Abstract
Background
The aim of this study was to explore the expression levels of micro ribonucleic acid-10b (miR-10b) and micro ribonucleic acid-181b (miR-181b) in gastric cancer tissues, as well as their application value in clinical diagnosis and treatment.
Material/Methods
A total of 120 patients with gastric cancer who were diagnosed and treated in the Department of Gastroenterology of our hospital were enrolled in this study. The gastric cancer tissues and paracancerous tissues were collected for measuring the expression of miR-10b and miR-181b by in situ hybridization and reverse transcription-polymerase chain reaction (RT-PCR). The 5-year survival rate was also analyzed.
Results
The expressions of miR-10b and miR-181b in gastric cancer tissues were both upregulated and were significantly higher than those in the paracancerous tissues (p<0.05). In addition, the expressions of miR-10b and miR-181b in gastric cancer tissues were correlated with tumor size, degree of pathological differentiation, depth of infiltration, tumor node metastasis (TNM) staging, and lymph node metastasis, as well as local lymph node and distant metastasis (p<0.05). For patients in stage II and III, the expressions of miR-10b and miR-181b were significantly correlated with the 5-year survival rate.
Conclusions
The high expressions of miR-10b and miR-181b are significantly correlated with poor prognosis in stage II and III patients with gastric cancer, suggesting that their expressions might be criteria for evaluating the prognosis of gastric cancer.
MeSH Keywords: Gastric Acid, Prognosis, Stomach Neoplasms
Background
Gastric cancer, the second most common malignant tumor that endangers human health, has an increasing incidence rate in recent years. More than 40% of the world’s gastric cancer cases occur in China [1]. Nearly 80% of the gastric cancer cases are already in the terminal stage at the time of first diagnosis, and surgical resection is widely used for these patients. However, the prognosis is poor, with the 5-year survival rate being less than 10% [2]. The occurrence and development of gastric cancer are complicated and involve multiple factors. With a length of 19–25 nt, the micro ribonucleic acids (microRNAs, miRNAs) are endogenous, single-stranded, non-coding, and small RNA molecules with a high degree of conservation and can inhibit the translation of messenger ribonucleic acid (mRNA) by combining with the untranslated region of the target gene [3]. Evidence shows that miRNAs are abnormally expressed in gastric cancer tissues [4]. A previous study showed [5] that the abnormal expressions of miRNAs are involved in the pathological processes of the occurrence and development of gastric cancer, such as abnormal proliferation, infiltration, and metastasis of gastric cancer cells. MicroRNA-10B (miR-10b) is located in the homeobox (HOX) gene cluster, and the HOX family is a special type of transcriptional regulator. miR-10b can play a unique regulatory role in proliferation and differentiation of normal cells as well as in the occurrence, development, infiltration, and metastasis of tumor cells [6,7]. Studies have shown that microRNA-181b (miR-181b) can inhibit the expression of tumor-suppressing genes in tissue cells and increase the activity of nuclear factor-kappa B (κB), ultimately resulting in the malignant transformation of tissue cells, indicating a regulatory role of miR-181b in the occurrence and development of tumor cells [8]. However, the association of miR-10b or miR-181b with gastric cancer remains poorly understood. In the present study, we assessed the expressions of miR-10b and miR-181b in gastric cancer tissues and their effect on the 5-year survival rate of gastric cancer patients, aiming to provide a reference for the prognosis of gastric cancer.
Material and Methods
General materials
A total of 120 patients with gastric cancer who were diagnosed and treated in the Department of Gastroenterology of our hospital were recruited in this study. There were 64 males and 56 females, aged from 30 to 82 years old with an average age of (58.2±27.4) years old. Gastric cancer tissues (n=120) and paracancerous tissues (n=120) were extracted for analysis of the expression levels of miR-10b and miR-181b. None of the patients were treated with chemotherapy or immunotherapy before the operation. Classification was performed by tumor diameters: 70 cases were less than 5 cm in diameter and 50 cases were greater than or equal to 5 cm. Classification by degrees of differentiation showed that 4 cases were well-differentiated, 35 cases were moderately differentiated, and 81 cases were poorly differentiated. Classification by depth of infiltration showed 16 cases of T1, 30 cases of T2, 67 cases of T3, and 7 cases of T4. Lymph node metastasis showed 74 cases with metastasis and 46 cases without metastasis. Tumor node metastasis (TNM) staging showed 25 cases in stage I, 29 in stage II, 48 in stage III, and 18 in stage IV. All patients and their families signed the informed consent and this study was approved by the Hospital Ethics Committee.
