Abstract
Abstract
Spinocerebellar ataxias (SCAs) are rare types of cerebellar ataxia with a dominant mode of inheritance. To date, 47 SCA subtypes have been identified, and the number of genes implicated in SCAs is continually increasing. Polyglutamine (polyQ) expansion diseases
( ATXN1/SCA1, ATXN2/SCA2, ATXN3/SCA3, CACNA1A/SCA6, ATXN7/SCA7, TBP/SCA17, and ATN1/DRPLA) are the most common group of SCAs. No preventive or curative treatments are currently available, but various therapeutic approaches, including RNA-targeting treatments, such as antisense oligonucleotides (ASOs), are being developed. Clinical trials of ASOs in SCA patients are already planned. There is, therefore, a need to identify valid outcome measures for such studies. In this review, we describe recent advances towards identifying appropriate biomarkers, which are essential for monitoring disease progression and treatment efficacy. Neuroimaging biomarkers are the most powerful markers identified to date, making it possible to reduce sample sizes for clinical trials. Changes on brain MRI are already evident at the premanifest stage in SCA1 and SCA2 carriers and are correlated with CAG repeat size. Other potential biomarkers have also been developed, based on neurological examination, oculomotor study, cognitive assessment, and blood and cerebrospinal fluid analysis. Longitudinal studies based on multimodal approaches are required to establish the relationships between parameters and to validate the biomarkers identified.
Keywords: spinocerebellar ataxias, biomarkers, antisense oligonucleotides, clinical trials, neuroimaging
Introduction
Spinocerebellar ataxias (SCAs) are a group of neurodegenerative diseases displaying autosomal dominant inheritance. To date, 47 SCA subtypes have been described, and 35 causal genes have been identified 1. SCAs are considered a rare group of cerebellar ataxias, with a mean prevalence of 2.7/100,000 2. Their most frequent forms are polyglutamine (polyQ) expansion diseases ( ATXN1/SCA1, ATXN2/SCA2, ATXN3/SCA3, CACNA1A/SCA6, ATXN7/SCA7, TBP/SCA17, and ATN1/DRPLA) 3. These diseases manifest above a threshold number of CAG repeats, which is different for each gene. The same mutational mechanism is found in Huntington disease 4 and in spinal bulbar muscular atrophy 5. Disease onset generally occurs between the ages of 20 and 40 years, and age at onset and CAG repeat expansion size are inversely correlated 6, 7. Age at onset also displays variability due to interactions between polyQ genes, i.e. between the expanded allele and the alleles of normal repeat size in the other genes 8, 9. The instability of expanded alleles results in genetic anticipation in successive generations 3. Unsteady gait and clumsiness are usually the first clinical symptoms at onset, followed by a progressive loss of the ability to walk. Cerebellar syndrome is often associated with extracerebellar signs (pyramidal syndrome, extrapyramidal syndrome, retinal degeneration, dementia, seizures, etc.) 3. There is currently no preventive or curative treatment, but different therapeutic approaches are being tested, including the use of disease-modifying compounds and therapeutic approaches, which are described below. However, in SCAs, the lack of sensitive biomarkers and the small number of patients to be included in clinical trials are a challenge for the statistical sample size estimation. Outcome measures providing information about treatment efficacy are vital, particularly for these genetic diseases that can be treated before the onset of symptoms. This review provides an overview of findings for biomarkers, recent results of clinical trials in SCA patients, and future perspectives for treatment.
Natural history of spinocerebellar ataxias (SCA1, 2, 3, and 6)
The natural history of SCA1, 2, 3, and 6 was established by the longitudinal European cohort study EUROSCA, which included 462 patients with at least one year of follow-up and a median of 49 months of observation 10. This study used the Scale for the Assessment and Rating of Ataxia (SARA), an accurate measurement of cerebellar dysfunction 11, 12 in SCA1, 2, 3, and 6 patients and premanifest individuals 13. The most severe disease turned out to be SCA1, with an annual progression of 2.11 on the 40-point SARA, followed by SCA3 (1.56 points), SCA2 (1.49 points), and SCA6 (0.8 points) 10. SCA1 has also been reported to have the most severe prognosis for progression in other studies, such as one in Europe 14 and another conducted by the North American consortium (the Clinical Research Consortium for Spinocerebellar Ataxias) 15. This may be because of motoneuron involvement, which, particularly in this form, leads to bulbar dysfunction and, thus, to respiratory and swallowing failure 16. These results are consistent with 10-year survival, which is lowest for SCA1 patients, intermediate for SCA2 and SCA3, and highest for SCA6 patients 17. A high SARA score has also been identified as one of the strongest risk factors for death in all subtypes. Other associated risk factors include dysphagia for SCA1, longer CAG length in pathologic allele for SCA2, and dystonia and interaction between age and CAG length in SCA3 17. The Composite Cerebellar Functional Severity (CCFS) score is another quantitative score for assessing cerebellar ataxia that has been validated in adults and children 18, 19. This score is calculated from scores for two tasks for the dominant hand—the nine-hole pegboard test and the click test—and is age dependent and available from an open source ( https://icm-institute.org/en/tutorial-for-making-ccfs-board/). In a study on Friedreich ataxia and SCA1, 2, 3, and 7 patients, SARA and CCFS scores were higher in SCAs than in Friedreich ataxia after adjustment for disease duration, revealing a slower progression in Friedreich ataxia than in SCAs. Considering each SCA subtype separately, SCA2 and SCA1 patients had higher CCFS scores than did patients with other subtypes, despite an absence of difference in SARA scores 20, probably because of the more accentuate cerebellar syndrome.
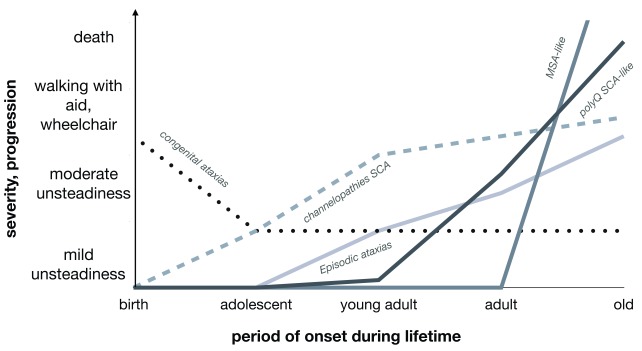
Based on SARA score progression, the EUROSCA study identified, for each genotype, the sample sizes required to detect a 50% decrease in SARA score progression in a two-arm clinical trial lasting one year: 142 patients for SCA1, 172 for SCA2, 202 for SCA3, and 602 for SCA6 10. These are large numbers of patients for such rare diseases, making it necessary to carry out clinical trials at an international level. Additional biomarkers are required to overcome this problem. Furthermore, these dominantly inherited cerebellar ataxias have very different progression profiles, as shown in Figure 1. Given the progressive nature of the disease and the sample sizes required, biomarkers are very important and essential.
