Version Changes
Revised. Amendments from Version 1
This version of the manuscript has been revised to consider the reviewers’ comments and suggestions based on the initial version1 of the paper. We have modified the main text, the figures and removed two tables that contained information or data that could be described in the main text. The main changes are: - Removed table 1 and table 2 from the main text and provided description of the same information in the main text. - Revised table 3 to reflect proposed corrections. - Revised figure 4 to reflect amended changes. - Added a new figure, Figure 5 as per reviewers’ suggestion. - Revised the initial R code to correct a bug that resulted in miscalculation of TP and TN for FreeBayes estimates. - Revised the supplementary materials to include explicit methods used in the variant calling procedures. These changes have been clarified further in responses to the reviewers’ specific comments.
Abstract
Background: High-throughput whole genome sequencing facilitates investigation of minority virus sub-populations from virus positive samples. Minority variants are useful in understanding within and between host diversity, population dynamics and can potentially assist in elucidating person-person transmission pathways. Several minority variant callers have been developed to describe low frequency sub-populations from whole genome sequence data. These callers differ based on bioinformatics and statistical methods used to discriminate sequencing errors from low-frequency variants.
Methods: We evaluated the diagnostic performance and concordance between published minority variant callers used in identifying minority variants from whole-genome sequence data from virus samples. We used the ART-Illumina read simulation tool to generate three artificial short-read datasets of varying coverage and error profiles from an RSV reference genome. The datasets were spiked with nucleotide variants at predetermined positions and frequencies. Variants were called using FreeBayes, LoFreq, Vardict, and VarScan2. The variant callers’ agreement in identifying known variants was quantified using two measures; concordance accuracy and the inter-caller concordance.
Results: The variant callers reported differences in identifying minority variants from the datasets. Concordance accuracy and inter-caller concordance were positively correlated with sample coverage. FreeBayes identified the majority of variants although it was characterised by variable sensitivity and precision in addition to a high false positive rate relative to the other minority variant callers and which varied with sample coverage. LoFreq was the most conservative caller.
Conclusions: We conducted a performance and concordance evaluation of four minority variant calling tools used to identify and quantify low frequency variants. Inconsistency in the quality of sequenced samples impacts on sensitivity and accuracy of minority variant callers. Our study suggests that combining at least three tools when identifying minority variants is useful in filtering errors when calling low frequency variants.
Keywords: variant calling, minority variants, concordance, performance, RSV
Introduction
RNA viruses have been described as a population of closely related sequences that arise from rapid genomic evolution coupled with a high replication and mutation rates ( Domingo et al., 2012; Eigen et al., 1988; Holland et al., 1992). Genetic changes in RNA viruses result from genetic drift, erroneous replication processes, mutagenic agents and upon which natural selection acts ( Moya et al., 2004). Rapid replication and mutations generate an ensemble of mutant genomes that are comprised of both dominant and low frequency variants. This diversity has been shown to affect virus fitness landscape, transmission, colonization and replication ( Henn et al., 2012; Stack et al., 2013; Vignuzzi et al., 2006).
Many recent studies ( Henn et al., 2012; Poon et al., 2016; Stack et al., 2013) have demonstrated the potential application of virus diversity to inform person-to-person transmission during virus outbreaks. A number of methods that incorporate both genomic and epidemiologic data to infer pathogen transmission have recently been developed ( Worby et al., 2017). These approaches rely partly on the accurate detection and quantification of minority variant populations from genomic samples.
Several tools have been developed to identify and quantify minority variants from short-read data ( Koboldt et al., 2009; Koboldt et al., 2012; Lai et al., 2016; Macalalad et al., 2012; Wilm et al., 2012; Yang et al., 2013). Nonetheless, these tools do not fully account for discrepancies that arise from sample collection, pre-processing and sequencing in addition to errors that are introduced during downstream bioinformatic analysis. Rigorous quality control in sample processing and analysis is often suggested to distinguish true biological variants from artefactual variants ( Zhang & Flaherty, 2017). In some cases, sequencing errors can be reduced by developing high-fidelity protocols and laboratory quality control measures ( Kinde et al., 2011; McCrone & Lauring, 2016; Watson et al., 2013). Additionally, the uncertainty resulting from random sequencing errors can be countered by sequencing larger populations at higher coverage ( Zukurov et al., 2016). A number of studies have extensively explored variants from somatic or tumour samples ( Hofmann et al., 2017; Koboldt et al., 2012; Krøigård et al., 2016; Lai et al., 2016; Pabinger et al., 2014) and their application in clinical genomics, but only a limited number of studies have explored the nature of variants from patient-derived samples that target viral populations ( Henn et al., 2012; Macalalad et al., 2012; Wilm et al., 2012; Yang et al., 2013; Zukurov et al., 2016) and especially when calling variants from respiratory viruses such as the respiratory syncytial virus (RSV).
