Version Changes
Revised. Amendments from Version 1
This revised version contains modifications to tables 1 & 2 and figures 1 & 7 to provide greater clarity on these data sets. We have highlighted how our study was undertaken across diverse malaria endemic countries beyond West Africa and the revised manuscript contains minor editing (including the addition of primer sequences) that was suggested by the reviewers. In addition, we have modified our discussion on the correlation between Plasmodium and Wolbachia prevalence in An. gambiae s.s. to provide a more balanced viewpoint on our data.
Abstract
Background: Wolbachia, a common insect endosymbiotic bacterium that can influence pathogen transmission and manipulate host reproduction, has historically been considered absent from the Anopheles (An.) genera, but has recently been found in An. gambiae s.l. populations in West Africa. As there are numerous Anopheles species that have the capacity to transmit malaria, we analysed a range of species across five malaria endemic countries to determine Wolbachia prevalence rates, characterise novel Wolbachia strains and determine any correlation between the presence of Plasmodium, Wolbachia and the competing bacterium Asaia.
Methods: Anopheles adult mosquitoes were collected from five malaria-endemic countries: Guinea, Democratic Republic of the Congo (DRC), Ghana, Uganda and Madagascar, between 2013 and 2017. Molecular analysis was undertaken using quantitative PCR, Sanger sequencing, Wolbachia multilocus sequence typing (MLST) and high-throughput amplicon sequencing of the bacterial 16S rRNA gene.
Results: Novel Wolbachia strains were discovered in five species: An. coluzzii, An. gambiae s.s., An. arabiensis, An. moucheti and An. species A, increasing the number of Anopheles species known to be naturally infected. Variable prevalence rates in different locations were observed and novel strains were phylogenetically diverse, clustering with Wolbachia supergroup B strains. We also provide evidence for resident strain variants within An. species A. Wolbachia is the dominant member of the microbiome in An. moucheti and An. species A but present at lower densities in An. coluzzii. Interestingly, no evidence of Wolbachia/Asaia co-infections was seen and Asaia infection densities were shown to be variable and location dependent.
Conclusions: The important discovery of novel Wolbachia strains in Anopheles provides greater insight into the prevalence of resident Wolbachia strains in diverse malaria vectors. Novel Wolbachia strains (particularly high-density strains) are ideal candidate strains for transinfection to create stable infections in other Anopheles mosquito species, which could be used for population replacement or suppression control strategies.
Keywords: Wolbachia, mosquitoes, malaria, Anopheles, Asaia, endosymbionts
Background
Malaria is a mosquito-borne disease caused by infection with Plasmodium ( P.) parasites, with transmission to humans occurring through the inoculation of Plasmodium sporozoites during blood-feeding of an infectious female Anopheles ( An.) mosquito. The genus Anopheles consists of 475 formally recognised species with ~40 vector species/species complexes responsible for the transmission of malaria at a level of public health concern 1. During the mosquito infection cycle, Plasmodium parasites encounter a variety of resident microbiota both in the mosquito midgut and other tissues. Numerous studies have shown that certain species of bacteria can inhibit Plasmodium development 2– 4. For example, Enterobacter bacteria that reside in the Anopheles midgut can inhibit the development of Plasmodium parasites prior to their invasion of the midgut epithelium 5, 6. Wolbachia endosymbiotic bacteria are estimated to naturally infect ~40% of insect species 7 including mosquito vector species that are responsible for transmission of human diseases, such as Culex (Cx.) quinquefasciatus 8– 10 and Aedes (Ae.) albopictus 11, 12. Although Wolbachia strains have been shown to have variable effects on arboviral infections in their native mosquito hosts 13– 15, transinfected Wolbachia strains have been considered for mosquito biocontrol strategies, due to observed arbovirus transmission blocking abilities and a variety of synergistic phenotypic effects. Transinfected strains in Ae. aegypti and Ae. albopictus provide strong inhibitory effects on arboviruses, with maternal transmission and cytoplasmic incompatibility enabling introduced strains to spread through populations 16– 22. Open releases of Wolbachia-transinfected Ae. aegypti populations have demonstrated the ability of the wMel Wolbachia strain to invade wild populations 23 and provide strong inhibitory effects on viruses from field populations 24, with releases currently occurring in arbovirus endemic countries such as Indonesia, Vietnam, Brazil and Colombia ( https://www.worldmosquitoprogram.org).
The prevalence of Wolbachia in Anopheles species has not been extensively studied, with most studies focused in Asia using classical PCR-based screening; up until 2014 there was no evidence of resident strains in mosquitoes from this genus 25– 29. Furthermore, significant efforts to establish artificially infected lines were, up until recently, also unsuccessful 30. Somatic, transient infections of the Wolbachia strains wMelPop and wAlbB in An. gambiae were shown to significantly inhibit P. falciparum 31, but the interference phenotype is variable with other Wolbachia strain-parasite combinations 32– 34. A stable line was established in An. stephensi, a vector of malaria in southern Asia, using the wAlbB strain and this was also shown to confer resistance to P. falciparum infection 35. One potential reason postulated for the absence of Wolbachia in Anopheles species was thought to be the presence of other bacteria, particularly from the genus Asaia 36. This acetic acid bacterium is stably associated with several Anopheles species and is often the dominant species in the mosquito microbiota 37. In laboratory studies, Asaia has been shown to impede the vertical transmission of Wolbachia in Anopheles 36 and was shown to have a negative correlation with Wolbachia in mosquito reproductive tissues 38.
Recently, resident Wolbachia strains (those naturally present in wild insect populations) have been discovered in the An. gambiae s.l. complex, which consists of multiple morphologically indistinguishable species including several major malaria vector species. Wolbachia strains (collectively named wAnga) were found in An. gambiae s.l. populations in Burkina Faso 39 and Mali 40, suggesting that Wolbachia may be more abundant in the An. gambiae complex across Sub-Saharan Africa. Globally, there is a large variety of Anopheles vector species (~70) that have the capacity to transmit malaria 41 and could potentially contain resident Wolbachia strains. Additionally, this number of malaria vector species may be an underestimate given that recent studies using molecular barcoding have also revealed a larger diversity of Anopheles species than would be identified using morphological identification alone 42, 43.
Investigating the prevalence and diversity of Wolbachia strains naturally present in Anopheles populations across diverse malaria endemic countries would allow a greater understanding of how this bacterium could be influencing malaria transmission in field populations and identify candidate strains for transinfection. In this study, we collected Anopheles mosquitoes from five malaria-endemic countries; Ghana, Democratic Republic of the Congo (DRC), Guinea, Uganda and Madagascar, from 2013–2017. Wild-caught adult female Anopheles were screened for P. falciparum malaria parasites, Wolbachia and Asaia bacteria. In total, we analysed mosquitoes from 17 Anopheles species that are known malaria vectors or implicated in transmission, and some unidentified species, discovering five species of Anopheles with resident Wolbachia strains; An. coluzzii from Ghana, An. gambiae s.s., An. arabiensis, An. moucheti and An. species A from DRC. Using Wolbachia gene sequencing, including multilocus sequence typing (MLST), we show that the resident strains in these malaria vectors are diverse, novel strains and quantitative PCR (qPCR) and 16S rRNA amplicon sequencing data suggests that the strains in An. moucheti and An. species A are higher density infections, compared to the strains found in the An. gambiae s.l. complex. We found no evidence for either Wolbachia- Asaia co-infections, or for either bacteria having any significant effect on the prevalence of Plasmodium in wild mosquito populations.
Methods
Study sites & collection methods
Anopheles adult mosquitoes were collected from five malaria-endemic countries in Sub-Saharan Africa (Guinea, Democratic Republic of the Congo (DRC), Ghana, Uganda and Madagascar) between 2013 and 2017 ( Figure 1). Human landing catches, Centers for Disease Control (CDC) light traps and pyrethrum spray catches were undertaken between April 2014 and February 2015 in 10 villages near four cities in Guinea; Foulayah (10.144633, -10.749717) and Balayani (10.1325, -10.7443) near Faranah; Djoumaya (10.836317, -14.2481) and Kaboye Amaraya (10.93435, -14.36995) near Boke; Tongbekoro (9.294295, -10.147953), Keredou (9.208919, -10.069525), and Gbangbadou (9.274363, -9.998639) near Kissidougou; and Makonon (10.291124, -9.363358), Balandou (10.407669, -9.219096), and Dalabani (10.463692, -9.451904) near Kankan. Human landing catches and pyrethrum spray catches were undertaken between January and September 2015 in seven sites of the DRC; Kinshasa (-4.415881, 15.412188), Mikalayi (-6.024184, 22.318251), Kisangani (0.516350, 25.221176), Katana (-2.225129, 28.831604), Kalemie (-5.919054, 29.186572), and Kapolowe (-10.939802, 26.952970). We also analysed a subset from collections obtained from Lwiro (-2.244097, 28.815232), a village near Katana, collected between September and October 2015. A combination of CDC light traps, pyrethrum spray catches and human landing catches were undertaken in Butemba, Kyankwanzi District in mid-western Uganda (1.1068444, 31.5910085) in August and September 2013, and June 2014. CDC light trap catches were undertaken in May 2017 in Dogo in Ada, Greater Accra, Ghana (5.874861111, 0.560611111). In Madagascar, sampling was undertaken in June 2016 at four sites: Anivorano Nord, located in the Northern domain, (-12.7645000, 49.2386944); Ambomiharina, Western domain, (-16.3672778, 46.9928889); Antafia, Western domain, (-17.0271667, 46.7671389); and Ambohimarina, Central domain, (-18.3329444, 47.1092500). Trapping consisted of CDC light traps and a net trap baited with Zebu (local species of cattle) to attract zoophilic species 44. Coordinate values for all locations are latitude and longitude respectively, in decimal degrees.
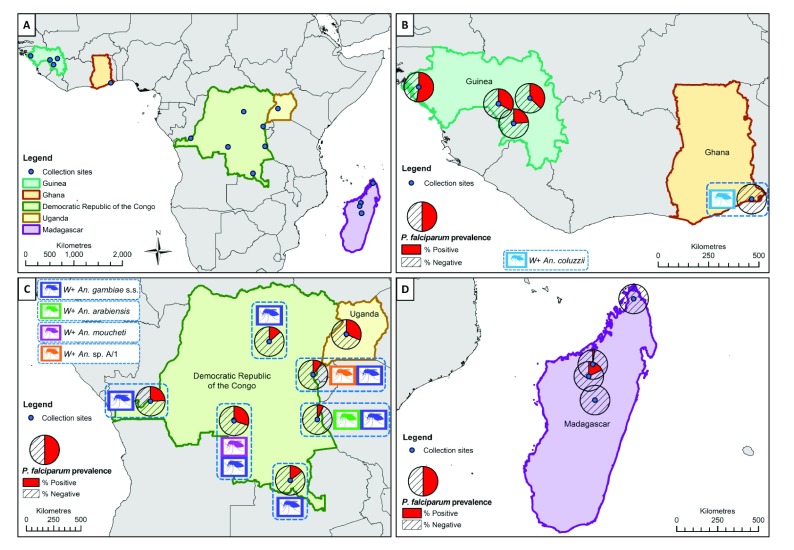
Figure 1. Locations of Anopheles species collections (including Wolbachia-infected species) and P. falciparum malaria prevalence rates in mosquitoes (across all species for each location).