Figure 1. Progression and severity profiles in cerebellar ataxias by underlying etiology.
Multisystem atrophy (MSA) is a rapidly progressing non-monogenic alpha-synucleinopathy. It is the most severe of these diseases, followed by polyglutamine (polyQ) spinocerebellar ataxias (SCAs), such as ATXN1/SCA1, ATXN2/SCA2, ATXN3/SCA3, CACNA1A/SCA6, ATXN7/SCA7, TBP/SCA17, and ATN1/DRPLA. Very different profiles are present in the forms because of missense mutations in channel genes ( CACNA1A, KCND3, KCNC3, and KCNA1).
Biomarkers
Neuroimaging biomarkers
The most powerful biomarkers identified to date are derived from neuroimaging examinations. Specific patterns of brain atrophy have been described in vivo in polyQ SCAs 21, 22 and confirmed in post-mortem studies 23, 24. Changes in brain MRI findings are already evident at the premanifest stage in SCA1 and SCA2 carriers in the form of losses of gray matter from the cerebellum and brainstem. In SCA2 carriers only, an additional decrease in brainstem volume relative to non-carriers has also been reported 13. A recent case-control study on a Cuban-German cohort of premanifest and manifest SCA2 carriers reported remarkable decreases in cerebellum and pontine volume and in the anteroposterior diameter of the pontine brainstem. A negative correlation was found between CAG repeat size and brainstem and cerebellum volume in premanifest individuals 25. Cerebellar and pons atrophy was more pronounced in manifest patients 25, representing a potential outcome measure. By contrast, midbrain and medulla volumes did not differ significantly between the preclinical and clinical stages 25. In SCA2, white matter alterations in the parietal lobe and anterior corona radiata (detected by fractional anisotropy), and in the cerebellum and middle cerebellar peduncle (detected by mean diffusivity), were found to be correlated with SARA scores, leading the authors of the report concerned to suggest that connections between the motor and sensory integration areas might be impaired 26. However, with the exception of rare follow-up studies 21, 27, only cross-sectional brain MRI studies have been performed to date. Longitudinal studies will be required to establish the rate of progression of cerebellar and brainstem atrophy.
Cervical spinal cord atrophy has been proposed as a possible biomarker for SCA1, as it is correlated with SARA score, CAG repeat length, and disease duration 28. Indeed, the clinical presentation of this subtype can include pyramidal signs and it may, at early stages, mimic spastic paraplegia 29. Cervical spinal cord atrophy has also been identified as a potential biomarker for SCA3 30.
Neurochemical abnormalities can be detected by MRI spectroscopy (MRS), as demonstrated by the group headed by Gulin Oz 31. Decreases in N-acetylaspartate and glutamate levels reflect a loss of neurons, whereas increases in myoinositol serve as a marker of gliosis. N-acetylaspartate and N-acetylaspartylglutamate levels are significantly lower in the vermis and pons of SCA1, 2, 3, and 7 patients than in controls. Myoinositol levels are higher in SCA1, 2, and 3 patients than in controls. This neurochemical profile is particularly evident in SCA2 and SCA3 patients and is correlated with SARA score 32. Interestingly, multicenter acquisition of this technique has been validated 32.
Such alterations have been demonstrated by ultra-high-field MRS, even at the premanifest stage, with SCA2 patients showing the most compromised profile, followed by SCA1, SCA3, and SCA6 variant carriers 31.
Another recent neuroimaging technique applied to SCAs is functional MRI (fMRI) at rest 33 or during a task 34. This technique can be used to characterize circuitry reorganization, making it possible to identify specific patterns of cerebellar activation 35, 36. Using this approach in the early phase of SCA3 disease, Duarte et al. discovered a reorganization of the motor network that could potentially serve as a biomarker 37. However, more detailed analyses of fMRI outcomes are required for future application.
Neuroimaging biomarkers are more powerful than clinical scores for the detection of disease progression. In a cohort of SCA1, 2, 3, and 7 patients, greater longitudinal effect size was detected for brain volumetry (>1.2) than SARA and CCFS scores (<0.8) 27. In conclusion, measures of pons and cerebellum atrophy seem to be the most promising biomarkers.
Oculomotor biomarkers
Oculomotor involvement is present at an early phase of SCA and is detectable even at premanifest stages as a higher rate of gaze-evoked nystagmus or other alterations, such as a square-wave jerk during central fixation, impaired vertical smooth pursuit, slow saccade, and a higher antisaccade error rate in SCA3 carriers than controls 13, 38, 39. These alterations have been studied in detail in SCA2 patients, all of whom present specific oculomotor abnormalities (slow horizontal saccades up to saccade paresis) 40. Preclinical SCA2 carriers present a reduced saccade velocity and antisaccade task errors 41.
Moreover, in manifest and premanifest SCA2 carriers, both the progression of oculomotor impairment and the slowing of horizontal saccade velocity are correlated, respectively, with CAG repeat size 42 and pontine atrophy 25. The annual decrease in saccade velocity and saccade accuracy and an increase in saccade latency may soon be used as biomarkers. This correlation between saccade measurements and other parameters, such as brain MRI and/or cognitive assessment, may make it possible to reduce sample size in future trials 42.
Biological biomarkers
No biological biomarkers for SCAs have yet been identified, a situation contrasting with other neurodegenerative diseases, such as Alzheimer’s disease and frontotemporal dementia. There is, therefore, a need to search for blood or cerebrospinal fluid (CSF) biomarkers of these diseases. In addition to guiding diagnostic process, biomarkers may be of use in clinical management if linked to disease progression.
