Abstract
Dysferlinopathy is a progressive myopathy caused by mutations in the dysferlin (DYSF) gene. Dysferlin protein plays a major role in plasma-membrane resealing. Some patients with DYSF deletion mutations exhibit mild symptoms, suggesting some regions of DYSF can be removed without significantly impacting protein function. Antisense-mediated exon-skipping therapy uses synthetic molecules called antisense oligonucleotides to modulate splicing, allowing exons harboring or near genetic mutations to be removed and the open reading frame corrected. Previous studies have focused on DYSF exon 32 skipping as a potential therapeutic approach, based on the association of a mild phenotype with the in-frame deletion of exon 32. To date, no other DYSF exon-skipping targets have been identified, and the relationship between DYSF exon deletion pattern and protein function remains largely uncharacterized. In this study, we utilized a membrane-wounding assay to evaluate the ability of plasmid constructs carrying mutant DYSF, as well as antisense oligonucleotides, to rescue membrane resealing in patient cells. We report that multi-exon skipping of DYSF exons 26–27 and 28–29 rescues plasma-membrane resealing. Successful translation of these findings into the development of clinical antisense drugs would establish new therapeutic approaches that would be applicable to ∼5%–7% (exons 26–27 skipping) and ∼8% (exons 28–29 skipping) of dysferlinopathy patients worldwide.
Keywords: exon skipping, antisense, morpholino, dysferlin, dysferlinopathy, limb-girdle muscular dystrophy type 2B, Miyoshi myopathy, distal myopathy, plasma membrane, membrane wounding
Introduction
The dysferlinopathies are a heterogeneous group of recessive myopathies caused by mutations in the dysferlin (DYSF) gene.1, 2 Characterized by progressive muscle weakness that typically begins during the second decade of life, the dysferlinopathies can be clinically divided into at least three types: Miyoshi myopathy (MM), limb-girdle muscular dystrophy type 2B (LGMD2B), and distal myopathy with anterior tibial onset (DMAT).1, 2, 3 The dysferlinopathies are clinically distinguished based on the initial pattern of muscle weakness, originating in either the proximal (shoulder and pelvic girdle; LGMD2B) or distal musculature (gastrocnemius and soleus; MM). The disease can also present initially and often advances to include both the proximal and distal muscles.4, 5, 6 While most patients experience a gradual decline over decades, several atypical phenotypes have been reported, including rapid loss of ambulation in less than 5 years.6 Variable age of onset has also been reported, ranging from 10 to 73 years.4, 6, 7
The DYSF gene codes for dysferlin protein, which is a large (∼240 kDa) type II transmembrane protein containing seven lipid- and protein-binding C2 domains, multiple Dysf and Fer domains, and a C-terminal transmembrane domain8, 9, 10, 11, 12, 13, 14, 15, 16 (Figure 1). Dysferlin is ubiquitously expressed but is most abundant in skeletal and cardiac muscle.15, 17 Dysferlin is predominantly localized to the plasma membrane but is also observed in cytoplasmic vesicles and is associated with the t-tubule network.15, 17, 18, 19, 20 Dysferlin protein plays an essential role in plasma membrane repair, and loss of dysferlin results in compromised membrane resealing and deterioration of muscle fibers.15, 21, 22 There is currently no cure for dysferlinopathy. Existing disease management consists mainly of physical therapy, orthopaedic surgery, and use of mechanical and respiratory aids.5, 23
Figure 1.
Human Dysferlin Domains Relative to Exons
There are 55 exons in the DYSF gene; dysferlin protein contains seven C2 domains (C2A–C2F), Ferlin family domains (F), an inner Dysf domain (D), and a second outer Dysf domain (N- and C-terminal), and a transmembrane domain (T). Also depicted are putative binding sites for various dysferlin interacting proteins, such as AHNAK (AK), Caveolin-3 (C), and Affixin (Beta-parvin; A). Predicted dysferlin domain positions and protein-protein interacting domains are based on coordinates available from the Universal Mutation Database (http://www.umd.be/DYSF/W_DYSF/Protein.html) and from Wein et al.53
A promising therapeutic strategy that has gained traction in recent years, especially with respect to neuromuscular disorders, is antisense-mediated exon skipping. Exon skipping utilizes synthetic nucleic acids called antisense oligonucleotides (AOs) to modulate pre-mRNA splicing and can be used to remove mutation-carrying exons and flanking exons to maintain an open reading frame.24, 25 In 2016, the United States Food and Drug Administration (FDA) approved eteplirsen (Exondys 51), the first-ever antisense drug for the treatment of Duchenne muscular dystrophy (DMD).26 Eteplirsen facilitates in-frame skipping of dystrophin exon 51 and utilizes the phosphorodiamidate morpholino oligomer (PMO) antisense chemistry. The rationale for exon skipping in dysferlinopathy is supported by reports of mildly affected patients harboring in-frame DYSF deletion mutations, such as exon 32.27 Even large deletions of DYSF have been associated with a milder disease course.28 Exon-skipping progress in dysferlinopathy has been limited to the in vitro skipping of DYSF exon 32 in human dysferlinopathy patient cells.29 To date, no other therapeutic exon-skipping targets have been identified for dysferlinopathy.
In this study, we undertook to identify novel exon-skipping targets in DYSF by characterizing the relationship between exon-deletion pattern and plasma-membrane-resealing ability. We created GFP-conjugated DYSF plasmid constructs lacking certain exons and transfected these into dysferlinopathy patient cells, then subjected the cells to a membrane-wounding assay. We demonstrate that DYSF exon combinations 19–21, 20–21, and 46–48 are required for proper plasma-membrane resealing, while exons 26–27 and 28–29 are dispensable for membrane resealing. After identifying which exons were not required for proper membrane resealing, we designed PMOs using a predictive software algorithm and tested the ability of PMO cocktails to facilitate multi-exon skipping and restore membrane-resealing ability in patient cells. We show that a PMO cocktail targeting DYSF exons 28–29 restores membrane resealing in patient cells. Our results provide a foundation for future in vivo investigations and possible clinical translation of DYSF exons 26–27 and 28–29 skipping approaches for treating dysferlinopathy.