( A) Overall map showing the five malaria-endemic countries where mosquito collections were undertaken. ( B) High P. falciparum prevalence rates in mosquitoes from Guinea, and Wolbachia-infected An. coluzzii from Ghana (no P. falciparum detected). ( C) Wolbachia strains in An. gambiae s.s., An. arabiensis, An. species A and An. moucheti from the Democratic Republic of the Congo (DRC) and variable P. falciparum prevalence rates in mosquitoes from DRC and Uganda. ( D) Low P. falciparum infection rates in mosquitoes from Madagascar and no evidence of resident Wolbachia strains. ( W+; Wolbachia detected in this species). Maps were generated using ArcMap ™ within the ArcGIS 10.5 software package (Esri ®, Redlands CA, USA, http://www.esri.com). Maps were constructed using country outline (level 0) data downloaded from the Database of Global Administrative Areas (GADM) ( http://www.gadm.org) (release number 2.8) for both the world, and each country of interest. The coloured mosquito icons were generated by the authors themselves (CLJ).
DNA extraction and mosquito species identification
DNA was extracted from individual whole mosquitoes or abdomens using QIAGEN DNeasy Blood and Tissue Kits according to manufacturer’s instructions. DNA extracts were eluted in a final volume of 100 μl and stored at −20°C. Mosquito species identification was initially undertaken using morphological keys followed by diagnostic species-specific PCR assays to distinguish between the morphologically indistinguishable sibling mosquito species of the An. gambiae 45– 47 and An. funestus complexes 48. To determine species identification for samples of interest and for samples that could not be identified by species-specific PCR, Sanger sequences were generated from ITS2 PCR products 49.
Detection of P. falciparum and Asaia
Detection of P. falciparum malaria was undertaken using qPCR targeting an 120-bp sequence of the P. falciparum cytochrome c oxidase subunit 1 ( Cox1) mitochondrial gene using primers 5’-TTACATCAGGAATGTTATTGC-3’ and 5’-ATATTGGATCTCCTGCAAAT-3’ 50. Positive controls from gDNA extracted from a cultured P. falciparum-infected blood sample (parasitaemia of ~10%) were serially diluted to determine the threshold limit of detection, in addition to the inclusion of no template controls (NTCs). Asaia detection was undertaken targeting the 16S rRNA gene using primers Asafor: 5’-GCGCGTAGGCGGTTTACAC-3’ and Asarev: 5’-AGCGTCAGTAATGAGCCAGGTT-3’ 37, 51. Ct values for both P. falciparum and Asaia assays in selected An. gambiae extracts were normalized to Ct values for a single copy An. gambiae rps17 housekeeping gene using primers 5’-GACGAAACCACTGCGTAACA-3’ and 5’-TGCTCCAGTGCTGAAACATC-3’ (accession no. AGAP004887 on www.vectorbase.org) 52, 53. As Ct values are inversely related to the amount of amplified DNA, a higher target gene Ct: host gene Ct ratio represented a lower estimated infection level. qPCR reactions were prepared using 5 μl of FastStart SYBR Green Master mix (Roche Diagnostics), a final concentration of 1 µM of each primer, 1 μl of PCR grade water and 2 μl template DNA, to a final reaction volume of 10 μl. Prepared reactions were run on a Roche LightCycler® 96 System and amplification was followed by a dissociation curve (95°C for 10 seconds, 65°C for 60 seconds and 97°C for 1 second) to ensure the correct target sequence was being amplified. PCR results were analysed using the LightCycler® 96 software (Roche Diagnostics). A sub-selection of PCR products from each assay was sequenced to confirm correct amplification of the target gene fragment.
Wolbachia detection
Wolbachia detection was first undertaken targeting three conserved Wolbachia genes previously shown to amplify a wide diversity of strains; 16S rRNA gene using primers W-Spec-16S-F: 5’-CATACCTATTCGAAGGGATA-3’ and W-Spec-16s-R: 5’-AGCTTCGAGTGAAACCAATTC-3’ 40, 54, Wolbachia surface protein ( wsp) gene using primers wsp81F: 5’-TGGTCCAATAAGTGATGAAGAAAC-3’ and wsp691R: 5’-AAAAATTAAACGCTACTCCA-3’ 55 and FtsZ cell cycle gene using primers ftsZqPCR F: 5’-GCATTGCAGAGCTTGGACTT-3’ and ftsZqPCR R: 5’-TCTTCTCCTTCTGCCTCTCC-3’ 56. DNA extracted from a Drosophila melanogaster fly (infected with the wMel strain of Wolbachia) was used as a positive control, in addition to no template controls (NTCs). The 16S rRNA 54 and wsp 55 gene PCR reactions were carried out in a Bio-Rad T100 Thermal Cycler using standard cycling conditions and PCR products were separated and visualised using 2% E-Gel EX agarose gels (Invitrogen) with SYBR safe and an Invitrogen E-Gel iBase Real-Time Transilluminator. FtsZ 56 and 16S rRNA 40 gene real time PCR reactions were prepared using 5 μl of FastStart SYBR Green Master mix (Roche Diagnostics), a final concentration of 1 µM of each primer, 1 μl of PCR grade water and 2 μl template DNA, to a final reaction volume of 10 μl. Prepared reactions were run on a Roche LightCycler® 96 System for 15 minutes at 95°C, followed by 40 cycles of 95°C for 15 seconds and 58°C for 30 seconds. Amplification was followed by a dissociation curve (95°C for 10 seconds, 65°C for 60 seconds and 97°C for 1 second) to ensure the correct target sequence was being amplified. PCR results were analysed using the LightCycler® 96 software (Roche Diagnostics). To estimate Wolbachia densities across multiple Anopheles mosquito species, ftsZ and 16S qPCR Ct values were compared to total dsDNA extracted, measured using an Invitrogen Qubit 4 fluorometer. A serial dilution series of a known Wolbachia-infected mosquito DNA extract was used to correlate Ct values and amount of amplified target product.
Wolbachia multilocus strain typing (MLST)
MLST was undertaken to characterize Wolbachia strains using the sequences of five conserved genes as molecular markers to genotype each strain. In brief, 450–500 base pair fragments of the gatB, coxA, hcpA, ftsZ and fbpA Wolbachia genes were amplified from individual Wolbachia-infected mosquitoes using previously optimised protocols 57. Primers used were as follows: gatB_F1: 5’-GAKTTAAAYCGYGCAGGBGTT-3’, gatB_R1: 5’-TGGYAAYTCRGGYAAAGATGA-3’, coxA_F1: 5’-TTGGRGCRATYAACTTTATAG-3’, coxA_R1: 5’-CTAAAGACTTTKACRCCAGT-3’, hcpA_F1: 5’-GAAATARCAGTTGCTGCAAA-3’, hcpA_R1: 5’-GAAAGTYRAGCAAGYTCTG-3’, ftsZ_F1: 5’-ATYATGGARCATATAAARGATAG-3’, ftsZ_R1: 5’-TCRAGYAATGGATTRGATAT-3’, fbpA_F1: 5’-GCTGCTCCRCTTGGYWTGAT-3’ and fbpA_R1: 5’-CCRCCAGARAAAAYYACTATTC-3’. A Cx. pipiens gDNA extraction (previously shown to be infected with the wPip strain of Wolbachia) was used as a positive control for each PCR run, in addition to no template controls (NTCs). If initial amplification with these primers was unsuccessful, the PCR was repeated using the standard primers but with the addition of M13 adaptors. If no amplification was detected using standard primers, further PCR analysis was undertaken using degenerate primer sets, with or without M13 adaptors, which for the hcpA gene of wAnga-Ghana allowed improved amplification (using hcpA_F3: 5’-ATTAGAGAAATARCAGTTGCTGC-3’, hcpA_R3: 5’-CATGAAAGACGAGCAARYTCTGG-3’ (no M13 adaptors)) 57. PCR products were separated and visualised using 2% E-Gel EX agarose gels (Invitrogen) with SYBR safe and an Invitrogen E-Gel iBase Real-Time Transilluminator. PCR products were submitted to Source BioScience (Source BioScience Plc, Nottingham, UK) for PCR reaction clean-up, followed by Sanger sequencing to generate both forward and reverse reads. Where PCR primers included M13 adaptors, just the M13 primers alone (M13_adaptor_F: 5’-TGTAAAACGACGGCCAGT-3’ and M13_adaptor_R: 5’-CAGGAAACAGCTATGACC-3’) were used for sequencing, otherwise the same primers as utilised for PCR were used. Sequencing analysis was carried out in MEGA7 58 as follows. Both chromatograms (forward and reverse traces) from each sample were manually checked, edited, and trimmed as required, followed by alignment with ClustalW and checking to produce consensus sequences. Consensus sequences were used to perform nucleotide BLAST (NCBI) database queries, and searches against the Wolbachia MLST database 59. If a sequence produced an exact match in the MLST database we assigned the appropriate allele number, otherwise we obtained a new allele number for each novel gene locus sequence through submission of the FASTA and raw trace files on the Wolbachia MLST website for new allele assignment and inclusion within the database. Full consensus sequences were also submitted to GenBank and assigned accession numbers. The Sanger sequencing traces from the wsp gene were also treated in the same way and analysed alongside the MLST gene locus scheme, as an additional marker for strain typing.
Phylogenetic analysis
Alignments were constructed in MEGA7 by ClustalW to include all relevant and available sequences highlighted through searches on the BLAST and Wolbachia MLST databases. Maximum Likelihood phylogenetic trees were constructed from Sanger sequences as follows. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model 60. The tree with the highest log likelihood in each case is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. The phylogeny test was by Bootstrap method with 1000 replications. Evolutionary analyses were conducted in MEGA7 58.
Microbiome analysis
The microbiomes of selected individual Anopheles were analysed using barcoded high-throughput amplicon sequencing of the bacterial 16S rRNA gene. Sequencing libraries for each isolate were generated using universal 16S rRNA V3-V4 region primers 61 in accordance with Illumina 16S rRNA metagenomic sequencing library protocols. The samples were barcoded for multiplexing using Nextera XT Index Kit v2. Sequencing was performed on an Illumina MiSeq instrument using a MiSeq Reagent Kit v2 (500-cycles). Quality control and taxonomical assignment of the resultant reads were performed using CLC Genomics Workbench 8.0.1 Microbial Genomics Module. Low quality reads containing nucleotides with quality threshold below 0.05 (using the modified Richard Mott algorithm), as well as reads with two or more unknown nucleotides were removed from analysis. Additionally, reads were trimmed to remove sequenced Nextera adapters. Reference-based operational taxonomic unit (OTU) picking was performed using the SILVA SSU v128 97% database 62. Sequences present in more than one copy but not clustered to the database were then placed into de novo OTUs (97% similarity) and aligned against the reference database with 80% similarity threshold to assign the “closest” taxonomical name where possible. Chimeras were removed from the dataset if the absolute crossover cost was 3 using a k-mer size of 6. Alpha diversity was measured using Shannon entropy (OTU level).