In this respect, several recent studies in patients with SCA3, the most frequent SCA subtype, have yielded promising results. SIRT1 encodes sirtuin-1, an NAD +-dependent deacetylase involved in several cellular functions, including chromatin modulation, the cell cycle, apoptosis, and autophagy regulation in response to DNA damage. SIRT1 mRNA levels are lower in SCA3 mice than in wild-type mice and are also low in the fibroblasts of SCA3 patients 43. In SCA3 mice, the rescue of SIRT1 by caloric restriction or the administration of resveratrol induces motor improvement and neuropathological changes, such as a decrease in neuroinflammation and reactive gliosis with an activation of autophagy 43. SIRT1 overexpression has been shown to activate autophagy and thus to induce higher levels of mutant protein clearance. For these reasons, resveratrol may be a useful neuroprotective drug, as shown in some assays in Drosophila models of SCA3 44 and in the rat 3-acetylpyridine-induced cerebellar ataxia model 45. Other studies have also demonstrated an imbalance between autophagy and apoptosis in SCA3. For example, an increase has been reported in the levels of BECN1, encoding the proautophagic Beclin 1 protein 46, and BCL2/BAX ratio has been shown to be lower in pre-ataxic SCA3 carriers than in controls 47, enhancing apoptotic processes.
Oxidative stress has been implicated in several neurodegenerative disorders, and SCA3 patients have been shown to produce abnormally large amounts of reactive oxygen species 48. This phenomenon results from a decrease in antioxidant capacity: both superoxide dismutase and glutathione peroxidase (GPx) activities are lower in symptomatic than in pre-symptomatic carriers 49. Moreover, the observed correlation of the decrease in GPx levels with disease severity suggests that GPx may be a reliable biomarker 49.
Cytokines have also been investigated as possible markers of SCA3. Indeed, enhanced inflammation has been linked to stronger staining for IL1B and IL6 and higher levels of activated microglia and reactive astrocytes in the brains of SCA3 patients 50. In a study on Brazilian SCA3 patients, no difference in cytokine levels was detected between 79 carriers and 43 controls. On the contrary, higher eotaxin levels were observed in asymptomatic carriers than in symptomatic carriers. It has been suggested that the levels of eotaxin released by astrocytes are inversely correlated with disease progression 51. In another cohort of SCA3 patients recruited in the Azores, lower IL6 mRNA levels, due to the presence of the IL6*C allele, were associated with an earlier age at onset than the presence of the IL6*G allele 52. In this study, the earlier age at onset (by about 10 years on average) resulted from the presence of the APOE*e2 allele in IL6*C carriers, probably because of additional decreases in the levels of other cytokines, such as IL1B and TNF, due to the presence of the APOE*e2 allele 53.
In a recent study, serum neurofilament light chain (Nfl) was identified as a powerful potential serum biomarker in polyQ SCAs 54. Neurofilaments are components of the neuron cytoskeleton, and their levels in the blood reflect damage to the axons of long fiber tracts 55. In Huntington disease, which is also a polyQ disease, a correlation between serum Nfl levels and disease severity (UHDRS, cognitive decline, brain atrophy) has been reported, even after adjustment for age and CAG repeat size 56. Nfl dosage is also useful for predicting disease onset in pre-symptomatic carriers and the progression of Huntington disease 56. In a study by Wilke et al., Nfl levels were higher in SCAs patients than in controls, particularly for SCA1 and SCA3 54. The CSF has been only briefly explored in SCAs. CSF Nfl levels are a potential diagnostic and prognostic biomarker for SCAs, as in amyotrophic lateral sclerosis 57. A study in SCA1, SCA2, and SCA6 patients evaluated α-synuclein, DJ-1, and glial fibrillary acidic protein levels in CSF. The levels of all of these proteins were higher in CSF from patients than in control CSF, but this difference was significant only for tau, the levels of which were significantly higher in SCA2 patients. No correlations were found for CAG repeat size, disease severity, and disease duration 58. It will be very interesting to determine polyQ protein levels in the CSF of SCA patients, as has been done already for Huntington disease.
Cognition in spinocerebellar ataxias
Cognitive impairment, of various degrees of severity, can occur in polyQ SCAs. It involves mostly the executive functions and verbal memory, as shown in one of the first cognitive studies to compare the profiles of SCA1, 2, and 3 patients 59. Cognitive decline was more prominent and rapid in SCA1 than in the other genotypes 59– 61 and was associated with an increase in the incidence of depression 61. In another study, SCA2 patients were found to have more visuospatial and visuoperceptive deficits than SCA1 patients 62. In a recent study focusing on cognitive dysfunction in SCA6 patients, mild impairments of executive functions, mental flexibility, and visuospatial skills were found to be correlated with decreased resting-state connectivity in the frontoparietal network 63. These data support a role for the cerebellum in cognitive processes, given that the cerebellum is the principal cerebral structure affected and neuronal loss is less severe in SCA6 than in other polyQ diseases 23. The scales most widely used to assess dementia, such as the Mini-Mental State Examination or Montreal Cognitive Assessment, are not appropriate for the detection of cognitive impairment in SCA patients, who present a cerebellar cognitive affective/Schmahmann’s syndrome (CCAS) 64. This syndrome includes executive dysfunctions, spatial cognition difficulties, language deficits, and personality changes. A CCAS scale has recently been validated in a large cohort of cerebellar patients with acquired, genetic, or idiopathic cause and shown to have a sensitivity of 95% and a selectivity of 78% for CCAS diagnosis 64. This scale, which is easy and fast to administer, can be used for cognitive evaluation, which should be performed in longitudinal studies of patients.
Genetic advances in spinocerebellar ataxias
The number of genes implicated in SCAs has steadily increased over time. The last causal gene to have been identified, in SCA47 patients, is PUM1, encoding Pumilio1, a member of the PUMILIO/FBF RNA-binding protein family. The loss of Pum1 led to an SCA1-like phenotype in mice by causing a 30–40% increase in wild-type Atxn1 protein levels in the cerebellum 65. These two proteins are functionally related: in SCA1 mice, the disease is more severe if one copy of Pum1 is removed, whereas the motor phenotype of Pum1-heterozygous mice is improved by the removal of one copy of Atxn1 65. Mutations of PUM1 were reported by Gennarino et al. in 15 patients, with different ages at disease onset (5 months to 50 years) and phenotypic presentations. A 50% loss of the protein resulted in a severe infantile disease and a developmental syndrome called Pumilio1-associated developmental disability, ataxia, and seizure (PADDAS), whereas the loss of 25% of the protein caused Pumilio1-related cerebellar ataxia (PRCA), with a later onset and incomplete penetrance 66.