Results
Mutation Analysis and Exon-Skipping Approach for Dysferlinopathy Cell Lines
We first assessed the mutation patterns present in two dysferlinopathy patient cell lines. The first cell line, MM-Pt1, was originally collected from a patient with MM. Sanger sequencing confirmed a homozygous missense mutation in DYSF exon 46 (Figure 2A). To remove the mutated exon while maintaining the reading frame would involve multi-exon skipping of DYSF exons 46–48 (Figure 2A). While exons 46 and 47 do not code for any known protein domain, exon 48 codes for a portion of the C2F domain (Figure 2A). The second cell line, MM-Pt2, was also collected from a patient with MM. Sanger sequencing confirmed that this cell line is transheterozygotic, carrying a frameshift-causing point mutation in exon 21 on one allele and a missense mutation in exon 28 on the other allele (Figure 2B). To remove the mutant exons while maintaining the reading frame would involve double exon skipping of exons 20 and 21, or 28 and 29 (Figure 2B). Exons 20 and 21 do not code for any known protein domain of dysferlin, while exons 28 and 29 code for portions of the Dysf-N and Dysf-C domains (Figure 2B).
Figure 2.
Dysferlinopathy Patient Cell Line Mutation Analysis
(A) Cell line MM-Pt1 was generated from a patient with Miyoshi myopathy and harbors a homozygous missense mutation in DYSF exon 46. (B) Cell line MM-Pt2 was generated from a patient with Miyoshi myopathy and contains two mutations: a frameshift mutation in exon 21 on one allele and a missense mutation in exon 28 on the second allele. Sanger sequencing was used to confirm the mutation patterns in both cell lines. Grey boxes represent exons, and ends of boxes denote corresponding phasing; colored boxes below represent predicted protein domains.
Determining the Feasibility of Exon-Skipping Approaches in Dysferlin through Transfection of Exon-Deleted Plasmid Constructs
Before proceeding with the application of an exon-skipping approach for our patient cell lines, we undertook to identify which DYSF exons could be removed without negatively impacting protein function. To do this, we mimicked the effect of exon skipping by using site-directed mutagenesis to generate DYSF plasmid constructs lacking exons corresponding to an exon-skipping approach amenable to each cell line. We generated GFP-fused DYSF plasmid constructs lacking DYSF exons 20–21, 28–29, and 46–48, all of which are in-frame deletions. We also generated an exon 19–21 deletion construct, as this pattern of exon skipping would also restore the reading frame in MM-Pt2 (Figure 2B). Additionally, we generated an exon 26–27 deletion construct, based on our later procurement of another dysferlinopathy patient cell line harboring a c.2875C > T missense mutation in exon 27. The deletion of DYSF exons 26–27 is in-frame (Figure 2B). As a control, we generated a GFP-only plasmid. To identify whether the exons deleted in the above constructs would have an impact on protein function, we transfected the constructs into cell line MM-Pt1, which exhibited impaired plasma-membrane-resealing ability compared to healthy cells (Figure 3) and observed whether any of these plasmids could rescue membrane-resealing ability. To assess membrane-resealing ability, we incubated cells in FM4-64 dye and generated focal lesions in the plasma membranes of GFP-positive cells using a two-photon laser microscope, as previously described.21 FM4-64 dye quickly fluoresces within the cytoplasm upon entering the cell through the lesion, and membrane resealing prevents entry of additional dye. We quantified the relative fluorescence values of FM dye within the cytoplasm over time.30 Our results show that Δ26–27 and Δ28–29 were able to rescue membrane-resealing ability to a degree similar to that of healthy cells and cells transfected with the full-length DYSF plasmid, as measured by changes in relative fluorescence intensity over time (Figures 3A and 3B). In contrast, Δ19–21, Δ20–21, and Δ46–48 were not able to rescue membrane-resealing ability (Figures 3A and 3B).
Figure 3.
Transfection with Δ26–27 or Δ28–29 DYSF Plasmid Restores Membrane Resealing Ability in Dysferlinopathy Patient Cells
(A) A membrane-wounding assay was performed on dysferlinopathy patient cells (MM-Pt1) transfected with various plasmids. Relative fluorescence intensity (ΔF/F0) over time is shown as means ± SE. (B) Graphical representation of (A) showing relative fluorescence intensity at final time point (t = 300 s). (C) Representative image of raw fluorescence values over time generated by membrane wounding assay. Line with double arrows depicts the final time point from which fluorescence values no longer significantly increase over time. Blue bar represents the time from laser wounding until the time point when fluorescence values peak, termed “steady state.” (D) Graphical representation of steady-state means ± SE. Statistics, one-way ANOVA and Tukey-Kramer multiple comparisons test. **p < 0.005, ***p < 0.0005, ****p < 0.0001 compared to GFP-only.
We next considered that, while great care is taken to ensure that all plasma-membrane-wounding parameters are consistent between experiments (e.g., laser power, wavelength, number of iterations, etc.) there can still be some disparity with respect to the degree of membrane damage following laser ablation. This might generate some small variation regarding quantification, in terms of the amount of fluorescent dye which infiltrates the cell. We utilized an additional measure of membrane-resealing ability, which we termed “time to steady state,” defining steady state as the point at which raw fluorescence values peak following laser wounding without any significant increase over time (Figure 3C). Time to steady state is calculated by subtracting the time prior to laser wounding from the time point when fluorescence values peak. We observed that the full-length as well as Δ26–27 and Δ28–29 constructs were able to rescue membrane resealing, as demonstrated by significantly shorter times to steady state when compared with GFP-only control; the Δ19–21, Δ20–21, and Δ46–48 plasmid constructs were not able to rescue membrane resealing, as their times to steady state were not statistically different from GFP-only control (Figure 3D). Taken together, these results show that DYSF exons 26–27 and 28–29 are dispensable for plasma-membrane resealing, suggesting that these exons are promising therapeutic targets for exon skipping, while exons 19–21, 20–21, and 46–48 are required for proper membrane resealing, suggesting that these are not promising exon-skipping targets.