In this study, we evaluated four published minority variant detection tools using artificial short-read data with different error profiles. We explored the tools’ ability to detect and quantify minority variants and assessed their overall agreement which we defined using two metrics, concordance accuracy, which measures the combined accuracy of the variant callers, and inter-caller concordance, which is the size of the largest set of variant callers that agree at each position. We show that concordance metrics are dependent on sample coverage and are influenced by the quality of input data.
Methods
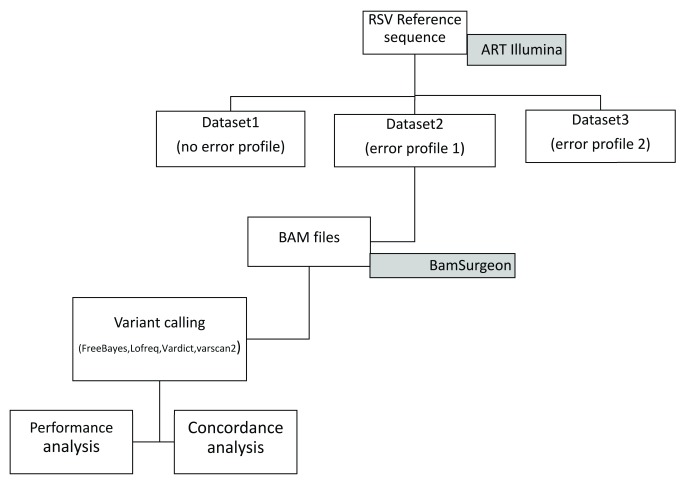
Overall, we considered ten published, open-source tools with presumed ability to call minority variants from virus deep sequence data. A number of callers were excluded from the analysis for various reasons, for example, the GATK HaplotypeCaller primarily targets germline calling from human and not variant calling from viral samples. We experienced technical difficulties in setting up the Platypus caller and even after setup, Platypus did not provide calls across all levels of coverage in our datasets. SAMtools mpileup did not provide direct allele frequencies while V-Phaser was superseded by V-Phaser 2 which has reported bugs and could not handle reads aligned with BWA-MEM. Therefore, the following four tools were evaluated, FreeBayes version 1.1.0-3-g961e5f3, LoFreq version 2.1.2, VarDict version 30.3.17 and VarScan version 2.4.2. A schematic diagram showing the overall approach is shown in Figure 1.
Figure 1. A schematic diagram showing the variant calling workflow.
The artificial datasets (BAM files) were generated using ART-Illumina based on an RSV reference genome. BAMSurgeon was used to spike the resulting BAM files by inserting known variants at known locations across the artificial BAM file.
Artificial datasets
Artificial datasets were generated based on an RSV reference sequence (GenBank accession number KX510245.1) using ART-Illumina version 2.5.8 ( Huang et al., 2012). ART-Illumina was took the reference RSV genome sequence as input and generated artificial reads using data derived error models to mimic sequence data. Each dataset comprised of eight samples with varying depth of coverage (20, 50, 100, 500, 1000, 2000, 5000, 10000) and was generated using the methods described in Supplementary File 1, section S1.1. The first dataset did not incorporate an error profile. Error profile models (empirical error models based on the distribution of base quality scores) were generated from uncompressed FastQC raw reads derived from sequenced RSV whole genome samples, referred to as “good” and a “bad” sample based on FastQC metrics ( Supplementary File 2 and Supplementary File 3) and used to generate artificial reads in dataset 2 and 3. ART-Illumina-generated artificial SAM files were converted to the BAM format, sorted and indexed using SAMtools version 1.3.1 for each dataset.
166 randomly and uniformly generated nucleotide mutations at different frequencies ( Supplementary Table 1) were inserted into each of the artificial datasets using BamSurgeon ( Ewing et al., 2015), such that a base change was made amongst the reads at each alignment position as described in Supplementary File 1, section S1.2. This process was repeated with a separate set of 155 positions that comprised a set of mutations with frequencies below 0.5 ( Supplementary Table 1).