Using whole-exome sequencing (WES), Nibbeling et al. identified the genes associated with SCA46 and SCA45: PLD3 and FAT2, respectively. These genes, which are strongly expressed in the cerebellum, cause pure adult-onset cerebellar ataxia. However, in some cases, SCA46 patients may present with a sensory neuropathy 67.
Genes encoding ion channels are frequently involved in dominant cerebellar ataxia: mutations of CACNA1A are the most frequent genetic cause of autosomal dominant cerebellar ataxia in patients negative for polyQ SCAs, followed by other channel-coding genes, such as KCND3, KCNC3, and KCNA1 68. These genes predispose the patient to an earlier disease onset, intellectual deficiency, and slower disease progression 68. CACNA1G mutations, underlying SCA42 69, lead to a slowly progressive pure 70 or complicated cerebellar ataxia associated with a spastic gait 69. Patients with earlier onset may also present facial dysmorphisms, microcephaly, digital abnormalities, and seizures 71.
Another channel gene implicated in SCA44 is GRM1, encoding the metabotropic glutamate receptor 1 (mGluR1) responsible for two phenotypic manifestations: adult-onset cerebellar ataxia in the presence of gain-of-function mutations and early onset ataxia with intellectual deficiency when associated with loss-of-function mutations 72.
The use of next-generation sequencing (NGS) techniques, such as WES, in everyday clinical practice has increased the diagnostic rate of complex neurological diseases 73, 74. The application of NGS in ataxia patients has revealed new SCA genes and improved our knowledge of the molecular pathways involved in cerebellar ataxia. It also affects the development of new therapeutic approaches: for example, elucidation of the role of mGluR1 in the excitability of Purkinje cells led to experiments modulating mGluR1 activity in SCA1 mice with drugs such as baclofen or the negative allosteric modulator JNJ16259685, both of which improved motor function 75, 76. As well, in a recent study by Bushart et al., the administration of potassium channel modulators induced improvement in SCA1 mice motor phenotype via electrophysiological changes 77; the authors also showed that chlorzoxazone and baclofen co-administration was well tolerated by SCA1 patients and should be considered as a promising approach for treating symptoms 77.
However, today, WES will not detect trinucleotide repeat disorders, mitochondrial DNA mutations, or large structural variants. Novel or de novo repeat expansions are still difficult to detect by short-read sequencing 78. Tandem repeats are usually discarded by NGS pipelines, but bioinformatics efforts will be made to detect them. Recently, a novel intronic expansion, a pentanucleotide-repeat in SAMD12, has been identified in familial myoclonic epilepsy 78. Long-read sequencing will probably allow one to identify more neurological repeat disorders.
Therapeutic approaches in spinocerebellar ataxias
Over the last few years, several disease-modifying treatments have been tested with different outcomes. The most beneficial molecules to date are riluzole and valproic acid, whereas other drugs, such as lithium carbonate, trimethoprim-sulfamethoxazole, or zinc, have no significant effect 79. A randomized placebo-controlled clinical trial in 40 SCA patients (SCA1, 2, 6, 8, and 10) and 20 Friedreich ataxia patients treated with riluzole (100 mg/day) for 12 months revealed a one-point improvement in SARA score in treated patients compared to the placebo group 80. The use of valproic acid (1,200 mg/day) in SCA3 patients resulted in a SARA score at 12 weeks lower than that obtained for patients on placebo 81. However, further trials are required to identify the genotype associated with a positive response to riluzole (ClinicalTrials.gov Identifier: NCT03347344).
Based on the hypothesis that one of the main omega-3 polyunsaturated fatty acids of the cerebellum, docosahexaenoic acid (DHA), is present at low levels in SCA38 patients owing to ELOVL5 mutations, a clinical trial of DHA (600 mg/day) was performed 82. This drug improved SARA score at 16 weeks in a small group of SCA38 patients and it also increased cerebellar metabolism, as shown by a brain 18-fluorodeoxyglucose positron emission tomography scan at 40 weeks 82.
However, the most encouraging and innovative strategies developed to date are RNA-targeting therapies for polyQ SCAs, which have been validated in several mouse models 83, 84. These approaches mostly use antisense oligonucleotides (ASOs) to downregulate levels of the pathological polyQ protein. SCA2 mice treated with intrathecal ASOs display improvements in motor abilities, a recovery of Purkinje cell firing frequency, and lower levels of mutated ATXN2 protein 85. ASOs have also been administered to SCA3 mice and were found to be well tolerated and to decrease the production of mutated protein 86. A correlation was also found with electrophysiological changes in Purkinje cells 87. These treatments may be used in patients with polyQ SCAs at early stages of the disease, even before the onset of symptoms. The long-term effects of decreasing levels of wild-type and mutated proteins and of decreasing the levels of other polyQ proteins remain unclear. The use of allele-specific ASOs may be required 88. A therapeutic approach based on ASOs is already at an advanced stage of development for Huntington disease 89, and a phase 1A clinical trial in patients has yielded promising results 90. ASO-based treatments are also being used for spinal muscular atrophy, in a different context, to retain exon 7 and produce a full-length SMN protein, with promising results 91, 92.
The advent of disease-modifying treatments for SCAs, including ASOs, will require the rapid identification and validation of robust biomarkers of disease progression or processes for the assessment of treatment response.
Conclusion
This review summarizes recent clinical and genetic advances in dominant SCAs, including discoveries of novel SCAs, development of biomarkers, and therapeutic progress. The identification of biomarkers will be essential to demonstrate treatment efficacy. Future longitudinal studies based on multimodal approaches to elucidate the relationships between parameters will be required for the establishment of valid biomarkers. To date, gene suppression therapies are the most promising in polyQ SCAs, and clinical trials in the next few years are justified.
Abbreviations
ASO, antisense oligonucleotide; CCAS, cerebellar cognitive affective/Schmahmann’s syndrome; CCFS, Composite Cerebellar Functional Severity; CSF, cerebrospinal fluid; DHA, docosahexaenoic acid; fMRI, functional MRI; GPx, glutathione peroxidase; mGluR1, metabotropic glutamate receptor 1; MRS, MRI spectroscopy; Nfl, neurofilament light; NGS, next-generation sequencing; polyQ, polyglutamine; SARA, Scale for the Assessment and Rating of Ataxia; SCA, spinocerebellar ataxia; WES, whole-exome sequencing.