Statistical analysis
Fisher’s exact post hoc test in Graphpad Prism 7 was used to compare infection rates. Normalised qPCR Ct ratios were compared using unpaired t-tests in GraphPad Prism 7.
Results
Mosquito species and resident Wolbachia strains
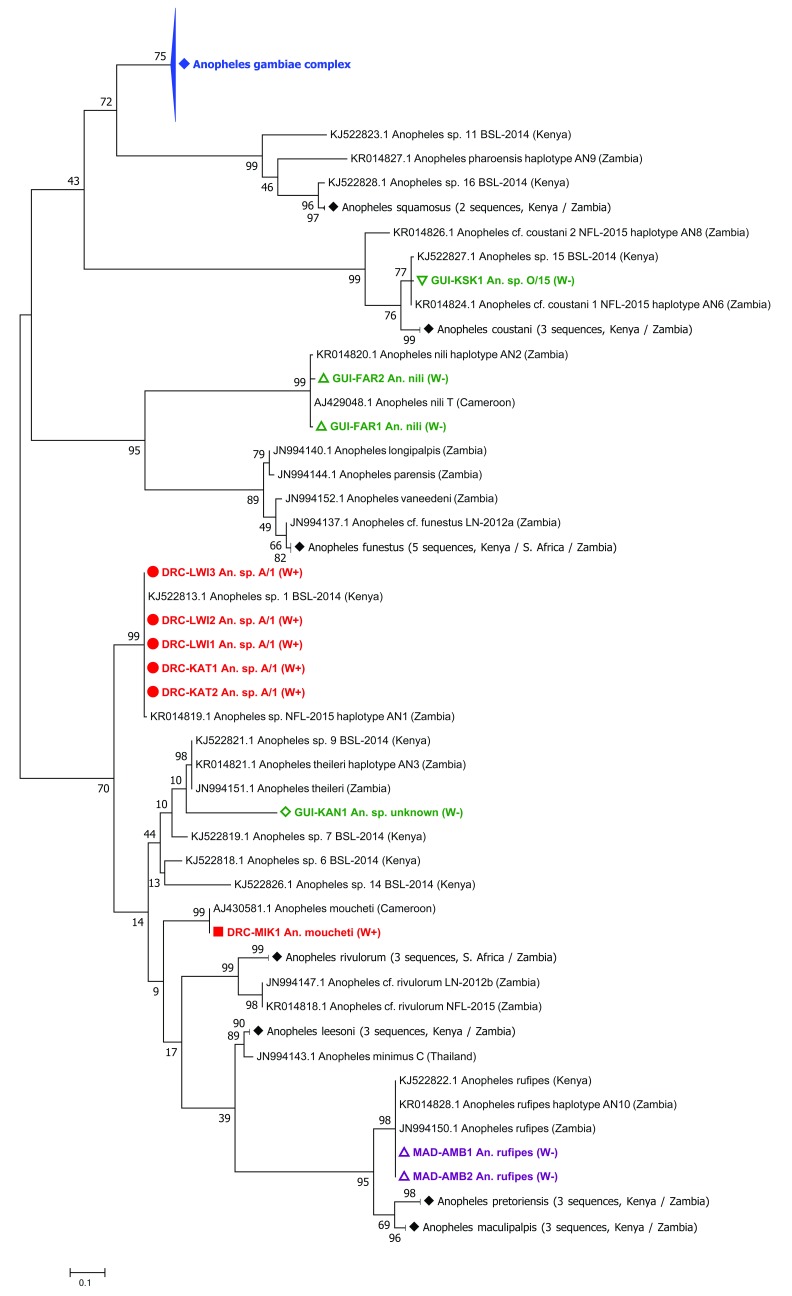
Anopheles species composition varied depending on country and mosquito collection sites ( Table 1). We detected Wolbachia in An. coluzzii mosquitoes from Ghana (prevalence of 4% - termed wAnga-Ghana) and An. gambiae s.s. from all six collection sites in DRC (prevalence range of 8–24%) in addition to a single infected An. arabiensis from Kalemie in DRC ( Figure 1 and Table 1). The molecular phylogeny of the ITS2 gene of Anopheles gambiae s.l. complex individuals (including both Wolbachia-infected and uninfected individuals analysed in our study) confirmed molecular species identifications made using species-specific PCR assays ( Figure 2). Novel resident Wolbachia infections were detected in two additional Anopheles species from DRC; An. moucheti (termed wAnM) from Mikalayi, and An. species A (termed wAnsA) from Katana. Additionally, we screened adult female mosquitoes of An. species A (collected as larvae and adults) from Lwiro, a village near Katana in DRC, and detected Wolbachia in 30/33 (91%), indicating this resident wAnsA strain has a high infection prevalence in populations in this region. The molecular phylogeny of the ITS2 gene revealed Wolbachia-infected individuals from Lwiro and Katana are the same An. species A ( Figure 3) previously collected in Eastern Zambia 43 and Western Kenya 63. All ITS2 sequences were deposited in GenBank (accession numbers MH598414–MH598445; listed in Supplementary Table 1).
Table 1. Anopheles mosquito species collected from locations within five malaria-endemic countries, including the infection status of individuals from each location.
Individuals were classified as having either single infections with Plasmodium ( Pla), Wolbachia ( Wol) or Asaia ( Asa), co-infections, or uninfected. Species containing Wolbachia-infected individuals are shown in bold.
| Country | Location | Species | Individuals with single
infections |
Individuals with
co-infections |
Uninfected
individuals |
Total | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pla | Wol | Asa |
Pla +
Wol |
Pla +
Asa |
Wol +
Asa |
|||||
| Guinea | Faranah | An. gambiae s.s. | 9 (18.8) | 0 (0) | 13 (27.1) | 0 (0) | 11 (22.9) | 0 (0) | 15 (31.3) | 48 |
| An. arabiensis | 0 (0) | 0 (0) | 7 (100.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7 | ||
| An. nili | 0 (0) | 0 (0) | 6 (75.0) | 0 (0) | 0 (0) | 0 (0) | 2 (25.0) | 8 | ||
| Kissidougou | An. gambiae s.s. | 0 (0) | 0 (0) | 26 (74.3) | 0 (0) | 9 (25.7) | 0 (0) | 0 (0) | 35 | |
| An. species O | 0 (0) | 0 (0) | 1(100.00) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 | ||
| Boke | An. gambiae s.s. | 7 (33.3) | 0 (0) | 3 (14.3) | 0 (0) | 3 (14.3) | 0 (0) | 8 (38.1) | 21 | |
| Kankan | An. gambiae s.s. | 10 (21.7) | 0 (0) | 15 (32.6) | 0 (0) | 9 (19.6) | 0 (0) | 12 (26.1) | 46 | |
| An. sp. unknown | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100.0) | 1 | ||
| DRC | Mikalayi | An. gambiae s.s. | 4 (25.0) | 1 (6.3) | 1 (6.3) | 1 (6.3) | 0 (0) | 0 (0) | 9 (56.3) | 16 |
| An. moucheti | 0 (0) | 1 (100.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 | ||
| An. funestus s.s. | 3 (23.1) | 0 (0) | 1 (7.7) | 0 (0) | 1 (7.7) | 0 (0) | 8 (61.5) | 13 | ||
| Kisangani | An. gambiae s.s. | 2 (8.0) | 2 (8.0) | 3 (12.0) | 0 (0) | 1 (4.0) | 0 (0) | 17 (68.0) | 25 | |
| An. arabiensis | 1 (25.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (75.0) | 4 | ||
| Katana | An. gambiae s.s. | 0 (0) | 2 (8.7) | 0 (0) | 0 (0) | 1 (4.3) | 0 (0) | 20 (87.0) | 23 | |
| An. funestus s.s. | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 5 (100.0) | 5 | ||
| An. species A | 0 (0) | 1 (50.0) | 0 (0) | 1 (50.0) | 0 (0) | 0 (0) | 0 (0) | 2 | ||
| Lwiro (Katana) | An. species A * | NT | 30 (91.0) | NT | NT | NT | NT | 3 (9.0) | 33 | |
| Kapolowe | An. gambiae s.s. | 1 (11.0) | 1 (11.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7 (78.0) | 9 | |
| An. funestus s.s. | 1 (20.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (80.0) | 5 | ||
| Kalemie | An. gambiae s.s. | 2 (7.1) | 6 (21.4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 20 (71.4) | 28 | |
| An. arabiensis | 0 (0) | 1 (50.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (50.0) | 2 | ||
| Kinshasa | An. gambiae s.s. | 5 (19.2) | 2 (7.7) | 1 (3.8) | 1 (3.8) | 0 (0) | 0 (0) | 17 (65.4) | 26 | |
| An. funestus s.s. | 1 (50.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (50.0) | 2 | ||
| Ghana | Dogo | An. coluzzii | 0 (0) | 12 (4.2) | 92 (32.1) | 0 (0) | 0 (0) | 0 (0) | 183 (63.8) | 287 |
| An. melas | 0 (0) | 0 (0) | 1 (100.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 | ||
| Uganda | Butemba
(2013) |
An. gambiae s.s. | 2 (3.5) | 0 (0) | 41 (71.9) | 0 (0) | 9 (15.8) | 0 (0) | 5 (8.8) | 57 |
| Butemba
(2014) |
An. gambiae s.s. | 23 (17.0) | 0 (0) | 38 (28.1) | 0 (0) | 27 (20.0) | 0 (0) | 47 (34.8) | 135 | |
| An. arabiensis | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100.00) | 1 | ||
| Madagascar | Anivorano
Nord |
An. funestus | 0 (0) | 0 (0) | 3 (37.5) | 0 (0) | 0 (0) | 0 (0) | 5 (62.5) | 8 |
| An. gambiae s.s. | 0 (0) | 0 (0) | 1 (33.3) | 0 (0) | 0 (0) | 0 (0) | 2 (66.6) | 3 | ||
| An. arabiensis | 0 (0) | 0 (0) | 2 (100.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 | ||
| An. mascarensis | 0 (0) | 0 (0) | 15 (44.1) | 0 (0) | 0 (0) | 0 (0) | 19 (55.9) | 34 | ||
| An. maculipalpis | 0 (0) | 0 (0) | 2 (15.4) | 0 (0) | 0 (0) | 0 (0) | 11 (84.6) | 13 | ||
| An. coustani | 0 (0) | 0 (0) | 6 (28.6) | 0 (0) | 0 (0) | 0 (0) | 15 (71.4) | 21 | ||
| An. rufipes | 0 (0) | 0 (0) | 3 (27.3) | 0 (0) | 0 (0) | 0 (0) | 8 (72.7) | 11 | ||
| Ambomiharina | An. funestus | 0 (0) | 0 (0) | 9 (81.8) | 0 (0) | 0 (0) | 0 (0) | 2 (18.2) | 11 | |
| An. pharoensis | 0 (0) | 0 (0) | 3 (42.9) | 0 (0) | 0 (0) | 0 (0) | 4 (57.1) | 7 | ||
| An. rufipes | 0 (0) | 0 (0) | 14 (66.7 | 0 (0) | 0 (0) | 0 (0) | 7 (33.3) | 21 | ||
| An. maculipalpis | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 9 (100.0) | 9 | ||
| An. gambiae s.s. | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 8 (100.0) | 8 | ||
| An. coustani | 0 (0) | 0 (0) | 6 (25.0) | 0 (0) | 0 (0) | 0 (0) | 18 (75.0) | 24 | ||
| An. squamosus | 0 (0) | 0 (0) | 2 (20.0) | 0 (0) | 0 (0) | 0 (0) | 8 (80.0) | 10 | ||
| An. mascarensis | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100.0) | 1 | ||
| An. pauliani | 0 (0) | 0 (0) | 3 (100.0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 | ||
| Antafia | An. gambiae s.s. | 1 (9.1) | 0 (0) | 3 (27.3) | 0 (0) | 2 (18.2) | 0 (0) | 5 (45.5) | 11 | |
| An. pauliani | 0 (0) | 0 (0) | 1 (50.0) | 0 (0) | 0 (0) | 0 (0) | 1 (50.0) | 2 | ||
| An. rufipes | 0 (0) | 0 (0) | 1 (50.0) | 0 (0) | 0 (0) | 0 (0) | 1 (50.0) | 2 | ||
| An. mascarensis | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (100.00) | 2 | ||
| Ambohimarina | An. funestus | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100.0) | 1 | |
| An. gambiae s.s. | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (100.0) | 1 | ||
| An. arabiensis | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (100.0) | 2 | ||
| An. rufipes | 0 (0) | 0 (0) | 3 (42.9) | 0 (0) | 0 (0) | 0 (0) | 4 (57.1) | 7 | ||
| An. coustani | 0 (0) | 0 (0) | 2 (11.1) | 0 (0) | 0 (0) | 0 (0) | 16 (88.9) | 18 | ||
| An. maculipalpis | 0 (0) | 0 (0) | 1 (12.5) | 0 (0) | 0 (0) | 0 (0) | 7 (87.5) | 8 | ||
| An. squamosus | 0 (0) | 0 (0) | 2 (4.3) | 0 (0) | 0 (0) | 0 (0) | 44 (95.7) | 46 | ||
| An. mascarensis | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 11 (100.0) | 11 | ||
*Adult individuals from Lwiro (Katana), DRC were collected as both larvae and adults so have been excluded from P. falciparum and Asaia prevalence analysis (NT; Not tested).