Experimental consumables
Tissue RNA extraction kit (Shanghai So-Fe Biomedical Co., Ltd.), real-time fluorescent quantitative polymerase chain reaction (PCR) reaction kit (Shanghai Kang Lang Biological Technology Co., Ltd.), primer (Shanghai Yanqi Biological Technology Co., Ltd.), hermle (Shanghai Lingcheng Biological Technology Co., Ltd.), PCR instrument [Application Binary Interface (ABI)], ultraviolet spectrophotometer (Hangzhou Noted Scientific Instrument Co., Ltd.), real-time fluorescent quantitative PCR software (Life Technologies), and sensitivity-enhanced in situ hybridization kit (Boster Biological Technology).
Methods
In situ hybridization
The expressions of miR-10b and miR-181b in the tissues were detected using an in situ hybridization kit according to the operation instructions. Digoxin-labeled miR-10b and miR-181b probes were provided by Exiqon. The proportion of positive tumor cells ≤5% was recorded as 0, 6~25% as 1, 26~50% as 2, and >50% as 3. Staining intensity: Non-stained was recorded as 0, canary yellow as 1, yellowish-brown as 2 and brown as 3. The product of scores of positive proportion and staining intensity was used as the judgment basis for the final evaluation. Product ≥4 was treated as high expression and <4 as low expression.
Reverse transcription-polymerase chain reaction (RT-PCR) detection
The reverse transcriptional complementary deoxyribonucleic acid (cDNA) system included: 5 μL 5×RT-PCR buffer solution, 1 μL Reverse transcriptase (RTase) Mix, 1 μL 2.5 U/μL polymerase, 2 μg sample RNA, and 25 μL 3-diethylamino-1-propyne (DEP) water.
Conditions of reverse transcription were 37°C for 60 min and 85°C for 5 min. In the end of the reaction, the products were diluted 5 times and stored at low temperature.
The RT-PCR amplification system consisted of 10 μL 2×All-in-One quantitative polymerase chain reaction (qPCR) Mix, 2 μL All-in-One miRNA qPCR Primer, 2 μL Universal Adaptor PCR Primer, 2 μL cDNA (Dilute 5 times), and 4 μL DEP water.
RT-PCR reaction conditions were 95°C for 10 min, 95°C for 15 s, 60°C for 30 s, and 72°C for 10 s, with 40 cycles.
MicroRNA calculation
The RT-PCR experiment was performed using U6 as the internal reference and 2-ΔCT method was used to calculate the relative expressions of miR-10b and miR-181b in gastric cancer tissues and adjacent tissues, among which ΔCT was the D-value between the CT target gene and CTU6.
Statistical analysis
Statistical Product and Service Solutions (SPSS) 20.0 software was used for statistical analysis. The non-normally distributed data are represented by the median and normally-distributed data are expressed by mean ± standard deviation (SD). The t test or Wilcoxon test was performed for comparison between groups. The Mann-Whitney U test was used for independent samples comparison between groups. P<0.05 was considered as statistically significant.