Acknowledgments
We thank the patients’ associations for their continuous support of our research: Association Connaitre les Syndromes Cérébelleux, Association Strumpell-Lorrain, Association Française de l’Ataxie de Friedreich, and the Tom Wahlig Foundation.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Vikram G Shakkottai, Department of Neurology, University of Michigan, Ann Arbor, MI, 48109-2200, USA
Bing-wen Soong, Department of Neurology, Shuang Ho Hospital, Taipei Medical University, Taipei, Taiwan
Harry Orr, Institute for Translational Neuroscience, University of Minnesota, Minneapolis, Minnesota, 55455, USA
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; referees: 3 approved]
References
- 1. Online Mendelian inheritance in man. OMIM Baltimore, MD: McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University. Reference Source [Google Scholar]
- 2. Ruano L, Melo C, Silva MC, et al. : The global epidemiology of hereditary ataxia and spastic paraplegia: a systematic review of prevalence studies. Neuroepidemiology. 2014;42(3):174–83. 10.1159/000358801 [DOI] [PubMed] [Google Scholar]
- 3. Durr A: Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol. 2010;9(9):885–94. 10.1016/S1474-4422(10)70183-6 [DOI] [PubMed] [Google Scholar]
- 4. MacDonald ME, Ambrose CM, Duyao MP, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72(6):971–83. 10.1016/0092-8674(93)90585-E [DOI] [PubMed] [Google Scholar]
- 5. La Spada AR, Wilson EM, Lubahn DB, et al. : Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352(6330):77–9. 10.1038/352077a0 [DOI] [PubMed] [Google Scholar]
- 6. Stevanin G, Dürr A, Brice A: Clinical and molecular advances in autosomal dominant cerebellar ataxias: From genotype to phenotype and physiopathology. Eur J Hum Genet. 2000;8(1):4–18. 10.1038/sj.ejhg.5200403 [DOI] [PubMed] [Google Scholar]
- 7. Orr HT, Zoghbi HY: Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. 10.1146/annurev.neuro.29.051605.113042 [DOI] [PubMed] [Google Scholar]
- 8. Tezenas du Montcel S, Durr A, Bauer P, et al. : Modulation of the age at onset in spinocerebellar ataxia by CAG tracts in various genes. Brain. 2014;137(Pt 9):2444–55. 10.1093/brain/awu174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tezenas du Montcel S, Durr A, Rakowicz M, et al. : Prediction of the age at onset in spinocerebellar ataxia type 1, 2, 3 and 6. J Med Genet. 2014;51(7):479–86. 10.1136/jmedgenet-2013-102200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jacobi H, Du Montcel ST, Bauer P, et al. : Long-term disease progression in spinocerebellar ataxia types 1, 2, 3, and 6: a longitudinal cohort study. Lancet Neurol. 2015;14(11):1101–8. 10.1016/S1474-4422(15)00202-1 [DOI] [PubMed] [Google Scholar]
- 11. Schmitz-Hübsch T, du Montcel ST, Baliko L, et al. : Scale for the assessment and rating of ataxia: development of a new clinical scale. Neurology. 2006;66(11):1717–20. 10.1212/01.wnl.0000219042.60538.92 [DOI] [PubMed] [Google Scholar]
- 12. Schmitz-Hübsch T, Fimmers R, Rakowicz M, et al. : Responsiveness of different rating instruments in spinocerebellar ataxia patients. Neurology. 2010;74(8):678–84. 10.1212/WNL.0b013e3181d1a6c9 [DOI] [PubMed] [Google Scholar]
- 13. Jacobi H, Reetz K, du Montcel ST, et al. : Biological and clinical characteristics of individuals at risk for spinocerebellar ataxia types 1, 2, 3, and 6 in the longitudinal RISCA study: analysis of baseline data. Lancet Neurol. 2013;12(7):650–8. 10.1016/S1474-4422(13)70104-2 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Monin ML, Tezenas du Montcel S, Marelli C, et al. : Survival and severity in dominant cerebellar ataxias. Ann Clin Transl Neurol. 2015;2(2):202–7. 10.1002/acn3.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ashizawa T, Figueroa KP, Perlman SL, et al. : Clinical characteristics of patients with spinocerebellar ataxias 1, 2, 3 and 6 in the US; a prospective observational study. Orphanet J Rare Dis. 2013;8:177. 10.1186/1750-1172-8-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Orengo JP, van der Heijden ME, Hao S, et al. : Motor neuron degeneration correlates with respiratory dysfunction in SCA1. Dis Model Mech. 2018;11(2): pii: dmm032623. 10.1242/dmm.032623 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Diallo A, Jacobi H, Cook A, et al. : Survival in patients with spinocerebellar ataxia types 1, 2, 3, and 6 (EUROSCA): a longitudinal cohort study. Lancet Neurol. 2018;17(4):327–34. 10.1016/S1474-4422(18)30042-5 [DOI] [PubMed] [Google Scholar]
- 18. du Montcel ST, Charles P, Ribai P, et al. : Composite cerebellar functional severity score: validation of a quantitative score of cerebellar impairment. Brain. 2008;131(Pt 5):1352–61. 10.1093/brain/awn059 [DOI] [PubMed] [Google Scholar]
- 19. Filipovic Pierucci A, Mariotti C, Panzeri M, et al. : Quantifiable evaluation of cerebellar signs in children. Neurology. 2015;84(12):1225–32. 10.1212/WNL.0000000000001403 [DOI] [PubMed] [Google Scholar]
- 20. Tanguy Melac A, Mariotti C, Filipovic Pierucci A, et al. : Friedreich and dominant ataxias: quantitative differences in cerebellar dysfunction measurements. J Neurol Neurosurg Psychiatry. 2018;89(6):559–65. 10.1136/jnnp-2017-316964 [DOI] [PubMed] [Google Scholar]
- 21. Reetz K, Costa AS, Mirzazade S, et al. : Genotype-specific patterns of atrophy progression are more sensitive than clinical decline in SCA1, SCA3 and SCA6. Brain. 2013;136(Pt 3):905–17. 10.1093/brain/aws369 [DOI] [PubMed] [Google Scholar]
- 22. Mascalchi M, Diciotti S, Giannelli M, et al. : Progression of brain atrophy in spinocerebellar ataxia type 2: a longitudinal tensor-based morphometry study. PLoS One. 2014;9(2):e89410. 10.1371/journal.pone.0089410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seidel K, Siswanto S, Brunt ERP, et al. : Brain pathology of spinocerebellar ataxias. Acta Neuropathol. 2012;124(1):1–21. 10.1007/s00401-012-1000-x [DOI] [PubMed] [Google Scholar]