In Silico and In Vitro Screening of PMOs for DYSF Exon 28–29 Skipping
Our exon-skipping predictive algorithm31 projected the expected exon-skipping efficiencies for 191 PMO sequences of 30-mer length, covering all possible target sites for exons 28 and 29. From these, the top three PMO sequences with the highest predicted exon-skipping efficiency that also met synthesis criteria (e.g., GC content, melting temperature [Tm], self-complementarity) were selected by us and produced by GeneTools (Philomath, Oregon) (Table 1). As measured by RT-PCR, all nine possible combinations of PMO cocktails were able to efficiently skip DYSF exons 28 and 29 using 10 μM each oligo in MM-Pt2 cells, suggesting that substantive multiple exon skipping can be achieved in DYSF through the use of PMO cocktails (Figure 4A). To examine rescue of dysferlin protein, we performed western blot analysis and found that there was no detectable change in protein levels between control and 10 μM each oligo treatment groups, while mutant cells also expressed detectable levels of dysferlin (Figure 4B).
Table 1.
Selected PMO Sequences for DYSF Exon 28–29 Skipping
| DYSF Exon to Skip | Oligo Name | Distance from Acceptor Splice Site | Oligo Sequence (5′ to 3′) | Predicted Skip (%) |
|---|---|---|---|---|
| 28 | Ac7 | 7 | ACTCAATGTCATCCTTGGGAAGCACCTTCT | 70.52 |
| 28 | Ac11 | 11 | GGGCACTCAATGTCATCCTTGGGAAGCACC | 68.21 |
| 28 | Ac41 | 41 | CATTCCTCATCTTCCCACTTCCAGCCCAGT | 61.01 |
| 29 | Ac36 | 36 | TCTTCTCAGCAGGGACCCAGTGCTTCGGCT | 86.38 |
| 29 | Ac32 | 32 | CTCAGCAGGGACCCAGTGCTTCGGCTTCCG | 79.7 |
| 29 | Ac40 | 40 | TACATCTTCTCAGCAGGGACCCAGTGCTTC | 62.06 |
Figure 4.
PMO Cocktails Facilitate Multi-exon Skipping of DYSF Exons 28–29 in Patient Cells
All transfections were performed using 10 μM each PMO. (A) Efficiency of exon 28–29 skipping as measured by one-step RT-PCR. Representative image shown. M, 100 bp marker. Values are shown as mean ± SE (n = 3). Statistics, one-way ANOVA and Tukey-Kramer multiple comparisons test. *p < 0.0001 compared to non-treated. (B) Western blot assessment of dysferlin protein expression following PMO cocktail transfection, with alpha-tubulin as a loading control.
Rescue of Plasma-Membrane Resealing in PMO Cocktail-Treated Dysferlinopathy Patient Cells
We selected PMO cocktail Ac11+32, which showed a high degree of DYSF exon 28–29 skipping, as measured by RT-PCR (Figure 4A), for transfection into patient cells (MM-Pt2). We observed that Ac11+32 was able to rescue plasma membrane resealing in dysferlinopathy patient fibroblasts, as measured by changes in relative fluorescence intensity over time (Figures 5A and 5B). Ac11+32 was also able to significantly reduce the time to membrane resealing, as measured by time to steady state (Figure 5C). These results show that functional recovery of membrane wounding in vitro is possible through antisense-mediated exon skipping of DYSF exons 28–29 via PMO cocktail, suggesting that these PMOs might be promising therapeutic agents for treating patients with mutations amenable to DYSF exon 28–29 skipping.
Figure 5.
Antisense-Mediated Skipping of DYSF Exons 28–29 via PMO Cocktail Rescues Membrane Resealing Ability in Dysferlinopathy Patient Cells
(A) Dysferlinopathy patient cells (MM-Pt2) transfected with PMO cocktail. Relative fluorescence intensity (ΔF/F0) over time is shown as means ± SE. (B) Graphical representation of (A) showing relative fluorescence intensity at the final time point (t = 300 s). (C) Graphical representation of time to steady state, means ± SE. Statistics, one-way ANOVA and Tukey-Kramer multiple comparisons test. ***p < 0.0005, ****p < 0.0001 compared to mock treated.
Discussion
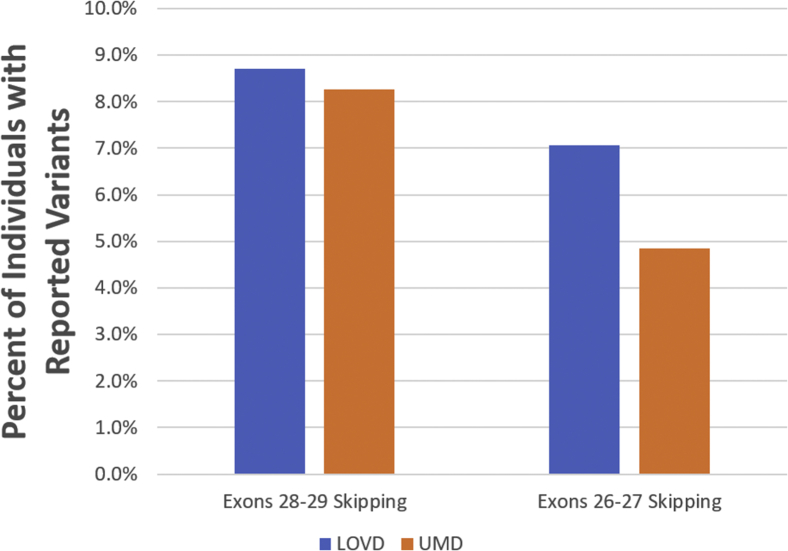
The first identification of a potential therapeutic exon skipping target in dysferlinopathy was DYSF exon 32, described by Sinnreich et al.27 in 2006. Since then, no new therapeutic exon-skipping targets have been described for DYSF. While some groups have attempted to identify redundant protein domains for the purpose of mini- or nano-dysferlin delivery via AAV vector,32 the relationship between exon-deletion pattern and protein functionality has gone largely uncharacterized. In this report, we described not only the first-ever success of multiple exon skipping in DYSF, but also the identification of two novel potential therapeutic exon-skipping targets for treating dysferlinopathy—DYSF exon 26–27 and 28–29 skipping. Notably, DYSF exons 26–29 were present in previously used nano-dysferlin constructs but not in mini-dysferlin constructs, both of which were able to ameliorate pathology and membrane-resealing defects in dysferlinopathy mouse models.32, 33 Successful translation of these findings into the development of clinical AO drugs would establish new therapeutic approaches that, combined, would be applicable to approximately 5%–7% (exon 26–27 skipping) and 8% (exon 28–29 skipping) of dysferlinopathy patients worldwide, according to reported variant data in the Leiden Open Variation Database (LOVD) (http://www.dmd.nl/) and Universal Mutation Database (UMD) (http://www.umd.be/DYSF/) (Figure 6).