Variant calling
The BAM files from each of the three datasets were used as input to each of the four variant callers (FreeBayes, Lofreq, VarDict and VarScan2). The default parameter options used in each tool are explicitly provided in Supplementary File 4. All output files were provided in the variant call format (VCF) or as a tabular file for the case of VarDict. The output from the VCF and tabular file was parsed and written as a comma separated (CSV) file.
Performance measures
To evaluate the performance of the variant calling algorithms, we compared the sequence generated by each variant caller vc, denoted to the gold standard "spiked" sequence, denoted S true, at each of N=15205 nucleotide positions. The accuracy of each variant caller is the normalized Hamming distance from the gold standard sequence, where d(x,y) is the standard discrete metric giving 1 when x=y, and 0 otherwise. By distinguishing between the sets of positions where variants did and did not occur in the gold standard sequence we calculated sensitivity, specificity, precision and accuracy ( Table 1).
Table 1. A breakdown of performance metrics of variant callers evaluated from first dataset that did not incorporate an error profile.
The samples represent simulated datasets of varying depth of coverage. True positive (TP), true negative (TN), false positive (FP) and false negatives (FN) were used to calculate performance metrics of each caller. FPR – False positive rate.
| Sample | Caller | TP | TN | FP | FN | Sensitivity | Specificity | Precision | FPR | Accuracy |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 (20X) | freebayes | 87 | 15018 | 21 | 79 | 0.5241 | 0.9986 | 0.8056 | 0.0014 | 0.9934 |
| lofreq | 29 | 15039 | 0 | 137 | 0.1747 | 1 | 1 | 0 | 0.991 | |
| vardict | 72 | 15039 | 0 | 94 | 0.4337 | 1 | 1 | 0 | 0.9938 | |
| varscan | 11 | 15039 | 0 | 155 | 0.0663 | 1 | 1 | 0 | 0.9898 | |
| 2 (50X) | freebayes | 118 | 14901 | 138 | 47 | 0.7152 | 0.9908 | 0.4609 | 0.00918 | 0.9878 |
| lofreq | 67 | 15039 | 0 | 99 | 0.40361 | 1 | 1 | 0 | 0.9935 | |
| vardict | 108 | 15039 | 0 | 58 | 0.6506 | 1 | 1 | 0 | 0.9962 | |
| varscan | 22 | 15039 | 0 | 144 | 0.13253 | 1 | 1 | 0 | 0.9905 | |
| 3 (100X) | freebayes | 127 | 14454 | 585 | 38 | 0.7697 | 0.9611 | 0.1784 | 0.0389 | 0.959 |
| lofreq | 57 | 15039 | 0 | 109 | 0.3434 | 1 | 1 | 0 | 0.9928 | |
| vardict | 104 | 15038 | 1 | 62 | 0.6265 | 0.9999 | 0.9905 | 6.65E-05 | 0.9959 | |
| varscan | 40 | 15039 | 0 | 126 | 0.241 | 1 | 1 | 0 | 0.9917 | |
| 4 (500X) | freebayes | 131 | 12559 | 2480 | 30 | 0.8137 | 0.8351 | 0.0502 | 0.1649 | 0.8349 |
| lofreq | 60 | 15039 | 0 | 106 | 0.3614 | 1 | 1 | 0 | 0.993 | |
| vardict | 110 | 15029 | 10 | 56 | 0.6627 | 0.9993 | 0.9167 | 6.65E-04 | 0.9957 | |
| varscan | 73 | 15039 | 0 | 93 | 0.4398 | 1 | 1 | 0 | 0.9939 | |
| 5 (1000X) | freebayes | 146 | 14414 | 625 | 20 | 0.8795 | 0.9584 | 0.1894 | 0.0416 | 0.9576 |
| lofreq | 57 | 15039 | 0 | 109 | 0.3434 | 1 | 1 | 0 | 0.9928 | |
| vardict | 109 | 15036 | 3 | 57 | 0.6567 | 0.9998 | 0.9732 | 1.99E-04 | 0.9961 | |
| varscan | 79 | 15039 | 0 | 87 | 0.4759 | 1 | 1 | 0 | 0.9943 | |
| 6 (2000X) | freebayes | 146 | 14923 | 116 | 20 | 0.8795 | 0.9923 | 0.5571 | 0.0077 | 0.9911 |
| lofreq | 70 | 15039 | 0 | 96 | 0.4217 | 1 | 1 | 0 | 0.9937 | |
| vardict | 120 | 15039 | 0 | 46 | 0.7229 | 1 | 1 | 0 | 0.997 | |
