenclature and classification of enzymes. (Published for the International Union of Biochemistry and Molecular Biology by Academic Press, 1992). [Google Scholar]
- 51.Ashburner M et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29, doi: 10.1038/75556 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M & Goto S KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fabregat A et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res 46, D649–D655, doi: 10.1093/nar/gkx1132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mi H et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45, D183–D189, doi: 10.1093/nar/gkw1138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slenter DN et al. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res 46, D661–D667, doi: 10.1093/nar/gkx1064 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57, doi: 10.1038/nprot.2008.211 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504, doi: 10.1101/gr.1239303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bindea G et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093, doi: 10.1093/bioinformatics/btp101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glaab E, Baudot A, Krasnogor N, Schneider R & Valencia A EnrichNet: network-based gene set enrichment analysis. Bioinformatics 28, i451–i457, doi: 10.1093/bioinformatics/bts389 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kramer A, Green J, Pollard J Jr. & Tugendreich S Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530, doi: 10.1093/bioinformatics/btt703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laukens K, Naulaerts S & Berghe WV Bioinformatics approaches for the functional interpretation of protein lists: from ontology term enrichment to network analysis. Proteomics 15, 981–996, doi: 10.1002/pmic.201400296 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Raghu G et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med 183, 788–824, doi: 10.1164/rccm.2009-040GL (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Dwyer DN, Ashley SL & Moore BB Influences of innate immunity, autophagy, and fibroblast activation in the pathogenesis of lung fibrosis. Am J Physiol Lung Cell Mol Physiol 311, L590–601, doi: 10.1152/ajplung.00221.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foster MW et al. Quantitative proteomics of bronchoalveolar lavage fluid in idiopathic pulmonary fibrosis. J Proteome Res 14, 1238–1249, doi: 10.1021/pr501149m (2015). [DOI] [PubMed] [Google Scholar]
- 65.Pardo A et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med 2, e251, doi: 10.1371/journal.pmed.0020251 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schiller HB et al. Deep Proteome Profiling Reveals Common Prevalence of MZB1-Positive Plasma B Cells in Human Lung and Skin Fibrosis. Am J Respir Crit Care Med 196, 1298–1310, doi: 10.1164/rccm.201611-2263OC (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Holgate ST et al. Asthma. Nat Rev Dis Primers 1, 15025, doi: 10.1038/nrdp.2015.25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teran LM et al. Eosinophil recruitment following allergen challenge is associated with the release of the chemokine RANTES into asthmatic airways. J Immunol 157, 1806–1812 (1996). [PubMed] [Google Scholar]
- 69.O’Neil SE et al. Network analysis of quantitative proteomics on asthmatic bronchi: effects of inhaled glucocorticoid treatment. Respir Res 12, 124, doi: 10.1186/1465-9921-12-124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hamsten C et al. Protein profiles of CCL5, HPGDS, and NPSR1 in plasma reveal association with childhood asthma. Allergy 71, 1357–1361, doi: 10.1111/all.12927 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Wu J et al. Differential proteomic analysis of bronchoalveolar lavage fluid in asthmatics following segmental antigen challenge. Mol Cell Proteomics 4, 1251–1264, doi: 10.1074/mcp.M500041-MCP200 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Cundall M et al. Neutrophil-derived matrix metalloproteinase-9 is increased in severe asthma and poorly inhibited by glucocorticoids. J Allergy Clin Immunol 112, 1064–1071, doi: 10.1016/j.jaci.2003.08.013 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Gharib SA et al. Induced sputum proteome in healthy subjects and asthmatic patients. J Allergy Clin Immunol 128, 1176–1184 e1176, doi: 10.1016/j.jaci.2011.07.053 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Loza M et al. Systemic corticosteroid-associated serum analyte profiles in the U-BIOPRED severe asthma cohort. European Respiratory Journal 46, doi: 10.1183/13993003.congress-2015.OA1774 (2015). [DOI] [Google Scholar]