- 24. Scherzed W, Brunt ER, Heinsen H, et al. : Pathoanatomy of cerebellar degeneration in spinocerebellar ataxia type 2 (SCA 2) and type 3 (SCA 3). Cerebellum. 2012;11(3):749–60. 10.1007/s12311-011-0340-8 [DOI] [PubMed] [Google Scholar]
- 25. Reetz K, Rodríguez-Labrada R, Dogan I, et al. : Brain atrophy measures in preclinical and manifest spinocerebellar ataxia type 2. Ann Clin Transl Neurol. 2018;5(2):128–37. 10.1002/acn3.504 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Hernandez-Castillo CR, Galvez V, Mercadillo R, et al. : Extensive White Matter Alterations and Its Correlations with Ataxia Severity in SCA 2 Patients. PLoS One. 2015;10(8):e0135449. 10.1371/journal.pone.0135449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adanyeguh IM, Perlbarg V, Henry PG, et al. : Autosomal dominant cerebellar ataxias: Imaging biomarkers with high effect sizes. Neuroimage Clin. 2018;19:858–67. 10.1016/j.nicl.2018.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martins CR, Jr, Martinez ARM, de Rezende TJR, et al. : Spinal Cord Damage in Spinocerebellar Ataxia Type 1. Cerebellum. 2017;16(4):792–6. 10.1007/s12311-017-0854-9 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Pedroso JL, de Souza PV, Pinto WB, et al. : SCA1 patients may present as hereditary spastic paraplegia and must be included in spastic-ataxias group. Parkinsonism Relat Disord. 2015;21(10):1243–6. 10.1016/j.parkreldis.2015.07.015 [DOI] [PubMed] [Google Scholar]
- 30. Fahl CN, Branco LM, Bergo FP, et al. : Spinal cord damage in Machado-Joseph disease. Cerebellum. 2015;14(2):128–32. 10.1007/s12311-014-0619-7 [DOI] [PubMed] [Google Scholar]
- 31. Joers JM, Deelchand DK, Lyu T, et al. : Neurochemical abnormalities in premanifest and early spinocerebellar ataxias. Ann Neurol. 2018;83(4):816–29. 10.1002/ana.25212 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Adanyeguh IM, Henry PG, Nguyen TM, et al. : In vivo neurometabolic profiling in patients with spinocerebellar ataxia types 1, 2, 3, and 7. Mov Disord. 2015;30(5):662–70. 10.1002/mds.26181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cocozza S, Saccà F, Cervo A, et al. : Modifications of resting state networks in spinocerebellar ataxia type 2. Mov Disord. 2015;30(10):1382–90. 10.1002/mds.26284 [DOI] [PubMed] [Google Scholar]
- 34. Guell X, Gabrieli JDE, Schmahmann JD: Triple representation of language, working memory, social and emotion processing in the cerebellum: convergent evidence from task and seed-based resting-state fMRI analyses in a single large cohort. Neuroimage. 2018;172:437–49. 10.1016/j.neuroimage.2018.01.082 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 35. Stefanescu MR, Dohnalek M, Maderwald S, et al. : Structural and functional MRI abnormalities of cerebellar cortex and nuclei in SCA3, SCA6 and Friedreich's ataxia. Brain. 2015;138(Pt 5):1182–97. 10.1093/brain/awv064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deistung A, Stefanescu MR, Ernst TM, et al. : Structural and Functional Magnetic Resonance Imaging of the Cerebellum: Considerations for Assessing Cerebellar Ataxias. Cerebellum. 2016;15(1):21–5. 10.1007/s12311-015-0738-9 [DOI] [PubMed] [Google Scholar]
- 37. Duarte JV, Faustino R, Lobo M, et al. : Parametric fMRI of paced motor responses uncovers novel whole-brain imaging biomarkers in spinocerebellar ataxia type 3. Hum Brain Mapp. 2016;37(10):3656–68. 10.1002/hbm.23266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu C, Chen DB, Feng L, et al. : Oculomotor deficits in spinocerebellar ataxia type 3: Potential biomarkers of preclinical detection and disease progression. CNS Neurosci Ther. 2017;23(4):321–8. 10.1111/cns.12676 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Raposo M, Vasconcelos J, Bettencourt C, et al. : Nystagmus as an early ocular alteration in Machado-Joseph disease (MJD/SCA3). BMC Neurol. 2014;14:17. 10.1186/1471-2377-14-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moscovich M, Okun MS, Favilla C, et al. : Clinical evaluation of eye movements in spinocerebellar ataxias: a prospective multicenter study. J Neuroophthalmol. 2015;35(1):16–21. 10.1097/WNO.0000000000000167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Velázquez-Pérez L, Rodríguez-Labrada R, Cruz-Rivas EM, et al. : Comprehensive study of early features in spinocerebellar ataxia 2: Delineating the prodromal stage of the disease. Cerebellum. 2014;13(5):568–79. 10.1007/s12311-014-0574-3 [DOI] [PubMed] [Google Scholar]
- 42. Rodríguez-Labrada R, Velázquez-Pérez L, Auburger G, et al. : Spinocerebellar ataxia type 2: Measures of saccade changes improve power for clinical trials. Mov Disord. 2016;31(4):570–8. 10.1002/mds.26532 [DOI] [PubMed] [Google Scholar]
- 43. Cunha-Santos J, Duarte-Neves J, Carmona V, et al. : Caloric restriction blocks neuropathology and motor deficits in Machado-Joseph disease mouse models through SIRT1 pathway. Nat Commun. 2016;7:11445. 10.1038/ncomms11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu YL, Chang JC, Lin WY, et al. : Caffeic acid and resveratrol ameliorate cellular damage in cell and Drosophila models of spinocerebellar ataxia type 3 through upregulation of Nrf2 pathway. Free Radic Biol Med. 2018;115:309–17. 10.1016/j.freeradbiomed.2017.12.011 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Ghorbani Z, Farahani RM, Aliaghaei A, et al. : Resveratrol Protects Purkinje Neurons and Restores Muscle Activity in Rat Model of Cerebellar Ataxia. J Mol Neurosci. 2018;65(1):35–42. 10.1007/s12031-018-1065-7 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Kazachkova N, Raposo M, Ramos A, et al. : Promoter Variant Alters Expression of the Autophagic BECN 1 Gene: Implications for Clinical Manifestations of Machado-Joseph Disease. Cerebellum. 2017;16(5–6):957–63. 10.1007/s12311-017-0875-4 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 47. Raposo M, Ramos A, Santos C, et al. : Accumulation of Mitochondrial DNA Common Deletion Since The Preataxic Stage of Machado-Joseph Disease. Mol Neurobiol. 2018;1–6. 10.1007/s12035-018-1069-x [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Pacheco LS, da Silveira AF, Trott A, et al. : Association between Machado-Joseph disease and oxidative stress biomarkers. Mutat Res. 2013;757(2):99–103. 10.1016/j.mrgentox.2013.06.023 [DOI] [PubMed] [Google Scholar]