| varscan | 83 | 15039 | 0 | 83 | 0.5 | 1 | 1 | 0 | 0.9945 | |
| 7 (5000X) | freebayes | 149 | 15020 | 19 | 17 | 0.8976 | 0.9987 | 0.8869 | 0.0013 | 0.9976 |
| lofreq | 67 | 15039 | 0 | 99 | 0.40366 | 1 | 1 | 0 | 0.9935 | |
| vardict | 117 | 15036 | 3 | 49 | 0.7048 | 0.9998 | 0.975 | 1.99E-04 | 0.9966 | |
| varscan | 78 | 15039 | 0 | 88 | 0.4699 | 1 | 1 | 0 | 0.9942 | |
| 8 (10000X) | freebayes | 145 | 15022 | 17 | 21 | 0.8735 | 0.9989 | 0.8951 | 0.0011 | 0.9975 |
| lofreq | 72 | 15039 | 0 | 94 | 0.4337 | 1 | 1 | 0 | 0.9938 | |
| vardict | 118 | 15038 | 1 | 48 | 0.7108 | 0.9999 | 0.9916 | 6.65E-05 | 0.9968 | |
| varscan | 97 | 15039 | 0 | 69 | 0.5843 | 1 | 1 | 0 | 0.9955 |
Concordance analysis
We defined two concordance metrics to present the level of agreement between different callers in detecting the same variant positions in the sequence. The first concordance metric is concordance accuracy, which measures the combined accuracy of the variant callers. At the true variant position i we then which can be either 0, 1, 2, 3, 4 for each true variant position. The second concordance metric is inter-caller concordance, which is the size of the largest set of variant callers that agree at each position i, without reference to any gold standard sequence. We used both bar plots and heat maps to visualize the effect of coverage on C acc. Visualization of inter-caller concordance for variant sets was achieved using a bar plot and expounded by UpSet plots ( Lex et al., 2014) in R version 3.4.2.
Results
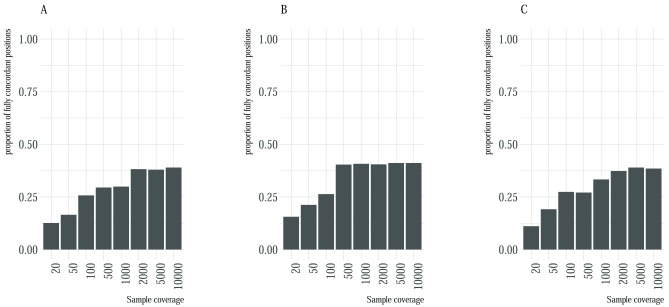
We used three artificial datasets of varying coverage and error profile to assess the concordance accuracy and inter-caller concordance for four minority variant callers. The first dataset comprised of artificial reads based on an RSV genome, the second dataset comprised of the similar simulated set of reads whilst incorporating an error profile from the set of reads used to assemble the reference genome, the third dataset was generated using an error profile from a poorly sequenced sample. Overall, concordance accuracy improved with increase in sample coverage ( Figure 2), and the proportion of positions that could not be identified by any variant caller decreased with increase in coverage ( Supplementary Figure 1). For all the three datasets, and at each coverage level, fully concordant variants were below 50% of the total variants suggesting that considering only fully concordant positions eliminated a substantial number of variant positions. There were marginal improvements in the number of concordant variants in the second dataset compared to the first and the third error profile. Across all datasets, there was little improvement at detecting fully concordant positions after a coverage of 2000 ( Figure 2). We utilized UpSetR plots to provide a visual summary of the combination of variant callers that contributed to the observed concordance accuracy (Supplementary Figure 4).
Figure 2. Proportion of fully concordant positions with respect to sample coverage.
Each plot A– C represents the proportion (y-axis) of fully concordant variants with respect to read coverage (x-axis) for the first, second and third dataset. Concordant positions were defined as positions that were identified by all the four variant callers.