Results
In situ hybridization analysis of the expressions of miR-10b and miR-181b
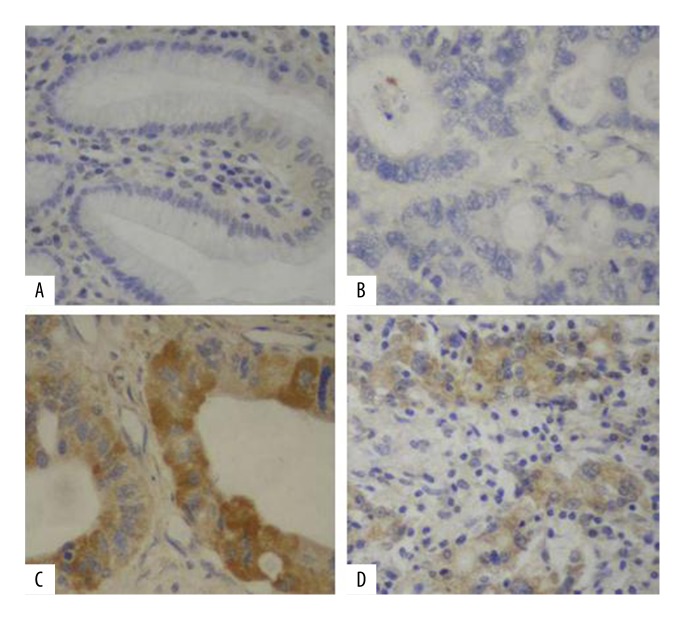
The positive-expression cells of miR-10b and miR-181b in gastric carcinoma were demonstrated to be stained in the cytoplasm under the light microscope, while the negative-expression cells were not stained (Figure 1).
Figure 1.
Expressions of miR-10b and miR-181b via in situ hybridization analysis. (A) Negative expression of miR-10b, (B) Negative expression of miR-181b, (C) Positive expression of miR-10b, (D) Positive expression of miR-181b.
Expressions of miR-10b in gastric cancer tissues and paracancerous tissues
The expressions of miR-10b in gastric cancer tissues in 109 patients were significantly higher than those in paracancerous tissues (p=0.017) (Figure 2).
Figure 2.
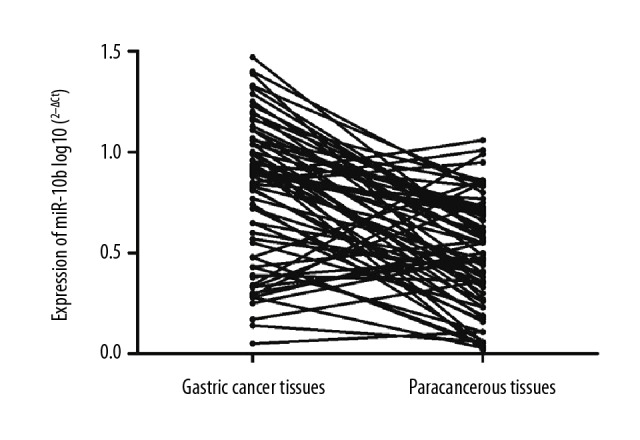
Expressions of miR-10b in gastric cancer tissues and paracancerous tissues.
Expressions of miR-181b in gastric cancer tissues and paracancerous tissues
The expressions of miR-181b in gastric cancer tissues in 97 patients were higher than those in paracancerous tissues, with a significant difference (p=0.0002) (Figure 3).
Figure 3.
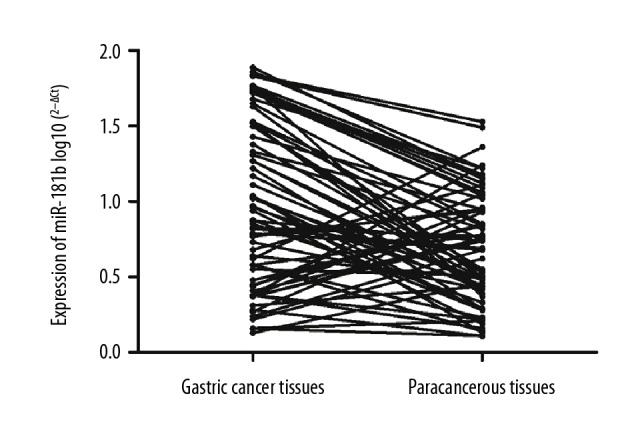
Expressions of miR-181b in gastric cancer tissues and paracancerous tissues.