- 49. de Assis AM, Saute JAM, Longoni A, et al. : Peripheral Oxidative Stress Biomarkers in Spinocerebellar Ataxia Type 3/Machado-Joseph Disease. Front Neurol. 2017;8:485. 10.3389/fneur.2017.00485 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Evert BO, Schelhaas J, Fleischer H, et al. : Neuronal intranuclear inclusions, dysregulation of cytokine expression and cell death in spinocerebellar ataxia type 3. Clin Neuropathol. 2006;25(6):272–81. [PubMed] [Google Scholar]
- 51. da Silva Carvalho G, Saute JAM, Haas CB, et al. : Cytokines in Machado Joseph Disease/Spinocerebellar Ataxia 3. Cerebellum. 2016;15(4):518–25. 10.1007/s12311-015-0719-z [DOI] [PubMed] [Google Scholar]
- 52. Raposo M, Bettencourt C, Ramos A, et al. : Promoter Variation and Expression Levels of Inflammatory Genes IL1A, IL1B, IL6 and TNF in Blood of Spinocerebellar Ataxia Type 3 (SCA 3) Patients. Neuromolecular Med. 2017;19(1):41–5. 10.1007/s12017-016-8416-8 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Zhang H, Wu LM, Wu J: Cross-talk between apolipoprotein E and cytokines. Mediators Inflamm. 2011;2011:949072. 10.1155/2011/949072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wilke C, Bender F, Hayer SN, et al. : Serum neurofilament light is increased in multiple system atrophy of cerebellar type and in repeat-expansion spinocerebellar ataxias: a pilot study. J Neurol. 2018;265(7):1618–24. 10.1007/s00415-018-8893-9 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Menke RA, Gray E, Lu CH, et al. : CSF neurofilament light chain reflects corticospinal tract degeneration in ALS. Ann Clin Transl Neurol. 2015;2(7):748–55. 10.1002/acn3.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Byrne LM, Rodrigues FB, Blennow K, et al. : Neurofilament light protein in blood as a potential biomarker of neurodegeneration in Huntington's disease: a retrospective cohort analysis. Lancet Neurol. 2017;16(8):601–9. 10.1016/S1474-4422(17)30124-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rossi D, Volanti P, Brambilla L, et al. : CSF neurofilament proteins as diagnostic and prognostic biomarkers for amyotrophic lateral sclerosis. J Neurol. 2018;265(3):510–21. 10.1007/s00415-017-8730-6 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Brouillette AM, Öz G, Gomez CM: Cerebrospinal Fluid Biomarkers in Spinocerebellar Ataxia: A Pilot Study. Dis Markers. 2015;2015:413098. 10.1155/2015/413098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bürk K, Globas C, Bösch S, et al. : Cognitive deficits in spinocerebellar ataxia type 1, 2, and 3. J Neurol. 2003;250(2):207–11. 10.1007/s00415-003-0976-5 [DOI] [PubMed] [Google Scholar]
- 60. Moriarty A, Cook A, Hunt H, et al. : A longitudinal investigation into cognition and disease progression in spinocerebellar ataxia types 1, 2, 3, 6, and 7. Orphanet J Rare Dis. 2016;11(1):82. 10.1186/s13023-016-0447-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jacobi H, du Montcel ST, Bauer P, et al. : Long-term evolution of patient-reported outcome measures in spinocerebellar ataxias. J Neurol. 2018;265(9):2040–51. 10.1007/s00415-018-8954-0 [DOI] [PubMed] [Google Scholar]
- 62. Fancellu R, Paridi D, Tomasello C, et al. : Longitudinal study of cognitive and psychiatric functions in spinocerebellar ataxia types 1 and 2. J Neurol. 2013;260(12):3134–43. 10.1007/s00415-013-7138-1 [DOI] [PubMed] [Google Scholar]
- 63. Pereira L, Airan RD, Fishman A, et al. : Resting-state functional connectivity and cognitive dysfunction correlations in spinocerebelellar ataxia type 6 (SCA6). Hum Brain Mapp. 2017;38(6):3001–10. 10.1002/hbm.23568 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Hoche F, Guell X, Vangel MG, et al. : The cerebellar cognitive affective/Schmahmann syndrome scale. Brain. 2018;141(1):248–70. 10.1093/brain/awx317 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Gennarino VA, Singh RK, White JJ, et al. : Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type Ataxin1 levels. Cell. 2015;160(6):1087–98. 10.1016/j.cell.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gennarino VA, Palmer EE, McDonell LM, et al. : A Mild PUM1 Mutation Is Associated with Adult-Onset Ataxia, whereas Haploinsufficiency Causes Developmental Delay and Seizures. Cell. 2018;172(5):924–936.e11. 10.1016/j.cell.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Nibbeling EAR, Duarri A, Verschuuren-Bemelmans CC, et al. : Exome sequencing and network analysis identifies shared mechanisms underlying spinocerebellar ataxia. Brain. 2017;140(11):2860–78. 10.1093/brain/awx251 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 68. Coutelier M, Coarelli G, Monin ML, et al. : A panel study on patients with dominant cerebellar ataxia highlights the frequency of channelopathies. Brain. 2017;140(6):1579–94. 10.1093/brain/awx081 [DOI] [PubMed] [Google Scholar]