Figure 6.
Projected Therapeutic Applicability of DYSF Exon 28–29 and 26–27 Skipping
Potential therapeutic applicability of DYSF exons 28–29 and 26–27 skipping is based on mutation variants reported in the Leiden Open Variation Database (LOVD) and Universal Mutation Database (UMD). Applicability was calculated by taking the number of reported variants within DYSF exons 28–29 (116 in LOVD and 97 in UMD) or 26–27 (94 in LOVD and 57 in UMD) and dividing by the total number of reported individuals with variants (1,332 total reported in LOVD and 1,174 total reported in UMD). LOVD data was obtained in May 2018, and UMD data available in May 2018 was taken from UMD-DYSF v1.4 data from June 16, 2015.
This work further supports the use of dysferlinopathy patient fibroblasts in screening novel AO sequences for the identification of therapeutic exon-skipping drugs, as fibroblasts expressed readily detectable amounts of DYSF mRNA at levels sufficient for in vitro assessment of exon-skipping efficiencies. While our AO sequences were able to facilitate robust exon skipping in fibroblasts, it remains to be seen whether the same sequences will be comparably effective when transfected into muscle cells and in vivo.
Our study also further validates the use of dysferlinopathy fibroblasts as an effective alternative to myoblasts or myotubes for the purpose of assessing plasma-membrane repair.30, 34 Here, healthy and dysferlinopathy patient fibroblasts displayed significant differences in their ability to reseal plasma membranes following two-photon laser wounding. Furthermore, this study highlights how the membrane-wounding assay can be used to validate the in vitro effectiveness of newly designed AOs at rescuing dysferlin protein function.
Since as little as 10% of wild-type protein has been associated with very mild pathology in dysferlinopathy27 and our cell line MM-Pt2 expresses dysferlin protein somewhere between 25% and 50% of wild-type (Figure 4B), it is reasonable to assume that while the endogenous missense mutation may not affect protein stability, there is an appreciable effect on protein functionality, as evidenced here by the significant difference in membrane-resealing ability between healthy and patient cells. The observation that our PMO cocktail was able to rescue plasma membrane resealing in patient cells despite no difference in protein expression between treated and non-treated cells is consistent with the idea that the proteins produced here via exon skipping could be more functional than non-treated proteins. Future studies aimed at characterizing the intracellular differences between native and exon-skipped proteins, such as their respective subcellular localization and interaction with other proteins, will help shed light on this issue. It would also be beneficial to test our PMO cocktail in a DYSF-null cell line with a mutation pattern amenable to exon 28–29 skipping in order to determine the degree of protein rescue following transfection.
The human dysferlin Dysf domains contain multiple positively charged and aromatic residues that exhibit a high degree of conservation in comparison to ferlin homologs myoferlin and fer-1.16 The secondary structure of the human inner Dysf domain consists of two antiparallel β strands, one at each terminus.35 This secondary structure is conserved in the inner Dysf domain of myoferlin.36 The solution structure of the myoferlin Dysf domain indicates the presence of stacked tryptophan/arginine (W/R) motifs, and mutations in this region are predicted to result in unfolding and protein degradation.36 Like myoferlin, the dysferlin inner Dysf domain is also held together by stacking of arginine and aromatic sidechains, and disruption of this region is also predicted to result in instability and unfolding.35 Notably, the majority of residues of this flat domain region contribute to the surface, suggesting that perhaps the Dysf domain is involved in protein-protein interactions.35 While this region is evidently susceptible to deleterious mutations according to the LOVD database,37 our demonstration that removal of exons 26–27 and 28–29 does not impact dysferlin protein function suggests that removal of portions of the Dysf domains is a possible therapeutic strategy. The hypothesis that this region of dysferlin may be superfluous is supported by the use of a mini-dysferlin that does not contain the Dysf domain but is able to rescue membrane resealing and dysferlin expression in a mouse model of dysferlinopathy.33
While dysferlin is known to have several binding partners,38, 39, 40, 41 the most likely interacting partner at the Dysf domains is caveolin-3 (CAV-3).13, 42 Mutations in CAV-3 are implicated in several forms of muscular dystrophy.43, 44, 45 CAV-3 is the only caveolin family member expressed in striated muscle and belongs to the dystrophin-glycoprotein complex, forming scaffolding along with t-tubules and caveolar membranes.46, 47, 48 CAV-3 is expressed primarily in muscle, where it plays a role in regulating sarcolemma stability, vesicular trafficking, signal transduction, and regulation of nitric-oxide-dependent functions.49, 50, 51, 52 Importantly, there exist two putative CAV-3 binding sites within the Dysf domains of dysferlin, with one found in the region of exons 25–26 and the other in the region of exons 28–29 (Figure 1).53 An additional CAV-3 binding site is located in a region that corresponds to exon 6. Our analyses were performed in fibroblasts, ruling out interactions with CAV-3 as influencing membrane resealing in this setting; however, it may be worth investigating possible Dysf domain and CAV-3 interactions in muscle cells.
It is also worth noting that patients with the exon 28 c.2997G > T missense mutation are reported to have significantly later disease onset (32.2 ± 4.8 years), and a patient homozygous for the c.2997G > T mutation did not use a cane at age 46.54, 55 Our results further underscore the therapeutic potential of a DYSF exon 28–29 skipping approach and suggest that whatever unknown function(s) the dysferlin Dysf domains serve, they are either not directly related to the process of plasma-membrane resealing or their function is redundant.
In conclusion, this study represents a significant achievement in the development of novel therapeutic strategies for treating dysferlinopathy. There are currently no ongoing or planned clinical trials involving exon skipping for dysferlinopathy, despite the successful translation of exon-skipping therapy into several clinical trials for other forms of muscular dystrophy, such as DMD (see ClinicalTrials.gov: NCT02958202, NCT02667483, NCT03375255, and NCT03167255). Our identification of two novel exon-skipping targets in vitro paves the way for future in vivo work that will help establish a foundation for the future clinical implementation of antisense-mediated exon skipping in dysferlinopathy.