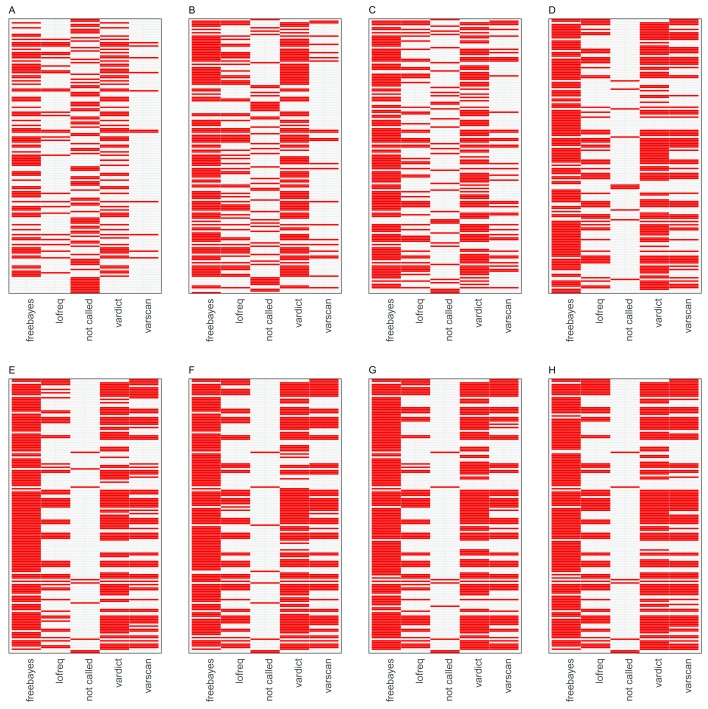
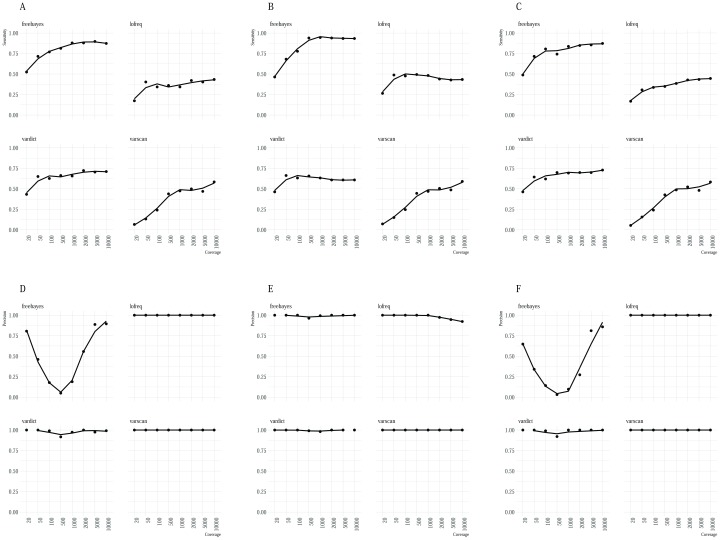
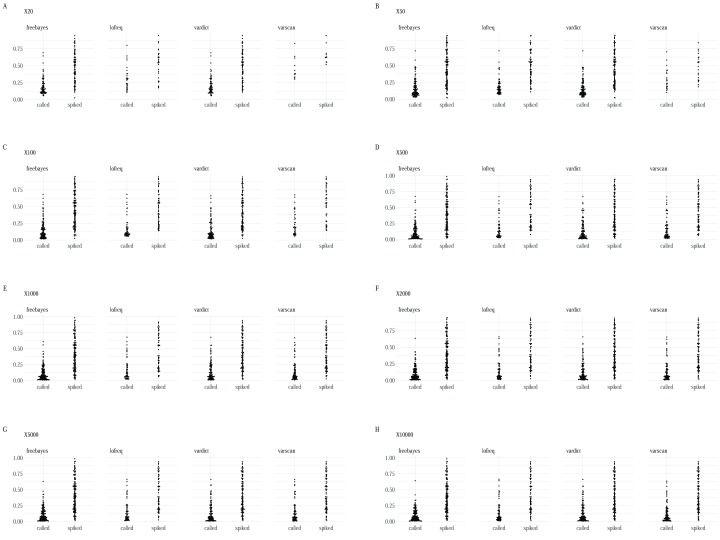
FreeBayes identified the majority of variants ( Figure 3) across all the datasets although it was characterised by a substantial trade-off between sensitivity and precision in artificial dataset 1 ( Figure 4) in addition to a high false positive rate relative to the other minority variant callers observed in datasets 1 and 3. Regardless, Freebayes reported comparatively better sensitivity relative to the rest of the tools. Lofreq was the most conservative of the evaluated callers and it missed majority of variants across all the three datasets. In addition, Lofreq’s sensitivity in coverages above 100 did not differ in a substantial way ( Figure 4). Vardict performance increased with read coverage but not by a great magnitude compared to FreeBayes and Varscan. Its performance across different datasets was more consistent relative to other tools. Overall, we observed lower reported frequency in called minority variants compared with spiked frequencies ( Figure 5). This observation was consistent in all the datasets from all the variant callers.