- 69. Coutelier M, Blesneac I, Monteil A, et al. : A Recurrent Mutation in CACNA1G Alters Cav3.1 T-Type Calcium-Channel Conduction and Causes Autosomal-Dominant Cerebellar Ataxia. Am J Hum Genet. 2015;97(5):726–37. 10.1016/j.ajhg.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kimura M, Yabe I, Hama Y, et al. : SCA42 mutation analysis in a case series of Japanese patients with spinocerebellar ataxia. J Hum Genet. 2017;62(9):857–9. 10.1038/jhg.2017.51 [DOI] [PubMed] [Google Scholar]
- 71. Chemin J, Siquier-Pernet K, Nicouleau M, et al. : De novo mutation screening in childhood-onset cerebellar atrophy identifies gain-of-function mutations in the CACNA1G calcium channel gene. Brain. 2018;141(7):1998–2013. 10.1093/brain/awy145 [DOI] [PubMed] [Google Scholar]
- 72. Watson LM, Bamber E, Schnekenberg RP, et al. : Dominant Mutations in GRM1 Cause Spinocerebellar Ataxia Type 44. Am J Hum Genet. 2017;101(3):451–458. 10.1016/j.ajhg.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 73. Jiang T, Tan MS, Tan L, et al. : Application of next-generation sequencing technologies in Neurology. Ann Transl Med. 2014;2(12):125. 10.3978/j.issn.2305-5839.2014.11.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee H, Deignan JL, Dorrani N, et al. : Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312(18):1880–7. 10.1001/jama.2014.14604 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Shuvaev AN, Hosoi N, Sato Y, et al. : Progressive impairment of cerebellar mGluR signalling and its therapeutic potential for cerebellar ataxia in spinocerebellar ataxia type 1 model mice. J Physiol. 2017;595(1):141–64. 10.1113/JP272950 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Power EM, Morales A, Empson RM: Prolonged Type 1 Metabotropic Glutamate Receptor Dependent Synaptic Signaling Contributes to Spino-Cerebellar Ataxia Type 1. J Neurosci. 2016;36(18):4910–6. 10.1523/JNEUROSCI.3953-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 77. Bushart DD, Chopra R, Singh V, et al. : Targeting potassium channels to treat cerebellar ataxia. Ann Clin Transl Neurol. 2018;5(3):297–314. 10.1002/acn3.527 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Ishiura H, Doi K, Mitsui J, et al. : Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat Genet. 2018;50(4):581–90. 10.1038/s41588-018-0067-2 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Zesiewicz TA, Wilmot G, Kuo SH, et al. : Comprehensive systematic review summary: Treatment of cerebellar motor dysfunction and ataxia: Report of the Guideline Development, Dissemination, and Implementation Subcommittee of the American Academy of Neurology. Neurology. 2018;90(10):464–71. 10.1212/WNL.0000000000005055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Romano S, Coarelli G, Marcotulli C, et al. : Riluzole in patients with hereditary cerebellar ataxia: a randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2015;14(10):985–91. 10.1016/S1474-4422(15)00201-X [DOI] [PubMed] [Google Scholar]
- 81. Lei LF, Yang GP, Wang JL, et al. : Safety and efficacy of valproic acid treatment in SCA 3/MJD patients. Parkinsonism Relat Disord. 2016;26:55–61. 10.1016/j.parkreldis.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 82. Manes M, Alberici A, Di Gregorio E, et al. : Docosahexaenoic acid is a beneficial replacement treatment for spinocerebellar ataxia 38. Ann Neurol. 2017;82(4):615–21. 10.1002/ana.25059 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 83. Keiser MS, Kordasiewicz HB, McBride JL: Gene suppression strategies for dominantly inherited neurodegenerative diseases: lessons from Huntington's disease and spinocerebellar ataxia. Hum Mol Genet. 2016;25(R1):R53–64. 10.1093/hmg/ddv442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Matos CA, de Almeida LP, Nóbrega C: Machado-Joseph disease/spinocerebellar ataxia type 3: lessons from disease pathogenesis and clues into therapy. J Neurochem. 2018. 10.1111/jnc.14541 [DOI] [PubMed] [Google Scholar]
- 85. Scoles DR, Meera P, Schneider MD, et al. : Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature. 2017;544(7650):362–6. 10.1038/nature22044 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 86. Moore LR, Rajpal G, Dillingham IT, et al. : Evaluation of Antisense Oligonucleotides Targeting ATXN3 in SCA3 Mouse Models. Mol Ther Nucleic Acids. 2017;7:200–10. 10.1016/j.omtn.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 87. McLoughlin HS, Moore LR, Chopra R, et al. : Oligonucleotide therapy mitigates disease in spinocerebellar ataxia type 3 mice. Ann Neurol. 2018;84(1):64–77. 10.1002/ana.25264 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 88. Toonen LJA, Rigo F, van Attikum H, et al. : Antisense Oligonucleotide-Mediated Removal of the Polyglutamine Repeat in Spinocerebellar Ataxia Type 3 Mice. Mol Ther Nucleic Acids. 2017;8:232–42. 10.1016/j.omtn.2017.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 89. Skotte NH, Southwell AL, Østergaard ME, et al. : Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all Huntington disease patients. PLoS One. 2014;9(9):e107434. 10.1371/journal.pone.0107434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wild EJ, Tabrizi SJ: Therapies targeting DNA and RNA in Huntington's disease. Lancet Neurol. 2017;16(10):837–47. 10.1016/S1474-4422(17)30280-6 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 91. Finkel RS, Mercuri E, Darras BT, et al. : Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N Engl J Med. 2017;377(18):1723–32. 10.1056/NEJMoa1702752 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 92. Mercuri E, Finkel R, Kirschner J, et al. : Efficacy and safety of nusinersen in children with later-onset spinal muscular atrophy (SMA): End of study results from the phase 3 CHERISH study. Neuromuscular Disorders. 2017;27(2):S210 10.1016/j.nmd.2017.06.418 [DOI] [Google Scholar]; F1000 Recommendation