Materials and Methods
Cell Culture
Human dysferlinopathy patient fibroblast cells harboring trans-heterozygous c.1958delG/c.2997G > T mutations (ID, MM-Pt2) and fibroblast cells harboring a homozygous c.5077C > T mutation (ID, MM-Pt1) were obtained from the induced pluripotent stem cell (iPSC) bank at Kyoto University. Healthy human fibroblasts (ID, GM23815) were obtained from the Coriell Institute for Medical Research (Camden, NJ, USA). Fibroblast cells were cultured in DMEM/F-12 growth media (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS) and 0.5% penicillin and streptomycin and incubated in a CO2 incubator at 37°C.
Plasmid Construct Design and Transfection
A plasmid construct containing N-terminally GFP-tagged DYSF (termed full-length DYSF plasmid) was generously provided by Dr. Katherine Bushby (Newcastle University). All other DYSF plasmid constructs investigated in this study (Δ19–21, Δ20–21, Δ26–27, Δ28–29, Δ46–48, GFP-only) were generated using a Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA) according to manufacturer’s instructions (for primers, see Table 2). Plasmid constructs were transfected into cells using a Lipofectamine LTX with Plus Reagent kit (Thermo Fisher) according to manufacturer’s instructions. In brief, fibroblast cells were seeded into 35-mm collagen-coated glass-bottom dishes (MatTek, Ashland, MA) and cultured in 2 mL of growth media for 24 hr. After incubation, media was changed to 2.3 mL of Opti-MEM (Thermo Fisher) containing 3 μL Lipofectamine LTX Transfection Reagent, 3.5 μL of PLUS Reagent, and 3.5 μg of plasmid. Cells were then incubated for 24 hr, after which media was changed to fresh growth media and cells were incubated an additional 24 hr before imaging and membrane-wounding analysis.
Table 2.
Primers Used in Site-Directed Mutagenesis and PCR
| Primer Name | Primer Sequence (5′ to 3′) |
|---|---|
| Site-Directed Mutagenesis | |
| hDYSF DSC-B GFP REV | CTACTACTTGTACAGCTCGTCCATGC |
| hDYSF DSC-B GFP FWD | GGCCGCTCGAGTCTAGAGGG |
| hDYSF DSC-B Ex49 FWD | GTTTTTCCTGCGTTGTATTATC |
| hDYSF DSC-B Ex45 REV | ACACAGTAGGTCTGTGGG |
| hDYSF DSC-B Ex22 FWD | GAAGCTGGCCTGGAGCAG |
| hDYSF DSC-B Ex18 REV | CTTGCCTGTGTTGAGCTCTG |
| hDYSF DSC-B Ex19 REV | CTCCACCCGGAGGATGTC |
| hDYSF DSC-B Ex28 FWD | AACGGGGAGAAGGTGCTT |
| hDYSF DSC-B Ex25 REV | GGTTTCAGCAAAGACAGACAG |
| hDYSF DSC-B Ex30 FWD | CACAGGCAGGCGGAGGCG |
| hDYSF DSC-B Ex27 REV | CACATCGGTGTAGTTGTCACTCATGTAGATCC |
| RT-PCR | |
| hDYSF Ex26 FWD | CGAGACTAAGTTGGCCCTTG |
| hDYSF Ex30 REV | GGCATCTGTCTTGCGGTACT |
PMO Design and Transfection
PMO sequences targeting DYSF exons were designed using a predictive software tool designed by our group that estimates expected exon-skipping efficiency.31 The top three PMOs per exon with the highest predicted exon-skipping ability and that met technical criteria for synthesis (e.g., GC content, Tm, self-complementarity) were supplied by Gene Tools (Philomath, Oregon). PMO sequences are shown in Table 1. For molecular analysis, PMO cocktails were transfected into 70%–80% confluent patient fibroblast cells using 6 μM Endo-Porter (Gene Tools) transfection reagent at a final concentration of 10 μM each PMO in DMEM (Invitrogen). Cells were incubated in PMOs for 48 hr before harvesting. For the two-photon membrane-wounding assay, 40%–50% confluent patient fibroblast cells were incubated with PMO in growth media for 48 hr before being subjected to the wounding assay.
RT-PCR Analysis
Total RNA was extracted from cells using Trizol (Invitrogen), and 200 ng of total RNA was used for assessing exon-skipping efficiency via SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Invitrogen). Primers are shown in Table 2. RT-PCR conditions are as follows: 1, 50°C (20 min); 2, 94°C (2 min); 3, 94°C (15 s); 4, 63°C (30 s); 5, 68°C (37 s); 6, go to 3, repeat 35×; 7, 68°C (5 min). PCR products were electrophoresed on a 1.5% agarose gel, and bands of interest were excised using PureLink Quick Gel Extraction Kit (Invitrogen) and sequenced via Sanger sequencing.
Immunoblotting
1.5 μg of protein from PMO-treated MM-Pt2 fibroblasts were loaded onto a NuPAGE Novex 3%–8% Tris-Acetate Midi Gel (Invitrogen) and separated by SDS-PAGE at 150 V for 70 min. Proteins were transferred onto an Immobilon polyvinylidene fluoride (PVDF) membrane (MilliporeSigma, Darmstadt, Germany) by semidry blotting using 20 V for 45 min. The membrane was blocked with PBS containing 0.05% Tween 20 and 5% skimmed milk, then incubated with the NCL Hamlet monoclonal antibody (Leica Biosystems, Wetzlar, Germany; 1:4,000 in blocking solution) or alpha-tubulin ab7291 antibody (Abcam, Cambridge, UK; 1:8,000 in blocking solution) at 4°C overnight. Membranes were then incubated with 1:10,000 goat anti-mouse immunoglobulin G (IgG) (H+L) secondary antibody, horseradish peroxidase (HRP) (Thermo Fisher), followed by incubation with ECL Select (GE Healthcare, Little Chalfont, UK). Bands were detected using a ChemiDoc system (Bio-Rad, Hercules, CA).