Figure 3. Heat maps illustrating tool specific concordance for the first artificial dataset.
The red tiles represent variants detected by each caller from the list of 166 variant positions. The panels are arranged left to right A– H in the order of increasing sample coverage (20,50,100,500,1000,2000,5000 and 10,000). The “not called” column in each panel represents the variants that were not identified by any of the variant callers.
Figure 4.
A summary of the relationship between sample coverage, sensitivity ( A– C) and sample coverage and precision ( D– F). The x-axis shows the sample coverage and the y-axis represents the sensitivity and precision respectively. Sensitivity of the callers rose gradually from low to high coverage samples. Again, precision was variable for FreeBayes calls while relatively high for the rest of the three callers.
Discussion
Detecting and reporting minority variant calls is challenging, given that low frequency calls occur at a frequency that is the same as error generated from sequencing and PCR reactions. Recent studies have linked the sharing of minority variants with transmission patterns, and hence it is important to distinguish actual minor variants from spurious variant calls. Several minority variants callers use different detection algorithms and statistics, each of which attempt to optimize an aspect of the variant calling process. Therefore, there could be disparities between what is reported by a given minority variant caller, given datasets of varying sequencing depths and error profiles.
This study aimed to identify the proportion of positions that were recognized as variants by a set of tools using three artificial datasets of varying coverage and error profiles. Concordance accuracy and inter-caller concordance measures were dependent on the sample coverage and error profile.
Sensitivity for the majority of the tools was positively correlated with depth of coverage and similarly observed previously ( Spencer et al., 2014) in a study that investigated performance in methods used to detect low-frequency variants. It is important to note that the tools provide different performance metrics depending on the variant’s threshold ( Supplementary File 5). In the first artificial dataset, VarDict detected true positive variants with comparably good performance (sensitivity 43.4% – 72.3%), though it was marginally invariant to changes in average coverage above 20. VarDict has in-built features that could contribute to its efficient performance. It is able to activate an “amplicon calling mode” that filters out amplicon biased variants and mispaired primers as PCR artefacts. A similar pattern was observed with LoFreq, where sensitivity was not significantly affected by depth of coverage. VarScan2 was more affected by coverage and maintained average sensitivity (6.6% – 58.4%). Applying filters in VarScan has been reported to improve sensitivity by reducing number of false positives ( Hofmann et al., 2017; Koboldt et al., 2013). FreeBayes’ trade-off between sensitivity and precision was also reported by other studies ( Hwang et al., 2015; Sandmann et al., 2017). The use of caller-specified filters could have enhanced the sensitivity of some callers, but the option to adopt default parameters allows equivalent assessment of tool performance. Moreover, all possible combinations of tuning parameters are challenging, time-consuming and sometimes impractical.
Based on artificial reads from the second dataset, FreeBayes performed comparatively better than the other tools with a very low false positive rate and better sensitivity (46.4% – 94.6%). This suggests that FreeBayes is potentially useful in identifying minority variants when sample data comes from reads with a low error profile. This implies the error rate results are outcome of tool performance.
Specificity of a caller is its ability to correctly predict the absence of a variant. The variant callers make use of a high specificity to minimize the number of false positive calls thereby reducing post-call filtering and consequently filter out true low-frequency variants. Moreover, high accuracy measures demonstrate the reliability of the variant caller in correctly identifying true variants.
All the minority variant callers reported slightly lower frequencies in called variants compared to the frequencies in the original spiked variants ( Figure 5). This could be explained by the fact that many of the callers are tuned to report lower differences in the calls owing to stringent pre-processing criteria. A thorough investigation of this observation is therefore required.
Figure 5.