Figure 2. Maximum Likelihood molecular phylogenetic analysis of Anopheles gambiae complex ITS2 sequences from field-collected mosquitoes.
The tree with the highest log likelihood (-785.65) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 42 nucleotide sequences. There were a total of 475 positions in the final dataset. Symbols, colours and codes used for the sequences generated in this study are as follows: W+; individual was Wolbachia positive (solid coloured symbol), W-; individual was Wolbachia negative (empty coloured symbol). DRC, Democratic Republic of the Congo (red); KAL, Kalemie; MIK, Mikalayi; KIN, Kinshasa; KAT, Katana. GHA, Ghana (blue); DOG, Dogo. GUI, Guinea (green); KSK, Kissidougou. MAD, Madagascar (purple); ANT, Antafia. UGA, Uganda (maroon); BUT, Butemba. Different shape coloured symbols are used to differentiate between the different mosquito species. GenBank sequences included (for comparison with sequences generated in this study) are in black with their accession numbers provided. Where GenBank sequence subtrees have been compressed, this is denoted by a solid black diamond symbol. GenBank accession numbers for sequences included in compressed subtrees are: GQ870318.1 and GQ870320.1 for Anopheles bwambae, and GQ870315.1, JN664146.1 and KR014832.1 for Anopheles quadriannulatus.
Figure 3. Maximum Likelihood molecular phylogenetic analysis of Anopheles ITS2 sequences from field-collected mosquitoes outside of the An. gambiae s.l. complex.
The tree with the highest log likelihood (-3084.12) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 118 nucleotide sequences. There were a total of 156 positions in the final dataset. Symbols, colours and codes used for sequences generated in this study are as follows: W+; individual was Wolbachia positive (solid coloured symbol), W-; individual was Wolbachia negative (empty coloured symbol). DRC, Democratic Republic of the Congo (red): KAT, Katana; LWI, Lwiro; MIK, Mikalayi. GUI, Guinea (green); FAR, Faranah; KAN, Kankan; KSK, Kissidougou. MAD, Madagascar (purple); AMB, Ambomiharina. Different shape coloured symbols are used to differentiate between different mosquito species. GenBank sequences included (for comparison with sequences generated in this study) are in black with their accession numbers provided. Where GenBank sequence subtrees have been compressed, this is denoted by a solid black diamond symbol. GenBank accession numbers for sequences included in compressed subtrees are as follows: Anopheles squamosus; KJ522825.1 and KR014825.1. Anopheles coustani; JN994134.1, KJ522815.1 and KR014823.1. Anopheles funestus; AF062512.1, JN994135.1, JN994136.1, KJ522816.1 and KR014830.1. Anopheles rivulorum; JN994148.1, JN994149.1 and KR014822.1. Anopheles lessoni; JN994139.1, KJ522824.1 and KR014834.1. Anopheles pretoriensis; JN994145.1, KJ522820.1 and KR014829.1. Anopheles maculipalpis; JN994142.1, KJ522817.1 and KR014835.1. (The blue Anopheles gambiae complex compressed subtree is shown in Figure 2.)
Wolbachia strain typing
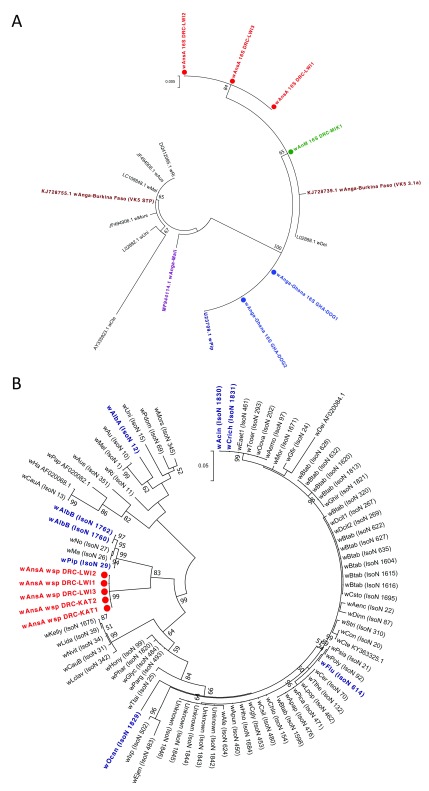
Phylogenetic analysis of the 16S rRNA gene demonstrated that the 16S sequences for these strains cluster with other Supergroup B strains such as wPip (99–100% nucleotide identity) ( Figure 4a). When compared to the resident Wolbachia strains in An. gambiae s.l. populations from Mali 40 and Burkina Faso 39, wAnga-Ghana is more closely related to the Supergroup B strain of wAnga from Burkina Faso. Although a resident strain was detected in An. gambiae s.s. and a single An. arabiensis from DRC through amplification of 16S rRNA fragments using two independent PCR assays 40, 54, we were unable to obtain 16S sequences of sufficient quality to allow further analysis. The Wolbachia wsp gene has been evolving at a faster rate and provides more informative strain phylogenies 55. As expected, however, and similar to Wolbachia-infected An. gambiae s.l. from Burkina Faso 39 and Mali 40, a fragment of the wsp gene was not amplified from Wolbachia-positive samples from An. gambiae s.s, An. arabiensis and An. coluzzii. Similarly, no wsp gene fragment amplification occurred from wAnM-infected An. moucheti. However, wsp sequences were obtained from both Wolbachia-infected individuals of An. species A from Katana. We also analysed the wsp sequences of 22 specimens of An. species A from Lwiro (near Katana) and found identical sequences to the two individuals from Katana. Phylogenetic analysis of the wsp sequences obtained for the wAnsA strain, for both individuals from Katana ( wAnsA wsp DRC-KAT1, wAnsA wsp DRC-KAT2) and three representative individuals from Lwiro ( wAnsA wsp DRC-LWI1, wAnsA wsp DRC-LWI2, wAnsA wsp DRC-LWI3) indicates wAnsA is most closely related to Wolbachia strains of Supergroup B (such as wPip, wAlbB, wMa and wNo), which is consistent with 16S rRNA phylogeny. However, the improved phylogenetic resolution provided by wsp indicates they cluster separately ( Figure 4b). Typing of the wAnsA wsp nucleotide sequences highlighted that there were no exact matches to wsp alleles currently in the Wolbachia MLST database and, in addition, wAnsA wsp sequences demonstrated novel amino acid motifs in three out of the four hypervariable regions (HVRs) when compared to those present in the MLST database ( Table 2). All Wolbachia 16S and wsp sequences of sufficient quality to generate a consensus were deposited into GenBank (accession numbers MH605275–MH605285; listed in Supplementary Table 2).
Figure 4. Resident Wolbachia strain phylogenetic analysis using 16S rRNA and wsp genes.
( A) Maximum Likelihood molecular phylogenetic analysis of the 16S rRNA gene for resident strains in An. coluzzii ( wAnga-Ghana; blue), An. moucheti ( wAnM; green) and An. species A ( wAnsA; red). The tree with the highest log likelihood (-660.03) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 17 nucleotide sequences. There were a total of 333 positions in the final dataset. Accession numbers of additional sequences obtained from GenBank are shown, including wPip (navy blue), wAnga-Mali (purple) and wAnga-Burkina Faso strains (maroon). ( B) Maximum Likelihood molecular phylogenetic analysis of the wsp gene for wAnsA-infected representative individuals from the DRC (red). The tree with the highest log likelihood (-3663.41) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 83 nucleotide sequences. There were a total of 443 positions in the final dataset. Reference numbers of additional sequences obtained from the MLST database (IsoN; Isolate number) or GenBank (accession number) are shown. Strains isolated from mosquitoes are highlighted in navy blue. KAT, Katana; LWI, Lwiro.
Table 2. Novel resident Wolbachia strain WSP typing and multilocus sequence typing (MLST) gene allelic profiles.
Novel allele numbers (in bold) assigned by the Wolbachia MLST database for strains from An. species A ( wAnsA) and An. moucheti ( wAnM) are shown, alongside the novel allelic profile from An. coluzzii ( wAnga-Ghana), comprising exact matches to existing alleles present in the database for each gene locus. (HVR; Hypervariable regions within the wsp sequence.).
| Mosquito species |
Wolbachia
strain |
WSP typing allele numbers | MLST gene allele numbers | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| wsp | HVR1 | HVR2 | HVR3 | HVR4 | gatB | coxA | hcpA | ftsZ | fbpA | ||
| An. species A | wAnsA | 728 | 254 | 288 | 284 | 23 | 279 | 274 | 302 | 240 | 445 |
| An. moucheti | wAnM | - | - | - | - | - | 280 | 275 | 303 | 241 | 446 |
| An. coluzzii | wAnga-Ghana | - | - | - | - | - | 9 | 64 | 3 * | 177 | 4 |
*Alternative degenerate primers (set 3) were used to generate sequence from another An. coluzzii individual from the same location to complete the full allelic profile.