Membrane-Wounding Assay
Human fibroblast plasma membranes were subjected to laser-induced injury using two-photon laser microscopy as described previously.30 In brief, cells in glass-bottom dishes were prepared for wounding by rinsing once with Tyrode’s salts solution (MilliporeSigma) followed by addition of 1 mL of Tyrode’s salts containing 2.5 mM FM4-64 dye (Invitrogen). Only GFP-positive cells were selected for membrane-wounding experiments. A minimum of 15 cells were wounded per treatment. Using a Zeiss LSM 710 inverted confocal laser scanning microscope and Zeiss ZEN software, a 0.2 μm × 2 μm target was placed at the edge of the cell membrane. A 5-min time series of sequential image scans was performed, with cells imaged every 5 s. Cells were ablated 25 s after the beginning of the time series using a two-photon laser set to 820 nm, using 15% laser power with 10 iterations. Fluorescence values at sites of injury were quantified using Zeiss ZEN software, and for each time point relative fluorescence values were determined by subtraction of the background value (mean of t = 0–25 s) and division of the net increase in fluorescence by the background fluorescence value.
Statistical Analysis
All data were reported as mean values ± SE. The statistical differences between treatment groups were assessed by one-way ANOVA with a Tukey-Kramer multiple comparison test. p < 0.05 was considered statistically significant.
Author Contributions
Conceptualization of Experiments, T.Y.; Investigation, J.J.A.L.; Formal Analysis, J.J.A.L., R.M., T.Y.; Supervision, R.M., T.Y.; Software, W.D.; Provision of Materials/Samples, H.S.; Writing: J.J.A.L., T.Y.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the University of Alberta Faculty of Medicine and Dentistry, The Friends of Garrett Cumming Research Chair Fund, the HM Toupin Neurological Science Research Chair Fund, Muscular Dystrophy Canada, the Canada Foundation for Innovation (CFI), Alberta Advanced Education and Technology (AET), the Canadian Institutes of Health Research (CIHR), Jesse’s Journey - The Foundation for Gene and Cell Therapy, the Women and Children's Health Research Institute (WCHRI), Alberta Innovates Health Solutions (AIHS), The Rare Disease Foundation, and the BC Children’s Hospital Foundation. We thank Dr. Katherine Bushby for supplying us with the full-length DYSF plasmid. We would also like to thank Dr. Katsuya Miyake for technical advice.
References
- 1.Liu J., Aoki M., Illa I., Wu C., Fardeau M., Angelini C., Serrano C., Urtizberea J.A., Hentati F., Hamida M.B. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 1998;20:31–36. doi: 10.1038/1682. [DOI] [PubMed] [Google Scholar]
- 2.Bashir R., Britton S., Strachan T., Keers S., Vafiadaki E., Lako M., Richard I., Marchand S., Bourg N., Argov Z. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat. Genet. 1998;20:37–42. doi: 10.1038/1689. [DOI] [PubMed] [Google Scholar]
- 3.Illa I., Serrano-Munuera C., Gallardo E., Lasa A., Rojas-García R., Palmer J., Gallano P., Baiget M., Matsuda C., Brown R.H. Distal anterior compartment myopathy: a dysferlin mutation causing a new muscular dystrophy phenotype. Ann. Neurol. 2001;49:130–134. [PubMed] [Google Scholar]
- 4.Klinge L., Dean A.F., Kress W., Dixon P., Charlton R., Müller J.S., Anderson L.V., Straub V., Barresi R., Lochmüller H., Bushby K. Late onset in dysferlinopathy widens the clinical spectrum. Neuromuscul. Disord. 2008;18:288–290. doi: 10.1016/j.nmd.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Aoki M. Dysferlinopathy. In: Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., editors. GeneReviews. University of Washington, Seattle; 1993. [Google Scholar]
- 6.Nguyen K., Bassez G., Krahn M., Bernard R., Laforêt P., Labelle V., Urtizberea J.A., Figarella-Branger D., Romero N., Attarian S. Phenotypic study in 40 patients with dysferlin gene mutations: high frequency of atypical phenotypes. Arch. Neurol. 2007;64:1176–1182. doi: 10.1001/archneur.64.8.1176. [DOI] [PubMed] [Google Scholar]
- 7.Jethwa H., Jacques T.S., Gunny R., Wedderburn L.R., Pilkington C., Manzur A.Y. Limb girdle muscular dystrophy type 2B masquerading as inflammatory myopathy: case report. Pediatr. Rheumatol. Online J. 2013;11:19. doi: 10.1186/1546-0096-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Therrien C., Di Fulvio S., Pickles S., Sinnreich M. Characterization of lipid binding specificities of dysferlin C2 domains reveals novel interactions with phosphoinositides. Biochemistry. 2009;48:2377–2384. doi: 10.1021/bi802242r. [DOI] [PubMed] [Google Scholar]
- 9.Davis D.B., Doherty K.R., Delmonte A.J., McNally E.M. Calcium-sensitive phospholipid binding properties of normal and mutant ferlin C2 domains. J. Biol. Chem. 2002;277:22883–22888. doi: 10.1074/jbc.M201858200. [DOI] [PubMed] [Google Scholar]
- 10.Lennon N.J., Kho A., Bacskai B.J., Perlmutter S.L., Hyman B.T., Brown R.H., Jr. Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J. Biol. Chem. 2003;278:50466–50473. doi: 10.1074/jbc.M307247200. [DOI] [PubMed] [Google Scholar]
- 11.Azakir B.A., Di Fulvio S., Therrien C., Sinnreich M. Dysferlin interacts with tubulin and microtubules in mouse skeletal muscle. PLoS ONE. 2010;5:e10122. doi: 10.1371/journal.pone.0010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai C., Weisleder N., Ko J.K., Komazaki S., Sunada Y., Nishi M., Takeshima H., Ma J. Membrane repair defects in muscular dystrophy are linked to altered interaction between MG53, caveolin-3, and dysferlin. J. Biol. Chem. 2009;284:15894–15902. doi: 10.1074/jbc.M109.009589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda C., Hayashi Y.K., Ogawa M., Aoki M., Murayama K., Nishino I., Nonaka I., Arahata K., Brown R.H., Jr. The sarcolemmal proteins dysferlin and caveolin-3 interact in skeletal muscle. Hum. Mol. Genet. 2001;10:1761–1766. doi: 10.1093/hmg/10.17.1761. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y., Laval S.H., van Remoortere A., Baudier J., Benaud C., Anderson L.V., Straub V., Deelder A., Frants R.R., den Dunnen J.T. AHNAK, a novel component of the dysferlin protein complex, redistributes to the cytoplasm with dysferlin during skeletal muscle regeneration. FASEB J. 2007;21:732–742. doi: 10.1096/fj.06-6628com. [DOI] [PubMed] [Google Scholar]