The associations of the expressions of miR-10b and miR-181b with pathological parameters of gastric cancer
The expressions of miR-10b and miR-181b in gastric cancer tissues were significantly associated with the tumor size, pathological differentiation, depth of infiltration, TNM staging, and lymph node metastasis, as well as with local lymph node and distant metastasis of the patients (p<0.05). However, they were not associated with age, sex, ot the location of gastric cancer (p>0.05) (Table 1).
Table 1.
The correlations of the expressions of miR-10b and miR-181b with pathological parameters of gastric cancer.
| miR-10b | t/χ2 | p | miR-181b | t/χ2 | p | |||
|---|---|---|---|---|---|---|---|---|
| High | Low | High | Low | |||||
| Age (years old) | 58.04±11.96 | 60.81±11.92 | 2.419 | 0.264 | 59.02±12.04 | 61.37±12.47 | 2.595 | 0.306 |
| Sex | 0.015 | 0.927 | 0.028 | 0.892 | ||||
| Male | 37 (57.81) | 27 (42.19) | 35 (54.69) | 29 (45.31) | ||||
| Female | 32 (57.14) | 24 (42.86) | 34 (60.71) | 22 (39.29) | ||||
| Location | 4.175 | 0.133 | 3.984 | 0.142 | ||||
| Near-end | 7 (46.67) | 8 (53.33) | 8 (53.33) | 7 (46.67) | ||||
| Mid-end | 27 (60.00) | 18 (40.00) | 25 (45.45) | 30 (54.55) | ||||
| Far-end | 35 (58.33) | 25 (41.67) | 36 (60.00) | 24 (40.00) | ||||
| Tumor size | 20.024 | 0.0001 | 24.054 | 0.0001 | ||||
| <5 cm | 47 (67.14) | 23 (32.86) | 49 (70.00) | 21 (30.00) | ||||
| ≥5 cm | 22 (44.00) | 28 (56.00) | 20 (40.00) | 30 (60.00) | ||||
| Pathological differentiation | 11.052 | 0.012 | 9.386 | 0.019 | ||||
| Well differentiated | 3 (75.00) | 1 (25.00) | 3 (75.00) | 1 (25.00) | ||||
| Moderately differentiated | 16 (45.71) | 19 (54.29) | 15 (42.86) | 20 (57.14) | ||||
| Poorly differentiated | 50 (61.73) | 31 (38.27) | 51 (62.96) | 30 (37.04) | ||||
| Depth of infiltration | 41.037 | 0.0001 | 38.048 | 0.0001 | ||||
| T1 | 14 (87.50) | 2 (12.50) | 11 (68.75) | 5 (31.25) | ||||
| T2 | 17 (56.67) | 13 (43.33) | 19 (63.33) | 11 (36.67) | ||||
| T3 | 35 (52.24) | 32 (47.76) | 34 (50.75) | 33 (49.25) | ||||