MLST was undertaken to provide more accurate strain phylogenies. This was done for the novel Wolbachia strains wAnM and wAnsA in addition to the resident wAnga-Ghana strain in An. coluzzii from Ghana. We were unable to amplify any of the five MLST genes from Wolbachia-infected An. gambiae s.s . and An. arabiensis from DRC (likely due to low infection densities). New alleles for all five MLST gene loci (sequences differed from those currently present in the MLST database) and novel allelic profiles confirm the diversity of these novel Wolbachia strains ( Table 2). The phylogeny of these three novel strains based on concatenated sequences of all five MLST gene loci confirms they cluster within Supergroup B ( Figure 5a). This also demonstrates the novelty as comparison with a wide range of strains (including all isolates highlighted through partial matching during typing of each locus) shows these strains are distinct from currently available sequences ( Figure 5a and Table 2). The concatenated phylogeny indicates that wAnM is most closely related to a Hemiptera strain: Isolate number 1616 found in Bemisia tabaci in Uganda, and a Coleoptera strain: Isolate number 20 found in Tribolium confusum. Concatenation of the MLST loci also indicates wAnsA is closest to a group containing various Lepidoptera and Hymenoptera strains from multiple countries in Asia, Europe and America, as well as two mosquito strains: Isolate numbers 1830 and 1831, found in Aedes cinereus and Coquillettidia richiardii in Russia. This highlights the lack of concordance between Wolbachia strain phylogeny and their insect hosts across diverse geographical regions.
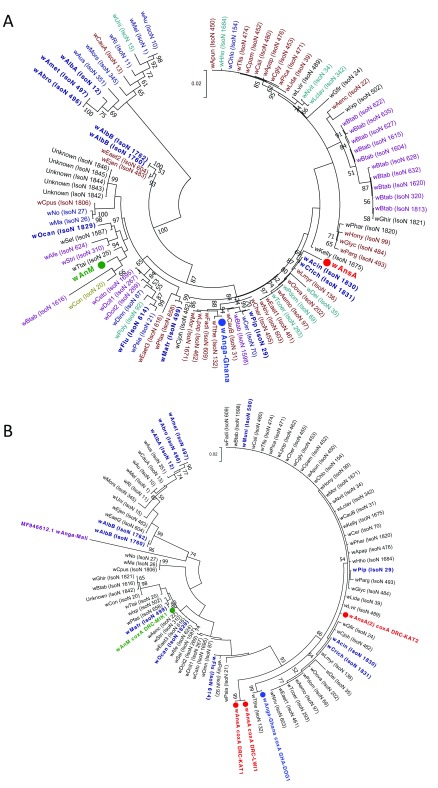
Figure 5. Wolbachia multilocus sequence typing (MLST) phylogenetic analysis of resident Wolbachia strains in An. coluzzii, An. moucheti and An. species A.
( A) Maximum Likelihood molecular phylogenetic analysis from concatenation of all five MLST gene loci for resident Wolbachia strains from An. coluzzii ( wAnga-Ghana; blue), An. moucheti ( wAnM; green) and An. species A ( wAnsA; red). The tree with the highest log likelihood (-10606.13) is shown and drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 94 nucleotide sequences. There were a total of 2067 positions in the final dataset. Concatenated sequence data from Wolbachia strains downloaded from MLST database for comparison are shown with isolate numbers in brackets (IsoN). Wolbachia strains isolated from mosquito species highlighted in navy blue, bold. Strains isolated from other Dipteran species are shown in navy blue, from Coleoptera in olive green, from Hemiptera in purple, from Hymenoptera in teal blue, from Lepidoptera in maroon and from other, or unknown orders in black. ( B) Maximum Likelihood molecular phylogenetic analysis for coxA gene locus for resident Wolbachia strains from An. coluzzii ( wAnga-Ghana; blue), An. moucheti ( wAnM; green) and An. species A ( wAnsA and wAnsA(2); red). The tree with the highest log likelihood (-1921.11) is shown and drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 84 nucleotide sequences. There were a total of 402 positions in the final dataset. Sequence data for the coxA locus from Wolbachia strains downloaded from MLST database for comparison are shown in black and navy blue with isolate numbers (IsoN) from the MLST database shown in brackets. Wolbachia strains isolated from mosquito species highlighted in navy blue. GenBank sequence for wAnga-Mali coxA shown in maroon with accession number.
We also found evidence of potential strain variants in wAnsA through variable MLST gene fragment amplification and resulting closest-match allele numbers. A second wAnsA-infected sample from Katana, An. sp. A/1 (W+) DRC-KAT2, only successfully amplified hcpA and coxA gene fragments and although identical sequences were obtained for wsp ( Figure 4b) and hcpA, genetic diversity was seen in the coxA sequences, with typing indicating a different, but still novel allele for the coxA sequence from this individual ( wAnsA(2) coxA DRC-KAT2) ( Figure 5b). Further analysis of the coxA sequence as part of MLST allele submission from this variant suggested the possibility of a double infection, where two differing strains of Wolbachia are present. MLST gene fragment amplification was also variable for wAnga-Ghana-infected An. coluzzii, requiring two individuals to generate the five MLST gene sequences, and for the hcpA locus, more degenerate primers (hcpA_F3/hcpA_R3) were required to generate sequence of sufficient quality for analysis. This is likely due to the low density of this strain potentially influencing the ability to successfully amplify all MLST genes, in addition to the possibility of genetic variation in primer binding regions. Despite the sequences generated for this strain producing exact matches with alleles in the database for each of the five gene loci, the resultant allelic profile, and therefore strain type, did not produce a match, showing this wAnga-Ghana strain is also a novel strain type. The closest matches to the wAnga-Ghana allelic profile were with strains from two Lepidopteran species: Isolate number 609 found in Fabriciana adippe from Russia, and Isolate number 658 found in Pammene fasciana from Greece, but each of these only produced a match for three out of the five loci. The concatenated phylogeny for this strain ( Figure 5a) indicates that across the 5 MLST loci, wAnga-Ghana is actually most closely related to a Lepidopteran strain found in Thersamonia thersamon in Russia (Isolate number 132). The phylogeny of Wolbachia strains based on the coxA gene ( Figure 5b) highlights the genetic diversity of both the wAnsA strain variants and also wAnga-Ghana, compared to the wAnga-Mali strain 40; coxA gene sequences are not available for wAnga strains from Burkina Faso 39. All Wolbachia MLST sequences were deposited into GenBank (accession numbers MH605286–MH605305; listed in Supplementary Table 3).
Resident strain densities and relative abundance
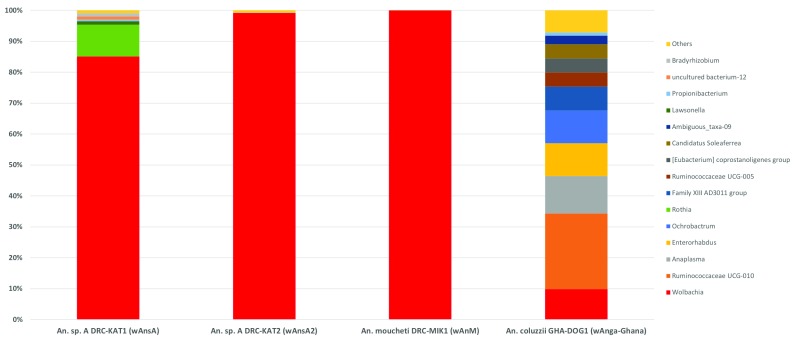
The relative densities of Wolbachia strains were estimated using qPCR targeting the ftsZ 56and 16S rRNA 40genes. qPCR analysis of ftsZ and 16S rRNA indicated the amount of Wolbachia detected in wAnsA-infected and wAnM-infected females was three orders of magnitude higher (Ct values 20–22) than Wolbachia-infected An. gambiae s.s. , An. arabiensis and wAnga-Ghana-infected An. coluzzii (Ct values 30–33). To account for variation in mosquito body size and DNA extraction efficiency, we compared the total amount of DNA for Wolbachia-infected mosquito extracts and conversely, we found less total DNA in the wAnsA-infected extract (1.36 ng/μl) and wAnM-infected extracts (5.85 ng/μl) compared to the mean of 6.64 ± 2.33 ng/μl for wAnga -Ghana-infected An. coluzzii. To estimate the relative abundance of resident Wolbachia strains in comparison to other bacterial species, we sequenced the bacterial microbiome using 16S rRNA amplicon sequencing on Wolbachia-infected individuals. We found wAnsA, wAnsA(2) and wAnM Wolbachia strains were the dominant OTUs of these mosquito species ( Figure 6). In contrast, the lower-density infection wAnga -Ghana strain represented only ~10% of the OTUs within the microbiome.
Figure 6. The relative abundance of resident Wolbachia strains in Anopheles.
Bacterial genus level taxonomy was assigned to operational taxonomic units clustered with a 97% cut-off using the SILVA SSU v128 97% database, and individual genera comprising less than 1% of total abundance was merged into “Others”.
P. falciparum, Wolbachia and Asaia prevalence
The prevalence of P. falciparum in female mosquitoes was extremely variable across countries and collection locations ( Figure 1 and Table 1) with very high prevalence recorded in An. gambiae s.s. from villages close to Boke (52%) and Faranah (44%) in Guinea. Despite the collection of other Anopheles species in Guinea, An. gambiae s.s. was the only species to have detectable malaria parasite infections. In contrast, P. falciparum was detected in multiple major vector species from DRC, including An. gambiae s.s, An. arabiensis and An. funestus s.s. A high prevalence of P. falciparum was also detected in An. gambiae s.s. from Uganda for both collection years; 19% for 2013 and 36% for 2014. In contrast, no P. falciparum infections were detected in any of the An. coluzzii or An. melas collected in Ghana. In Madagascar, P. falciparum was detected in only two species; An. gambiae s.s. and An. rufipes. We compared the overall P. falciparum infection rates in An. gambiae s.s. mosquitoes collected across all locations from DRC to determine if there was any correlation with the presence of the low density wAnga-DRC Wolbachia resident strain. Overall, of the 128 mosquitoes collected, only 1.56% (n=2) had detectable Wolbachia- Plasmodium co-infections, compared to 10.16% (n=13) where we only detected Wolbachia. A further 11.72% (n=15) were only PCR-positive for P. falciparum. As expected, for the vast majority of mosquitoes (76.56%, n=98) we found no evidence of Wolbachia or P. falciparum present, resulting in no correlation across all samples (Fisher’s exact post hoc test on unnormalized data, two-tailed, P=0.999). Interestingly, one An. species A female from Katana, DRC (infected with wAnsA) was co-infected with P. falciparum.