- 15.Bansal D., Campbell K.P. Dysferlin and the plasma membrane repair in muscular dystrophy. Trends Cell Biol. 2004;14:206–213. doi: 10.1016/j.tcb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Therrien C., Dodig D., Karpati G., Sinnreich M. Mutation impact on dysferlin inferred from database analysis and computer-based structural predictions. J. Neurol. Sci. 2006;250:71–78. doi: 10.1016/j.jns.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 17.Anderson L.V., Davison K., Moss J.A., Young C., Cullen M.J., Walsh J., Johnson M.A., Bashir R., Britton S., Keers S. Dysferlin is a plasma membrane protein and is expressed early in human development. Hum. Mol. Genet. 1999;8:855–861. doi: 10.1093/hmg/8.5.855. [DOI] [PubMed] [Google Scholar]
- 18.Piccolo F., Moore S.A., Ford G.C., Campbell K.P. Intracellular accumulation and reduced sarcolemmal expression of dysferlin in limb—girdle muscular dystrophies. Ann. Neurol. 2000;48:902–912. [PubMed] [Google Scholar]
- 19.Kerr J.P., Ziman A.P., Mueller A.L., Muriel J.M., Kleinhans-Welte E., Gumerson J.D., Vogel S.S., Ward C.W., Roche J.A., Bloch R.J. Dysferlin stabilizes stress-induced Ca2+ signaling in the transverse tubule membrane. Proc. Natl. Acad. Sci. USA. 2013;110:20831–20836. doi: 10.1073/pnas.1307960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klinge L., Laval S., Keers S., Haldane F., Straub V., Barresi R., Bushby K. From T-tubule to sarcolemma: damage-induced dysferlin translocation in early myogenesis. FASEB J. 2007;21:1768–1776. doi: 10.1096/fj.06-7659com. [DOI] [PubMed] [Google Scholar]
- 21.Bansal D., Miyake K., Vogel S.S., Groh S., Chen C.C., Williamson R., McNeil P.L., Campbell K.P. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 2003;423:168–172. doi: 10.1038/nature01573. [DOI] [PubMed] [Google Scholar]
- 22.Roche J.A., Lovering R.M., Roche R., Ru L.W., Reed P.W., Bloch R.J. Extensive mononuclear infiltration and myogenesis characterize recovery of dysferlin-null skeletal muscle from contraction-induced injuries. Am. J. Physiol. Cell Physiol. 2010;298:C298–C312. doi: 10.1152/ajpcell.00122.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi K., Izawa T., Kuwamura M., Yamate J. Dysferlin and animal models for dysferlinopathy. J. Toxicol. Pathol. 2012;25:135–147. doi: 10.1293/tox.25.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J.J., Yokota T. Antisense therapy in neurology. J. Pers. Med. 2013;3:144–176. doi: 10.3390/jpm3030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niks E.H., Aartsma-Rus A. Exon skipping: a first in class strategy for Duchenne muscular dystrophy. Expert Opin. Biol. Ther. 2017;17:225–236. doi: 10.1080/14712598.2017.1271872. [DOI] [PubMed] [Google Scholar]
- 26.Aartsma-Rus A., Krieg A.M. FDA Approves Eteplirsen for Duchenne Muscular Dystrophy: The Next Chapter in the Eteplirsen Saga. Nucleic Acid Ther. 2017;27:1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinnreich M., Therrien C., Karpati G. Lariat branch point mutation in the dysferlin gene with mild limb-girdle muscular dystrophy. Neurology. 2006;66:1114–1116. doi: 10.1212/01.wnl.0000204358.89303.81. [DOI] [PubMed] [Google Scholar]
- 28.Krahn M., Wein N., Lostal W., Bourg-Alibert N., Nguyen K., Courrier S., Vial C., Labelle V., De Petris D., Borges A. G.O.5 Partial functionality of a Mini-dysferlin molecule identified in a patient affected with moderately severe primary dysferlinopathy. Neuromuscul. Disord. 2008;18:781. [Google Scholar]
- 29.Barthélémy F., Blouin C., Wein N., Mouly V., Courrier S., Dionnet E., Kergourlay V., Mathieu Y., Garcia L., Butler-Browne G. Exon 32 Skipping of Dysferlin Rescues Membrane Repair in Patients’ Cells. J. Neuromuscul. Dis. 2015;2:281–290. doi: 10.3233/JND-150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J.J.A., Maruyama R., Sakurai H., Yokota T. Cell Membrane Repair Assay Using a Two-photon Laser Microscope. J. Vis. Exp. 2018;131:e56999. doi: 10.3791/56999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Echigoya Y., Mouly V., Garcia L., Yokota T., Duddy W. In silico screening based on predictive algorithms as a design tool for exon skipping oligonucleotides in Duchenne muscular dystrophy. PLoS ONE. 2015;10:e0120058. doi: 10.1371/journal.pone.0120058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Llanga T., Nagy N., Conatser L., Dial C., Sutton R.B., Hirsch M.L. Structure-Based Designed Nano-Dysferlin Significantly Improves Dysferlinopathy in BLA/J Mice. Mol. Ther. 2017;25:2150–2162. doi: 10.1016/j.ymthe.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krahn M., Wein N., Bartoli M., Lostal W., Courrier S., Bourg-Alibert N., Nguyen K., Vial C., Streichenberger N., Labelle V. A naturally occurring human minidysferlin protein repairs sarcolemmal lesions in a mouse model of dysferlinopathy. Sci. Transl. Med. 2010;2:50ra69. doi: 10.1126/scitranslmed.3000951. [DOI] [PubMed] [Google Scholar]