| T4 | 3 (42.86) | 4 (57.14) | 5 (71.43) | 2 (28.57) | ||||
| TNM staging | 89.017 | 0.0001 | 77.045 | 0.0001 | ||||
| I | 22 (88.00) | 3 (12.00) | 15 (60.00) | 10 (40.00) | ||||
| II | 23 (79.31) | 6 (20.69) | 21 (72.41) | 8 (27.59) | ||||
| III | 19 (39.58) | 29 (60.42) | 25 (52.08) | 23 (47.92) | ||||
| IV | 5 (27.78) | 13 (72.22) | 8 (44.44) | 10 (55.56) | ||||
| Lymph node metastasis | 57.231 | 0.0001 | 68.551 | 0.0001 | ||||
| No | 37 (80.43) | 9 (19.57) | 39 (84.78) | 7 (15.22) | ||||
| Yes | 32 (43.24) | 42 (56.76) | 30 (40.54) | 44 (59.46) | ||||
| Local lymphatic node | 66.048 | 0.0001 | 52.375 | 0.0001 | ||||
| pN0 | 36 (78.26) | 10 (21.74) | 32 (69.57) | 14 (30.43) | ||||
| pN1 | 20 (54.05) | 17 (45.95) | 22 (59.46) | 15 (40.54) | ||||
| pN2 | 9 (33.33) | 18 (66.67) | 10 (37.04) | 17 (62.96) | ||||
| pN3 | 4 (40.00) | 6 (60.00) | 5 (50.00) | 5 (50.00) | ||||
| Distant metastasis | 15.926 | 0.0001 | 12.049 | 0.0001 | ||||
| No | 63 (61.17) | 40 (38.83) | 60 (58.25) | 43 (41.75) | ||||
| Yes | 6 (35.29) | 11 (64.71) | 9 (52.94) | 8 (47.06) | ||||
Prognostic analysis
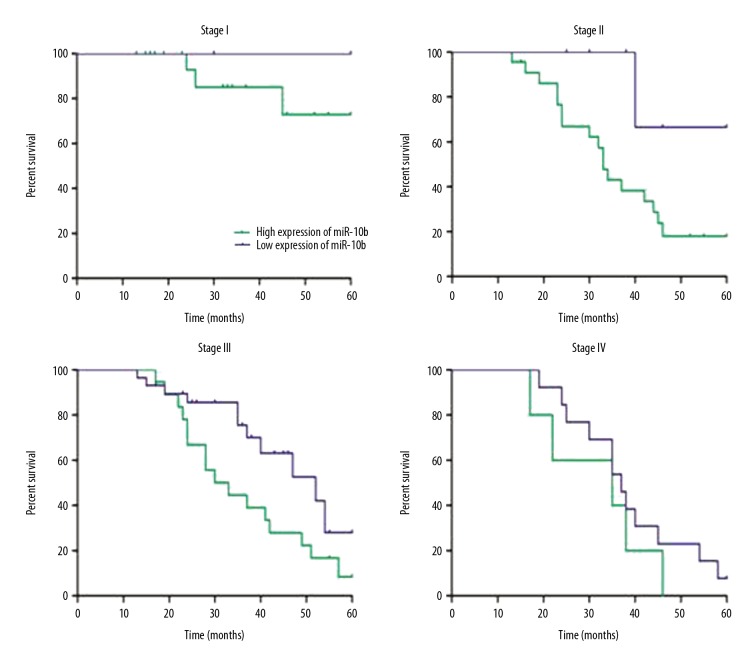
In patients in stage I, II, and III, the 5-year survival rates for patients with low expression of miR-10b were significantly higher than those of patients with high expression (p<0.05) (Figure 4). However, in patients in stage IV, the 5-year survival rates of patients with high expression and low expression of miR-10b were not statistically significant (p>0.05) (Figure 4).
Figure 4.
Prognostic analyses of the expressions of miR-10b and the 5-year survival rate of gastric cancer patients.
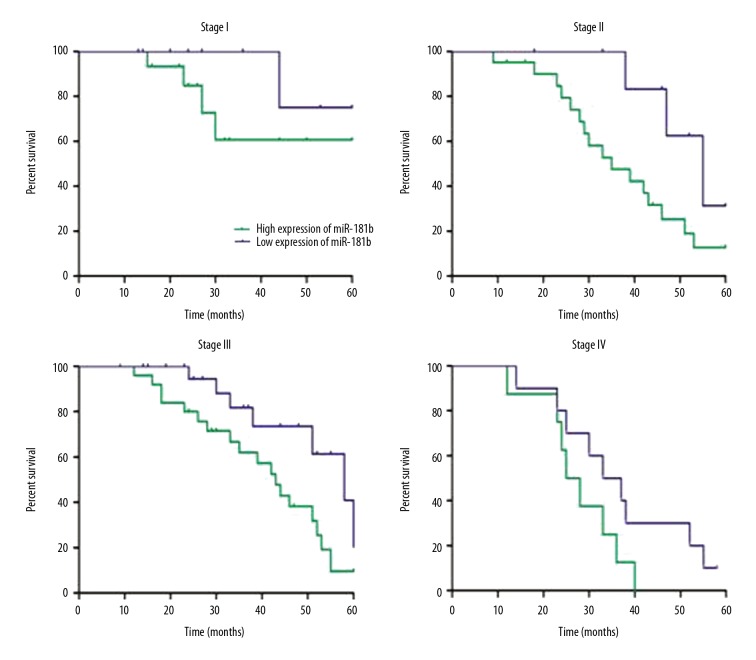
In stage II and III, the 5-year survival rates of patients with low expression of miR-181b were significantly higher than those in patients with high expression (p<0.05) (Figure 5). Interestingly, in patients in stage I and IV, no statistically significant difference in the 5-year survival rates was found in patients with high expression and low expression of miR-181b (p>0.05) (Figure 5).