For all Wolbachia-infected females collected in our study (including An. coluzzii from Ghana and novel resident strains in An. moucheti and An. species A), we did not detect the presence of Asaia. No resident Wolbachia strain infections were detected in Anopheles mosquitoes from Guinea, Uganda or Madagascar. However, high Asaia and malaria parasite prevalence rates were present in Anopheles mosquitoes from Uganda and Guinea (including in multiple species in all four sites in Guinea). We compared the overall P. falciparum infection rates in An. gambiae s.s. collected across all locations from Guinea, with and without Asaia bacteria, and found no overall correlation (Fisher’s exact post hoc test on unnormalized data, two-tailed, P=0.4902). There was also no overall correlation between Asaia and P. falciparum infections in An. gambiae s.s. from Uganda for both 2013 (Fisher’s exact post hoc test on unnormalized data, two-tailed, P=0.601) and 2014 (Fisher’s exact post hoc test on unnormalized data, two-tailed, P=0.282).
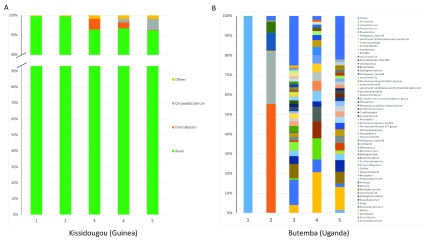
Asaia can be environmentally acquired at all life stages but can also have the potential to be vertically and horizontally transmitted between individual mosquitoes. Therefore, we performed 16S microbiome analysis on a sub-sample of Asaia-infected An. gambiae s.s. from Kissidougou (Guinea), a location in which high levels of Asaia were detected by qPCR (mean Asaia Ct = 17.84 ± 2.27) 64. Asaia in these individuals is the dominant bacterial species present ( Figure 7a) but in Uganda we detected much lower levels of Asaia (qPCR mean Ct = 33.33 ± 0.19) and this was reflected in Asaia not being a dominant species in microbiome analysis ( Figure 7b). The alpha and beta diversity of An. gambiae s.s. from Kissidougou, Guinea and Butemba, Uganda shows much more overall diversity in the microbiome for Uganda individuals ( Supplementary Figure 1). Interestingly, 2/5 of these individuals from Kissidougou (Guinea) were P. falciparum-infected compared to 3/5 individuals from Uganda. To determine if the presence of Asaia had a quantifiable effect on the level of P. falciparum detected, we normalized P. falciparum Ct values from qPCR (n = 61) ( Supplementary Figure 2a) and compared gene ratios for An. gambiae s.s. mosquitoes from Guinea, with or without Asaia ( Supplementary Figure 2b). Statistical analysis using student’s t-tests revealed no significant difference between normalized P. falciparum gene ratios between the Asaia positive (n = 33) and negative (n = 28) groups (p = 0.51, df = 59). Larger variation of Ct values was seen for Asaia (n = 90) ( Supplementary Figure 2c) suggesting the bacterial densities in individual mosquitoes were more variable than P. falciparum parasite infection levels.
Figure 7. The relative abundance of bacteria in An. gambiae s.s. comparing two locations with contrasting Asaia infection densities.
Bacterial genus level taxonomy was assigned to operational taxonomic units clustered with a 97% cut-off using the SILVA SSU v128 97% database, and individual genera comprising less than 1% of total abundance was merged into “Others”.
Discussion
Malaria transmission in Sub-Saharan Africa is highly dependent on the local Anopheles vector species, but the primary vector complexes recognised are An. gambiae s.l. , An. funestus s.l. An. nili s.l. and An. moucheti s.l. 41, 65. An. gambiae s.s. and An. coluzzii sibling species are considered the most important malaria vectors in Sub-Saharan Africa and recent studies indicate that An. coluzzii extends further north, and closer to the coast than An. gambiae s.s. within West Africa 66. In our study, high Plasmodium prevalence rates in An. gambiae s.s . across Guinea would be consistent with high malaria parasite prevalence in humans (measured by rapid diagnostic tests) in Guéckédou prefecture, and the overall national malaria prevalence, estimated to be 44% in 2013 67. However, malaria prevalence has decreased in the past few years with an overall prevalence across Guinea estimated at 15% for 2016. Although our P. falciparum infection prevalence rates were also high in DRC, recent studies have shown comparable levels of infection with 35% of An. gambiae s.l. mosquitoes infected from Kinshasa 68. We detected P. falciparum in An. gambiae s.s, An. arabiensis, An. funestus s.s. and An. species A from DRC. Morphological differences have been widely used for identification of malaria vectors but species complexes (such as An. gambiae s.l. and An. funestus s.l.) require species-diagnostic PCR assays. Historically, malaria parasite entomology studies in Africa have focused predominantly on species from these complexes, likely due to the fact that mosquitoes from these complexes dominate the collections 43. In our study, we used ITS2 sequencing to confirm secondary vector species that were P. falciparum-infected given the difficulties of morphological identification and recent studies demonstrating the inaccuracy of diagnostic species PCR-based molecular identification 69. Our study is the first to report the detection of P. falciparum in An. rufipes from Madagascar; previously this species was considered a vector of Plasmodium species of non-human origin and has only very recently been implicated in human malaria transmission 70. However, detection of P. falciparum parasites in whole body mosquitoes does not confirm that the species plays a significant role in transmission. Detection could represent infected bloodmeal stages or oocysts present in the midgut wall so further studies are warranted to determine the ability of this species to transmit human malaria parasites.
The mosquito microbiota can modulate the mosquito immune response and bacteria present in wild Anopheles populations can influence malaria vector competence 4, 5. Endosymbiotic Wolbachia bacteria are particularly widespread through insect populations, but they were commonly thought to be absent from Anopheles mosquitoes. However, the recent discovery of Wolbachia strains in An. gambiae s.l. in Burkina Faso and Mali 39, 40, in addition to our study showing infection in Anopheles from Ghana and DRC, suggest resident strains could be widespread across Sub-Saharan Africa. The discovery of resident strains in Burkina Faso resulted from sequencing of the 16S rRNA gene identifying Wolbachia sequences rather than screening using Wolbachia-specific genes 39. Intriguingly, Wolbachia infections in these mosquitoes could not be detected using conventional PCR targeting the wsp gene. As the wsp gene has often been used in previous studies to detect strains in Anopheles species 25, 27, this could explain why resident strains in the An. gambiae complex have gone undetected until very recently. Recent similar methods using 16S rRNA amplicon sequencing to determine the overall microbiota in wild mosquito populations has provided evidence for Wolbachia infections in An. gambiae s.l. in additional villages in Burkina Faso 71 and Anopheles species collected in Illinois, USA 72. Our study describing resident Wolbachia strains in numerous species of Anopheles malaria vectors also highlights the potential for Wolbachia to be influencing malaria transmission, as postulated by previous studies 39, 40, 73. No significant correlation was present in our study for Plasmodium and Wolbachia prevalence in the 128 An. gambiae s.s. individuals from DRC. As the majority (77%) of samples had neither detectable Wolbachia resident strains or P. falciparum, a larger sample size would provide a more comprehensive assessment factoring in the Plasmodium parasite life stages. Although there is evidence from previous studies that Wolbachia is negatively correlated with Plasmodium in both Burkina Faso 73 and Mali 40, our infection prevalence rates for resident Wolbachia strains in An. coluzzii from Ghana (4%) and An. gambiae s.s. from the DRC were variable but low (8–24%). These results are more aligned to infection prevalence rates in An. gambiae s.l. from Burkina Faso (11%) 39 but much lower than those reported in Mali (60–80%) 40 where infection was associated with reduced prevalence and intensity of sporozoite infection in field-collected females.
The discovery of a resident Wolbachia strain in An. moucheti, a highly anthropophilic and efficient malaria vector found in the forested areas of Western and Central Africa 41, suggests further studies are warranted that utilize large sample sizes to examine the influence of the wAnM Wolbachia strain on Plasmodium infection dynamics in this malaria vector. An. moucheti is often the most abundant vector, breeding in slow moving streams and rivers, contributing to year round malaria transmission in these regions 74, 75. This species has also been implicated as a main bridge vector species in the transmission of ape Plasmodium malaria in Gabon 76. There is thought to be high genetic diversity in An. moucheti populations 77, 78, which may either influence the prevalence of Wolbachia resident strains, or Wolbachia could be contributing to genetic diversity through its effect on host reproduction. A novel Wolbachia strain in An. species A, present at high infection frequencies in Lwiro (close to Katana in DRC), also suggests more Anopheles species, including unidentified and potentially new species, could be infected with this widespread endosymbiotic bacterium. An. species A should be further investigated to determine if this species is a potential malaria vector, given our study demonstrated P. falciparum infection in one of two individuals screened and ELISA-positive samples of this species were reported from the Western Highlands of Kenya 42.
The variability of Wolbachia prevalence rates in An. gambiae complex from locations within DRC and Ghana and previous studies in Burkina Faso 39 and Mali 40 suggest the environment is one factor that influences the presence or absence of resident strains. In our study we found no evidence of Wolbachia- Asaia co-infections across all countries, supporting laboratory studies that have shown these two bacterial species demonstrate competitive exclusion in Anopheles species 36, 38. We also found that Asaia infection densities (whole body mosquitoes) were variable and location dependent which would correlate with this bacterium being environmentally acquired at all life stages, but also having the potential for both vertical and horizontal transmission 37. Significant variations in overall Asaia prevalence and density across different Anopheles species and locations in our study would also correlate with our data indicating no evidence of an association with P. falciparum prevalence in both Guinea and Uganda populations. Further studies are needed to determine the complex interaction between these two bacterial species and malaria in diverse Anopheles malaria vector species. Horizontal transfer of Wolbachia strains between species (even over large phylogenetic differences) has shaped the evolutionary history of this endosymbiont in insects, and there is evidence for loss of infection in host lineages over evolutionary time 79. Our results showing a novel strain present in An. coluzzii from Ghana (phylogenetically different to strains present in An. gambiae s.l. mosquitoes from both Burkina Faso and Mali), strain variants observed in An. species A, and the concatenated grouping of the novel Anopheles strains with strains found in different Orders of insects, support the lack of congruence between insect host and Wolbachia strain phylogenies 80.
Our qPCR and 16S microbiome analysis indicates the densities of wAnM and wAnsA strains are significantly higher than resident Wolbachia strains in An. gambiae s.l. However, caution must be taken as we were only able to analyse selected individuals, and larger collections of wild populations would be required to confirm these results. Native Wolbachia strains dominating the microbiome of An. species A and An. moucheti is consistent with other studies of resident strains in mosquitoes showing Wolbachia 16S rRNA gene amplicons vastly outnumber sequences from other bacteria in Ae. albopictus and Cx. quinquefasciatus 81, 82. The discovery of novel Wolbachia strains provides the rationale to undertake vector competence experiments to determine what effect these strains are having on malaria transmission. The tissue tropism of novel Wolbachia strains in malaria vectors will be particularly important to characterise given this will determine if these endosymbiotic bacteria are proximal to malaria parasites within the mosquito. It would also be important to determine the additional phenotypic effects novel resident Wolbachia strains have on their mosquito hosts. Some Wolbachia strains induce a reproductive phenotype termed cytoplasmic incompatibility (CI) that results in inviable offspring when an uninfected female mates with a Wolbachia-infected male. In contrast, Wolbachia-infected females produce viable progeny when they mate with both infected and uninfected male mosquitoes. This reproductive advantage over uninfected females allows Wolbachia to spread within mosquito populations.