- 34.Matsuda C., Kiyosue K., Nishino I., Goto Y., Hayashi Y.K. Dysferlinopathy Fibroblasts Are Defective in Plasma Membrane Repair. PLoS Curr. 2015;7 doi: 10.1371/currents.md.5865add2d766f39a0e0411d38a7ba09c. ecurrents.md.5865add2d766f39a0e0411d38a7ba09c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sula A., Cole A.R., Yeats C., Orengo C., Keep N.H. Crystal structures of the human Dysferlin inner DysF domain. BMC Struct. Biol. 2014;14:3. doi: 10.1186/1472-6807-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel P., Harris R., Geddes S.M., Strehle E.M., Watson J.D., Bashir R., Bushby K., Driscoll P.C., Keep N.H. Solution structure of the inner DysF domain of myoferlin and implications for limb girdle muscular dystrophy type 2b. J. Mol. Biol. 2008;379:981–990. doi: 10.1016/j.jmb.2008.04.046. [DOI] [PubMed] [Google Scholar]
- 37.Fokkema I.F., Taschner P.E., Schaafsma G.C., Celli J., Laros J.F., den Dunnen J.T. LOVD v.2.0: the next generation in gene variant databases. Hum. Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 38.Cacciottolo M., Belcastro V., Laval S., Bushby K., di Bernardo D., Nigro V. Reverse engineering gene network identifies new dysferlin-interacting proteins. J. Biol. Chem. 2011;286:5404–5413. doi: 10.1074/jbc.M110.173559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han R., Campbell K.P. Dysferlin and muscle membrane repair. Curr. Opin. Cell Biol. 2007;19:409–416. doi: 10.1016/j.ceb.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ono H., Suzuki N., Kanno S., Izumi R., Takahashi T., Kitajima Y., Osana S., Akiyama T., Ikeda K., Shijo T. Novel binding partner of dysferlin is required for plasma-membrane repair. Neuromuscul. Disord. 2017;27(Suppl 2):S146. [Google Scholar]
- 41.Matsuda C., Kameyama K., Tagawa K., Ogawa M., Suzuki A., Yamaji S., Okamoto H., Nishino I., Hayashi Y.K. Dysferlin interacts with affixin (beta-parvin) at the sarcolemma. J. Neuropathol. Exp. Neurol. 2005;64:334–340. doi: 10.1093/jnen/64.4.334. [DOI] [PubMed] [Google Scholar]
- 42.Couet J., Li S., Okamoto T., Ikezu T., Lisanti M.P. Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J. Biol. Chem. 1997;272:6525–6533. doi: 10.1074/jbc.272.10.6525. [DOI] [PubMed] [Google Scholar]
- 43.Galbiati F., Volonte D., Minetti C., Bregman D.B., Lisanti M.P. Limb-girdle muscular dystrophy (LGMD-1C) mutants of caveolin-3 undergo ubiquitination and proteasomal degradation. Treatment with proteasomal inhibitors blocks the dominant negative effect of LGMD-1C mutanta and rescues wild-type caveolin-3. J. Biol. Chem. 2000;275:37702–37711. doi: 10.1074/jbc.M006657200. [DOI] [PubMed] [Google Scholar]
- 44.Galbiati F., Volonte D., Minetti C., Chu J.B., Lisanti M.P. Phenotypic behavior of caveolin-3 mutations that cause autosomal dominant limb girdle muscular dystrophy (LGMD-1C). Retention of LGMD-1C caveolin-3 mutants within the golgi complex. J. Biol. Chem. 1999;274:25632–25641. doi: 10.1074/jbc.274.36.25632. [DOI] [PubMed] [Google Scholar]
- 45.Minetti C., Sotgia F., Bruno C., Scartezzini P., Broda P., Bado M., Masetti E., Mazzocco M., Egeo A., Donati M.A. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat. Genet. 1998;18:365–368. doi: 10.1038/ng0498-365. [DOI] [PubMed] [Google Scholar]
- 46.Rothberg K.G., Heuser J.E., Donzell W.C., Ying Y.S., Glenney J.R., Anderson R.G. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- 47.Sargiacomo M., Scherer P.E., Tang Z., Kübler E., Song K.S., Sanders M.C., Lisanti M.P. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc. Natl. Acad. Sci. USA. 1995;92:9407–9411. doi: 10.1073/pnas.92.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parton R.G., Way M., Zorzi N., Stang E. Caveolin-3 associates with developing T-tubules during muscle differentiation. J. Cell Biol. 1997;136:137–154. doi: 10.1083/jcb.136.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song K.S., Scherer P.E., Tang Z., Okamoto T., Li S., Chafel M., Chu C., Kohtz D.S., Lisanti M.P. Expression of caveolin-3 in skeletal, cardiac, and smooth muscle cells. Caveolin-3 is a component of the sarcolemma and co-fractionates with dystrophin and dystrophin-associated glycoproteins. J. Biol. Chem. 1996;271:15160–15165. doi: 10.1074/jbc.271.25.15160. [DOI] [PubMed] [Google Scholar]
- 50.Scherer P.E., Lisanti M.P. Association of phosphofructokinase-M with caveolin-3 in differentiated skeletal myotubes. Dynamic regulation by extracellular glucose and intracellular metabolites. J. Biol. Chem. 1997;272:20698–20705. doi: 10.1074/jbc.272.33.20698. [DOI] [PubMed] [Google Scholar]
- 51.Aravamudan B., Volonte D., Ramani R., Gursoy E., Lisanti M.P., London B., Galbiati F. Transgenic overexpression of caveolin-3 in the heart induces a cardiomyopathic phenotype. Hum. Mol. Genet. 2003;12:2777–2788. doi: 10.1093/hmg/ddg313. [DOI] [PubMed] [Google Scholar]
- 52.Williams T.M., Lisanti M.P. The caveolin proteins. Genome Biol. 2004;5:214. doi: 10.1186/gb-2004-5-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wein N., Avril A., Bartoli M., Beley C., Chaouch S., Laforêt P., Behin A., Butler-Browne G., Mouly V., Krahn M. Efficient bypass of mutations in dysferlin deficient patient cells by antisense-induced exon skipping. Hum. Mutat. 2010;31:136–142. doi: 10.1002/humu.21160. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi T., Aoki M., Tateyama M., Kondo E., Mizuno T., Onodera Y., Takano R., Kawai H., Kamakura K., Mochizuki H. Dysferlin mutations in Japanese Miyoshi myopathy: relationship to phenotype. Neurology. 2003;60:1799–1804. doi: 10.1212/01.wnl.0000068333.43005.12. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi T., Aoki M., Suzuki N., Tateyama M., Yaginuma C., Sato H., Hayasaka M., Sugawara H., Ito M., Abe-Kondo E. Clinical features and a mutation with late onset of limb girdle muscular dystrophy 2B. J. Neurol. Neurosurg. Psychiatry. 2013;84:433–440. doi: 10.1136/jnnp-2011-301339. [DOI] [PMC free article] [PubMed] [Google Scholar]