Box plots showing the distribution of frequencies between the spiked variants and the corresponding called variant for each variant caller at each coverage ( A– H) for the first dataset.
In absence of an explicit error model from samples of heterogeneous sequencing quality, combining at least three tools when identifying minority variants could potentially assist in filtering out errors from low frequency variants. Given that there are no definitive data and next generation sequencing pipeline standards for variant calling approaches that are specific for viruses, there are opportunities to develop robust methods and tools that strike a balance between detecting errors and true minority variants from field virus samples that present with different sequencing quality.
Data availability
The data analysis scripts and datasets used in analysis are available from our institutional Dataverse repository: http://dx.doi.org/10.7910/DVN/ZIO43M ( Mohammed & Githinji, 2018)
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Funding Statement
The work was funded by the Wellcome Trust Senior Investigator Award to Prof D. James Nokes [102975] in addition, this work was supported through the DELTAS Africa Initiative [DEL-15-003]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)'s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa's Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [107769] and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 2 approved]
Supplementary material
Supplementary File 1: A description of data generation methods and command line tools used to create the artificial datasets.Supplementary File 5: Figures showing the performance of variant calling for minority variants that were inserted at different thresholds, standard (>25%), moderate (5-25%) and low (<5%).
Supplementary Figure 1: Proportion of fully concordant positions with respect to sample coverage in dataset1. (A) Represents the proportion (y-axis) of fully concordant variants with respect to read coverage (x-axis) and (B) shows the proportion of variants positions that could not be identified by any minority caller at each level of sample coverage.
Supplementary Figure 2: UpSetR plots showing the concordance between called variants and the respective variant callers using the first artificial dataset. The intersection size illustrates the number of variants in each intersection set. The horizontal axis shows the combination matrix identifying the intersections. A single filled circle represents a unique set of variants. Connected lines depict shared variants (intersections) among the variant callers. Intersection size was positively correlated with average coverage at (a) 20X, (b) 50X, (c) 100X, (d) 500X, (e) 1000X, (f) 2000X, (g) 5000X and (h) 10,000X.
Supplementary Tables 1-4.
Supplementary Table 1: 166 artificially generated nucleotide mutations with frequencies from 0-1.
Supplementary Table 2: 156 artificially generated nucleotide mutations with frequencies below 0.5.
Supplementary Table 3: A breakdown of performance metrics of variant callers evaluated using the second dataset incorporated with an error profile derived from the set of reads used to assemble the reference genome. The samples represent simulated dataset of varying average depth of coverage. True positive (TP), true negative (TN), false positive (FP) and false negatives (FN) were used to calculate performance metrics of each caller. FPR – False positive rate.
Supplementary Table 4: A breakdown of performance metrics of variant callers evaluated using the third dataset generated with an error profile from a poorly sequenced sample. The samples represent simulated dataset of varying average depth of coverage. True positive (TP), true negative (TN), false positive (FP) and false negatives (FN) were used to calculate performance metrics of each caller. FPR – False positive rate.
References
- Domingo E, Sheldon J, Perales C: Viral quasispecies evolution. Microbiol Mol Biol Rev. 2012;76(2):159–216. 10.1128/mmbr.05023-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigen M, McCaskill J, Schuster P: Molecular Quasi-Species. J Phys Chem. 1988;92(24):6881–6891. 10.1021/j100335a010 [DOI] [Google Scholar]
- Ewing AD, Houlahan KE, Hu Y, et al. : Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat Methods. 2015;12(7):623–630. 10.1038/nmeth.3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn MR, Boutwell CL, Charlebois P, et al. : Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 2012;8(3):e1002529. 10.1371/journal.ppat.1002529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AL, Behr J, Singer J, et al. : Detailed simulation of cancer exome sequencing data reveals differences and common limitations of variant callers. BMC Bioinformatics. 2017;18(1):8. 10.1186/s12859-016-1417-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JJ, De La Torre JC, Steinhauer DA: RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. 10.1007/978-3-642-77011-1_1 [DOI] [PubMed] [Google Scholar]