Conclusions
Wolbachia has been the focus of recent biocontrol strategies in which Wolbachia strains transferred into naïve mosquito species provide strong inhibitory effects on arboviruses 16, 18– 20, 83, 84 and malaria parasites 31, 35. The discovery of two novel Wolbachia strains in Anopheles mosquitoes that are potentially present at much higher density than resident strains in the An. gambiae complex, also suggests the potential for these strains to be transinfected into other Anopheles species to produce inhibitory effects on Plasmodium parasites. Wolbachia transinfection success is partly attributed to the relatedness of donor and recipient host so the transfer of high density Wolbachia strains between Anopheles species may result in stable infections (or co-infections) that have strong inhibitory effects on Plasmodium development. Finally, if the resident strain present in An. moucheti is at low infection frequencies in wild populations, an alternative strategy known as the incompatible insect technique (IIT) could be implemented where Wolbachia-infected males are released to suppress the wild populations through CI (reviewed by 22). In summary, the important discovery of diverse novel Wolbachia strains in Anopheles species will help our understanding of how Wolbachia strains can potentially impact malaria transmission, through natural associations or being used as candidate strains for transinfection to create stable infections in other species.
Data availability
ITS2 GenBank accession numbers are listed in Supplementary Table 1; Wolbachia 16S and wsp gene GenBank accession numbers are listed in Supplementary Table 2; Wolbachia MLST gene GenBank accession numbers are listed in Supplementary Table 3.
Raw PCR screening data is available at Open Science Framework: DOI: https://doi.org/10.17605/OSF.IO/MW6XZ 64.
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
Acknowledgements
We would like to thank all the mosquito collectors and residents of the villages where collections took place. We would also like to thank John Gimnig, Bill Hawley and Barb Marston for reviewing our manuscript. This publication made use of the PubMLST website ( https://pubmlst.org/wolbachia/) sited at the University of Oxford (Jolley & Maiden 2010, BMC Bioinformatics, 11:595). The development and maintenance of this site has been funded by the Wellcome Trust.
Funding Statement
CLJ and TW were supported by a Wellcome Trust /Royal Society grant awarded to TW (101285): http://www.wellcome.ac.uk; https://royalsociety.org. GLH is supported by NIH grants (R21AI124452 and R21AI129507), a University of Texas Rising Star award, the John S. Dunn Foundation Collaborative Research Award, the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation, and the Centers for Disease Control and Prevention (CDC) (Cooperative Agreement Number U01CK000512). The papers contents are solely the responsibility of the authors and do not necessarily represent the official views of the CDC or the Department of Health and Human Services. This work was also supported by a James W. McLaughlin postdoctoral fellowship at the University of Texas Medical Branch to SH. Field work in Uganda was funded by UK aid (through the Programme Partnership Arrangement grant to Malaria Consortium). YAA and ARM were supported by a NIH grant R01AI123074. SRI was funded by the U.S. President’s Malaria Initiative.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; referees: 3 approved]
Supplementary material
Supplementary Table 1. Additional sample details and ITS2 GenBank accession numbers.References
- 1. Hay SI, Sinka ME, Okara RM, et al. : Developing global maps of the dominant anopheles vectors of human malaria. PLoS Med. 2010;7(2):e1000209. 10.1371/journal.pmed.1000209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cirimotich CM, Dong Y, Clayton AM, et al. : Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332(6031):855–8. 10.1126/science.1201618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cirimotich CM, Ramirez JL, Dimopoulos G: Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe. 2011;10(4):307–10. 10.1016/j.chom.2011.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dennison NJ, Jupatanakul N, Dimopoulos G: The mosquito microbiota influences vector competence for human pathogens. Curr Opin Insect Sci. 2014;3:6–13. 10.1016/j.cois.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boissière A, Tchioffo MT, Bachar D, et al. : Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 2012;8(5):e1002742. 10.1371/journal.ppat.1002742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dennison NJ, Saraiva RG, Cirimotich CM, et al. : Functional genomic analyses of Enterobacter, Anopheles and Plasmodium reciprocal interactions that impact vector competence. Malar J. 2016;15(1):425. 10.1186/s12936-016-1468-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zug R, Hammerstein P: Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One. 2012;7(6):e38544. 10.1371/journal.pone.0038544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klasson L, Walker T, Sebaihia M, et al. : Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol Biol Evol. 2008;25(9):1877–87. 10.1093/molbev/msn133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laven H: Speciation by cytoplasmic isolation in the Culex pipiens-complex. Cold Spring Harb Symp Quant Biol. 1959;24:166–73. 10.1101/SQB.1959.024.01.017 [DOI] [PubMed] [Google Scholar]
- 10. Sinkins SP, Walker T, Lynd AR, et al. : Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature. 2005;436(7048):257–60. 10.1038/nature03629 [DOI] [PubMed] [Google Scholar]
- 11. Dutton TJ, Sinkins SP: Strain-specific quantification of Wolbachia density in Aedes albopictus and effects of larval rearing conditions. Insect Mol Biol. 2004;13(3):317–22. 10.1111/j.0962-1075.2004.00490.x [DOI] [PubMed] [Google Scholar]
- 12. Sinkins SP, Braig HR, O’Neill SL: Wolbachia pipientis: bacterial density and unidirectional cytoplasmic incompatibility between infected populations of Aedes albopictus. Exp Parasitol. 1995;81(3):284–91. 10.1006/expr.1995.1119 [DOI] [PubMed] [Google Scholar]
- 13. Glaser RL, Meola MA: The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One. 2010;5(8):e11977. 10.1371/journal.pone.0011977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mousson L, Zouache K, Arias-Goeta C, et al. : The native Wolbachia symbionts limit transmission of dengue virus in Aedes albopictus. PLoS Negl Trop Dis. 2012;6(12):e1989. 10.1371/journal.pntd.0001989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Silva JBL, Magalhães Alves D, Bottino-Rojas V, et al. : Wolbachia and dengue virus infection in the mosquito Aedes fluviatilis (Diptera: Culicidae). PLoS One. 2017;12(7):e0181678. 10.1371/journal.pone.0181678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walker T, Johnson PH, Moreira LA, et al. : The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476(7361):450–3. 10.1038/nature10355 [DOI] [PubMed] [Google Scholar]
- 17. Iturbe-Ormaetxe I, Walker T, O' Neill SL: Wolbachia and the biological control of mosquito-borne disease. EMBO Rep. 2011;12(6):508–18. 10.1038/embor.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Joubert DA, Walker T, Carrington LB, et al. : Establishment of a Wolbachia Superinfection in Aedes aegypti Mosquitoes as a Potential Approach for Future Resistance Management. PLoS Pathog. 2016;12(2):e1005434. 10.1371/journal.ppat.1005434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, et al. : A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139(7):1268–78. 10.1016/j.cell.2009.11.042 [DOI] [PubMed] [Google Scholar]
- 20. Bian G, Xu Y, Lu P, et al. : The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 2010;6(4):e1000833. 10.1371/journal.ppat.1000833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blagrove MS, Arias-Goeta C, Failloux AB, et al. : Wolbachia strain wMel induces cytoplasmic incompatibility and blocks dengue transmission in Aedes albopictus. Proc Natl Acad Sci U S A. 2012;109(1):255–60. 10.1073/pnas.1112021108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bourtzis K, Dobson SL, Xi Z, et al. : Harnessing mosquito- Wolbachia symbiosis for vector and disease control. Acta Trop. 2014;132 Suppl:S150–63. 10.1016/j.actatropica.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 23. Hoffmann AA, Montgomery BL, Popovici J, et al. : Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476(7361):454–7. 10.1038/nature10356 [DOI] [PubMed] [Google Scholar]
- 24. Frentiu FD, Zakir T, Walker T, et al. : Limited dengue virus replication in field-collected Aedes aegypti mosquitoes infected with Wolbachia. PLoS Negl Trop Dis. 2014;8(2):e2688. 10.1371/journal.pntd.0002688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kittayapong P, Baisley KJ, Baimai V, et al. : Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae). J Med Entomol. 2000;37(3):340–5. 10.1093/jmedent/37.3.340 [DOI] [PubMed] [Google Scholar]
- 26. Tiawsirisup S, Sripatranusorn S, Oraveerakul K, et al. : Distribution of mosquito (Diptera: Culicidae) species and Wolbachia (Rickettsiales: Rickettsiaceae) infections during the bird immigration season in Pathumthani province, central Thailand. Parasitol Res. 2008;102(4):731–5. 10.1007/s00436-007-0825-z [DOI] [PubMed] [Google Scholar]
- 27. Wiwatanaratanabutr I: Geographic distribution of wolbachial infections in mosquitoes from Thailand. J Invertebr Pathol. 2013;114(3):337–40. 10.1016/j.jip.2013.04.011 [DOI] [PubMed] [Google Scholar]
- 28. Ricci I, Cancrini G, Gabrielli S, et al. : Searching for Wolbachia (Rickettsiales: Rickettsiaceae) in mosquitoes (Diptera: Culicidae): large polymerase chain reaction survey and new identifications. J Med Entomol. 2002;39(4):562–7. 10.1603/0022-2585-39.4.562 [DOI] [PubMed] [Google Scholar]
- 29. Rasgon JL, Scott TW: An initial survey for Wolbachia (Rickettsiales: Rickettsiaceae) infections in selected California mosquitoes (Diptera: Culicidae). J Med Entomol. 2004;41(2):255–7. 10.1603/0022-2585-41.2.255 [DOI] [PubMed] [Google Scholar]
- 30. Walker T, Moreira LA: Can Wolbachia be used to control malaria? Mem Inst Oswaldo Cruz. 2011;106 Suppl 1:212–7. 10.1590/S0074-02762011000900026 [DOI] [PubMed] [Google Scholar]
- 31. Hughes GL, Koga R, Xue P, et al. : Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 2011;7(5):e1002043. 10.1371/journal.ppat.1002043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hughes GL, Vega-Rodriguez J, Xue P, et al. : Wolbachia strain wAlbB enhances infection by the rodent malaria parasite Plasmodium berghei in Anopheles gambiae mosquitoes. Appl Environ Microbiol. 2012;78(5):1491–5. 10.1128/AEM.06751-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murdock CC, Blanford S, Hughes GL, et al. : Temperature alters Plasmodium blocking by Wolbachia. Sci Rep. 2014;4: 3932. 10.1038/srep03932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hughes GL, Rivero A, Rasgon JL: Wolbachia can enhance Plasmodium infection in mosquitoes: implications for malaria control? PLoS Pathog. 2014;10(9):e1004182. 10.1371/journal.ppat.1004182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bian G, Joshi D, Dong Y, et al. : Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340(6133):748–51. 10.1126/science.1236192 [DOI] [PubMed] [Google Scholar]