Figure 5.
Prognostic analyses of the expressions of miR-181b and the 5-year survival rate of gastric cancer patients.
Discussion
At present, most cancers in humans are the results of a variety of factors, and gastric cancer is no exception. Several factors are involved in the occurrence and development of cancer, such as mutations of oncogene and proto-oncogenes, formation of DNA methylation, and abnormal expression of non-coding RNA [9–11]. miRNAs are a kind of small-molecule RNA and nearly 2000 such small-molecule RNAs have been reported. Studies have suggested that small-molecule RNAs regulate 40% of gene expressions in humans and abnormal expressions during pathological processes have extremely important effects on the development of disease and prognosis through regulating the expressions of other genes [12,13].
At present the prognosis of gastric cancer is poor and the 5-year survival rate is low. Therefore, it is urgent to find a marker that can predict the prognosis of gastric cancer. Previous studies [14,15] found that compared with serum or tissue proteins as the markers, miRNAs are more sensitive markers, which can improve the prognosis of patients. In this study, we showed that the expression of miR-10b in gastric cancer was increased significantly, suggesting that it may be involved in the occurrence and development of gastric cancer, which is consistent with a previous study [16]. In addition, the expressions of miR-181b and miR-200c have also been demonstrated to be upregulated in gastric cancer tissues [17], indicating that multiple miRNAs might be involved in the occurrence and development of gastric cancer.
Several studies [18–20] found that miRNAs can effectively regulate the posttranscriptional expression of the gene through binding to the target gene via base pairing, and the effect of regulation is closely associated with the prognosis. The present study found that the expressions of miR-10b and miR-181b in gastric cancer tissues were significantly associated with tumor size, pathological differentiation, depth of infiltration, TNM staging, and lymph node metastasis, as well as with local lymph node and distant metastasis (p<0.05), while the expressions were not associated with age, sex, or the location of gastric cancer. We also showed that in stage II and III patients, the expressions of miR-10b and miR-181b were significantly associated with the prognosis of the 5-year survival rate of gastric cancer patients. In stage I, the expression of miR-10b was correlated with the 5-year survival rate, while the expression of miR-181b was not correlated with the 5-year survival rate, suggesting that attention should be paid to the expressions of miR-10b and miR-181b in patients in stage II and III, which may be used as potential prognostic indicators for gastric cancer.
Conclusions
High expressions of miR-10b and miR-181b in the tissues of patients with gastric cancer have significant associations with the poor prognoses of patients in stage I and II, and detection of their expressions can be used as potential criteria for evaluating the prognosis of gastric cancer.