- 75.Qaseem A et al. Diagnosis and management of stable chronic obstructive pulmonary disease: a clinical practice guideline update from the American College of Physicians, American College of Chest Physicians, American Thoracic Society, and European Respiratory Society. Ann Intern Med 155, 179–191, doi: 10.7326/0003-4819-155-3-201108020-00008 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Barnes PJ et al. Chronic obstructive pulmonary disease. Nat Rev Dis Primers 1, 15076, doi: 10.1038/nrdp.2015.76 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Tu C et al. Large-scale, ion-current-based proteomics investigation of bronchoalveolar lavage fluid in chronic obstructive pulmonary disease patients. J Proteome Res 13, 627–639, doi: 10.1021/pr4007602 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Franciosi L et al. Proteomic analysis of human epithelial lining fluid by microfluidics-based nanoLC-MS/MS: a feasibility study. Electrophoresis 34, 2683–2694, doi: 10.1002/elps.201300020 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Baraniuk JN et al. Protein networks in induced sputum from smokers and COPD patients. Int J Chron Obstruct Pulmon Dis 10, 1957–1975, doi: 10.2147/COPD.S75978 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ohlmeier S et al. Lung tissue proteomics identifies elevated transglutaminase 2 levels in stable chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol 310, L1155–1165, doi: 10.1152/ajplung.00021.2016 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Herbst RS, Heymach JV & Lippman SM Lung cancer. N Engl J Med 359, 1367–1380, doi: 10.1056/NEJMra0802714 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fujii K, Nakamura H & Nishimura T Recent mass spectrometry-based proteomics for biomarker discovery in lung cancer, COPD, and asthma. Expert Rev Proteomics 14, 373–386, doi: 10.1080/14789450.2017.1304215 (2017). [DOI] [PubMed] [Google Scholar]
- 83.Wu HY et al. Qualification and Verification of Serological Biomarker Candidates for Lung Adenocarcinoma by Targeted Mass Spectrometry. J Proteome Res 14, 3039–3050, doi: 10.1021/pr501195t (2015). [DOI] [PubMed] [Google Scholar]
- 84.Rice SJ et al. Proteomic profiling of human plasma identifies apolipoprotein E as being associated with smoking and a marker for squamous metaplasia of the lung. Proteomics 15, 3267–3277, doi: 10.1002/pmic.201500029 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Pernemalm M et al. Quantitative proteomics profiling of primary lung adenocarcinoma tumors reveals functional perturbations in tumor metabolism. J Proteome Res 12, 3934–3943, doi: 10.1021/pr4002096 (2013). [DOI] [PubMed] [Google Scholar]
- 86.Kikuchi T et al. In-depth proteomic analysis of nonsmall cell lung cancer to discover molecular targets and candidate biomarkers. Mol Cell Proteomics 11, 916–932, doi: 10.1074/mcp.M111.015370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ganz P et al. Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients With Stable Coronary Heart Disease. JAMA 315, 2532–2541, doi: 10.1001/jama.2016.5951 (2016). [DOI] [PubMed] [Google Scholar]
- 88.Loffredo FS et al. Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell 153, 828–839, doi: 10.1016/j.cell.2013.04.015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ren X, Gelinas AD, von Carlowitz I, Janjic N & Pyle AM Structural basis for IL-1alpha recognition by a modified DNA aptamer that specifically inhibits IL-1alpha signaling. Nat Commun 8, 810, doi: 10.1038/s41467-017-00864-2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sullivan KD et al. Trisomy 21 causes changes in the circulating proteome indicative of chronic autoinflammation. Sci Rep 7, 14818, doi: 10.1038/s41598-017-13858-3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Williams SA et al. Improving Assessment of Drug Safety Through Proteomics: Early Detection and Mechanistic Characterization of the Unforeseen Harmful Effects of Torcetrapib. Circulation 137, 999–1010, doi: 10.1161/CIRCULATIONAHA.117.028213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun BB et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79, doi: 10.1038/s41586-018-0175-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]