- 36. Hughes GL, Dodson BL, Johnson RM, et al. : Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc Natl Acad Sci U S A. 2014;111(34):12498–503. 10.1073/pnas.1408888111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Favia G, Ricci I, Damiani C, et al. : Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc Natl Acad Sci U S A. 2007;104(21):9047–51. 10.1073/pnas.0610451104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rossi P, Ricci I, Cappelli A, et al. : Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasit Vectors. 2015;8:278. 10.1186/s13071-015-0888-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baldini F, Segata N, Pompon J, et al. : Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nat Commun. 2014;5: 3985. 10.1038/ncomms4985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gomes FM, Hixson BL, Tyner MDW, et al. : Effect of naturally occurring Wolbachia in Anopheles gambiae s.l. mosquitoes from Mali on Plasmodium falciparum malaria transmission. Proc Natl Acad Sci U S A. 2017;114(47):12566–12571. 10.1073/pnas.1716181114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sinka ME, Bangs MJ, Manguin S, et al. : The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic précis. Parasit Vectors. 2010;3:117. 10.1186/1756-3305-3-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stevenson J, St Laurent B, Lobo NF, et al. : Novel vectors of malaria parasites in the western highlands of Kenya. Emerg Infect Dis. 2012;18(9):1547–9. 10.3201/eid1809.120283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lobo NF, St Laurent B, Sikaala CH, et al. : Unexpected diversity of Anopheles species in Eastern Zambia: implications for evaluating vector behavior and interventions using molecular tools. Sci Rep. 2015;5: 17952. 10.1038/srep17952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tantely ML, Rakotoniaina JC, Tata E, et al. : Biology of mosquitoes that are potential vectors of Rift Valley Fever virus in different biotopes of the central highlands of Madagascar. J Med Entomol. 2013;50(3):603–10. 10.1603/ME12069 [DOI] [PubMed] [Google Scholar]
- 45. Scott JA, Brogdon WG, Collins FH: Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49(4):520–9. 10.4269/ajtmh.1993.49.520 [DOI] [PubMed] [Google Scholar]
- 46. Santolamazza F, Mancini E, Simard F, et al. : Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J. 2008;7:163. 10.1186/1475-2875-7-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bass C, Williamson MS, Field LM: Development of a multiplex real-time PCR assay for identification of members of the Anopheles gambiae species complex. Acta Trop. 2008;107(1):50–3. 10.1016/j.actatropica.2008.04.009 [DOI] [PubMed] [Google Scholar]
- 48. Cohuet A, Simard F, Toto JC, et al. : Species identification within the Anopheles funestus group of malaria vectors in Cameroon and evidence for a new species. Am J Trop Med Hyg. 2003;69(2):200–5. 10.4269/ajtmh.2003.69.200 [DOI] [PubMed] [Google Scholar]
- 49. Beebe NW, Saul A: Discrimination of all members of the Anopheles punctulatus complex by polymerase chain reaction--restriction fragment length polymorphism analysis. Am J Trop Med Hyg. 1995;53(5):478–81. 10.4269/ajtmh.1995.53.478 [DOI] [PubMed] [Google Scholar]
- 50. Marie A, Boissière A, Tsapi MT, et al. : Evaluation of a real-time quantitative PCR to measure the wild Plasmodium falciparum infectivity rate in salivary glands of Anopheles gambiae. Malar J. 2013;12:224. 10.1186/1475-2875-12-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yamada Y, Katsura K, Kawasaki H, et al. : Asaia bogorensis gen. nov., sp. nov., an unusual acetic acid bacterium in the alpha- Proteobacteria. Int J Syst Evol Microbiol. 2000;50 Pt 2:823–9. 10.1099/00207713-50-2-823 [DOI] [PubMed] [Google Scholar]
- 52. Holt RA, Subramanian GM, Halpern A, et al. : The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298(5591):129–49. 10.1126/science.1076181 [DOI] [PubMed] [Google Scholar]
- 53. Coggins SA, Estévez-Lao TY, Hillyer JF: Increased survivorship following bacterial infection by the mosquito Aedes aegypti as compared to Anopheles gambiae correlates with increased transcriptional induction of antimicrobial peptides. Dev Comp Immunol. 2012;37(3–4):390–401. 10.1016/j.dci.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 54. Werren JH, Windsor DM: Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc Biol Sci. 2000;267(1450):1277–85. 10.1098/rspb.2000.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou W, Rousset F, O'Neil S: Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc Biol Sci. 1998;265(1395):509–15. 10.1098/rspb.1998.0324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. de Oliveira CD, Gonçalves DS, Baton LA, et al. : Broader prevalence of Wolbachia in insects including potential human disease vectors. Bull Entomol Res. 2015;105(3):305–15. 10.1017/S0007485315000085 [DOI] [PubMed] [Google Scholar]
- 57. Baldo L, Bordenstein S, Wernegreen JJ, et al. : Widespread recombination throughout Wolbachia genomes. Mol Biol Evol. 2006;23(2):437–49. 10.1093/molbev/msj049 [DOI] [PubMed] [Google Scholar]
- 58. Kumar S, Stecher G, Tamura K: MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jolley KA, Chan MS, Maiden MC: mlstdbNet - distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 2004;5:86. 10.1186/1471-2105-5-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tamura K, Nei M: Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–26. 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- 61. Klindworth A, Pruesse E, Schweer T, et al. : Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41(1):e1. 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Quast C, Pruesse E, Yilmaz P, et al. : The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–6. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. St Laurent B, Cooke M, Krishnankutty SM, et al. : Molecular Characterization Reveals Diverse and Unknown Malaria Vectors in the Western Kenyan Highlands. Am J Trop Med Hyg. 2016;94(2):327–35. 10.4269/ajtmh.15-0562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Walker T: Novel Wolbachia Strains in Anopheles Malaria Vectors from Sub-Saharan Africa. Open Science Framework. 2018. Web. 10.17605/OSF.IO/MW6XZ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kyalo D, Amratia P, Mundia CW, et al. : A geo-coded inventory of anophelines in the Afrotropical Region south of the Sahara: 1898-2016 [version 1; referees: 3 approved]. Wellcome Open Res. 2017;2:57. 10.12688/wellcomeopenres.12187.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wiebe A, Longbottom J, Gleave K, et al. : Geographical distributions of African malaria vector sibling species and evidence for insecticide resistance. Malar J. 2017;16(1):85. 10.1186/s12936-017-1734-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tiffany A, Moundekeno FP, Traoré A, et al. : Encouraging impact following 2.5 years of reinforced malaria control interventions in a hyperendemic region of the Republic of Guinea. Malar J. 2016;15(1):298. 10.1186/s12936-016-1353-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Riveron JM, Watsenga F, Irving H, et al. : High Plasmodium Infection Rate and Reduced Bed Net Efficacy in Multiple Insecticide-Resistant Malaria Vectors in Kinshasa, Democratic Republic of Congo. J Infect Dis. 2018;217(2):320–328. 10.1093/infdis/jix570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Erlank E, Koekemoer LL, Coetzee M: The importance of morphological identification of African anopheline mosquitoes (Diptera: Culicidae) for malaria control programmes. Malar J. 2018;17(1):43. 10.1186/s12936-018-2189-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tabue RN, Awono-Ambene P, Etang J, et al. : Role of Anopheles (Cellia) rufipes (Gough, 1910) and other local anophelines in human malaria transmission in the northern savannah of Cameroon: a cross-sectional survey. Parasit Vectors. 2017;10(1):22. 10.1186/s13071-016-1933-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Buck M, Nilsson LK, Brunius C, et al. : Bacterial associations reveal spatial population dynamics in Anopheles gambiae mosquitoes. Sci Rep. 2016;6:22806. 10.1038/srep22806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Muturi EJ, Ramirez JL, Rooney AP, et al. : Comparative analysis of gut microbiota of mosquito communities in central Illinois. PLoS Negl Trop Dis. 2017;11(2):e0005377. 10.1371/journal.pntd.0005377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shaw WR, Marcenac P, Childs LM, et al. : Wolbachia infections in natural Anopheles populations affect egg laying and negatively correlate with Plasmodium development. Nat Commun. 2016;7:11772. 10.1038/ncomms11772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Antonio-Nkondjio C, Simard F, Awono-Ambene P, et al. : Malaria vectors and urbanization in the equatorial forest region of south Cameroon. Trans R Soc Trop Med Hyg. 2005;99(5):347–54. 10.1016/j.trstmh.2004.07.003 [DOI] [PubMed] [Google Scholar]
- 75. Cano J, Descalzo MA, Moreno M, et al. : Spatial variability in the density, distribution and vectorial capacity of anopheline species in a high transmission village (Equatorial Guinea). Malar J. 2006;5:21. 10.1186/1475-2875-5-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Makanga B, Yangari P, Rahola N, et al. : Ape malaria transmission and potential for ape-to-human transfers in Africa. Proc Natl Acad Sci U S A. 2016;113(19):5329–34. 10.1073/pnas.1603008113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Antonio-Nkondjio C, Ndo C, Kengne P, et al. : Population structure of the malaria vector Anopheles moucheti in the equatorial forest region of Africa. Malar J. 2008;7:120. 10.1186/1475-2875-7-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fouet C, Kamdem C, Gamez S, et al. : Extensive genetic diversity among populations of the malaria mosquito Anopheles moucheti revealed by population genomics. Infect Genet Evol. 2017;48:27–33. 10.1016/j.meegid.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 79. Zug R, Koehncke A, Hammerstein P: Epidemiology in evolutionary time: the case of Wolbachia horizontal transmission between arthropod host species. J Evol Biol. 2012;25(11):2149–60. 10.1111/j.1420-9101.2012.02601.x [DOI] [PubMed] [Google Scholar]
- 80. Bailly-Bechet M, Martins-Simões P, Szöllosi GJ, et al. : How Long Does Wolbachia Remain on Board? Mol Biol Evol. 2017;34(5):1183–1193. 10.1093/molbev/msx073 [DOI] [PubMed] [Google Scholar]
- 81. Minard G, Tran FH, Dubost A, et al. : Pyrosequencing 16S rRNA genes of bacteria associated with wild tiger mosquito Aedes albopictus: a pilot study. Front Cell Infect Microbiol. 2014;4:59. 10.3389/fcimb.2014.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hegde S, Khanipov K, Albayrak L, et al. : Microbiome Interaction Networks and Community Structure From Laboratory-Reared and Field-Collected Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus Mosquito Vectors. Front Microbiol. 2018;9:2160. 10.3389/fmicb.2018.02160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dutra HL, Rocha MN, Dias FB, et al. : Wolbachia Blocks Currently Circulating Zika Virus Isolates in Brazilian Aedes aegypti Mosquitoes. Cell Host Microbe. 2016;19(6):771–4. 10.1016/j.chom.2016.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. van den Hurk AF, Hall-Mendelin S, Pyke AT, et al. : Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PLoS Negl Trop Dis. 2012;6(11):e1892. 10.1371/journal.pntd.0001892 [DOI] [PMC free article] [PubMed] [Google Scholar]