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Duell EJ, Lujan-Barroso L, Sala N, et al. Plasma microRNAs as biomarkers of pancreatic cancer risk in a prospective cohort study. Int J Cancer. 2017;141(5):905–15. doi: 10.1002/ijc.30790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saito M, Okayama H, Saito K, et al. CDX2 is involved in microRNA-associated inflammatory carcinogenesis in gastric cancer. Oncol Lett. 2017;14(5):6184–90. doi: 10.3892/ol.2017.6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Q, Song Q, Zhong W, et al. MicroRNA-10b and the clinical outcomes of various cancers: A systematic review and meta-analysis. Clin Chim Acta. 2017;474:14–22. doi: 10.1016/j.cca.2017.08.034. [DOI] [PubMed] [Google Scholar]
- 4.Li FQ, Xu B, Wu YJ, et al. Differential microRNA expression in signet-ring cell carcinoma compared with tubular adenocarcinoma of human gastric cancer. Genet Mol Res. 2015;14(1):739–47. doi: 10.4238/2015.January.30.17. [DOI] [PubMed] [Google Scholar]
- 5.Wang YY, Ye ZY, Zhao ZS, et al. Clinicopathologic significance of miR-10b expression in gastric carcinoma. Hum Pathol. 2013;44(7):1278–85. doi: 10.1016/j.humpath.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Ebrahimi S, Hosseini M, Ghasemi F, et al. Circulating microRNAs as potential diagnostic, prognostic and therapeutic targets in pancreatic cancer. Curr Pharm Des. 2016;22(42):6444–50. doi: 10.2174/1381612822666160817095047. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Yang Q, Kong WQ, et al. MicroRNA-181b targets cAMP responsive element binding protein 1 in gastric adenocarcinomas. IUBMB Life. 2012;64(7):628–35. doi: 10.1002/iub.1030. [DOI] [PubMed] [Google Scholar]
- 8.Krishnan AR, Zheng H, Kwok JG, et al. A comprehensive study of smoking-specific microRNA alterations in head and neck squamous cell carcinoma. Oral Oncol. 2017;72:56–64. doi: 10.1016/j.oraloncology.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou D, Zhou Q, Wang D, et al. The downregulation of microRNA-10b and its role in cervical cancer. Oncol Res. 2016;24(2):99–108. doi: 10.3727/096504016X14611963142173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Chen X, Meng Q, et al. MiR-181b regulates cisplatin chemosensitivity and metastasis by targeting TGFβR1/Smad signaling pathway in NSCLC. Sci Rep. 2015;5:17618. doi: 10.1038/srep17618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge YZ, Xu LW, Zhou CC, et al. A BAP1 mutation-specific microRNA signature predicts clinical outcomes in clear cell renal cell carcinoma patients with wild-type BAP1. J Cancer. 2017;8(13):2643–52. doi: 10.7150/jca.20234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YY, Li L, Ye ZY, et al. MicroRNA-10b promotes migration and invasion through Hoxd10 in human gastric cancer. World J Surg Oncol. 2015;13:259. doi: 10.1186/s12957-015-0673-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin X, Chen Y, Chen H, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017;23(17):5311–19. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 14.Tian F, Shen Y, Chen Z, et al. Aberrant miR-181b-5p and miR-486-5p expression in serum and tissue of non-small cell lung cancer. Gene. 2016;591(2):338–43. doi: 10.1016/j.gene.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Guan Y, Wang L, et al. MicroRNA-10b expression in node-negative breast cancer-correlation with metastasis and angiogenesis. Oncol Lett. 2017;14(5):5845–52. doi: 10.3892/ol.2017.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Z, Chen Y, Min L, et al. Augmented miR-10b expression associated with depressed expression of its target gene KLF4 involved in gastric carcinoma. Int J Clin Exp Pathol. 2015;8(5):5071–79. [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, Jian W, Wei C, et al. Down-regulation of miR-181b promotes apoptosis by targeting CYLD in thyroid papillary cancer. Int J Clin Exp Pathol. 2014;7(11):7672–80. [PMC free article] [PubMed] [Google Scholar]
- 18.Karsy M, Arslan E, Moy F. Current progress on understanding MicroRNAs in glioblastoma multiforme. Genes Cancer. 2012;3(1):3–15. doi: 10.1177/1947601912448068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouyang M, Li Y, Ye S, et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One. 2014;9(5):e96228. doi: 10.1371/journal.pone.0096228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tezcan G, Tunca, Bekar A, et al. microRNA expression pattern modulates temozolomide response in GBM tumors with cancer stem cells. Cell Mol Neurobiol. 2014;34(5):679–92. doi: 10.1007/s10571-014-0050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]