- Huang HW, NISC Comparative Sequencing Program, Mullikin JC, et al. : Evaluation of variant detection software for pooled next-generation sequence data. BMC Bioinformatics. 2015;16:235. 10.1186/s12859-015-0624-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Li L, Myers JR, et al. : ART: a next-generation sequencing read simulator. Bioinformatics. 2012;28(4):593–594. 10.1093/bioinformatics/btr708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Kim E, Lee I, et al. : Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep. 2015;5:17875. 10.1038/srep17875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde I, Wu J, Papadopoulos N, et al. : Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108(23):9530–9535. 10.1073/pnas.1105422108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, et al. : VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25(17):2283–2285. 10.1093/bioinformatics/btp373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Larson DE, Wilson RK: Using VarScan 2 for Germline Variant Calling and Somatic Mutation Detection. Curr Protoc Bioinformatics. 2013;44:15.4.1–17. 10.1002/0471250953.bi1504s44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, et al. : VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krøigård AB, Thomassen M, Laenkholm AV, et al. : Evaluation of Nine Somatic Variant Callers for Detection of Somatic Mutations in Exome and Targeted Deep Sequencing Data. PLoS One. 2016;11(3):e0151664. 10.1371/journal.pone.0151664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Markovets A, Ahdesmaki M, et al. : VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44(11):e108. 10.1093/nar/gkw227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A, Gehlenborg N, Strobelt H, et al. : UpSet: Visualization of Intersecting Sets. IEEE Trans Vis Comput Graph. 2014;20(12):1983–1992. 10.1109/tvcg.2014.2346248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macalalad AR, Zody MC, Charlebois P, et al. : Highly sensitive and specific detection of rare variants in mixed viral populations from massively parallel sequence data. PLoS Comput Biol. 2012;8(3):e1002417. 10.1371/journal.pcbi.1002417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrone JT, Lauring AS: Measurements of Intrahost Viral Diversity Are Extremely Sensitive to Systematic Errors in Variant Calling. J Virol. 2016;90(15):6884–6895. 10.1128/jvi.00667-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed KS, Githinji G: Replication Data for: Evaluating the Performance of Tools Used to Call Minority Variants from Whole Genome Short-Read Data. Harvard Dataverse, V3. 2018. 10.7910/DVN/ZIO43M [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Holmes EC, González-Candelas F: The population genetics and evolutionary epidemiology of RNA viruses. Nat Rev Microbiol. 2004;2(4):279–288. 10.1038/nrmicro863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabinger S, Dander A, Fischer M, et al. : A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform. 2014;15(2):256–278. 10.1093/bib/bbs086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon LL, Song T, Rosenfeld R, et al. : Quantifying influenza virus diversity and transmission in humans. Nat Genet. 2016;48(2):195–200. 10.1038/ng.3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann S, de Graaf AO, Karimi M, et al. : Evaluating Variant Calling Tools for Non-Matched Next-Generation Sequencing Data. Sci Rep. 2017;7:43169. 10.1038/srep43169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer DH, Tyagi M, Vallania F, et al. : Performance of common analysis methods for detecting low-frequency single nucleotide variants in targeted next-generation sequence data. J Mol Diagn. 2014;16(1):75–88. 10.1016/j.jmoldx.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JC, Murcia PR, Grenfell BT, et al. : Inferring the inter-host transmission of influenza A virus using patterns of intra-host genetic variation. Proc Biol Sci. 2013;280(1750):20122173. 10.1098/rspb.2012.2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, et al. : Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439(7074):344–348. 10.1038/nature04388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson SJ, Welkers MR, Depledge DP, et al. : Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos Trans R Soc Lond B Biol Sci. 2013;368(1614):20120205. 10.1098/rstb.2012.0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm A, Aw PP, Bertrand D, et al. : LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40(22):11189–11201. 10.1093/nar/gks918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CJ, Lipsitch M, Hanage WP: Shared Genomic Variants: Identification of Transmission Routes Using Pathogen Deep-Sequence Data. Am J Epidemiol. 2017;186(10):1209–1216. 10.1093/aje/kwx182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Charlebois P, Macalalad A, et al. : V-Phaser 2: variant inference for viral populations. BMC Genomics. 2013;14:674. 10.1186/1471-2164-14-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Flaherty P: Variational inference for rare variant detection in deep, heterogeneous next-generation sequencing data. BMC Bioinformatics. 2017;18(1):45. 10.1186/s12859-016-1451-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukurov JP, do Nascimento-Brito S, Volpini AC, et al. : Estimation of genetic diversity in viral populations from next generation sequencing data with extremely deep coverage. Algorithms Mol Biol. 2016;11:2. 10.1186/s13015-016-0064-x [DOI] [PMC free article] [PubMed] [Google Scholar]